Abstract
Insertional mutagenesis by retroviral vectors has led to the discovery of many oncogenes associated with leukemia. Here we investigated the role of HOXC6, identified by proximal retrovirus insertion a large animal stem cell gene therapy study, for a potential involvement in hematopoietic stem cell activity and hematopoietic fate decision. HOXC6 was overexpressed in the murine bone marrow transplantation model and tested in a competitive repopulation assay in comparison to the known hematopoietic stem cell expansion factor, HOXB4. We have identified HOXC6 as a factor that enhances competitive repopulation capacity in vivo and colony formation in vitro. Ectopic HOXC6 expression also induced strong myeloid differentiation and expansion of granulocyte-macrophage progenitors/common myeloid progenitors (GMPs/CMPs) in vivo, resulting in myeloid malignancies with low penetrance (3 out of 17 mice), likely in collaboration with Meis1 due to a provirus integration mapped to the 3' region in the malignant clone. We characterized the molecular basis of HOXC6-induced myeloid differentiation and malignant cell transformation with complementary DNA microarray analysis. Overexpression of HOXC6 induced a gene expression signature similar to several acute myeloid leukemia subtypes when compared to normal GMPs/CMPs. These results demonstrate HOXC6 as a regulator in hematopoiesis and its involvement in malignant transformation.
Keywords: HOXC6, hematopoiesis, leukemogenesis
Introduction
Retroviral insertional mutagenesis, the activation or inactivation of a gene by an integrating retrovirus, is a consequence of retrovirus-based gene therapy and also a powerful tool for the identification of genes that influence cell survival or cell fate decisions.
Gamma retroviral vectors integrate in a semi-random manner with a propensity to integrate in proximity of transcription start sites of actively transcribed genes [1]. If a vector with an intact long terminal repeat (LTR), harboring a strong enhancer, is used, the LTR-enhancer can stimulate the transcription of proximal genes. This effect has induced clonal dominance in multiple animal studies before [2] and was partially responsible for adverse events in gene therapy trials after proximal integration and activation of the proto-oncogenes Lim-domain-only 2 (LMO2) or Cyclin D2 (CCND2) [3].
Besides being a consequence of retroviral gene therapy, the study of retroviral integration sites has led to the discovery of many important oncogenes and cellular transcription factors (e.g. EVI1, PIM1, MEIS1, PU.1) and is still frequently used as a discovery tool [4]. Genes proximal to the retroviral integrations are often enriched for genes that increase self-renewal [5].
In our study we took advantage of the property that retroviral integrations can mark genes in pathways responsible for expansion and self-renewal of hematopoietic stem and progenitor cells (HSPCs) or mark important cell fate regulators that might play a role in normal and malignant hematopoiesis. We focused on a gene, HOXC6, identified proximally to a retrovirus integration site in bone marrow colony forming units (CFUs), years after gene-modified transplantation in a large animal model. Ectopic expression of HOXC6 was studied in vitro and in vivo for HSPC activity-enhancing effects and its influence on hematopoietic cell fate decision as shown before [6].
Homeobox (HOX) genes play an important role in the patterning of the embryo and take over essential functions in the adult organisms. In murine adult hematopoietic stem cells, 22 of 39 HOX genes are expressed, but tightly regulated during hematopoietic differentiation [7]. Ectopic expression of HOXB4, for example, leads to an expansion of CD34+ cells in vivo and in vitro, indicating the important role of HOXB4 as regulator of the self-renewal of HSCs [8,9]. Overexpression of other HOX genes had similar effects on hematopoietic stem cells; the ectopic expression of them, however, resulted in myeloid malignancies [10-12]. For example, HOXA10-overexpression leads to a selective expansion of megakaryocytes and leads to an AML in mice within 5-8 months [10]. HOXA9 does similarly when expressed together with Meis1 or when translocated to Nup98. In addition, Meis1 is essential for all MLL-rearranged leukemias [11] and for maintenance of the hematopoietic stem cell pool [13]. The role of many other HOX genes in normal or malignant hematopoiesis, however, remains to be elucidated. Here we describe HOXC6 as a regulator of early hematopoiesis and myeloid differentiation and describe its potential involvement in myeloid malignancies.
Materials and Methods
Integration site analysis
Integration site detection and genomic localization was carried out as previously described [14].
Retroviral vectors and virus production
A self-inactivating lentiviral vector with an internal spleen focus-forming virus (SFFV) promoter, which drives the candidate genes, was used. For detection of gene expression, the GFP reporter gene is co-expressed from an internal ribosomal entry site (IRES). Human cDNA of the following reference sequences were obtained from the Dana Farber/Harvard Cancer Center DNA Resource Core (http://plasmid.med.harvard.edu/PLASMID) (Boston, MA, USA): HOXC6 BC074844, HOXB4 BC049204, LPXN BC019035.2, ZFP91 BC051743, TRIM44 BC013166, SMAD7 BC074819, and EIF2S2 BC000934. All genes were PCR amplified with primers to include a 5’ AgeI and 3’ NdeI or MluI restriction enzyme site and cloned into the lentiviral pRRL.cPPT.SFFV.IRES.eGFP.PRE vector backbone. Virus production was performed as previously described [15].
Animals
All mice experiments and manipulations were conducted in accordance with and approved by the Institutional Animal Care and Use Committee (IACUC) of the Fred Hutchinson Cancer Research Center (FHCRC). Mice were housed at the FHCRC Animal Health Resources.
Two mouse strains were used for these experiments: C57BL/6J-CD45.1-Pep3b (CD45.1) mice as stem cell donors and C57BL/6J-CD45.2 (CD45.2) mice as recipients. Mice were obtained from Jackson Laboratory (Bar Harbor, ME, USA).
Hematopoietic stem cell purification and transduction
Bone marrow cells were harvested from CD45.1 mice from the femur, tibia and pelvis. Lineage-positive cells were depleted according to manufacturing instructions (BD Bioscience, San Jose, CA) Lineage-negative (Lin−) cells were stained after depletion with Streptavidine-PE-Cy7, cKit-APC and Sca-PE antibodies sorted for Lin−, cKit+ and Sca+ cells (LSK). Cells were plated on CH296 coated plates (Retronectin, Takara Bio Inc., Otsu, Shiga, Japan) with a density of 25,000 cells per 48-well plate. Cells were cultured in IMDM with 15% FCS and cytokines (mTPO, 20 ng/mL; mSCF, 10 ng/mL; mIL-3, 5 ng/mL; and hIL-6, 10 ng/mL) as previously described [6]. Transduction was performed 16 and 24 hours after plating. Cells were cultured for 6 days; half of the medium was exchanged every second day. The prolonged culture in the given cytokine conditions has been shown to decrease stem cell potential in unmodified cells, which led to an increased signal-to-noise ratio for detection of stem cell activity influenced by the associated genes [6].
Transplantation
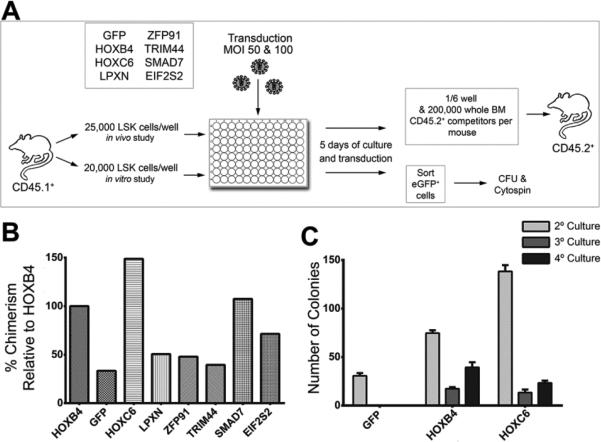
Recipient mice were lethally irradiated (1050 cGy) on the morning of transplantation and received 1/6 of the progeny of the 25,000 transduced cells along with 200,000 whole bone marrow competitor cells from C57Bl/6J-CD45.2 mice. Schematic overview of the transplantation setting is shown in Figure 1A.
Figure 1. Experimental design for screening candidate proto-oncogenes.
(A) Schematic overview of the experimental design used to test genes identified as common insertions sites for their influence on hematopoietic progenitor activity. (B) Results of the competitive repopulation assays. Shown is BM chimerism after 18 weeks normalized to the chimerism induced by HOXB4 expression. (C) Replanting frequency of transduced and sorted LSK cells in CFU assays. Five hundred cells per ml MethoCult were plated in all three rounds of replating (n=3).
In vitro colony-forming assays
Lin− cells from C57Bl/6 mice were transduced as described above. Forty-eight hours post-transduction, GFP-positive cells were sorted by fluorescent-activated cell-sorting (FACS-Aria, BD Bioscience, San Jose, CA, USA). Myeloid colony forming assays were performed in methylcellulose-based medium (MethoCult GF M3434, STEMCELL Technologies, Vancouver, British Columbia, Canada). Five hundred cells were plated per ml MethoCult. Plates were incubated at 37°C for seven days and colonies counted. For replanting, colonies were washed, counted and replated at 500 cells per ml.
Flow cytometry analysis of hematopoiesis
Mouse peripheral blood (PB) was evaluated at weeks 6, 12 and 16 after transplantation. PB, bone marrow (BM) and spleen (SP) cells were analyzed at week 18 or when mice were symptomatic of declining health. Cells were stained for CD45.1, CD45R, CD11b (all from eBioscience, San Diego, CA, USA), CD3ε, and GR-1 (BD Bioscience, San Jose, CA). BM cells were stained also for Sca1 (BD Bioscience, San Jose, CA, USA), cKit, CD34 and CD16/32 (eBioscience, San Diego, CA, USA).
B-cell differentiation
Lin− cells were transduced two times with a multiplicity of infection (MOI) of 10 and cultivated in alpha-MEM with 20% fetal bovine serum (FBS), 1% penicillin / streptomycin (P/S), and supplemented with 100 ng/ml mouse stem cell factor (mSCF), 25 ng/ml mouse Flt3L, 12.5 ng/ml interleukin (IL) 6 and 25 ng/ml IL-7 (all from PeproTech, Rocky Hill, NJ, USA). Four days after transduction, cells were sorted for GFP expression and 200,000 cells were co-cultured with OP9 stromal cells (American Type Culture Collection (ATCC), Manassas, VA). Cytokines were supplemented to the medium as follows: mFlt3L (10 ng/ml) and IL-7 (10 ng/ml). In 4-day intervals, cells were transferred to fresh OP9 stromal cells, and B-cell differentiation was assessed by flow cytometry for CD45R and CD19.
Histopathological evaluation
All necropsies were fixed in formalin solution for at least 24 hours and embedded in paraffin wax after decalcification of the BM specimens by ethylene-diamine tetra-acetic acid. From all necropsies, 3 μm thick sections were cut and stained with hematoxylin and eosin (H&E). Blood smears were also stained with H&E.
Microarray
GFP-positive GMP and CMP cells were isolated by FACS sorting from mice. Six weeks after transplantation, bone marrow was collected and stained as described above and sorted for GFP-positive cells and for the myeloid progenitor (GMP/CMP) population. RNA from sorted cells was isolated with an RNeasy Micro Kit (Qiagen, Hilden, Germany), and RNA amplification and cDNA labelling was performed with an Ovation PicoSL WTA System 2 (NuGen, San Carlos, CA, USA). For RNA expression analysis, a MouseWG-6 v2 Expression BeadChip (Illumina, San Diego, CA, USA) was used. Microarray data was analysed using GenePattern Package [16].
Quantitative (q)RT-PCR expression studies
For gene expression profiling, cells were isolated from the peripheral blood and bone marrow of untransplanted mice. RNA isolation was performed with an RNerasy Micro Kit (Qiagen, Hilden, Germany) according to the manufacturing guide. cDNA synthesis was performed with a high-capacity cDNA Kit (Applied Biosystem, Foster City, CA, USA). The HOXC6, Meis1 and Gapdh Quantitec primers were used (Qiagen, Hilden, Germany) in combination with the Fast Cyber Green Master Mix (Applied Biosystem, Foster City, CA, USA), and the cycling conditions were as follows: 50°C for 2 minutes, followed by 95°C for 10 minutes, then 40 cycles of 95°C for 15 seconds and 60°C for 1 minute.
Results
Ectopic expression of HOXC6 increases HSPC activity in vitro and in vivo
As part of ongoing retroviral integration site analyses in our large animal model of HSC gene therapy, we identified several interesting genes located close to retroviral integration sites in long-term repopulating cells (Figure 1A). Common integration sites detected were ranked according to following criteria: 1) the clones showed up late after transplantation indicating integration in long-term engrafting cells, 2) genes were identified in different species, and 3) whether several integrations were found in close proximity indicating clusters of integration. For an initial testing, genes were over-expressed in HSPCs (Lineage-negative, Sca1-positive, cKit-positive cells) and the cells were used in a competitive repopulation assay as described above [6]. We included a group with GFP-transduced cells only as a negative and HOXB4 as positive control [9]. As expected, HOXB4-overexpressing cells resulted in increased donor chimerism compared to the control GFP-transduced cells, but HOXC6 resulted in an even higher donor chimerism (10.5% GFP vs. 35.7% HOXB4, p=0.005; 53.0% HOXC6 vs. 35.74% HOXB4, p=0.03) (Figure 1B). In addition, CFU assays showed that HOXC6-expressing cells formed more CFUs than cells expressing HOXB4 in primary CFU plating (Figure 1C) and that ectopic HOXC6 expression allowed for similar serial replating results of CFUs when compared to HOXB4. GFP-transduced cells yielded no CFUs upon replating (Figure 1C). Both in vitro and in vivo data indicated that HOXC6 expression induced sustained self-renewal in HSPCs. These results clearly indicated an impact of HOXC6 on HSC activity.
HOXC6 induces myeloid differentiation and blocks B-cell development
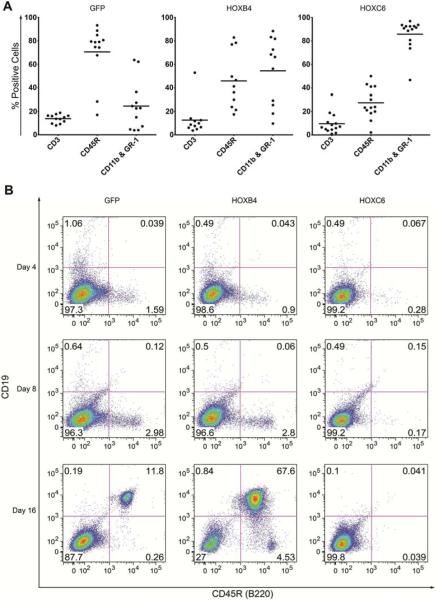
Based on this initial screen, we further analyzed the role of HOXC6 in hematopoiesis. Peripheral blood of HOXC6 over-expressing mice was analyzed at weeks 6, 12 and 16 after transplantation. Contribution of gene-marked cells to the T-cell (CD3), B-cell (CD45R) and myeloid lineage (CD11b/Gr1) was analyzed (Figure 2A, week 16). Mice transplanted with HOXC6-expressing cells (detected by GFP) showed a differentiation bias into the myeloid lineage (Figure 2A). It was shown before that HOXB4 expression biases differentiation into the myeloid lineage [8], but HOXC6 expression induced a significantly (p=0.0013) stronger differentiation towards the myeloid lineage compared to HOXB4. Conversely, the number of B-cells of HOXC6-expressing cells were also significantly reduced (p=0.037) compared to HOXB4-expressing cells. The number of T-cells was not significantly different. Compared to mice that received GFP-only expressing cells, both cell types were also significantly deregulated (Figure 2A).
Figure 2. HOXC6 induces myeloid differentiation bias.
(A) Lineage distribution of cells positive for GFP (control), HOXB4, and HOXC6 16 weeks after bone marrow transplantation in the peripheral blood. Shown are the results from three independent transplantation experiments. (B) In vitro differentiation of GFP, HOXB4, and HOXC6 Lin– transduced and sorted cells on OP9 stroma into the B-cell lineage. Differentiation progression was analysed at days 4, 8 and 16 with a CD19 and CD45R (B220) double staining.
To further investigate the phenomenon of B-cell suppression in HOXC6-expressing cells, we performed in vitro B-cell differentiation by co-culture of hematopoietic progenitor cells with OP9 stroma cells. B-cell differentiation of co-cultured cells that either overexpressed GFP, HOXB4, or HOXC6 was analyzed on days 4, 8 and 16 of differentiation (Figure 2B). GFP- and HOXB4- transduced cells showed B220 expression from day 4 onwards. At day 16, GFP- and HOXB4- expressing cells showed cells double-positive for CD19 and B220. HOXC6 inhibited early (B220+) and late (CD19+) B-cell differentiation at all analyzed time points. These experiments confirmed that HOXC6 overexpression inhibits the differentiation into the B-cell lineage (Figure 2B).
HOXC6 overexpression in vivo expands a GMP/CMP-like population
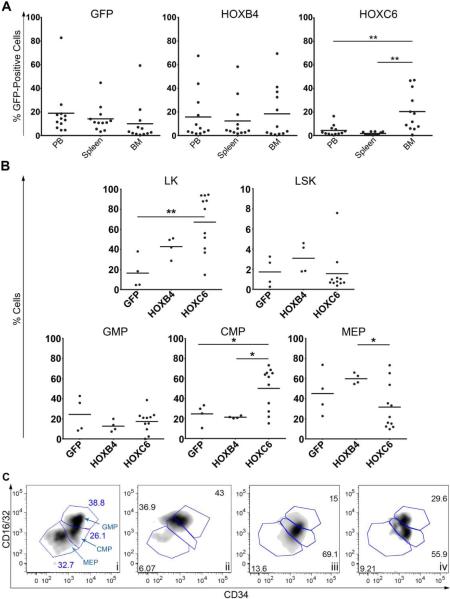
Peripheral blood, bone marrow and spleen cells of each mouse were analyzed by flow cytometry at week 18 after transplantation. Mice that developed leukemia were excluded from this analysis. The contribution of transgenic cells to each cell compartment was analyzed separately (Figure 3A) Cells over-expressing HOXC6 were retained in the bone marrow as indicated by increased percentages of gene-marked cells in the BM compared to spleen (p=0.0029) and peripheral blood (p=0.0079). Since retention to the bone marrow is a trait almost uniquely found in progenitor cells, our observation indicates that HOXC6-overexpression induces a differentiation block in the cells. To further investigate this observation, bone marrow cells were stained for the expression of hematopoietic progenitor markers. We focused on the compartment of hematopoietic stem cells (Lineage-negative/Kit-positive/Sca-positive (LSK)) as well as myeloid progenitors (Lineage-negative/Kit-positive (LK)) and their subsets as shown in Figure 3B. The myeloid progenitor fraction (LK) was significantly increased (p=0.0051) in the HOXC6 over-expressing cells in comparison to GFP-expressing cells (Figure 3B). Detailed analyses of the LK cell fraction revealed that physiological differentiation into GMPs, CMPs and megakaryocyteerythroid progenitors (MEPs) was disturbed by HOXC6 expression, and a significant shift towards the CMP-like cells was observed in comparison to GFP- and HOXB4-overexpressing cells (GMP/CMP/MEP flow cytometry GFP ctrl Figure 3Ci; disturbed GMP/CMP/MEP HOXC6, Figure 3C ii-iv).
Figure 3. HOXC6 overexpressing cells are retained in the bone marrow and show preferential differentiation toward myeloid progenitors.
(A) Gene marking of GFP; HOXB4 and HOXC6-transduced cells 18 weeks after transplantation in different hematopoietic organs (i.e. peripheral blood (PB), bone marrow (BM) and spleen). Shown are the results from three independent transplantation experiments; mice that developed leukemia were excluded. HOXC6-transduced cells were significantly less represented in the peripheral blood compared to the bone marrow (p=0.0079, t-test (Welch's corrected), unpaired, two-sided). The number of HOXC6-positive cells in the spleen was also significantly reduced compared to the BM (p-=0.0029; t test (Welch's’ corrected, unpaired, two-sided). (B) LK, LSK, GMP, CMP and MEP progenitor cell fractions from GFP-positive and HOXB4-positive cells 18 weeks after transplantation (mice that developed leukemia were excluded). The LK cell fraction in the HOXC6-transduced cells is significantly (p=0.0051, t test, unpaired, two-sided) enhanced in comparison to the GFP control group. The CMP cell fraction of HOXC6-transduced cells is significantly higher compared to GFP (p=0.04, t test, unpaired, two-sided) and to HOXB4-overexpressing cells (p=0.011, t test (Welch's corrected, unpaired, two sided). The MEP cell fraction was significantly reduced in HOXC6 expressing cells compared to HOXB4 expressing cells (p=0.025, t test (Welch's corrected, unpaired, two-sided). (C) The LK fraction was used to identify the different myeloid progenitor GMP, MEP and CMP as shown for a GFP control in i. Screwed myeloid progenitor differentiation of HOXC6 positive LK cells is shown in plot ii-iv. Plots show LK cells fractions of three different mice with a physiological blood cell counts out of two independent experiments.
Leukemic transformation of HOXC6-expressing cells
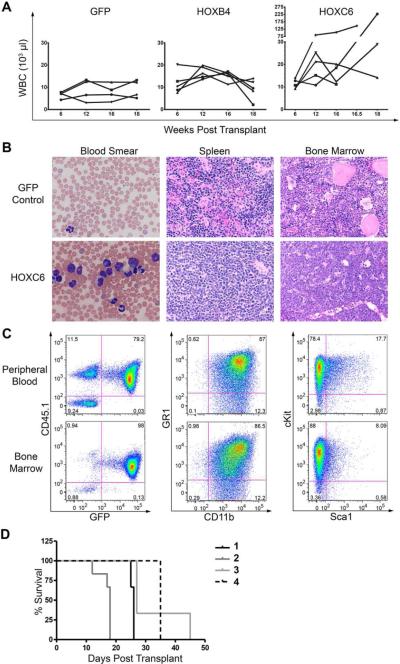
In three independent experiments, 3 out of 17 mice (approximately 20%) developed leukemia with high leukocytosis (WBC counts were 227, 146 and 29 × 103/μl; Figure 4A). Blood smears from mice with leukemia showed enhanced white blood cell numbers and immature myeloid cells (Figure 4B). The histology of all mice with leukemia revealed excessive myeloid progenitor cells in the bone marrow and also strong infiltration in the spleen with loss of normal spleen architecture (Figure 4B). Induction of disease with the same histological features in secondary animals supported leukemic phenotype (data not shown). Analysis of cells from BM and PB showed that they were donor derived (CD45.1+) and expressed high levels HOXC6 (indicated by GFP expression) (Figure 4C). Furthermore, the majority of cells were positive for GR1 and CD11b (~85%). The leukemic cells were further analysed for cKit and Sca1 expression as progenitor markers. The majority of cells expressed cKit both in peripheral blood or bone marrow (Figure 4C). The cell surface marker expression pattern of the cells revealed a myelomonocytic phenotype with partial maturation for the leukemic cells which is consistent with the histological analysis.
Figure 4. HOXC6 mice developed leukemia with an AML phenotype.
(A) White blood cell counts from GFP, HOXB4 and HOXC6 transplanted mice. (B) Blood smears from leukemic HOXC6 mice show increased white blood cell counts, left shifted myelopoiesis in the peripheral blood and cells with blast morphology (Hematoxylin-Eosin staining; x60). Spleen and bone marrow is overgrown with myeloid progenitor cells (Hematoxylin-Eosin staining; x40). (C) Flow cytometry analysis of the peripheral blood and bone marrow revealed that the cells of leukemic mice have an immature myeloid phenotype as indicated by high cKit expression. No phenotypic difference was detectable between cells from peripheral blood or bone marrow. (D) Survival of secondary transplants of mice from four different primary mice. One mouse with a myeloproliferative disorder diagnosed by histology (Number 4) showed evolution to leukemia in secondary transplant.
Leukemic phenotype was verified by secondary transplantations (Figure 4D). One additional primary mouse was diagnosed with a myeloproliferative disorder. This diagnosis was based on excessive expansion of mature myeloid lineage cells in the bone marrow but the absence of excessive blasts, which may have been indicative for overt leukemia (supplemental Figure 1; histological diagnosis of the bone marrow/liver). Secondary transplantation of cells from this mouse and re-initiation of disease in secondary animals indicates that the primary myeloproliferation disorder either transformed to overt leukemia or was a low-grade leukemia. All secondary mice became leukemic between days 12 and 45 post-transplantation in sublethally irradiated mice (Figure 4D). Retrovirus integration site data in mice that developed leukemia showed a dominant gene-modified clone with likely two provirus integration sites (Supplemental Table 1). The first lentiviral integration was close to Meis1, a key regulator of early hematopoietic progenitors that is also frequently involved in the pathogenesis of hematologic malignancies[17]. The second detected lentiviral integration site was close to Zfp606, which has no described role in hematopoiesis.
Characterization of HOXC6 overexpression GMP/CMP-like progenitor cells
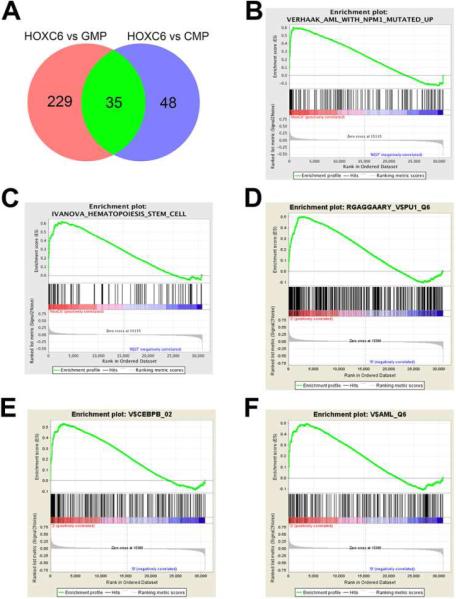
To elucidate the molecular mechanism underlying the blocked myeloid differentiation and progenitor expansion by HOXC6, mice transplanted with HOXC6 or GFP alone were sacrificed 6 weeks after transplantation. At 6 weeks post transplantation, full reconstitution of hematopoiesis is achieved and cells with blocked myeloid differentiation are expected. This early time point was also chosen to avoid heterogeneity from advanced malignant development and acquisition of additional abnormalities. Myeloid progenitor cells were FACS sorted (GMPs/CMPs from control mice and corresponding cells from HOXC6 mice) and a gene expression microarray was performed (normalized heatmap, see Supplemental Figure 2B). Principle component analysis showed independent grouping of GMPs, CMPs and HOXC6-expressing GMPs/CMPs (Supplemental Figure 2A). After adjusting for multiple testing, 229 genes and 48 genes were differentially expressed between HOXC6 and GMPs or HOXC6 and CMPs, respectively (Figure 5A; Supplemental Table 2: see list of genes). Out of these differentially regulated genes we could isolate a list of 35 genes that were differentially regulated between HOXC6 expressing early myeloid progenitors and normal GMPs and CMPs (p<0.05 after adjustment for multiple testing; gene list and fold change shown in Table 1). Since differentially expressed genes did not yield a clear explanation for the blocked differentiation, gene set enrichment analysis was applied to investigate deregulated pathways or common gene expression signatures. We identified that HOXC6 expression induced a gene expression signature similar to acute myeloid leukemia with nucleophosmin-1 mutations (nominal-enrichment score (NES) 2.07, FDR=0.0) (Figure 5B) [18]. In addition, other AML gene expression signatures were found enriched [19] in the HOXC6-overexpressing progenitors compared to their individual wild type counterparts and merged gene expression of CMPs and GMPs (NES 2.02, FDR=0.0; Supplemental Figure 2C). We could further identify an established hematopoietic stem cell signature to be enriched in HOXC6-expressing cells over GMPs/CMPs (NES 1.94, FDR=0.001; Figure 5CC) [20]. The enrichment of hematopoietic stem cell genes may explain the growth advantage observed in our experiments. In further Gene Set Enrichment Analysis (GSEAs) enrichment of target genes of the essential myeloid transcription factors PU.1, CEBPß and Runx1 (AML1) was identified (Figure 5 D-FF) [21]. No enrichment of the signature was detected between GMPs and CMPs, making a differentiation-induced phenomenon unlikely [21].
Figure 5. HOXC6 myeloid progenitor gene expression pattern is similar to nucleophosmin mutated AML. cDNA microarray analysis was performed on flow cytometry purified GMPs and CMPs from GFP transduced control mice and abnormal GMP/CMP-like progenitors from HOXC6 transduced mice.
(A) Differentially regulated genes between HOXC6 GMP/CMP-like progenitors and normal GMPs or CMPs were filtered and analysed (p<0.05 after adjustment for multiple testing). Data is presented as a Venn diagram. List of individual comparisons is given in Supplementary Table 1. (B) Gene expression data was analysed for gene set enrichment. HOXC6 overexpressing GMP/CMP-like progenitors presented with expression signature resembling NPM1c mutated AML (NES 2.07, FDR=0.0). (C) GSEA plots further isolate a formerly established stem cell signature by Ivanova et al. (NES 1.69, FDR=0.03) [20]. Furthermore GSEA plots evaluated HOXC6 driven changes for (D) PU.1 (NES 1.93, FDR=0.003), (E) CEBPβ (NES 1.92, FDR=0.003) and for (F) RUNX1 (AML-1) (NES 1.77, FDR=0.006). Those set of myeloid transcription factors confirm the findings of the myeloid differentiation into the myeloid lineage.
Table 1. Genes that are differentially regulated by HOXC6 compared to both normal CMPs and GMPs (p<0.05 after adjustment for multiple testing).
Fold changes in gene expression levels are shown.
| Overlap | HOXC6 vs GMP | HOXC6 vs CMP |
|---|---|---|
| GPNMB | 16.07 | 15.46 |
| PRSS34 | 11.93 | 31.43 |
| CPA3 | 10.75 | 8.1 |
| FRZB | 10.37 | 10.51 |
| GLYAT | 8.74 | 8.3 |
| ST8SIA6 | 7.39 | 7.26 |
| CDH1 | 6.37 | 21.86 |
| ST6GAL1 | 6.32 | 8.25 |
| FYB | 5.7 | 4.05 |
| SOCS2 | 4.84 | 4.92 |
| BC023892 | 4.8 | 5.49 |
| JEREMYREITER_BGALACTOSIDASE_40 | 4.54 | 4.17 |
| BTNL9 | 4.25 | 2.64 |
| SIGLEC5 | 3.94 | 3.44 |
| AI115600 | 3.79 | 3.86 |
| TCF7L2 | 3.51 | 2.79 |
| RBP1 | 2.99 | 2.67 |
| S100A6 | 2.75 | 3.51 |
| DNAJC10 | 2.57 | 4.63 |
| LOC675709 | 2.44 | 5.8 |
| VLDLR | 2.43 | 2.08 |
| SUSD5 | 2.34 | 2.32 |
| CD63 | 2.31 | 5 |
| LRRFIP2 | 2.3 | 2.62 |
| AHNAK | 2.3 | 2.71 |
| ITM2B | 1.94 | 1.9 |
| LYZ | 1.89 | 5.82 |
| SCHIP1 | 1.77 | 23.12 |
| SPATA13 | 1.67 | 2.87 |
| PILRA | 1.41 | 1.46 |
| RPL4 | 0.64 | 0.64 |
| MPO | 0.62 | 1.34 |
| EG666231 | 0.53 | 0.62 |
| MIF | 0.46 | 0.47 |
| CCND1 | 0.17 | 0.09 |
Gene expression analysis of the HOXC6-overexpressing aberrant myeloid progenitors thus showed induction of a stemness phenotype and AML-like traits [18-20] that were accompanied by signatures of myeloid transcription factors (Supplemental Figure 2B).
Finally, we sought to investigate the physiological role of HOXC6 in hematopoiesis. We performed quantitative real-time PCRs on RNA from different hematopoietic cell lineages to analyse physiological HOXC6 expression in hematopoiesis (Figure 6). HOXC6 expression was compared to Meis1, a gene mainly expressed in early hematopoietic progenitors and myeloid progenitors. There were very low levels of HOXC6 expression in mature cells (T-/B-/myeloid cells), but a strong increase of expression in myeloid progenitors (LK) and hematopoietic stem and progenitor cells (LSK).
Figure 6. HOXC6 Expression in Different Hematopoietic Cell Lineages.
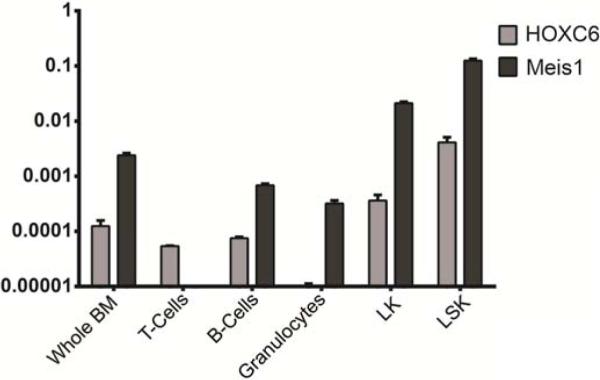
HOXC6 expression in flow cytometry purified hematopoietic cell populations. Expression was compared to Meis1 expression, a gene mainly expressed in the early hematopoietic progenitor compartment. Very low levels of HOXC6 expression were detected in mature cells (T-/B-/myeloid-cells) but an increase of expression in myeloid progenitors (LK) and hematopoietic stem and progenitor cells (LSK).
These data indicate that HOXC6 plays an important role in both AML and hematopoiesis that has not been described previously.
Discussion
In this study, we identified HOXC6 as a novel gene with an important role in hematopoietic cell fate. We demonstrate that HOXC6 affects self-renewal and cellular differentiation of hematopoietic progenitor cells.
HOXC6 overexpression in hematopoietic progenitor cells led to a higher CFU frequency compared to progenitor cells overexpressing HOXB4 or GFP, with HOXB4 shown before to be a positive regulator of HSC self-renewal [22]. Thus, we provide evidence that HOXC6 positively regulates the self-renewal or induces long-term proliferation of hematopoietic progenitor cells. HOXC6 overexpression further allowed serial replating of hematopoietic cells able to form CFUs, which supports the hypothesis that a self-renewal was induced in the transduced cells. In accordance with the in vitro data, HOXC6-expressing cells showed a significantly higher chimerism in mice compared to HOXB4-expressing LSK cells. Both HOXB4-expressing and HOXC6-expressing cells resulted in increased donor chimerism about 3–5-fold compared to GFP-expressing cells. The in vivo experiments further revealed that HOXC6 is a potent inducer of myeloid differentiation at the expense of lymphoid differentiation. The finding of a blocked B-lymphoid differentiation by ectopic expression of HOXC6 was further verified in OP9 co-culture experiments for induction of B-cell differentiation.
In experiments employing cDNA microarrays on myeloid progenitors from mice transplanted with cells expressing either HOXC6 or GFP, we could identify increased activity of the myeloid transcription factor Runx1 (AML1), PU.1, and CEBPβ by GSEA [21,23,24]. For PU.1 and CEBPβ they have been shown to synergistically act in granulopoisis [25] and our observed enrichment of target genes of these transcription factors may thus be a potential mechanism by which HOXC6 induces the strong bias towards myeloid differentiation. Although none of these transcription factors were significantly upregulated by itself, our data suggest that HOXC6 may induce the myeloid differentiation bias via these known regulators or their upstream regulators. A similar bias towards myeloid differentiation has been shown before for HOXB4 with unknown mechanisms [8]. Induction of downstream signatures of these myeloid transcription factors might also explain why HOXC6 blocks the lymphoid differentiation in vivo. More studies have to be performed to elucidate if HOXC6 directly regulates Pu.1 and Runx1. Studies showing HOXC6 to be overexpressed in non-Hodgkin lymphomas, malignant lymphoid tissue and lymphoid human cell lines support a potential role of HOXC6 in lymphocyte development [26-29].
Since ectopic expression of a transcription factor might induce non-physiological effects, we tested to see if HOXC6 is expressed in the BM. Our quantitative, real-time expression data confirmed a former report that HOXC6 is expressed in healthy bone morrow [30]. Our study further refines this expression signature by investigating HOXC6 expression in the different mature and immature cell lineages of the BM, and, therefore, we could show that HOXC6 is almost exclusively expressed in hematopoietic stem and progenitor cells and within the early myeloid progenitor compartment. These data, together with our functional studies, strongly argue for a role of HOXC6 in early hematopoietic lineage development.
Deeper analysis of our mouse data further revealed that HOXC6 not only induced myeloid differentiation but, also resulted in a partially-penetrant block of differentiation as indicated by accumulation of HOXC6 expressing myeloid progenitors in the BM and the decrease of HOXC6 expressing cells in the peripheral blood of the animals. Besides activation of a myeloid transcriptional signature as mentioned above, our microarray analysis further showed that HOXC6 myeloid progenitors have a gene expression signature highly correlated with different AMLs. In accordance with a leukemic- or pre-leukemic-phenotype of blocked differentiation and BM retention, self-renewal of these progenitors was upregulated in comparison to their physiologic phenotypical correlates (GMPs and CMPs) as shown in our microarray data. Our findings confirm previous studies which also indicated upregulation of HOXC6 in human AML and acute lymphoid leukemia (ALL) bone marrow [31]. Thus, we provide the first functional evidence for these previous studies that HOXC6 truly regulates myeloid differentiation, blocks differentiation, and induces self-renewal or partial transformation. Further evidence for this hypothesis was shown by the development of acute myeloid leukemia, or a myeloproliferative disorder in approximately 20% of mice that received HOXC6-expressing cells and the recapitulation of disease in a secondary mouse. Leukemic transformation was associated with provirus integration in Meis1 that has been previously identified as contributing to leukemic transformation in murine gene therapy studies [17].
Until now, HOXC6 was mainly known as a transcription factor during embryogenesis and neuronal differentiation [29]. Its role in the hematopoietic system was not well described. Our data suggest that HOXC6 is involved in the regulation of myeloid and B-lymphoid cell fate, but further studies will be necessary to clarify the molecular basis which we suggest to be a direct or indirect regulation of the transcription factors Runx1 and Pu.1. Regulation of Runx1 by HOXC6 has been shown before in non-hematopoietic tissue [32]. We could further show that HOXC6 is mainly expressed in early hematopoietic progenitors and myeloid progenitors. This finding was important since ectopic overexpression of a Homeobox gene could have caused non-physiological effects. Overall, our study provides functional evidence for a role of HOXC6 in the hematopoietic cell differentiation and during development of hematopoietic malignancies.
Supplementary Material
Acknowledgements
We thank Grace Choi for help with preparing the manuscript. We also thank the Fred Hutchinson Cancer Research Center Animal Health Resources staff for care of the mice. This work was supported by the Dr. Mildred Scheel Scholarship of the German Cancer Foundation (Deutsche Krebshilfe, Bonn, Germany) and in part by the National Institutes of Health (Bethesda, MD) grants HL053750, HL036444, HL084345, and DK056465. H.-P.K. is a Markey Molecular Medicine Investigator and the recipient of the José Carreras/E.D. Thomas Endowed Chair for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
H.-P.K designed experiments, analyzed data and wrote the manuscript. M.W. performed experiments, performed analyses and wrote the manuscript. J.K. performed experiments and analyses. D.H. performed analyses. X.-B.Z. performed analyses. V.N. performed experiments. B.B. performed experiments and analyses.
Conflicts of interest disclosure
The authors confirm no conflicts of interest.
References
- 1.Liu M, Li CL, Stamatoyannopoulos G, et al. Gammaretroviral vector integration occurs overwhelmingly within and near DNase hypersensitive sites. Hum Gene Ther. 2012;23:231–237. doi: 10.1089/hum.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kustikova OS, Schiedlmeier B, Brugman MH, et al. Cell-intrinsic and vector-related properties cooperate to determine the incidence and consequences of insertional mutagenesis. Mol Ther. 2009;17:1537–1547. doi: 10.1038/mt.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hacein-Bey-Abina S, Garrigue A, Wang GP, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baron BW, Anastasi J, Hyjek EM, et al. PIM1 gene cooperates with human BCL6 gene to promote the development of lymphomas. Proc Natl Acad Sci USA. 2012;109:5735–5739. doi: 10.1073/pnas.1201168109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kustikova OS, Geiger H, Li Z, et al. Retroviral vector insertion sites associated with dominant hematopoietic clones mark “stemness” pathways. Blood. 2007;109:1897–1907. doi: 10.1182/blood-2006-08-044156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deneault E, Cellot S, Faubert A, et al. A functional screen to identify novel effectors of hematopoietic stem cell activity. Cell. 2009;137:369–379. doi: 10.1016/j.cell.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sauvageau G, Lansdorp PM, Eaves CJ, et al. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc Natl Acad Sci USA. 1994;91:12223–12227. doi: 10.1073/pnas.91.25.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schiedlmeier B, Klump H, Will E, et al. High-level ectopic HOXB4 expression confers a profound in vivo competitive growth advantage on human cord blood CD34+ cells, but impairs lymphomyeloid differentiation. Blood. 2003;101:1759–1768. doi: 10.1182/blood-2002-03-0767. [DOI] [PubMed] [Google Scholar]
- 9.Sauvageau G, Thorsteinsdottir U, Eaves CJ, et al. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev. 1995;9:1753–1765. doi: 10.1101/gad.9.14.1753. [DOI] [PubMed] [Google Scholar]
- 10.Thorsteinsdottir U, Sauvageau G, Hough MR, et al. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol Cell Biol. 1997;17:495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pineault N, Buske C, Feuring-Buske M, et al. Induction of acute myeloid leukemia in mice by the human leukemia-specific fusion gene NUP98-HOXD13 in concert with Meis1. Blood. 2003;101:4529–4538. doi: 10.1182/blood-2002-08-2484. [DOI] [PubMed] [Google Scholar]
- 13.Unnisa Z, Clark JP, Roychoudhury J, et al. Meis1 preserves hematopoietic stem cells in mice by limiting oxidative stress. Blood. 2012;120:4973–4981. doi: 10.1182/blood-2012-06-435800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neff T, Beard BC, Peterson LJ, Anandakumar P, Thompson J, Kiem H-P. Polyclonal chemoprotection against temozolomide in a large-animal model of drug resistance gene therapy. Blood. 2005;105:997–1002. doi: 10.1182/blood-2004-08-3169. [DOI] [PubMed] [Google Scholar]
- 15.Beard BC, Trobridge GD, Ironside C, McCune JS, Adair JE, Kiem H-P. Efficient and stable MGMT-mediated selection of long-term repopulating stem cells in nonhuman primates. J Clin Invest. 2010;120:2345–2354. doi: 10.1172/JCI40767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura T, Largaespada DA, Shaughnessy JD, Jr., Jenkins NA, Copeland NG. Cooperative activation of Hoxa and Pbx1-related genes in murine myeloid leukaemias. Nat Genet. 1996;12:149–153. doi: 10.1038/ng0296-149. [DOI] [PubMed] [Google Scholar]
- 18.Verhaak RG, Goudswaard CS, van Putten W, et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005;106:3747–3754. doi: 10.1182/blood-2005-05-2168. [DOI] [PubMed] [Google Scholar]
- 19.Valk PJ, Verhaak RG, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 20.Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 21.Popernack PM, Truong LT, Kamphuis M, Henderson AJ. Ectopic expression of CCAAT/enhancer binding protein beta (C/EBPbeta) in long-term bone marrow cultures induces granulopoiesis and alters stromal cell function. J Hematother Stem Cell Res. 2001;10:631–642. doi: 10.1089/152581601753193841. [DOI] [PubMed] [Google Scholar]
- 22.Antonchuk J, Sauvageau G, Humphries RK. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell. 2002;109:39–45. doi: 10.1016/s0092-8674(02)00697-9. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka T, Tanaka K, Ogawa S, et al. An acute myeloid leukemia gene, AML1, regulates transcriptional activation and hemopoietic myeloid cell differentiation antagonistically by two alternative spliced forms. Leukemia. 1997;11(Suppl 3):299–302. [PubMed] [Google Scholar]
- 24.Nerlov C, Graf T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 1998;12:2403–2412. doi: 10.1101/gad.12.15.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ford AM, Bennett CA, Healy LE, Towatari M, Greaves MF, Enver T. Regulation of the myeloperoxidase enhancer binding proteins Pu1, C-EBP alpha, -beta, and -delta during granulocyte-lineage specification. Proc Natl Acad Sci USA. 1996;93:10838–10843. doi: 10.1073/pnas.93.20.10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bijl J, van Oostveen JW, Kreike M, et al. Expression of HOXC4, HOXC5, and HOXC6 in human lymphoid cell lines, leukemias, and benign and malignant lymphoid tissue. Blood. 1996;87:1737–1745. [PubMed] [Google Scholar]
- 27.Bijl JJ, Rieger E, van Oostveen JW, et al. HOXC4, HOXC5, and HOXC6 expression in primary cutaneous lymphoid lesions. High expression of HOXC5 in anaplastic large-cell lymphomas. Am J Pathol. 1997;151:1067–1074. [PMC free article] [PubMed] [Google Scholar]
- 28.Bijl JJ, van Oostveen JW, Walboomers JM, et al. HOXC4, HOXC5, and HOXC6 expression in non-Hodgkin's lymphoma: preferential expression of the HOXC5 gene in primary cutaneous anaplastic T-cell and oro-gastrointestinal tract mucosa-associated B-cell lymphomas. Blood. 1997;90:4116–4125. [PubMed] [Google Scholar]
- 29.Peljto M, Dasen JS, Mazzoni EO, Jessell TM, Wichterle H. Functional diversity of ESC-derived motor neuron subtypes revealed through intraspinal transplantation. Cell Stem Cell. 2010;7:355–366. doi: 10.1016/j.stem.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi Y, Hamada J, Murakawa K, et al. Expression profiles of 39 HOX genes in normal human adult organs and anaplastic thyroid cancer cell lines by quantitative real-time RT-PCR system. Exp Cell Res. 2004;293:144–153. doi: 10.1016/j.yexcr.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 31.Bijl JJ, van Oostveen JW, Walboomers JM, et al. Differentiation and cell-type-restricted expression of HOXC4, HOXC5 and HOXC6 in myeloid leukemias and normal myeloid cells. Leukemia. 1998;12:1724–1732. doi: 10.1038/sj.leu.2401106. [DOI] [PubMed] [Google Scholar]
- 32.McCabe CD, Spyropoulos DD, Martin D, Moreno CS. Genome-wide analysis of the homeobox C6 transcriptional network in prostate cancer. Cancer Res. 2008;68:1988–1996. doi: 10.1158/0008-5472.CAN-07-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.