During the course of a viral or bacterial infection, an innate immune response is activated upon the detection of pathogen-associated molecular patterns (PAMPs) by host pattern recognition receptors (PRRs). Three major classes of PRRs have been well characterized to date: the Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), and the nucleotide binding domain and leucine-rich-repeat-containing proteins (NLRs). PAMP ligation to these PRRs triggers the activation of interferon regulator factor 3 (IRF3) and/or NF-κB transcription factors, which facilitate the production of antiviral and proinflammatory cytokines including IFN-β and IL-6, respectively. These cytokines play an essential role in orchestrating an antimicrobial immune response leading to pathogen clearance and, ultimately, immunological memory (Kumar et al., 2011). Regulation of antiviral and proinflammatory cytokines, however, must be tightly controlled because their excessive production has been linked to autoimmune and chronic inflammatory disorders (Karin et al., 2006). In this issue of Immunity, two groups demonstrate that NLRX1 functions as a bona fide in vivo negative regulator of RLR and TLR signaling to IRF3 and NF-κB ( Allen et al., 2011) and go on to provide the mechanisms by which NLRX1 exerts its inhibitory effect (Xia et al., 2011).
NLRX1 is a member of the NLR family of PRRs and was previously shown to function as a negative regulator of RLR signaling, not via ligand competition, but instead by targeting the RLR downstream mitochondrial antiviral signaling adaptor, MAVS (also known as IPS-1, Cardif, and VISA). As such, ectopic expression of NLRX1 in cells resulted in reduction of MAVS-dependent signaling to IRF3 and NF-κB. Conversely, depletion of NLRX1 via small interfering RNA (siRNA) led to enhanced activation of these transcription factors and correspondingly, elevated levels of antiviral and proinflammatory cytokines (Moore et al., 2008). In an effort to better understand how NLRX1 functions physiologically, Allen et al. (2011) undertook a genetic approach and generated NLRX1-deficient mice to examine antiviral and proinflammatory cytokine production in an in vivo model of virus infection. Consistent with published NLRX1 knockdown data, they found that fibroblasts derived from Nlrx1−/− mice produced markedly higher amounts of IFN-β and IL-6 upon infection with viruses that activate RIG-I signaling. In vivo, Nlrx1−/− mice had decreased viral titers in lung homogenates after influenza infection because of an increase in IFN-β production compared to wild-type animals. At the same time, however, Nlrx1−/− mice also presented with severe lung pathology, displaying enhanced denuding of airway epithelial cells and airway occlusion, probably because of exacerbated production of proinflammatory cytokines. Indeed, IL-6 concentrations were markedly higher in the serum and lungs of virally infected mice lacking NLRX1; however, it is currently unclear which cytokine(s) are responsible for the increased lung pathology observed in these mice. In the context of virus infection, RLRs undergo ligand-dependent interaction with MAVS where signal bifurcation toward IRF3 and NF-κB is thought to occur at the level of MAVS ( Figure 1). Interestingly in NLRX1-deficient cells, Allen et al. (2011) found that MAVS was persistently associated with RIG-I (i.e., even in the absence of virus infection). However, the RIG-I-MAVS interaction was insufficient to activate IRF3 or NF-κB; basal amounts of IFN-β or IL-6 were found to be similar between WT and Nlrx1−/− fibroblasts. It is therefore unclear whether other immediate factors are required to facilitate downstream signaling after RIG-IMAVS complex formation, and therefore future analysis of this signaling module will be necessary to determine the mechanism of its activation.
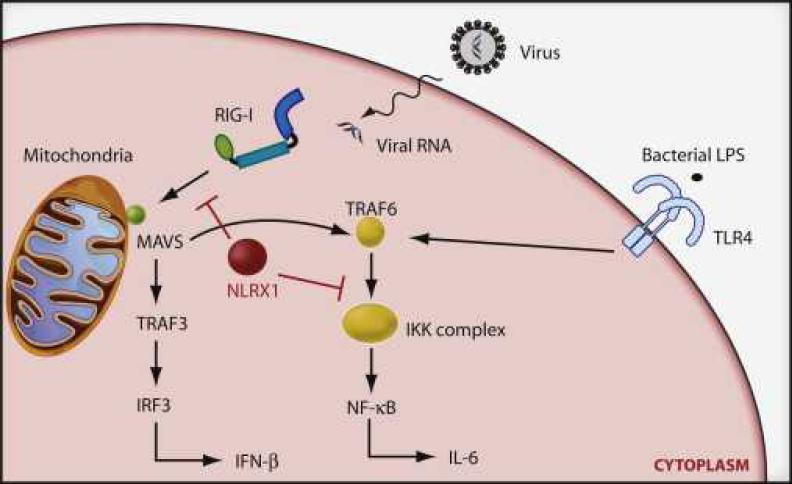
Figure 1. NLRX1 Blocks RIG-I and NF-κB Signaling.
Upon detection of viral- or bacterial-derived PAMPs, RIG-I and TLR4 facilitate the production of antiviral and/or proinflammatory cytokines via IRF3 and NF-κB, respectively. In RIG-I signaling, NLRX1 targets the MAVS adaptor to terminate downstream signal transmission to the IRF3 and NF-κB transcription factors. Alternatively, in TLR4 signaling pathways, NLRX1 undergoes ligand-dependent Lys63 polyubiquitination and translocation from TRAF6 to the IKK complex, blocking its kinase function and consequently NF-κB activation.
Extending their studies toward other PRRs, Allen et al. (2011) further evaluated the role of NLRX1 on TLR signaling. They found that stimulation of Nlrx1−/− macrophages with the bacterial PAMP lipopolysaccharide (LPS) (a TLR4 agonist) resulted in elevated amounts of IFN-β and IL-6 compared to WT stimulated cells, suggesting that NLRX1 additionally functions as a negative regulator of TLR signaling. Indeed, NLRX1-deficient mice that were given an in vivo airway challenge with LPS displayed increased IL-6 production that correlated with a marked increase in lung inflammation and pathology. TLR-mediated signaling occurs in a MAVS-independent fashion, where instead, the TRAF6 adaptor molecule is utilized to activate NF-κB. As such, Allen et al. (2011) found that NLRX1 could further target TRAF6 and inhibit its capacity to signal to NF-κB. Because TRAF6 is also downstream of MAVS in RLR signaling, it will be of future interest to determine whether NLRX1 additionally targets TRAF6 in this pathway as part of a MAVS-containing complex or whether multiple pools of NLRX1 exist to target different substrates.
In an attempt to gain further insight on the role of NLRX1 in TLR biology, Xia et al. (2011) employed a biochemical approach that led to a substantial understanding in the mechanism by which NLRX1 inhibits NF-κB. As latent transcription factors, NF-κB consists of prototypical RelA-p50 heterodimers that are sequestered in the cytoplasm by an inhibitory κB protein IκBα, which masks a nuclear localization signal on RelA. An upstream IκB kinase (IKK) complex, consisting of two kinases (IKKα and IKKβ) and a regulatory subunit IKKγ (also known as NEMO), upon activation, phosphorylates two highly conserved serine residues on IκBα, which in turn leads to phosphorylation-dependent ubiquitination and proteasomal degradation of IκBα. The freed NF-κB then translocates to the nucleus and binds specific promoter or enhancer sequences of NF-κB target genes to activate transcription (Hayden and Ghosh, 2004). In a canonical pathway of activation, it is the IKK complex that has the capacity to integrate a wide variety of signals transduced from diverse receptors including the TLRs. Interestingly, in cells stimulated with LPS, Xia et al. (2011) found that NLRX1 translocated from a TRAF6 module to the IKK complex. Further analysis indicated that NLRX1 underwent ligand-dependent Lys63 polyubiquitination, a posttranslational modification that modulates protein function in a proteasome-independent manner. Because NEMO contains a Lys63-ubiquitin binding domain (UBD), and deletion mutants of NEMO lacking the UBD showed a reduction in NLRX1 interactions, it may be that Lys63-ubiquitinated NLRX1 is targeted to NEMO in an initial phase of signal inhibition. Interestingly, the UBD of NEMO has previously been shown to be essential for downstream signaling to both NF-κB and IRF3 (Ea et al., 2006, Wu et al., 2006 and Zeng et al., 2009). As such, the antagonism of the UBD on NEMO by NLRX1 could prove to be an attractive mechanism of inhibition of both transcription factors. Future studies employing NLRX1 point mutants lacking ubiquitin acceptor sites would be beneficial in determining the exact function of NLRX1 Lys63 ubiquitination. Nevertheless, Xia et al. (2011) went on to demonstrate that IKK targeting by NLRX1 resulted in defective IKK function in that IKKα/β autophosphorylation and IκBα phosphorylation was impaired when NLRX1 was ectopically expressed or in NLRX1-containing IKK complexes after LPS stimulation. Conversely, both IKKα/β and IκBα phosphorylation was significantly enhanced in LPS-stimulated cells when NLRX1 was depleted via siRNA. Lastly, and in line with the results of Allen et al., 2011 and Xia et al., 2011 examined the in vivo role of NLRX1 on TLR signaling in a mouse model of LPS-induced septic shock. Intraperitoneal administration of LPS into NLRX1-knockdown mice resulted in significantly higher rates of death (compared to LPS-injected WT mice) resulting from LPS-mediated toxicity. Indeed, higher levels of serum IL-6 were present in NLRX1- depleted mice, giving further credence that NLRX1 serves as an in vivo negative regulator of inflammation.
Together, Allen et al. (2011) and Xia et al. (2011) provide a complementary and fairly comprehensive analysis on how NLRX1 operates to shut down antiviral and proinflammatory cytokine production upon engagement of PRRs. Although some variability in cytokine profiles and mechanism are observed between the reports, it is important to note that an additional group generated NLRX1-deficient mice that displayed no significant defects in RLR signaling (Rebsamen et al., 2011). Further clarification for these discrepancies will be essential in determining a conclusive function for NLRX1. Nevertheless, the presented works provide significant insight into the understanding of how NLRs pervade key PRR pathways. Future studies examining how NLRX1 is regulated in PRR signaling, particularly in the context of other negative regulators, and the role of NLRX1 in modulating other signaling programs will be beneficial in obtaining a more complete and refined analysis of NLRX1 function.
References
- 1.Allen IC, Moore CB, Schneider M, Lei Y, Davis BK, Scull MA, Gris D, Roney KE, Zimmermann AG, Bowzard JB, et al. Immunity. 2011;34:854–865. doi: 10.1016/j.immuni.2011.03.026. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Mol. Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 3.Hayden MS, Ghosh S. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 4.Karin M, Lawrence T, Nizet V. Cell. 2006;124:823–835. doi: 10.1016/j.cell.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 5.Kumar H, Kawai T, Akira S. Int. Rev. Immunol. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- 6.Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmermann AG, Accavitti-Loper MA, Madden VJ, Sun L, Ye Z, et al. Nature. 2008;451:573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- 7.Rebsamen M, Vazquez J, Tardivel A, Guarda G, Curran J, Tschopp J. Cell Death Differ. 2011 doi: 10.1038/cdd.2011.64. in press. Published online May 27, 2011. 1010.1038/cdd.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Nat. Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 9.Xia X, Cui J, Wang HY, Zhu L, Matsueda S, Wang Q, Yang X, Hong J, Songyang Z, Chen ZJ, Wang R-F. Immunity. 2011;34:843–853. doi: 10.1016/j.immuni.2011.02.022. his issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng W, Xu M, Liu S, Sun L, Chen ZJ. Mol. Cell. 2009;36:315–325. doi: 10.1016/j.molcel.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]