Abstract
Purpose
FGFR1 gene copy number (GCN) is being evaluated as a biomarker for FGFR tyrosine kinase inhibitor (TKI) response in squamous-cell lung cancers (SCC). The exclusive use of FGFR1 GCN for predicting FGFR TKI sensitivity assumes increased GCN is the only mechanism for biologically-relevant increases in FGFR1 signaling. Herein, we tested whether FGFR1 mRNA and protein expression may serve as better biomarkers of FGFR TKI sensitivity in lung cancer.
Experimental Design
Histologically diverse lung cancer cell lines were submitted to assays for ponatinib sensitivity, a potent FGFR TKI. A tissue microarray comprised of resected lung tumors was submitted to FGFR1 GCN and mRNA analyses and the results were validated with TCGA lung cancer data.
Results
14/58 cell lines exhibited ponatinib sensitivity (IC50 values ≤ 50 nM) that correlated with FGFR1 mRNA and protein expression, but not with FGFR1 GCN or histology. Moreover, ponatinib sensitivity associated with mRNA expression of the ligands, FGF2 and FGF9. In resected tumors, 22% of adenocarcinomas and 28% of SCCs expressed high FGFR1 mRNA. Importantly, only 46% of SCCs with increased FGFR1 GCN expressed high mRNA. Lung cancer TCGA data validated these findings and unveiled overlap of FGFR1 mRNA positivity with KRAS and PIK3CA mutations.
Conclusions
FGFR1 dependency is frequent across various lung cancer histologies and FGFR1 mRNA may serve as a better biomarker of FGFR TKI response in lung cancer than FGFR1 GCN. The study provides important and timely insight into clinical testing of FGFR TKIs in lung cancer and other solid tumor types.
INTRODUCTION
FGFR1 gene amplification in lung cancers, especially of squamous cell carcinoma (SCC) histology is well established in the literature (1–3) and increased FGFR1 gene copy number (GCN) is currently used as the predictive biomarker for prescreening SCC patients for entry into clinical trials of the FGFR-specific TKIs, BGJ398 (NCT01004224) and AZD4547 (NCT00979134). The rationale for use of this biomarker derives from studies showing that sensitivity of lung cancer cell lines to FGFR inhibitors correlates with increased FGFR1 GCN (1–3). Using ponatinib, another potent FGFR inhibitor, Gozgit et al (4) demonstrated growth inhibition of three FGFR1 gene amplified lung cancer cell lines. Similarly, Zhang et al (5) identified only two AZD4547-sensitive cell lines within a panel of 78 lung cancer cell lines, both were FGFR1 gene amplified.
While genomic amplification is a mechanism accounting for increased gene expression in cancer cells that is useful as a surrogate for oncogene activity, it is likely that transcriptional and translational control mechanisms may also mediate increased expression of proteins driving aberrant signal transduction. In support, our previous investigation of FGFR-dependent autocrine signaling in lung cancer cell lines (6) identified several as FGFR-dependent that have not been found to exhibit FGFR1 gene amplification (1, 2). We therefore screened a large panel of lung cancer cell lines including all histological subtypes for sensitivity to ponatinib and show that FGFR1 mRNA and protein function as superior biomarkers for response relative to FGFR1 GCN. While others have noted the association between FGFR1 gene amplification and SCC histology, we used assays applicable to tumor biopsy samples and observe that FGFR1 mRNA is more broadly increased across lung cancers of all histologies and that expression is not significantly correlated with FGFR1 GCN. The hypothesis that measurements of FGFR1 expression, not GCN, will provide more accurate markers of FGFR1-dependent lung cancers is being tested in an ongoing clinical trial.
MATERIALS AND METHODS
Cell Culture
All cell lines were cultured in RPMI-1640 growth medium supplemented with 10% fetal bovine serum at 37°C in an humidified 5% CO2 incubator. The following cell lines were available in our laboratories and submitted to DNA fingerprint analysis for authentication; H1703, HCC95, NE-18, DMS-114, SK-MES-1, H460, SW1573, H520, H661, H125, HCC44, H1299, H157, Colo699, H1581, HCC15, H2126, H1869, H1435 and H441. The remaining 38 cell lines were obtained directly from the University of Colorado Cancer Center Tissue Culture Core and were cultured fewer than 6 months after receipt. The core laboratory routinely performs DNA fingerprint analyses on all banked cell lines to ensure their authenticity.
Quantitative Real-Time PCR (RT-PCR)
Total RNA was purified from cells using RNeasy mini prep kits (Qiagen, Valencia, CA) and aliquots (5 μg) were reverse transcribed in a volume of 20 μL using Maxima First Strand cDNA Synthesis Kit (Thermo Scientific, Pittsburgh, PA). Aliquots (5 μL) of a 1:25 dilution of the reverse transcription reactions were submitted to quantitative RT-PCR in 25 μL reactions with SYBR green Jumpstart Taq Readymix (Sigma) with GAPDH, FGF2, FGF7, FGF9, FGFR1, FGFR2, FGFR3, FGFR4 primers previously described (6–8) using a My iQ real time-PCR detection system (BioRad, Hercules, CA). GAPDH mRNA levels were measured as a housekeeper gene for normalization of the different mRNA expression values.
Immunoblot Analysis
For immunoblot analysis of FGFR1 and the α-subunit of the NaK-ATPase, cells were collected in phosphate-buffered saline, centrifuged (3min at 3000 rpm) and suspended in lysis buffer. Aliquots of the cell lysates containing 150 μg of protein were submitted to SDS-PAGE and immunoblotted for FGFR1 (Origene, Rockville, MD) and NaK-ATPase α-subunit (sc-21712) (Santa Cruz Biotechnology, Santa Cruz, CA). Densitometry was performed using Quantity One® for FGFR1 and NaK-ATPase immunoblots where NaK-ATPase was measured as a loading control.
For immunoblot analysis of phospho-ERK and total ERK, cells were plated at 25,000 cells/well in a 6-well plate. 24 hours later, cells were switched to HITES media for 2 hours and subsequently treated with concentrations of ponatinib (0–1μM) for 2 hours. Cells were rinsed in phosphate-buffered saline, lysed in 250 μl lysis buffer and aliquots were submitted to SDS-PAGE and immunoblotted for phospho- ERK (P-p44/42 MAPK) (Cell Signaling Technology, Danvers, MA) and ERK 1 (sc-93) and ERK 2 (sc-154) (Santa Cruz Biotechnology, Santa Cruz, CA).
Anchorage Independent Growth Assay
For measurement of anchorage-independent cell growth in soft agar, 15,000 cells were suspended in 1.5 ml RPMI-1640 containing 10% fetal bovine serum and 0.35% Difco agar noble and overlaid on base layers containing 1.5mL RPMI-1640 containing 10% fetal bovine serum and 0.5% agar noble in six-well plates. The wells were overlayed with 2 ml growth media containing test agents or DMSO as a diluent control. Drugs were replaced in fresh growth medium once per week. The plates were incubated for 14 days and viable colonies were stained for 24 hours with 200 μl of 1mg/ml nitroblue tetrazolium. The wells were photographed and submitted to quantification using the MetaMorph (Molecular Devices, Downingtown, PA) imaging software program. Data are the total colony area (colony number × colony size) and presented as a percent of the DMSO control.
Clonogenic Growth Assay
To measure the effect of ponatinib on single cell colony formation in a clonogenic assay, lung cancer cells were seeded in 6-well plates at 100 to 200 cells/well in full growth media. Twenty-four hours later, cells were treated with ponatinib (0 to 300 nM) and cultured for 14 additional days. Wells were rinsed with phosphate-buffered saline (PBS) and stained and fixed with 0.5% (wt/vol) crystal violet in 6.0% (vol/vol) gluteraldehyde solution (Fisher Scientific, Fair Lawn, NJ). Following destaining with distilled water, the plates were photographed and total colony area was quantified using the MetaMorph imaging software program.
Cell Proliferation Assay
Cell lines were plated at 100 cells per well in 96-well tissue culture plates and treated in triplicate with test agents. Cell number per well was estimated after 10 days of culture using a CyQUANT Direct Cell Proliferation Assay (Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions.
FGFR1 Silencing with shRNAs
Distinct FGFR1-targeting shRNAs (clone IDs TRCN0000121185 and TRCN0000000417) in the pLKO.1 lentiviral vector were obtained from University of Colorado Cancer Center Functional Genomics shared resource. The pLKO.1 constructs encoding the FGFR1 shRNAs as well as pLKO.1 encoding a control shRNA targeting GFP were packaged in 293T cells with packaging component vectors pCMV-VSV-G and pΔ8.9. The lentiviruses released into the medium were filtered using a 0.45 μm filter and used to transduce the cell lines shown in Figure S2 in 6-well plates at 100,000 cells/well. Transfected cells were selected with puromycin (1 μg/ml) for 8 days after which the plates were rinsed with phosphate-buffered saline (PBS) and fixed and stained with 0.5% (wt/vol) crystal violet in 6.0% (vol/vol) glutaraldehyde solution for 30 minutes at room temperature. Plates were rinsed in distilled H2O and photographed. The MetaMorph imaging software program (Molecular Devices, Downingtown, PA) was used to quantify percent threshold area. RNA was purified from replicate wells and submitted to quantitative RT-PCR analysis to confirm knockdown.
Measurement of FGFR1 gene copy number in cell lines
Selected lung cancer cell lines were submitted to the University of Colorado Cancer Center Molecular Pathology shared resource for evaluation of FGFR1 gene copy number by fluorescence in situ hybridization (FISH) analysis. Also, publicly available SNP data archived in the Cancer Cell Line Encyclopedia (http://www.broadinstitute.org/ccle/home) were interrogated for FGFR1 gene copy number status in 49 of the lung cancer cell lines used in this study. Finally, total genomic DNA was purified from cells using Quick-gDNA MiniPrep kits (Zymo Research, Irvine, CA). Dilutions (1:100 and 1:20) of the genomic DNA were submitted to qRT-PCR in 25 μL reactions with SYBR green Jumpstart Taq Readymix (Sigma) and Line-1 primers (8) or FGFR1 primers (6, 8) using a My iQ real time-PCR detection system (BioRad). FGFR1 genomic levels were normalized to Line-1 gene copy levels as previously described (8). The normalized value for H157 cells was adjusted to 1 as this cell line exhibited an FGFR1 gene copy number of 1 (2 FGFR1 signals and 2 CEP8 signals) as determined by FISH assay. All other values were normalized accordingly.
Surgical lung cancer cohort and preparation of a tissue microarray
The tissue microarray (TMA) was generated from lung tumors obtained from a previously described series of patients who underwent pulmonary resection at the Medical University of Gdansk, Poland (9). In brief, the TMA is comprised of 3 cores of 1.5 mm in diameter from each tumor. The TMA slides were H-E stained for histology verification and quality control. Clinical data were derived from patient records. Tissue banking and the present studies were approved by the Institutional Review Boards at the University of Colorado and the Medical University of Gdansk.
Silver in situ hybridization of formalin-fixed, paraffin-embedded tissues for FGFR1 gene copy number
FGFR1 copy number was analyzed using bright-field microscopy and the silver in situ hybridization (SISH) technology. Probing was carried out using an FGFR1-specific probe according to protocols from the manufacturer (Ventana Medical Systems, Tucson, AZ). The scoring was carried out in 50 non-overlapping nuclei per core in regions identified by optical analysis of tissue sections as having higher gene copy number. The reading was completed for each core in the TMA and the core with the highest average FGFR1 gene copy number per cell was selected for each patient as described previously (9, 10).
In situ hybridization of formalin-fixed, paraffin-embedded tissues for FGFR1 mRNA
The mRNA in situ hybridization (ISH) assay was performed using the RNA scope® 2.0 assay system and the FGFR1 probe provided by Advanced Cell Diagnostics, Inc (Hayward, CA) according to the protocol provided by the company. ISH scores are generated and recorded using the following algorithm with a 400 X magnification setting on the microscope: 0, no staining; 1+, 1–3 dots/cell in >1% but <50% of the tumor cells; 2+, 1–3 dots per cell in >50% of the cells; 3+, clusters in <10% or 3–5 dots in >50% or >5 dots in <10% of tumor cells; 4+, clusters in >10% of the tumor cells or >5 dots in >10% of tumor cells.
Statistics
Spearman correlation coefficients and corresponding p-values were used to evaluate association between two continuous variables. A fixed effects ANOVA model was used to examine the potential association between histology (adenocarcinoma (AC), SCC, large cell carcinoma (LCC), small cell lung cancer (SCLC) and others) and IC50. ROC curves were generated to display the performance of dichotomized SISH and ISH in discriminating sensitive and non-sensitive cell lines based on ponatinib IC50.
RESULTS
Identification of ponatinib-sensitive lung cancer cell lines
We tested a panel of 58 well-characterized lung cancer cell lines (Table S1) comprised of 30 AC cell lines, 14 SCC cell lines, 4 large cell carcinoma (LCC) cell lines, 5 SCLC cell lines and 5 non-small cell lung cancer (NSCLC)/mixed histology cell lines for growth sensitivity to the multi-kinase TKI, ponatinib (4). No cell lines bearing EGFR mutations were included as we have recently reported their baseline insensitivity to FGFR-specific TKIs (11). The specificity of ponatinib (AP24534) has been previously defined (12) and although designed as a next generation ABL inhibitor, ponatinib is also a potent inhibitor of FGFR1, 2 and 4 (IC50s = 2, 2 and 8 nM, respectively) and a weaker inhibitor of FGFR3 (18 nM). PDGFRA, PDGFRB, RET and VEGFR1-3 are inhibited by ponatinib at IC50s ranging from 0.2–8 nM. Among the 58 cell lines, none contain RET gene rearrangements. Cancer Cell Line Encyclopedia data (CCLE; http://www.broadinstitute.org/ccle/home) were queried for VEGFR1-3 and RET mRNA expression, revealing that H1734 cells express detectable VEGFR1/FLT1 while H441 and H2170 cells express VEGFR2/KDR. VEGFR3/FLT4 was not expressed in any of the cell lines as assessed by CCLE gene expression data. RET mRNA was detected in SW1573 and H1385 cells, although no gain-of-function mutations are noted in CCLE. Also, consistent with published reports, H1703 cells expressed high levels of PDGFRA mRNA mediated by gene amplification (13, 14).
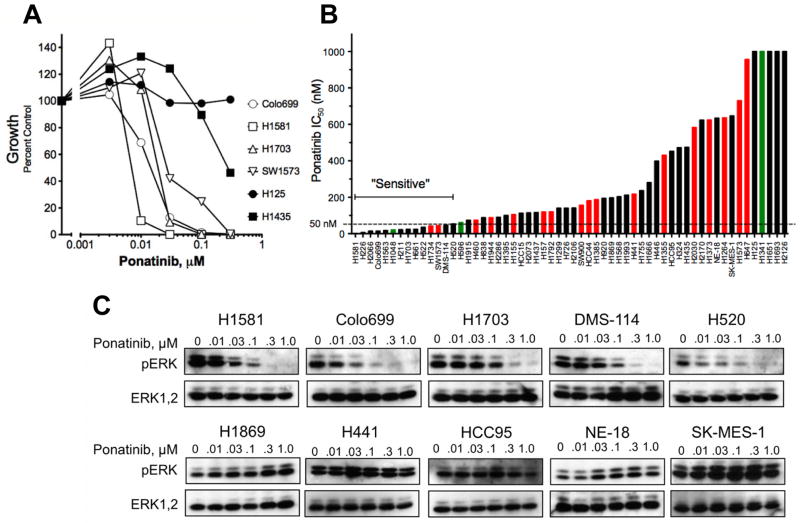
Representative ponatinib growth inhibitory curves for 6 cell lines are shown in Figure 1A. The calculated IC50 values for the entire panel (Table S1, Figure 1B) ranged from 1 nM to greater than 1 μM. The recent clinical trial in BCR-ABL positive leukemia revealed a serum trough level of ~60 nM ponatinib at the maximum tolerated dose (15). As a conservative cut-point, we defined sensitivity to ponatinib as IC50 ≤ 50 nM, thereby including 14 ponatinib sensitive lung cancer cell lines (Figure 1B, Table S1). In addition to inhibition of in vitro growth, ponatinib inhibited signaling through the ERK pathway in sensitive, but not insensitive cell lines (Figure 1C). The phosphorylation of AKT on T308 or S473 was not affected (data not shown), consistent with the reported lack of effect of the FGFR inhibitor, PD173074, on AKT phosphorylation (2). The cell lines with IC50s ≤ 50 nM for ponatinib were derived from ACs (5 of 30, 17%), LCCs (2 of 4, 50%), SCCs (3 of 14, 21%) and SCLCs (3 of 5, 60%) (Figure 1B (inset), Table S1). Also, 1 of 5 cell lines of NSCLC/mixed histology was sensitive to ponatinib. Using a fixed effects ANOVA model, no significant correlation of ponatinib IC50 to cell line histology was observed (p=0.16). Thus, ponatinib sensitivity in lung cancer cell lines distributes across all histological subtypes.
Figure 1. Ponatinib sensitivity of lung cancer cell lines of all histologies.
Lung cancer cell lines were submitted to anchorage-independent or clonogenic growth assays in the presence of 0 – 300 nM ponatinib. Some cell lines failed to grow in these assays and the effect of TKI on cell proliferation was determined instead. A, Dose-response curves for lung cancer cell lines representative of the full panel are shown. B, The IC50 values were calculated from the data and plotted from the lowest to highest. Green and red bars indicate the presence of mutations in PIK3CA and KRAS, respectively. C, Selected cell lines were treated for 2 hrs with ponatinib and cell extracts were immunoblotted for phospho-ERK and total ERK.
Sensitivity of cell lines to ponatinib correlates with FGFR1 mRNA and protein expression, but not gene amplification
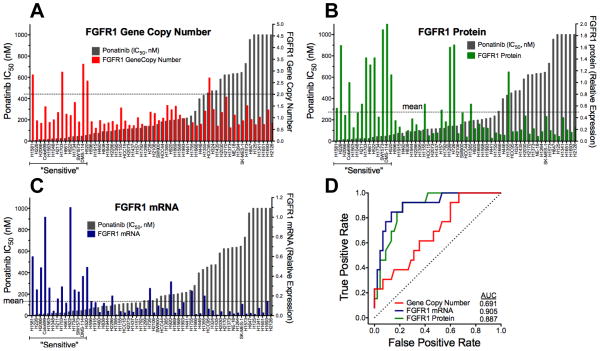
Ponatinib has previously been shown to potently inhibit in vitro and in vivo growth of three lung cancer cell lines, H1581, DMS-114 and H520, all of which bear FGFR1 gene amplification (4, 16). To further explore the association of ponatinib sensitivity with increased FGFR1 gene copy levels, we assessed FGFR1 GCN in 58 lung cancer cell lines using multiple techniques (see Materials and Methods) including quantitative PCR on genomic DNA (8), an FGFR1 FISH assay and SNP array data from the CCLE. In addition to H1581, H520 and DMS-114, increased FGFR1 GCN relative to a chromosome 8 centromeric (CEP8) probe was observed in H1703 and HCC95 cells (Table S1). Only 4/14 ponatinib-sensitive lung cancer cell lines exhibited a ratio of FGFR1:CEP8 signals > 2 (Table S1, Figure 2A). Also, FGFR1-amplified HCC95 cells are not sensitive to ponatinib (Figure 1B) or PD173074 (2). Moreover, Weiss et al (2) noted that H2444 NSCLC cells exhibited FGFR1 amplification, but are insensitive to PD173074. When the FGFR1 target:CEP8 ratio was considered as a continuous variable, there was no statistical association with ponatinib IC50 (Table S1).
Figure 2. Association of FGFR1 GCN, protein and mRNA expression with ponatinib IC50.
The FGFR1:CEP8 ratio (A), FGFR1 protein from immunoblot analysis (B) and FGFR1 mRNA measured by quantitative RT-PCR (C) were determined in the panel of cell lines and plotted as shown. The data are graphically overlayed on the ponatinib IC50 values from Figure 1B. The resulting datasets were submitted to ROC analysis (D) and the area under the curve (AUC) values are shown.
In addition to FGFR1 GCN, FGFR1 protein and mRNA levels were measured (Figure 2B and C, Table S1) by immunoblotting with an antibody against the carboxyl-terminus of FGFR1 and quantitative PCR with primers annealing to sequences within the invariant second immunoglobulin domain. Figure S1 shows representative immunoblots of FGFR1 protein in 34 of the lung cancer cell lines. Multiple FGFR1 protein species were identified by immunoblotting and is related to the known precursor/product relationship dictated by the degree of glycosylation as well as by alternative mRNA splicing as reviewed by Turner and Grose (17). Expression of FGFR1 protein (r=−0.52, <0.0001) and mRNA (r=−0.62, p<0.0001) were highly associated with ponatinib IC50 (Figure 2B and C, Table S1) by Spearman correlation analysis. Both markers were correlated with each other (r=0.466, P=0.00023). FGFR3 mRNA was weakly associated (r=−0.231, p=0.081), but not with FGFR2 or FGFR4 mRNA expression (Table S1). Using a cut-point of ≥ mean expression of the 58 cell lines, 12/14 and 10/14 ponatinib-sensitive lung cancer cell lines expressed FGFR1 mRNA and protein, respectively (Table S1). By contrast, 5/44 and 6/44 ponatinib-insensitive cell lines expressed FGFR1 mRNA and protein, respectively. To independently validate the association of FGFR1 mRNA levels with ponatinib sensitivity, we queried Affymetrix array data derived from 49 of the 58 lung cancer cell lines deposited in the CCLE. Spearman correlation revealed an association of ponatinib IC50 with FGFR1 (r=−0.50, p=0.0002), the FGFR signaling scaffold, FRS2 (r=−0.54, p<0.0001) and FGF2 (−0.43, p=0.002), but not FGFR2-4. It was recently shown that MYC was coexpressed in 40% of FGFR1-amplified tumors and that tumor cells coexpressing MYC were more sensitive to FGFR inhibition (18), However, MYC mRNA levels in the 49 lung cancer cell lines assessed by CCLE were not significantly associated with ponatinib sensitivity (r=0.958). The FGFR1 GCN and expression data from the 58 cell lines were submitted to receiver operating characteristic (ROC) curve analysis and FGFR1 mRNA and protein expression levels greatly outperformed FGFR1 GCN as predictors of ponatinib sensitivity (Figure 2D).
Targets distinct from FGFR1 may contribute to growth inhibition by ponatinib in specific cell lines due to its multi-kinase activity. In addition to amplification and high-level expression of FGFR1, H1703 cells also bear amplified PDGFRA and are dependent on a PDGFC-PDGFRA pathway for growth (13, 14). We have previously reported the sensitivity of H1703 cells to the FGFR/PDGFR/VEGFR inhibitor, RO4383596 (6). Thus, the dual activity of ponatinib on FGFR1 and PDGFRA likely contributes to full growth inhibition in this cell line.
Molecular confirmation of FGFR1 as a driver in an amplified and non-amplified cell line
We molecularly tested FGFR1 as a driver in non-amplified cell lines (Colo699, H1048, H596) as well as amplified cell lines (H1581, H520, H1703). Demonstration of H1581 cell growth dependence on FGFR1 through RNAi techniques has been previously published (1, 2), but not for any of the non-FGFR1 amplified cell lines. The cell lines were transduced with lentivirus encoding a control shRNA targeting GFP or two independent shRNAs targeting FGFR1 (Figure S2). Relative to the equal colony growth among the 3 lentiviruses following transduction of mouse embryo fibroblast cells where murine FGFR1 is not predicted to be silenced, or two independent lung cancer cell lines lacking FGFR1 expression and insensitive to ponatinib (HCC15, NE-18), the FGFR1-targeting shRNAs induced marked suppression of colony outgrowth in H1581, H520, Colo699, H1048 and H596 cells (Figure S2). Clonogenic growth of H1703 cells exhibiting amplified FGFR1, PDGFRA and WHSC1L1 (14, 19) was partially decreased upon FGFR1 silencing (Figure S2). Also noteworthy, H596 cells expressing lower levels of FGFR1 and exhibiting a ponatinib IC50 = 61 nM showed a marked decrease in growth following FGFR1 silencing. Thus, growth of representative FGFR1 amplified and non-amplified cell lines are dependent upon FGFR1 for growth, thereby supporting this RTK as a primary target mediating ponatinib sensitivity.
Requirement for autocrine FGFs in FGFR1-dependent lung cancer cell lines and association of FGF2 and FGF9 expression with ponatinib sensitivity
While increased expression and activity of FGFR1 has been associated with gene amplification, no gain-of-function mutations have been identified in lung cancer cell lines. To test for dependency on autocrine FGFs, growth of selected cell lines was measured in the presence of the FGF ligand trap, FP-1039 (20). As shown in Figure S3, FP-1039 inhibited growth of Colo699 cells with an IC50 of 3 nM. Consistent with a recent report (20), we observed potent growth inhibition of FGFR1 gene amplified H1581 cells (IC50 = 9 nM) and less potent inhibition of H1703 cells (IC50 = 33 nM) in which both FGFR1 and PDGFRA genes are amplified (14). NE18 cells that express no FGFR1, FGF2 or FGF9 (Table S1) were ~50 fold less sensitive (IC50 = 162 nM) to FP-1039 compared to Colo699 cells. Thus, the sensitivity to FP-1039 supports the autocrine activity of FGFs, at least within a subset of the ponatinib-sensitive lung cancer cell lines as previously shown (20).
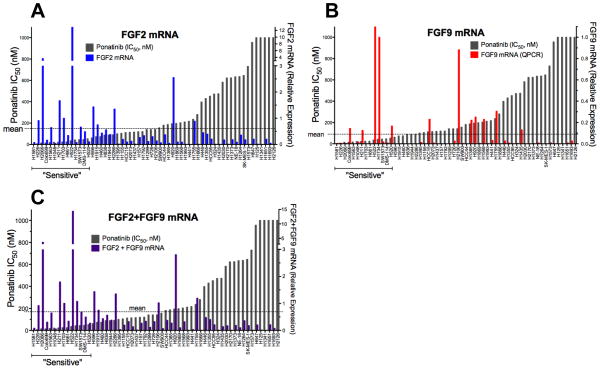
We measured the expression of FGF2 and FGF9, two FGFs previously shown to be expressed in FGFR-dependent lung cancer cell lines (6), and tested the association of expression with ponatinib sensitivity. FGF2 (r=−0.44, p=0.005), but not FGF9 mRNA expression (r=−0.21, p=0.11), was significantly associated with the ponatinib IC50 (Figure 3, Table S1). Because the majority (10/14) of the ponatinib-sensitive cell lines express FGF2 and/or FGF9 (Table S1, Figure S4), we also tested the correlation of their combined measurement and observed a stronger association (r=−0.55, p<0.0001) than FGF2 alone (Figure 3C). It is noteworthy that two sensitive cell lines, H1581 and H1048, express little or no FGF2 or FGF9 mRNA (Figure 3, Table S1), although the CCLE database reveals high expression of FGF10 and FGF20 mRNA in H1581 cells and high FGF7 mRNA in H1048 cells (Table S2). While the FGFR1 expressed in H1048 cells exists as the “c” isoform (data not shown) and is not predicted to bind FGF7, these cells also express significant levels of FGFR2 (Table S1) exclusively spliced to the “b” isoform (Figure S4B) and are predicted to bind FGF7. Moreover, both exogenous FGF2 (via FGFR1) and FGF7 (via FGFR2b) stimulate ERK activity in H1048 cells (Figure S4C). CCLE data reveal FGF10 and FGF20 mRNA expression, but not FGF2 or FGF9 mRNA expression in H1581 cells, consistent with our data (Table S1). While FGF10 binding, like FGF7, is restricted to FGFR “b” isoforms, FGF20 is an FGF9 subfamily member predicted to bind the FGFR “c” polypeptides (21). H1581 cells also express significant levels of FGFR2 (Table S1), but as the “c” isoform (Figure S4B) and elicit ERK activation in response to FGF2 and FGF9, but not FGF7 (Figure S4C). Thus, our findings support the requirement for autocrine FGFs as activators of FGFR1 in ponatinib-sensitive lung cancer cell lines.
Figure 3. Association of FGF2 and FGF9 mRNA expression with ponatinib IC50.
FGF2 (A) and FGF9 (B) mRNA levels were determined by quantitative RT-PCR and plotted as shown. In (C), the sum of the FGF2 and FGF9 mRNA relative expression values are plotted.
FGFR1 GCN status and mRNA expression in primary lung tumors
A TMA comprised of FFPE-processed lung cancer cell lines was generated and used to develop and optimize assays for FGFR1 GCN and mRNA in fixed tumor tissues. FGFR1 GCN was measured with silver in situ hybridization (SISH) and FISH assays on the lung cancer cell line TMA and the respective measurements were highly correlated (r=0.68, p<0.0001), indicating the two assays are essentially equivalent for detection of FGFR1 GCN. Moreover, analysis of the lung cancer cell line TMA with the RNAscope in situ hybridization (ISH) assay detected FGFR1 mRNA to a degree that is in excellent agreement (r=0.853, p<0.0001) with mRNA levels measured by quantitative RT-PCR. Next, we tested the FGFR1 SISH and ISH assays for their ability to identify the TKI sensitive cell lines within the lung cancer cell line TMA. Choosing the IC50 cut point of ≤50 nM for ponatinib sensitivity, ROC curve analyses were performed and FGFR1 SISH at GCN ≥ 4 or ISH = 4 provides reasonable combinations of sensitivity and specificity in discriminating ponatinib sensitive vs. resistant cell lines (Figure S5), with ISH being higher in both sensitivity and specificity than SISH
A TMA containing a cohort of surgically resected lung cancers was submitted to FGFR1 GCN analysis by SISH. Of the 182 tumors assessed by SISH, 14 (8%) exhibited FGFR1 GCN ≥ 4 and 11 of these showed FGFR1 amplification (FGFR1 target:CEP8; ratio ≥ 2; Table S3). All tumors with increased FGFR1 GCN or amplification were of SCC, mixed or not otherwise specified histology. Thus, the frequency of FGFR1 gene amplification as measured by SISH assay in lung SCC (11 of 100 tumors, ~11%) is similar to that reported by others (1–3). Measurement of FGFR1 mRNA by ISH revealed 46 of 158 tumors evaluated (29%) as positive at a score of 4+ (Table S4). FGFR1 mRNA was positive in 25 of the 89 SCCs, (28%) and in 10 of 45 ACs (22%). Among the 89 squamous cell carcinomas, 25 (~28%) were positive for FGFR1 mRNA while 10 of 45 (22%) adenocarcinomas were positive. Thus, expression of FGFR1 mRNA is more frequent than increased GCN and is not restricted to SCC histology. Finally, there was no prognostic association, either overall survival (Figure S6) or disease-free survival (data not shown), with increased FGFR1 GCN or mRNA expression.
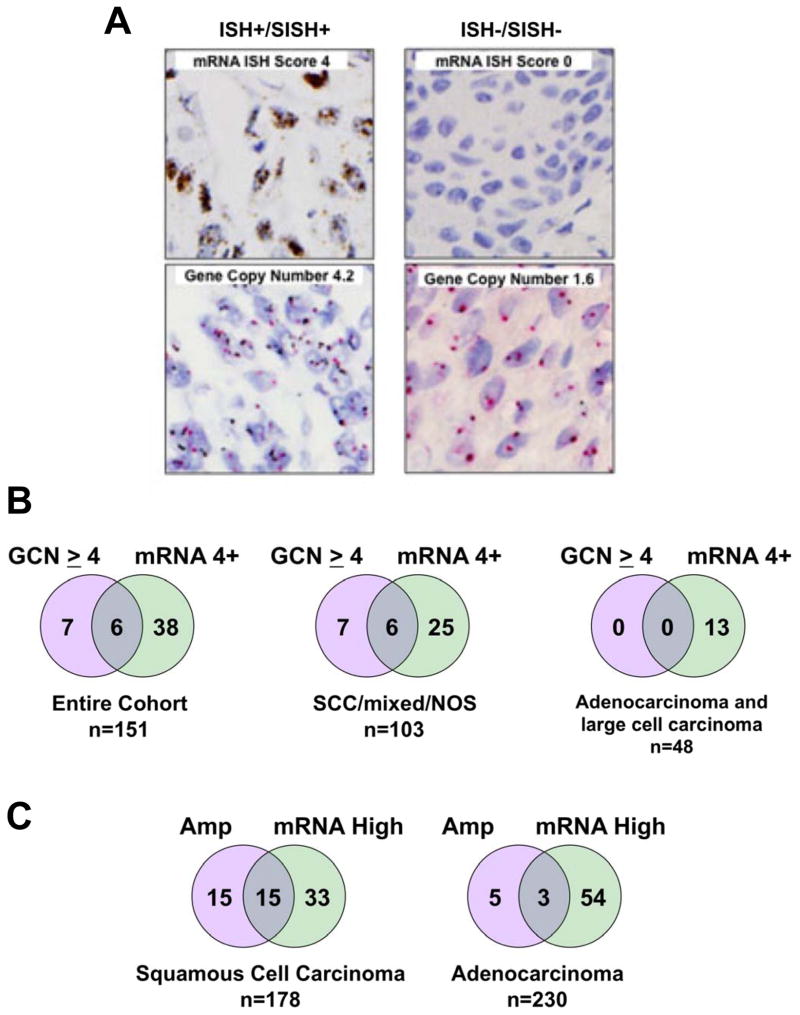
Of the primary tumors, 151 were assayed for both FGFR1 GCN and mRNA. Figure 4A shows SISH and ISH staining on a lung tumor exhibiting increased FGFR1 GCN and high mRNA as well as a tumor negative for both markers. In Figure 4B, the degree of overlap between positivity for FGFR1 GCN and mRNA is shown for all histologies as well as in subsets comprised of SCC/mixed/NOS or AC/LC. Of the 44 (29%) tumors with 4+ FGFR1 mRNA levels, only 6 were also positive for FGFR1 GCN ≥ 4. The findings indicate that use of FGFR1 GCN as a predictive biomarker in SCC for FGFR TKIs will select some lung tumors analogous to HCC95 cells that bear amplified FGFR1, but fail to express the mRNA and protein and are also insensitive to FGFR inhibitors. In addition, increased FGFR1 GCN is largely observed in lung SCC such that FGFR1 GCN alone will fail to detect a significant fraction of mRNA positive lung SCC tumors as well as 27% of the AC/LC subset. The Cancer Genome Atlas project has characterized a set of ACs (n=230) and SCCs (n=178) by SNP array and RNAseq analyses, thereby permitting an independent assessment of the FGFR1 GCN and mRNA expression in primary lung tumors. Among the lung SCCs, increased FGFR1 GCN was observed in 30 (17%) while only 8 of 230 ACs (~4%) were found to bear increased FGFR1 GCN (Figure 4C, Table S5). Of the 30 SCCs exhibiting increased FGFR1 GCN, only 15 (50%) also expressed FGFR1 mRNA. Likewise, only 3 of the 8 ACs (38%) bearing increased FGFR1 GCN highly expressed FGFR1 mRNA. Overall, 48 of 178 (27%) SCCs and 57 of 230 (25%) ACs expressed elevated FGFR1 mRNA, validating the lung tumor cohort findings.
Figure 4. Overlap of FGFR1 gene amplification and FGFR1 mRNA expression in primary lung tumors.
A, FGFR1 GCN (SISH) and mRNA (ISH) staining of a representative double-positive tumor and a tumor negative for both markers is shown. B, The overlap in positivity for FGFR1 GCN (>4) and mRNA (4+) in the surgical lung cancer cohort is shown for the entire cohort as well as for the SCC/mixed histology/not otherwise specified (NOS) or the ACs and LCCs. C, Lung SCC and ACs from TCGA cohorts were queried for the overlap between FGFR1 amplification as specified in the extracted datasets and high FGFR1 mRNA (≥ mean RNAseq expression value in each dataset).
Co-expression of FGF2 and/or FGF9 in FGFR1 mRNA positive lung cancers
Among the 14 lung cancer cell lines with ponatinib IC50s ≤ 50 nM, 12 expressed FGFR1 mRNA at a level ≥ to the mean value among the 58 cell lines determined by quantitative RT-PCR (Table S1). By this criterion, H1048 and H661 cells were defined as negative for FGFR1 mRNA expression, although H661 cells are positive for FGFR1 protein (Table S1) and H1048 cells exhibit detectable FGFR1 protein as well (Figure S1). Among the 12 FGFR1 mRNA positive cell lines, 10 also co-express significant levels of FGF2 and/or FGF9 (Figure S4). H1581 cells are negative for both of these ligands (Table S1), but highly express FGF10 and FGF20 (Table S2). Our data (Figure S3) and published findings (20) support the requirement for autocrine FGFs in these lung cancer cell lines, suggesting that co-expression of FGFs and FGFR1 may present a more accurate biomarker of FGFR1 dependency in lung tumors. When the overlap of FGF2 and FGF9 mRNA expression with FGFR1 mRNA was queried in TCGA RNAseq data, 28/48 (58%) of lung SCCs and 34/57 (60%) of ACs were also positive for FGF2 and/or FGF9 (Figure S7, Table S5). In total, FGFR1 and ligand double-positive tumors accounted for ~15% of all lung SCC and ACs.
Overlap of FGFR1 mRNA expression and oncogene mutations
No EGFR mutated cell lines were included in the cell line panel, but we have previously reported the insensitivity of EGFR mutant lung cancer cell lines to FGFR TKIs and their lack of FGFR1 expression (6, 11). Of the 17 EGFR mutant lung cancers in the lung TCGA data (15 ACs, 2 SCCs), 2 ACs were positive for FGFR1 mRNA (Table S5) and none of the 3 ALK fusion positive adenocarcinomas were positive for FGFR1 mRNA. The AC-derived cell lines, H1734 and SW1573, bear KRAS mutations, and the SCLC line, H1048, bears mutated PIK3CA, yet all are sensitive to ponatinib (Figure 1B, Table S1), suggesting that mutations in these oncogenes may not be exclusive of FGFR TKI sensitivity. We queried TCGA data for the overlap of FGFR1 mRNA positivity and KRAS and PIK3CA mutations (Table S5). Overall, 1 lung SCC and 35 ACs bore defined gain-of-function mutations in KRAS and 11 (31%) of these were positive for FGFR1 mRNA. Similarly, 28 SCCs and 5 ACs were mutated for PIK3CA and 7 (21%) of these also expressed FGFR1 mRNA. Thus, FGFR1 mRNA positivity is largely non-overlapping with EGFR and ALK oncogenes, but does overlap with KRAS and PIK3CA mutations.
DISCUSSION
Herein, we present the novel finding that elevated FGFR1 mRNA and/or protein expression occurs frequently in lung cancer cell lines and primary lung tumors and is often independent of increased FGFR1 GCN. Thus, FGFR1 mRNA and protein expression predict ponatinib sensitivity in lung cancer cell lines more accurately than GCN. The data support the hypothesis that ~ 20% of all lung cancers, regardless of histology, may be FGFR1 dependent and thereby ponatinib sensitive. If confirmed by clinical testing, the FGFR1-driven subset of lung cancers would be equal in size to the combined fraction of lung tumors driven by oncogenic EGFR and ALK.
Lung SCCs are presently the highlighted tumors in which to target FGFR1 due to the frequency of increased FGFR1 GCN in this histological subset (1–3). A prediction based on our findings is that FGFR1 GCN will identify a significant fraction of lung SCCs that fail to express FGFR1 mRNA or protein and are therefore insensitive to FGFR TKIs. HCC95 cells with high FGFR1 GCN, little or no FGFR1 expression and insensitivity to ponatinib serve as an example (Figure 1). There is published precedent for PDGFRA gene amplification not being associated with increased expression (13, 14). Our findings in resected lung SCCs and recent TCGA data indicate that ~50% of lung SCCs with increased FGFR1 GCN express high FGFR1 mRNA (Figure 4). In support of this prediction, only 3 of 6 patient-derived primary lung SCC xenografts exhibiting increased FGFR1 GCN exhibited shrinkage in response to AZD4547 treatment, although TKI response associated closely with high FGFR1 protein expression (5). The ongoing use of FGFR1 GCN, alone, as a biomarker in clinical trials of AZD4547 (NCT00979134) and BGJ398 (NCT01004224) is also predicted to overlook FGFR1-dependent lung ACs where increased FGFR1 GCN is infrequent (2) and more than half of FGFR1-dependent lung SCCs in which increased FGFR1 GCN is not observed. While increased FGFR1 GCN is also observed in ~6% of primary SCLC (22), our cell line studies indicate that increased FGFR1 expression and dependence will also occur independently of increased GCN in SCLC as well.
A study defining co-expressed FGFs as autocrine factors in FGFR1-dependent lung cancer cell lines (20) is consistent with our results. Because FGFR1 does not exhibit frequent somatic mutations in lung cancer cell lines (CCLE database) or in primary lung cancers (22–24), we propose that co-expression of FGFR1 and one or more ligands represent the oncogenic driving event in these tumors. Our previous publication (6) and the present findings indicate that FGF2 and FGF9 are the most common ligands involved in FGFR1 autocrine signaling. Overall, 10 of the 14 ponatinib-sensitive lung cancer cell lines expressed FGF2 or FGF9 mRNA. Among the 4 sensitive cell lines that were “negative” for FGF2 or FGF9, 2 cell lines highly expressed alternative FGFs, FGF7 (H1048) or FGF10 and FGF20 (H1581). Moreover, H1581 cell growth is inhibited by the ligand trap, FP-1039 (see Results and (20)). Inspection of lung SCC and AC TCGA RNAseq datasets reveals significant (SCC, 58%; AC, 60%), but not complete overlap of high FGFR1 mRNA expression and FGF2 and/or FGF9 mRNA expression (Figure S7). While autocrine FGFs may be enriched in cancer cell lines grown in vitro, it is likely that paracrine action of FGFs from the tumor microenvironment may serve as a distinct means to activate cancer cell-expressed FGFR1 in situ. If, in fact, co-expression of FGF2 or FGF9 is a requirement for FGFR1 dependency and ponatinib sensitivity, FGFR1 and ligand positive double positivity may represent a more accurate biomarker of FGFR1 dependent lung cancers. Importantly, TCGA lung cancers that satisfy this criterion still represent ~15% of the total (Figure S7), a frequency similar to that for EGFR, ALK, ROS1 and RET-driven lung cancers combined. The ability of co-expressed FGF2 or FGF9 to strengthen FGFR1 mRNA as a biomarker in our ponatinib clinical trial (described below) will be explored retrospectively.
Our findings indicate a partial overlap of FGFR1 dependency with distinct oncogene drivers. While no lung cancer cell lines bearing EGFR mutations or other re-arranged RTK drivers (ALK, ROS1, RET) were included in our panel of 58 cell lines, we have previously tested many of the known EGFR mutant lung cancer cell lines and found no overlap with FGFR1 dependence except following acquisition of resistance to EGFR-specific TKIs (6, 11). TCGA data support a general lack of overlap between EGFR and ALK oncogenes and FGFR1 mRNA expression (Table S5). By contrast, mutations in PIK3CA and KRAS were frequent in the panel of cell lines (see Figure 1B) and partially overlapped with ponatinib sensitivity. Inspection of the TCGA datasets for lung SCC and AC identifies clear overlap between high FGFR1 mRNA expression and the presence of KRAS or PIK3CA mutations (Table S5). Thus, unlike the mutual exclusivity of EGFR mutations and KRAS, FGFR1 dependency may overlap to a certain extent with PIK3CA and KRAS mutations and their presence will not be a basis for exclusion of such lung cancer patients from our clinical trial of ponatinib in lung cancer. Singh et al (25) reported that lung cancer cell lines bearing mutated KRAS can be defined as dependent or independent of this oncogene for growth and survival. It is noteworthy that KRAS-mutant SW1573 cells (identified as ponatinib-sensitive in our study) were classified as KRAS-independent. It seems plausible that autocrine FGFR1 activation has supplanted KRAS as the driving event in this particular lung cancer cell line.
When these issues are taken into consideration, it will be interesting to observe the response rates in the ongoing AZD4547 and BGJ398 trials relative to a trial now opened at our institution entitled “A Phase II Study of Ponatinib in Cohorts of Patients With Lung Cancer Preselected Using Different Candidate Predictive Biomarkers” (NCT01935336). In this trial, lung cancer patients of all histologies are enrolled for ponatinib treatment based on tumor positivity for FGFR1 mRNA assessed by ISH (4+), GCN by SISH ≥ 4 or positivity for both FGFR1 mRNA and GCN. The presence of KRAS or PIK3CA mutations will not exclude patients from enrollment. The goal is to test the hypothesis that FGFR1 mRNA expression levels will identify greater numbers of ponatinib-sensitive lung cancer patients in a more accurate fashion than FGFR1 GCN, alone.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
While only squamous cell lung cancer patients are being tested and enrolled in FGFR TKI clinical trials based on elevated FGFR1 gene copy number, our findings demonstrate that FGFR1 mRNA expression is a more biologically relevant marker of FGFR1 TKI sensitivity in lung cancers of all histologies including squamous, adenocarcinoma, and small cell lung cancer. The findings provide support for a clinical trial with an FGFR TKI in lung cancer patients enrolled by FGFR1 mRNA positivity and call for a reassessment of the use of FGFR1 gene copy number as a sole biomarker in ongoing FGFR TKI trials.
Acknowledgments
The authors acknowledge technical contributions of Kim Ellison (Hirsch lab) for staining and preparation of slides and publically available Cancer Genome Atlas data from lung adenocarcinomas and squamous cell carcinomas.
Grant Support
The studies were supported by the NIH (CA127105, Lung SPORE P50 CA58187, UC Cancer Center Support Grant P30 CA046934) and a research contract from ARIAD Pharmaceuticals to LH.
Footnotes
Disclosure of Potential Conflicts of Interest: L. Heasley is the recipient of a research contract from ARIAD Pharmaceuticals, Inc.
Author’s Contributions
Conception and design: L. Heasley, F. Hirsch, D.R. Camidge
Development of methodology: D. Boehm, S. Perner, F. Hirsch, M. Wynes
Acquisition of data: T. Hinz, L. Marek, M. Martini, M. Wynes, A.C. Tan, K. Ware
Analysis and interpretation of data: L. Heasley, Dexiang Gao, M. Wynes, M. Edwards, A.C. Tan
Writing, review and/or revision of the manuscript: L. Heasley, S. Perner, F. Hirsch, M. Wynes, T. Hinz, L. Marek, D.R. Camidge, P. Bunn, B. Helfrich, D. Gao, J. Jassem, R. Dziadziuszko
Technical or material support: D. Boehm, B. Helfrich, P. Bunn, J. Jassem, R. Dziadziuszko, S. Wojtylak, A. Sejda, J. Gozgit
References
- 1.Dutt A, Ramos AH, Hammerman PS, Mermel C, Cho J, Sharifnia T, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One. 2011;6:e20351. doi: 10.1371/journal.pone.0020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goke F, Franzen A, Menon R, Goltz D, Kirsten R, Boehm D, et al. Rationale for treatment of metastatic squamous cell carcinoma of the lung using fibroblast growth factor receptor inhibitors. Chest. 2012;142:1020–6. doi: 10.1378/chest.11-2943. [DOI] [PubMed] [Google Scholar]
- 4.Gozgit JM, Wong MJ, Moran L, Wardwell S, Mohemmad QK, Narasimhan NI, et al. Ponatinib (AP24534), a multitargeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Mol Cancer Ther. 2012;11:690–9. doi: 10.1158/1535-7163.MCT-11-0450. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, Zhang L, Su X, Li M, Xie L, Malchers F, et al. Translating the therapeutic potential of AZD4547 in FGFR1-amplified non-small cell lung cancer through the use of patient-derived tumor xenograft models. Clin Cancer Res. 2012;18:6658–67. doi: 10.1158/1078-0432.CCR-12-2694. [DOI] [PubMed] [Google Scholar]
- 6.Marek L, Ware KE, Fritzsche A, Hercule P, Helton WR, Smith JE, et al. Fibroblast growth factor (FGF) and FGF receptor-mediated autocrine signaling in non-small-cell lung cancer cells. Mol Pharmacol. 2009;75:196–207. doi: 10.1124/mol.108.049544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marshall ME, Hinz TK, Kono SA, Singleton KR, Bichon B, Ware KE, et al. Fibroblast growth factor receptors are components of autocrine signaling networks in head and neck squamous cell carcinoma cells. Clin Cancer Res. 2011;17:5016–25. doi: 10.1158/1078-0432.CCR-11-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soh J, Okumura N, Lockwood WW, Yamamoto H, Shigematsu H, Zhang W, et al. Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS One. 2009;4:e7464. doi: 10.1371/journal.pone.0007464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dziadziuszko R, Wynes MW, Singh S, Asuncion BR, Ranger-Moore J, Konopa K, et al. Correlation between MET gene copy number by silver in situ hybridization and protein expression by immunohistochemistry in non-small cell lung cancer. J Thorac Oncol. 2012;7:340–7. doi: 10.1097/JTO.0b013e318240ca0d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Badzio A, Wynes MW, Dziadziuszko R, Merrick DT, Pardo M, Rzyman W, et al. Increased insulin-like growth factor 1 receptor protein expression and gene copy number in small cell lung cancer. J Thorac Oncol. 2010;5:1905–11. doi: 10.1097/JTO.0b013e3181f38f57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ware KE, Hinz TK, Kleczko E, Singleton KR, Marek LA, Helfrich BA, et al. A mechanism of resistance to gefitinib mediated by cellular reprogramming and the acquisition of an FGF2-FGFR1 autocrine growth loop. Oncogenesis. 2013;2:e39. doi: 10.1038/oncsis.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, Wang F, et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009;16:401–12. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDermott U, Ames RY, Iafrate AJ, Maheswaran S, Stubbs H, Greninger P, et al. Ligand-dependent platelet-derived growth factor receptor (PDGFR)-alpha activation sensitizes rare lung cancer and sarcoma cells to PDGFR kinase inhibitors. Cancer Res. 2009;69:3937–46. doi: 10.1158/0008-5472.CAN-08-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramos AH, Dutt A, Mermel C, Perner S, Cho J, Lafargue CJ, et al. Amplification of chromosomal segment 4q12 in non-small cell lung cancer. Cancer biology & therapy. 2009;8:2042–50. doi: 10.4161/cbt.8.21.9764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cortes JE, Kantarjian H, Shah NP, Bixby D, Mauro MJ, Flinn I, et al. Ponatinib in refractory Philadelphia chromosome-positive leukemias. N Engl J Med. 2012;367:2075–88. doi: 10.1056/NEJMoa1205127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren M, Hong M, Liu G, Wang H, Patel V, Biddinger P, et al. Novel FGFR inhibitor ponatinib suppresses the growth of non-small cell lung cancer cells overexpressing FGFR1. Oncol Rep. 2013;29:2181–90. doi: 10.3892/or.2013.2386. [DOI] [PubMed] [Google Scholar]
- 17.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–29. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 18.Malchers F, Dietlein F, Schottle J, Lu X, Nogova L, Albus K, et al. Cell-Autonomous and Non-Cell-Autonomous Mechanisms of Transformation by Amplified FGFR1 in Lung Cancer. Cancer Discov. 2014;4:246–57. doi: 10.1158/2159-8290.CD-13-0323. [DOI] [PubMed] [Google Scholar]
- 19.Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci U S A. 2005;102:9625–30. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harding TC, Long L, Palencia S, Zhang H, Sadra A, Hestir K, et al. Blockade of nonhormonal fibroblast growth factors by FP-1039 inhibits growth of multiple types of cancer. Sci Transl Med. 2013;5:178ra39. doi: 10.1126/scitranslmed.3005414. [DOI] [PubMed] [Google Scholar]
- 21.Belov AA, Mohammadi M. Molecular mechanisms of fibroblast growth factor signaling in physiology and pathology. Cold Spring Harbor perspectives in biology. 2013:5. doi: 10.1101/cshperspect.a015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–10. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Network CGAR. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.