Abstract
Human health risks of exposure to low dose ionizing radiation remain ambiguous and are the subject of intense debate. A wide variety of biological effects are induced after cellular exposure to ionizing radiation, but the underlying molecular mechanism(s) remain to be completely understood. We hypothesized that low dose c-radiation-induced effects are controlled by the modulation of micro RNA (miRNA) that participate in the control of gene expression at the posttranscriptional level and are involved in many cellular processes. We monitored the expression of several miRNA in human cells exposed to acute or chronic low doses of 10 cGy or a moderate dose of 400 cGy of 137Cs γ-rays. Dose, dose rate and time dependent differences in the relative expression of several miRNA were investigated. The expression patterns of many miRNA differed after exposure to either chronic or acute 10 cGy. The expression of miRNA let-7e, a negative regulator of RAS oncogene, and the c-MYC miRNA cluster were upregulated after 10 cGy chronic dose but were downregulated after 3 h of acute 10 cGy. The miR-21 was upregulated in chronic or acute low dose and moderate dose treated cells and its target genes hPDCD4, hPTEN, hSPRY2, and hTPM1 were found to be downregulated. These findings provide evidence that low dose and dose rate c-irradiation dictate the modulation of miRNA, which can result in a differential cellular response than occurs at high doses. This information will contribute to understanding the risks to human health after exposure to low dose radiation.
Keywords: Micro-RNA, Gene expression, Low dose ionizing radiation, Radiation effects
Introduction
The health risks of occupational or environmental exposure to low dose ionizing radiation (IR) in humans have not been fully understood [1, 2]. Several types of cellular responses to IR, such as the adaptive response or the bystander effect, suggest that low-dose radiation has different characteristics than high-dose radiation. Based on current evidence, it is becoming increasingly clear that the biological effects of IR are not linear. Different responses are induced at high doses than at low doses [3].
Non-targeted and delayed effects of low dose radiation exposure include the bystander effect, genomic instability, hormesis, adaptive responses, hypersensitivity and transgenerational responses [4, 5]. Radiation-induced genomic instability (RIGI) manifests itself by an increase in genomic alterations in the progeny of irradiated cells [6]. Chromosomal rearrangements, micronuclei formation, aneuploidy, delayed mutagenesis with different mutation spectra, gene amplification and cell death have been identified as RIGI [7]. On the other hand, radiation-induced adaptive responses protect against endogenous damage from normal metabolism and attenuate the damage due to a subsequent challenge dose of radiation [8, 9]. Adaptive responses can alter radiation-induced DNA damage, mutagenesis, the frequency of chromosomal aberrations, micronuclei and cell transformants [10–12]. Low-dose radiation exposures can also affect unirradiated neighboring cells. This phenomenon has been termed the bystander effect and has been mainly observed after exposure of cell cultures to low fluences of high linear energy transfer (LET) radiations. Sister-chromatid exchanges (SCEs) and mutations have been observed in bystander cells neighboring a-particle-irradiated cells. The stress-responsive protein p21Waf1 was induced in bystander cells within a 100-micron radius (corresponding to a set of approximately 30 cells) from an irradiated cell [13]. Both gap-junction intercellular communication (GJIC) and endogenous oxidative metabolism mediate ‘adaptive’ and ‘bystander’ effects [9]. While adaptive responses are thought to mitigate the harmful effects of IR, bystander effects have been suggested to amplify its consequences. The exact molecular steps by which radiation-induced adaptive and bystander effects are elicited have not been defined.
It has been suggested that perturbations in intracellular metabolic oxidation/reduction reactions contribute to the biological effects of radiation exposure. These processes include redox sensitive signaling pathways, transcription factor activation, gene expression, and metabolic activities that govern the formation of intracellular oxidants and reductants [14]. The goal of this study was to understand the molecular processes by which exposure to low dose IR may elicit biological effects. The purpose was to analyze micro RNA (miRNA) expression after exposure of normal human fibroblasts to 10 cGy of γ rays delivered as either an acute or a chronic dose. miRNA are an abundant class of small non-protein-coding single-stranded RNA of ~22 nucleotides that function as negative gene regulators. Recent studies have established that miRNA control a wide range of biological functions, including cellular proliferation, differentiation and apoptosis. miRNA can control hundreds of gene targets and are critical in cell development, differentiation and communication [15]. miRNA negatively regulate gene expression by means of mRNA destabilization, through the RNAi mechanism, or translation repression [16]. Most miRNAs do not cleave their mRNA targets as a mechanism of gene regulation, but rather bind to imperfect complementary sites within the 3′ untranslated regions (UTRs) of their mRNA targets. In this case, the target-gene repression occurs post-transcriptionally at the level of translation [17] resulting in reduced protein levels, while the mRNA levels remain unaffected [18]. A single miRNA has the capability to bind to hundreds of gene targets ranging from transcription factors, secreted factors, receptors, and transporters [18, 19]. The miRNAome in its entirety has the capacity to potentially control the expression of more than half of all human mRNAs.
We recently published on the impact of radiation dose, cellular sensitivity to radiation, and DNA repair capability of the cell on the modulation of miRNA in human cells exposed to low LET radiation [21, 22]. In this study, we asked whether the miRNA are modulated in low dose/ dose rate γirradiated cells. If so, are there differences or similarities in the miRNA responses? It is possible that the function of most miRNA is to alter gene expression to provide cells more flexibility and the ability to quickly respond to environmental changes. Based on the growing body of evidence suggesting the involvement of miRNA in diverse cellular processes, it is reasonable to hypothesize that radiation-induced effects could be influenced by alterations in miRNA expression. We argued that the stress induced by ionizing radiation can alter the expression of miRNA in a dose and dose rate dependent manner.
Materials and methods
Growth and maintenance of cells
AG1522 normal human skin fibroblasts were obtained from the Genetic Cell Repository at the Coriell Institute for Medical Research (Camden, NJ). Passage 10–11 cells were seeded at a density of ~1.5 × 105 cells/mL in Eagle's Minimal Essential Medium (EMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS). The cells were maintained in a 37°C humidified incubator in an atmosphere of 5% CO2 in air. The cells were grown in three-dimensional (3-D) architecture on carbon scaffold Cytomatrices™ (Cell Science Pte, Woburn, MA) coated with FNC solution (AthenaES, Baltimore, MD) consisting of fibronectin and collagen to facilitate cell attachment. The cytomatrices were maintained in six well plates. The Cytomatrix™ is a highly porous and biocompatible scaffold; it is organized as an array of continuously, interconnected dodecahedrons with no dead space. By growing cells in 3-D architecture, the intercellular communication via gap junctions is enhanced [12, 13], and cells do not show signs of differentiation when grown in 3-D after growth in 2-D for many generations. Their morphology does not change as observed by light microscopy examination, and their rate of growth is not altered. Growth of AG1522 cells on FNC-coated surfaces does not lead to their differentiation; however, other cells (e.g. WB-F344) may become larger and stop growing.
Ionizing radiation treatment
A patented 137Cs irradiator that delivers dose rates ranging from about 30 cGy/h down to less than 0.01 cGy/h in the low dose range has been designed [23, 24] and was used for this project. The Model 28-8 irradiator (J.L. Shepherd & Associates, San Fernando, CA) contains an 18 Ci 137Cs source and is fitted with a beam shaper and a computer controlled mercury attenuator system that is capable of either maintaining a constant dose rate or modulating the dose rate as a function of time. For the high dose exposure, a 137Cs source (J.L. Shepherd Mark I, San Fernando, CA) that delivers γ rays at 130 cGy/min was used.
As cellular radiation sensitivity changes at different phases of the cell cycle, the cells were synchronized in G0/G1. The cells were fed with medium containing 0.5% FCS at 24 h prior to irradiation. When irradiated, ~ 95% of the cells were in G0/G1 as determined by [3H]-thymidine uptake. Irradiation of 5 × 106 cells was performed at various dose rates. For chronic 10 cGy exposure,the dose rate was 0.3 cGy/h. Acute 10 cGy dose was delivered at 3 cGy/min and acute 400 cGy was delivered at 120 cGy/min. The irradiations were administered at 37°C in an atmosphere of 5% CO2 in air. The control cells did not receive any radiation and were mock irradiated. The 10 and 400 cGy acutely exposed cells were incubated at 37°C for 3 and 8 h prior to harvesting and isolating RNA. The low dose rate treated cells were harvested within 10 min after the completion of irradiation. The experiment was repeated for each dose in triplicate. Table 1 summarizes the radiation doses, dose rates, analysis time after irradiation, and the designation of doses as “chronic” or “acute”.
Table 1.
Radiation doses, dose rates and miRNA analysis time in AG1522 cells
| Dose designation | Dose (cGy) | Dose rate | Time needed to deliver the dose | Analysis time after irradiation |
|---|---|---|---|---|
| Chronic | 10 | 0.3 cGy/h | 33.33 h | Within 10 min |
| Acute | 10 | 3 cGy/min | 3.33 min | 3 h |
| Acute | 10 | 3 cGy/min | 3.33 min | 8 h |
| Acute | 400 | 120 cGy/min | 3.33 min | 3 h |
| Acute | 400 | 120 cGy/min | 3.33 min | 8 h |
RNA isolation
The total RNA and miRNA were isolated from the harvested cells. The control (mock irradiated) and irradiated cells were counted with a hemocytometer. Approximately 5 × 106 cells were pelleted by centrifugation at 1,500 rpm for 5 min, and washed with 1 mL Dulbecco's phosphate-buffered saline (PBS) without MgCl2 and CaCl2 (Invitrogen, Carlsbad, CA, USA). Small RNA less than 200 nucleotides were isolated from cells using the mirVana™ miRNA isolation kit following the enrichment procedure for small RNA recovery (Applied Biosystems; Foster City, CA, USA). Total RNA was isolated with RNAeasy™ kit (Qiagen, Valencia, CA, USA). The quantity and quality of the total RNA and miRNA was measured on the NanoDrop 2000 (Thermo Scientific; West Palm Beach, FL, USA) and by analysis on 2% agarose gels stained with ethidium bromide on BioSpectrum® Imaging System (UVP, Upland, CA).
miRNA and gene targets
The miRNA investigated in this study were hsa-let-7a, hsa-let-7b, hsa-let-7c, hsa-let-7d, hsa-let-7e, hsa-let-7f, hsa-let 7i, hsa-miR-142-3p, hsa-miR-142-5p, hsa-miR-143, hsa-miR-145, hsa-miR-155, hsa-miR-15a, hsa-miR-16, hsa-miR-17-3p, hsa-miR-17-5p, hsa-miR-18a, hsa-miR-19a, hsa-miR-19b, and hsa-miR-21. The RNA, U6 small nuclear (RNU6B) and small nucleolar RNA, C/D box 48 (RNU48) were used for normalization.
These miRNA genes were selected from the Sanger Center miRNA Registry at http://www.sanger.ac.uk/Software/Rfam/mirna/index.shtml. All TaqMan assays and endogenous controls for miRNA analysis and assays on demand for gene expression analysis were purchased from Applied Biosystems. Standard TaqMan assays have been designed with PrimerExpress software (Applied Biosystems). All RNA samples for miRNA analysis were normalized based on the TaqMan Gene Expression Assays for human endogenous RNU6B or RNU48 controls as we have previously described [21].
Assays-on-demand for programmed cell death factor 4 (hPDCD4), phosphatase and tensin homolog (hPTEN), sprouty homolog 2 (hSPRY2), tropomyosin 1 (hTPM1) RAS, and c-MYC genes were purchased from Applied Biosystems. RNA samples for gene expression analysis were normalized based on the TaqMan Gene Expression Assays for human endogenous hypoxanthine phosphor-ibosyltransferase (HPRT) gene as previously described [21].
Reverse transcription and cDNA synthesis reactions
Reverse transcriptase reactions on miRNA-enriched RNA fractions were set up using a commercial kit from Applied Biosystems. The reaction contained RNA samples, 50 nM stem loop reverse transcriptase (RT) primer, 1 × RT buffer, 0.25 mM each of dNTPs, 3.33 U/μL MultiScribe™ reverse transcriptase and 0.25 U/μL RNase inhibitor. The 15μL reactions were incubated in Techne TC-312 thermocycler (Burlington, NJ, USA) for 30 min at 16°C, 30 min at 42°C, 5 min at 85°C and then cooled at 4°C. All reverse transcriptase reactions, including no-template controls and RT minus controls, were run in duplicate.
For gene expression analysis, total RNA was treated with DNAse prior to reverse transcription in order to avoid genomic DNA contamination. The cDNA were generated from total RNA with random hexamer primers using a cDNA synthesis Kit from Applied Biosystems, following the recommendations of the manufacturer.
Quantitative real-time polymerase chain reaction (QPCR) and data analysis
QPCR was performed on an Applied Biosystems 7900HT Sequence Detection System by using a standard TaqMan PCR kit protocol. The 10μL PCR contained 0.67μL RT product, 19 TaqMan Universal PCR Master Mix, 0.2 μM TaqMan probe, 1.5 μM forward primer and 0.7 μM reverse primer. The reactions were incubated in a 384-well plate at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The relative expression values of cycle thresholds were calculated by using the comparative delta cycle threshold, ΔΔCT method [25] by normalization to the control miRNA RNU6B or RNU48 and to the control nonirradiated sample. The mock irradiated control sample was used as calibrator to calculate the relative expression and Log2 values. The threshold cycle (CT) is defined as the fractional cycle number at which the fluorescence passes the fixed threshold. The statistics and data analysis were performed with ABI prism and GraphPad Prism 5 software, both licensed to the University of Vermont. CT values of the 20-target miRNA in each cell line were statistically evaluated using a one-way t-test (p < 0.05). The experiments were repeated three times.
Results
Assessment of the expression of miRNA in low dose γ irradiated AG1522 cells
We investigated the expression of several miRNA in AG1522 cells exposed to low dose γ-rays delivered at different dose rates by quantitative real-time RT-PCR (QPCR). The AG1522 cells were exposed to acute 10 cGy (3 cGy/min) and harvested after 3 and 8 h for miRNA expression analysis. In parallel experiments, AG1522 cells were also irradiated at low dose rate (10 cGy, 0.3 cGy/h) and harvested for analyses within 10 min after the exposure. The miRNA expression was also monitored in AG1522 cells after an acute dose of 400 cGy delivered at 120 cGy/min, and the irradiated cells were harvested at 3 and 8 h following irradiation for miRNA expression studies. We chose to examine miRNA expression at 3 h when the repair activity and/or commitment for permanent arrest in the cell cycle occurred, and at 8 h when most of the DNA damage had been repaired [12, 13]. We monitored the expression of miRNA which had been shown to regulate a range of genes in diverse cellular pathways particularly in carcinogenesis [20]. Relative to the control unirradiated cells, several miRNA were modulated after 400 cGy dose or 10 cGy delivered acutely or chronically.
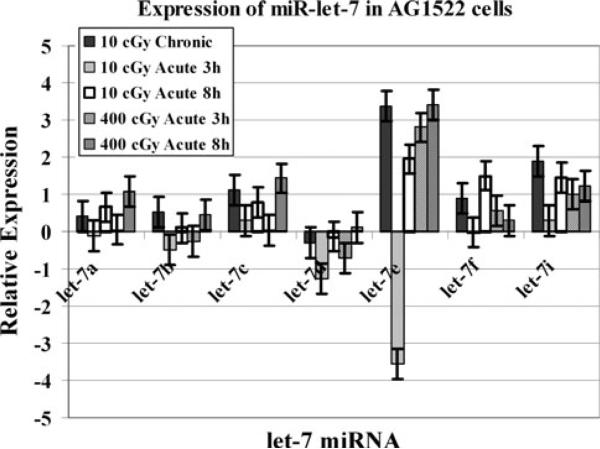
We first monitored the expression of seven miRNA belonging to the miR-let-7 family. AG1522 cells were irradiated with various doses from a 137Cs source. A time course experiment was conducted where irradiated cells were harvested at 3 and 8 h post-exposure for molecular analysis. The relative expression as determined by real-time QPCR for miR-let-7a, miR-let-7b, miR-let-7c, miR-let-7d, miR-let-7e, miR-let-7f, and miR-let-7i is shown in Fig. 1. Micro RNA miR-let-7e exhibited the highest degree of modulation in irradiated AG1522 cells. The miR-let-7e was found to be induced when examined in 400 cGy irradiated cells. The miR-let-7e was also upregulated in the cells that were treated with 10 cGy γradiation dose delivered at 0.3 cGy/h. When the 10 cGy dose was delivered at a dose rate of 3 cGy/min, the miR-let-7e was initially down regulated at 3 h but was induced at the 8 h time point. The dose rate dependent differences in the miR-let-7e expression were statistically significant (p < 0.05). The induction of miR-let-7c in 400 cGy acutely treated cells was significantly higher after 8 h of post irradiation as compared to the 3 h time point (p < 0.05).
Fig. 1.
The expression analysis of let-7 miRNA in AG1522 cells harvested for analyses at 3 or 8 h after exposure to acute 10 cGy (3 cGy/min) or 400 cGy (120 cGy/min), or within 10 min after chronic exposure to 10 cGy (0.3 cGy/h). The data are expressed as Log2. The error bars indicate the standard error of the mean (SEM) for three independent experiments
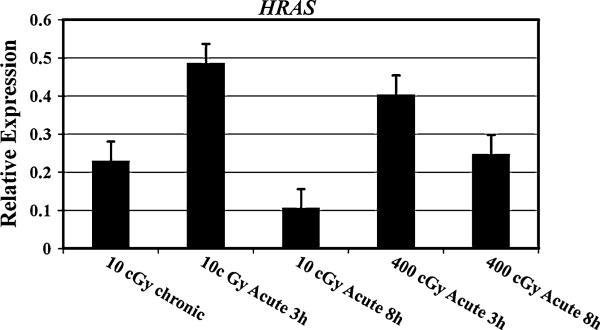
The let-7 family of miRNA negatively regulates the RAS oncogene. We monitored the expression of the RAS oncogene in AG1522 cells irradiated with various doses of γ-radiation. The mRNA levels of RAS were significantly higher (p = 0.023) in 10 cGy (3 cGy/min) treated cells examined at 3 h after irradiation as compared to the cells investigated at 8 h and the cells treated with chronic 10 cGy (0.3 cGy/h) (Fig. 2).
Fig. 2.
The relative expression of RAS mRNA in AG1522 cells harvested for analyses at 3 or 8 h after exposure to acute 10 cGy (3 cGy/min) or acute 400 cGy (120 cGy/min), or within 10 min after chronic exposure to 10 cGy (0.3 cGy/h). The data are expressed as Log2. The error bars indicate the standard error of the mean (SEM) for three independent experiments
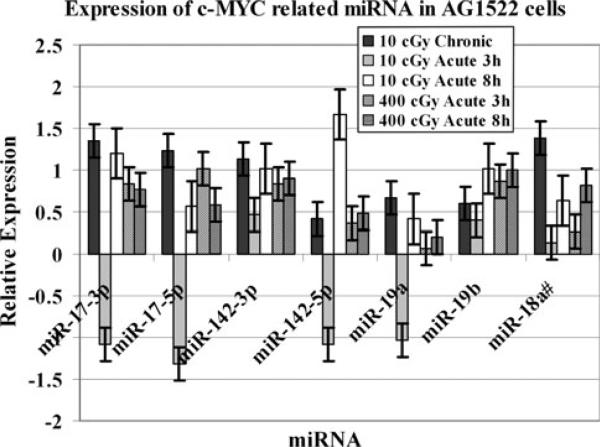
We next examined the modulation of several miRNA, shown to be involved in c-MYC gene regulation in low or moderate dose irradiated AG1522 cells. The expression of miR-17-3p, miR-17-5p, miR-142-3p, miR-142-5p, miR-18a, and miR-19b was significantly upregulated (p = 0.04) in AG1522cells after 10 cGy of γ rays delivered chronically at 0.3 cGy/h (Fig. 3). Interestingly, when the 10 cGy dose was given acutely at 3 cGy/min., miR-17-3p, miR-17-5p, miR-142-5p, and miR-19a were down regulated at 3 h post irradiation followed by an induction in their expression at 8 h. All of these miRNA were significantly upregulated (p = 0.05) after acute 400 cGy γ ray dose.
Fig. 3.
Relative expression of c-MYC-related miRNA in AG1522 cells after exposure to 10 or 400 cGy doses of γ-rays. The miRNA expression was assessed at 10 min, 3 h, and 8 h post irradiation. The data are shown as Log2 values. The error bars indicate the standard error of the mean (SEM) for three independent experiments
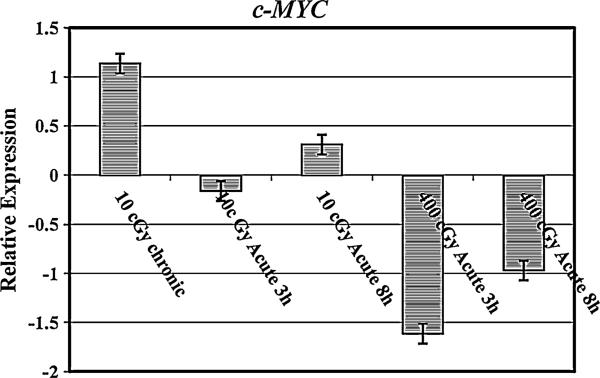
We further examined the expression levels of c-MYC miRNAin irradiated AG1522 cells. The c-MYC expression was upregulated after 10 cGy γ-radiation dose when delivered at a low dose rate of 0.3 cGy/h (Fig. 4). The expression of c-MYC miRNA remained repressed after 3 and 8 h of exposure to 10 cGy dose delivered at high dose rate of 3 cGy/min. This dose rate dependent difference in the expression of c-MYC was significantly different (p < 0.05). When AG1522 cells were exposed to 400 cGy (dose rate 120 cGy/min), c-MYC was downregulated at the 3 h time point (Fig. 6). Interestingly, most of the miRNA targeting c-MYC were upregulated in 400 cGy treated cells (Fig. 4).
Fig. 4.
Relative expression of c-MYC mRNA in AG1522 cells after exposure to 10 or 400 cGy doses of γ-rays. The mRNA expression was assessed at 10 min, 3 h, and 8 h post irradiation. The data are shown as Log2 values. The error bars indicate the standard error of the mean (SEM) for three independent experiments
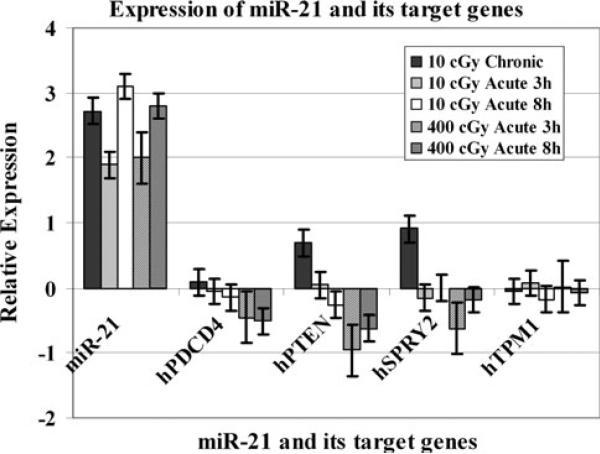
Fig. 6.
mRNA expression analyses of the miR-21 target genes, hPDCD4, hPTEN and hSPRY2 in 10 or 400 cGy irradiated AG1522 cells. The expression pattern of miR-21 is included for comparison. The data are plotted as Log2 values and indicate relative mRNA expression measured at 10 min, 3 h, and 8 h time points after radiation treatment. The error bars indicate the standard error of the mean (SEM) for three independent experiments
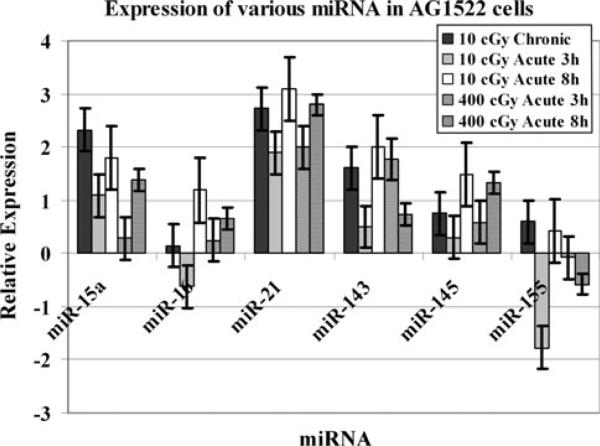
Finally, we examined the modulation of miR-15a, miR-16, miR-21, miR-143, miR-145 and miR-155 in AG1522 cells. Most of these miRNA were upregulated in AG1522 cells after radiation treatment (Fig. 5). The miR-15a, miR-21, and miR-143 were significantly upregulated (p = 0.02) in cells exposed to chronic 10 cGy. For miR-16, the acute 10 cGy treatment resulted in reduced expression levels at 3 h post irradiation and an upregulation of expression at 8 h time point (Fig. 5). Although miR-21, miR-143, and miR-145 were not down regulated at the 3 h time point following an acute 10 cGy dose, their expression levels were lower and increased to a higher level when examined at the 8 h time point. When the miRNA modulation was monitored in acute 400 cGy treated AG1522 cells, it was observed that miR-15a, miR-21, and miR-145 were gradually upregulated during the 3 h to 8 h time window. miR-143 was first induced at 3 h post irradiation and its expression levels declined at the 8 h time point (Fig. 5).
Fig. 5.
Modulation of various miRNA in irradiated AG1522 cells. The cells were treated with 10 or 400 cGy acute γ-rays doses. The relative expression, shown as Log2 values, was computed at 10 min, 3 h, and 8 h time points post-irradiation. The error bars indicate the standard error of the mean (SEM) for three independent experiments
Modulation of miR-21 target genes in irradiated AG1522 cells
In order to determine the correlation of miRNA expression levels to the modulation of target genes that they control, we investigated the relative expression of four miR-21 target genes: programmed cell death 4 (hPDCD4), PTEN tumor suppressor gene (hPTEN), sprouty 2 (hSPRY2), and the tumor suppressor gene tropomyosin 1 (hTPM1) in 10 or 400 cGy irradiated AG1522 cells by employing QPCR. The miR-21 target genes were selected from the published literature. The impact of miR-21 upregulation on the modulation of its target genes was observed in AG1522 cells exposed to chronic 10 cGy or acute 10 or 400 cGy of 0γ rays (Fig. 6). The miR-21 was upregulated in acute or chronic 10 cGy and in acute 400 cGy treated AG1522 cells (Fig. 6). The relative mRNA expression of miR-21 target genes hPDCD4 (p = 0.004), hPTEN (p = 0.001), hSPRY2 (p = 0.006) and hTPM1 (p = 0.008) was also significantly downregulated in these cells (Fig. 6). The mRNA levels of hPTEN and hSPRY2 were induced in chronic 10 cGy irradiated cells.
Discussion
To investigate low-dose/low-dose-rate effects of low-linear energy transfer (LET) ionizing radiation, we used cells adapted to grow in a 3-D architecture that mimics cell growth in vivo. Quiescent normal human fibroblasts were irradiated with single acute or chronic 10 cGy doses of 137Cs γ rays (Table 1). Cell cultures maintained in 3-D were used as they are ideal to investigate radiation-induced biological effects. The Cytomatrix™ allows cells to grow in 3-D, as they do in the body [26, 27]. Its inert material is not coated with metal, hence no complications with radiation dosimetry would be anticipated. We have characterized the IR response of several human cell strains growing on the Cytomatrix™. We have shown that the expression of connexin 43, a constitutive protein of gap junctions, and the G1 checkpoint were more sensitive to regulation by crays in cells maintained in a 3-D than in a 2-D configuration [12]. The IR-induced modulation of connexin proteins and the expression of adaptive responses were greater in a 3-D setup than in cells cultured in 2-D [12]. Hence, this platform provides in vivo-like culture conditions to monitor the biological effects of radiation.
To achieve significant reductions in the uncertainties associated with risk projections for cancer and other health effects caused by low dose IR, progress in the understanding of fundamental biological processes is necessary. We explored the role of miRNA in the cellular response to low dose radiation delivered at different dose rates. Exposure of cells to IR results in the formation of free radicals. It has been assumed that the subsequent alterations in multiple intracellular processes following irradiation are mainly due to the initial oxidative damage caused by these free radicals. The initial damage sustained by the irradiated cells is processed by a range of constitutive and induced repair, signaling, and communication processes involving the control of a large number of gene products [8, 9]. How all these processes are coordinated and controlled remain unknown. We asked whether there are differences in the modulation of miRNA in acute or chronic 10 cGy low dose γ-irradiated cells. We also wanted to investigate the changes in the levels of miRNA in cells after exposure to a moderate dose of 400 cGy of γ-radiation. The data presented here demonstrates that exposure to low dose ionizing radiation has different miRNA responses than high dose irradiation.
The selected miRNA examined in this investigation are regulators of many genes related to various diseases, including cancer. The monitoring of miRNA expression in 10 cGy γ-ray-treated cells revealed differences in the relative expression of the let-7 family of miRNA when the dose was given at different rates mimicking the acute or chronic low dose human radiation exposures. The expression patterns of miR-let-7 differed after exposure to either chronic or acute 10 cGy doses of γ-rays. Most of these miRNA were upregulated after 10 cGy chronic dose. In case of acute 10 cGy, they were first repressed and then induced (Fig. 1). The differences in the expression of let-7 miRNA among cells irradiated with low doses delivered at different rates might have a role in defining the biological effects of such exposures. The let-7 family of miRNA is a negative regulator of the RAS oncogene [28]. Ras proteins are membrane-associated GTPase signaling proteins that regulate cellular growth and differentiation [29]. Down regulation of the RAS gene is strongly associated with a poor prognosis of lung, pancreatic, and colon cancers [30]. The examination of RAS expression in these cells indicated that RAS was moderately induced after 10 cGy chronic dose. In contrast to the let-7 miRNA expression in AG1522 cells treated with 10 cGy dose delivered at a high dose rate, RAS was first induced and then repressed (Fig. 2). Previous studies have shown that the expression levels of the let-7 miRNA family were modulated in a cell type specific fashion [20]. We have previously examined the modulation of miRNA in cells that differed in DNA-dependent protein kinase (DNA-PK) activity. The let-7 family miRNA were upregulated in irradiated M059K cells with normal DNA-PK activity but were downregulated in irradiated DNA-PK deficient M059J cells [22].
The miR-17-3p, miR-17-5p, miR-142-3p, miR-142-5p, miR-18a, miR-19a, and miR-19b were shown to be involved in c-MYC gene regulation. These miRNA were upregulated in AG1522 cells after 10 cGy chronic dose. The miR-17-3p, miR-17-5p, miR-142-5p, and miR-19a were first down regulated at 3 h post 10 cGy acute dose followed by an induction in their expression at 8 h. Investigation of the modulation of c-MYC in irradiated AG1522 indicated that while the pattern of c-MYC mRNA expression was not different in cells treated with 10 cGy delivered either at 0.3 cG/h or 3 cGy/min, c-MYC was found to be significantly downregulated in cells after 400 cGy acute exposure. This observation indicates that the regulation of c-MYC by miRNA after radiation exposure is dose dependent.
Previous studies have shown that the expression of miRNA associated with c-MYC translocation was upregulated in TK6 cells exposed to γ-radiation [20]. The protooncogene c-MYC regulates the expression of genes involved in cell division, cell growth, and apoptosis [31]. The miR-17-3p, miR-17-5p, miR-19a, miR-19b, miR-142-3p, and miR-142-5p were upregulated in both M059K and M059J cells following exposure to IR [22]. c-MYC induces a cluster of miRNA known as miR-17-92 including miR-17-5p, miR-17-3p, miR-18a, miR-19a, miR-20a, miR-19b, and miR-92-1. This cluster is amplified in B cell lymphomas and other cancers [32]. Induced expression of c-MYC has been correlated with accelerated tumor development and many studies support the miR-17-92 cluster as a potential human oncogene. Over-expressed miR-17-3p, miR-18a, and miR-19a have been found in lung cancers and B-cell lymphomas, and were shown to participate in the process of angiogenesis [32]. The c-MYC oncogene, which encodes a basic helix-loop-helix transcription factor, functions as a regulator of cell growth owing to its ability to induce both cell proliferation and apoptosis [33].
The miR-15a and miR-16 negatively regulate B-cell lymphoma 2 (BCL2), an anti-apoptotic gene overexpressed in leukaemias and lymphomas [34]. These miRNA are down-regulated in B-cell chronic lymphocytic leukemia (CLL) [34, 35]. The deletion or downregulation of miR-15a and miR-16-1 result in increased expression of BCL2, which promotes leukaemogenesis and lymphomagenesis in haematopoietic cells. Importantly, BCL2 has been implicated in the cellular radiation response [36]. miR-15a and miR-16 also target MYB, that is involved in myeloid, lymphoid, or mixed-lineage leukemias [37]. miR-16 was initially repressed after acute 10 cGy treatment followed by an upregulation in its expression (Fig. 5). The miR-15a, miR-21, miR-143, miR-145 and miR-155 were upregulated in chronic 10 cGy exposed cells (Fig. 5). The miR-155 expression is upregulated in pediatric Burkitt's and Hodgkin's lymphomas [38]. The miR-15a, miR-16, miR-21, miR-143 and miR-145 were upregulated and miR-155x was down regulated in 400 cGy acutely-irradiated AG1522 cells (Fig. 5). These results indicate the importance of radiation dose, and dose rate, in determining the cellular response to ionizing radiation. The miR-143 regulates connective tissue growth factor and its expression correlates with adipocyte differentiation and carcinogenesis [39]. The expression of miR-145 and miR-155 is altered in lung cancers and has been correlated with patient's survival [40]. The miR-143 and miR-145 levels are reduced in colorectal tumors [41] while the upregulation of miR-21 in glioblastoma has been reported [42]. Similar expression patterns of multiple miRNA observed in the present study indicate common functions in the ionizing radiation induced pathways. Previous studies from our laboratory examined the impact of radiation dose on the modulation of miRNA in low LET irradiated human cells. The expression patterns of many miRNA differed markedly within the same cell line after exposure to either low or high doses of radiation [21]. Expression profiles of miRNAs analyzed with microarrays after exposure to low or high dose radiation revealed modulation of miRNA in a dose-dependent manner [43]. These studies have provided evidence that miRNA are regulated in response to IR exposure.
Why are miRNA repressed at 3 h after acute 10 cGy? One possible explanation is that the processing of DNA damage is perhaps different after a low dose delivered at high rather than low dose rate and the damage may require longer time to repair. The repression of certain miRNA could be necessary for the cell to accomplish that task. Previous studies have revealed differences in the extent of DNA damage induced by 10 cGy delivered at different dose rates. The protraction of the dose over 48 h reduced micronucleus frequency to below the spontaneous frequency. The DNA damage by acute 10 cGy was protected by the up-regulation of MnSOD, catalase or glutathione peroxidase and by the inhibition of superoxide anion generation by flavin-containing oxidases [12].
A number of published studies have reported target genes for many miRNA. Most of the target genes for the miRNA investigated in this study are involved in cellular proliferation [21]. We selected a few target genes for miR-21 from the published literature. The miR-21 is an anti-apoptotic factor expressed in glioblastoma cells, breast cancers, and is located at the genomic region amplified in lung cancer [40]. High expression of miR-21 is correlated with a poor prognosis of adenocarcinoma [40]. The miR-21 target genes include programmed cell death 4 (hPDCD4) [44–46], tumor suppressor gene tropomyosin 1 (hTPM1) [47], hPTEN tumor suppressor gene [48], and sprouty 2 (SPRY2) [49]. In order to determine the correlation of miRNA expression levels to the modulation of target genes that they control, we investigated the relative expression of miR-21 target genes hPDCD4, hPTEN, hSPRY2 and hTPM1 in irradiated AG1522 cells. The miR-21 was upregulated in 10 cGy treated cells and its target genes hPDCD4, hPTEN, hSPRY2 were found to be downregulated in these cells (Fig. 6). These results establish a direct role of miR-21 in controlling the expression of hPDCD4, hPTEN, hTPM1, and hSPRY2 in irradiated cells. Other studies have shown that while miR-21 is upregulated in irradiated cells, its target genes PDCD4, hPTEN, and hSPRY2 are downregulated [21].
The relationship between low dose IR exposure and health effects is not understood. The understanding of the process(es) underlying low dose radiation effects would contribute to better understanding of system responses and estimates of health risks from environmental and occupational exposures. The results reported here show that miRNA are modulated in low dose irradiated cells. There were dose, dose rate, and time dependent variations in the miRNA expression patterns. The relationship between miRNA expression and target gene regulation is complicated because mRNAs are typically regulated by many different miRNAs and that miRNAs frequently have hundreds of mRNA targets. It is not feasible to use the expression levels of a few target genes to discover physiologically relevant miRNA-mRNA interactions. A large number of studies have investigated the global gene expression profiles of a variety of cell types exposed to low doses of IR [50–53]. It is clear from these studies that the gene expression is different in low dose treated cells as compared to high dose IR exposed cells. Furthermore, many studies have identified low dose rate radiation-induced gene expression [54–56]. Our study shows that dose and dose rate dependent modulations of miRNA occur in the exposed cells. Whether these alterations are responsible for large-scale gene expression alterations that lead to radiation-induced cellular effects remains to be seen. Future studies to dissect the direct impact of miRNA modulation after radiation exposure to the subsequent multiple gene regulation could explain the global gene expression alterations.
Acknowledgments
The authors are grateful to the DNA Analysis Facility, University of Vermont for assistance with the real-time PCR experiments. This work was funded by a grant to MAC from College of Nursing and Health Sciences, University of Vermont. Grants from the US Department of Energy Low Dose Radiation Research Program, NASA and the NIH supported research in EIA laboratory.
Contributor Information
M. Ahmad Chaudhry, Department of Medical Laboratory and Radiation Sciences, University of Vermont, 302 Rowell Building, Burlington, VT 05405, USA.
Romaica A. Omaruddin, Department of Medical Laboratory and Radiation Sciences, University of Vermont, 302 Rowell Building, Burlington, VT 05405, USA
Bridget Kreger, Department of Medical Laboratory and Radiation Sciences, University of Vermont, 302 Rowell Building, Burlington, VT 05405, USA.
Sonia M. de Toledo, Department of Radiology, UMDNJ-New Jersey Medical School, Newark, NJ 07103, USA
Edouard I. Azzam, Department of Radiology, UMDNJ-New Jersey Medical School, Newark, NJ 07103, USA
References
- 1.Preston RJ. The LNT model is the best we can do—today. J Radiol Prot. 2003;23(3):263–268. doi: 10.1088/0952-4746/23/3/303. [DOI] [PubMed] [Google Scholar]
- 2.Little MP, Muirhead CR. Derivation of low-dose extrapolation factors from analysis of curvature in the cancer incidence dose response in Japanese atomic bomb survivors. Int J Radiat Biol. 2000;76(7):939–953. doi: 10.1080/09553000050050954. [DOI] [PubMed] [Google Scholar]
- 3.Chaudhry MA, Kreger B, Omaruddin RA. Transcriptional modulation of micro-RNA in human cells differing in radiation sensitivity. Int J Radiat Biol. 2010;86(7):569–583. doi: 10.3109/09553001003734568. [DOI] [PubMed] [Google Scholar]
- 4.Tubiana M, Aurengo A, Averbeck D, Masse R. Recent reports on the effect of low doses of ionizing radiation and its dose-effect relationship. Radiat Environ Biophys. 2006;44(4):245–251. doi: 10.1007/s00411-006-0032-9. [DOI] [PubMed] [Google Scholar]
- 5.Averbeck D. Non-targeted effects as a paradigm breaking evidence. Mutat Res. 2010;687(1–2):7–12. doi: 10.1016/j.mrfmmm.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Huang L, Kim PM, Nickoloff JA, Morgan WF. Targeted and nontargeted effects of low-dose ionizing radiation on delayed genomic instability in human cells. Cancer Res. 2007;67(3):1099–1104. doi: 10.1158/0008-5472.CAN-06-3697. [DOI] [PubMed] [Google Scholar]
- 7.Murnane JP, Sabatier L. Chromosome rearrangements resulting from telomere dysfunction and their role in cancer. Bioessays. 2004;26(11):1164–1174. doi: 10.1002/bies.20125. [DOI] [PubMed] [Google Scholar]
- 8.Wolff S. The adaptive response in radiobiology: evolving insights and implications. Environ Health Perspect. 1998;106(Suppl 1):277–283. doi: 10.1289/ehp.98106s1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azzam EI, Little JB. The radiation-induced bystander effect: evidence and significance. Hum Exp Toxicol. 2004;23(2):61–65. doi: 10.1191/0960327104ht418oa. [DOI] [PubMed] [Google Scholar]
- 10.Rigaud O, Moustacchi E. Radioadaptation for gene mutation and the possible molecular mechanisms of the adaptive response. Mutat Res. 1996;358(2):127–134. doi: 10.1016/s0027-5107(96)00113-3. [DOI] [PubMed] [Google Scholar]
- 11.de Toledo SM, Azzam EI. Adaptive and bystander responses in human and rodent cell cultures exposed to low level ionizing radiation: the impact of linear energy transfer. Dose Response. 2006;4(4):291–301. doi: 10.2203/dose-response.06-103.deToledo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Toledo SM, Asaad N, Venkatachalam P, Li L, Howell RW, Spitz DR, Azzam EI. Adaptive responses to low-dose/low-dose-rate gamma rays in normal human fibroblasts: the role of growth architecture and oxidative metabolism. Radiat Res. 2006;166(6):849–857. doi: 10.1667/RR0640.1. [DOI] [PubMed] [Google Scholar]
- 13.Gaillard S, Pusset D, de Toledo SM, Fromm M, Azzam EI. Propagation distance of the alpha-particle-induced bystander effect: the role of nuclear traversal and gap junction communication. Radiat Res. 2009;171(5):513–520. doi: 10.1667/RR1658.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spitz DR, Azzam EI, Li JJ, Gius D. Metabolic oxidation/reduction reactions and cellular responses to ionizing radiation: a unifying concept in stress response biology. Cancer Metastasis Rev. 2004;23(3–4):311–322. doi: 10.1023/B:CANC.0000031769.14728.bc. [DOI] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 16.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304(5670):594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 17.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309(5740):1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 18.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 19.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaudhry MA. Real-time PCR analysis of micro-RNA expression in ionizing radiation-treated cells. Cancer Biother Radiopharm. 2009;24(1):49–56. doi: 10.1089/cbr.2008.0513. [DOI] [PubMed] [Google Scholar]
- 21.Chaudhry MA, Kreger B, Omaruddin RA. Transcriptional modulation of micro-RNA in human cells differing in radiation sensitivity. Int J Radiat Biol. 2010;86:569–583. doi: 10.3109/09553001003734568. [DOI] [PubMed] [Google Scholar]
- 22.Chaudhry MA, Sachdeva H, Omaruddin RA. Radiation-induced Micro-RNA modulation in glioblastoma cells differing in DNA-repair pathways. DNA Cell Biol. 2010;29(9):553–561. doi: 10.1089/dna.2009.0978. [DOI] [PubMed] [Google Scholar]
- 23.Goddu SM, Howell RW, Rao DV. Method and means for variably attenuating radiation. 2001:B1. United States Patent US 6,201,852.
- 24.Howell RW, Goddu SM, Rao DV. Design and performance characteristics of an experimental Cs-137 irradiator to simulate internal radionuclide dose rate patterns. J Nucl Med. 1997;38:727–731. [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Bagley J, Rosenzweig M, Marks DF, Pykett MJ. Extended culture of multipotent hematopoietic progenitors without cytokine augmentation in a novel three-dimensional device. Exp Hematol. 1999;27(3):496–504. doi: 10.1016/s0301-472x(98)00053-8. [DOI] [PubMed] [Google Scholar]
- 27.Poznansky MC, Evans RH, Foxall RB, Olszak IT, Piascik AH, Hartman KE, Brander C, Meyer TH, Pykett MJ, Chabner KT, Kalams SA, Rosenzweig M, Scadden DT. Efficient generation of human T cells from a tissue-engineered thymic organoid. Nat Biotechnol. 2000;18(7):729–734. doi: 10.1038/77288. [DOI] [PubMed] [Google Scholar]
- 28.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 29.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64(11):3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 30.Goodsell DS. The molecular perspective: the ras oncogene. Oncologist. 1999;4(3):263–264. [PubMed] [Google Scholar]
- 31.Yaguang Xi JRE, Jingfang Ju. Investigation of miRNA Biology by bioinformatic tools and impact of miRNAs in colorectal cancer—regulatory relationship of c-Myc and p53 with miRNAs. Cancer Inform. 2007;3:245–253. [PMC free article] [PubMed] [Google Scholar]
- 32.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109(3):321–334. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- 34.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akerman GS, Rosenzweig BA, Domon OE, Tsai CA, Bishop ME, McGarrity LJ, Macgregor JT, Sistare FD, Chen JJ, Morris SM. Alterations in gene expression profiles and the DNA-damage response in ionizing radiation-exposed TK6 cells. Environ Mol Mutagen. 2005;45(2–3):188–205. doi: 10.1002/em.20091. [DOI] [PubMed] [Google Scholar]
- 37.Chung EY, Dews M, Cozma D, Yu D, Wentzel EA, Chang TC, Schelter JM, Cleary MA, Mendell JT, Thomas-Tikhonenko A. c-Myb oncoprotein is an essential target of the dleu2 tumor suppressor microRNA cluster. Cancer Biol Ther. 2008;7(11):1758–1764. doi: 10.4161/cbt.7.11.6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102(10):3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shahi P, Loukianiouk S, Bohne-Lang A, Kenzelmann M, Kuffer S, Maertens S, Eils R, Grone HJ, Gretz N, Brors B. Argonaute—a database for gene regulation by mammalian microRNAs. Nucleic Acids Res. 2006;34(Database issue):D115–D118. doi: 10.1093/nar/gkj093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 41.Michael MZ, O'Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1(12):882–891. [PubMed] [Google Scholar]
- 42.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 43.Cha HJ, Seong KM, Bae S, Jung JH, Kim CS, Yang KH, Jin YW, An S. Identification of specific microRNAs responding to low and high dose gamma-irradiation in the human lymphoblast line IM9. Oncol Rep. 2009;22(4):863–868. [PubMed] [Google Scholar]
- 44.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27(15):2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 45.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283(2):1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 46.Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, Li Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PubMed] [Google Scholar]
- 47.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem. 2007;282(19):14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 48.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sayed D, Rane S, Lypowy J, He M, Chen IY, Vashistha H, Yan L, Malhotra A, Vatner D, Abdellatif M. MicroRNA-21 targets sprouty2 and promotes cellular outgrowths. Mol Biol Cell. 2008 doi: 10.1091/mbc.E08-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding LH, Shingyoji M, Chen F, Hwang JJ, Burma S, Lee C, Cheng JF, Chen DJ. Gene expression profiles of normal human fibroblasts after exposure to ionizing radiation: a comparative study of low and high doses. Radiat Res. 2005;164(1):17–26. doi: 10.1667/rr3354. [DOI] [PubMed] [Google Scholar]
- 51.Long XH, Zhao ZQ, He XP, Wang HP, Xu QZ, An J, Bai B, Sui JL, Zhou PK. Dose-dependent expression changes of early response genes to ionizing radiation in human lymphoblastoid cells. Int J Mol Med. 2007;19(4):607–615. [PubMed] [Google Scholar]
- 52.Fujimori A, Okayasu R, Ishihara H, Yoshida S, Eguchi-Kasai K, Nojima K, Ebisawa S, Takahashi S. Extremely low dose ionizing radiation up-regulates CXC chemokines in normal human fibroblasts. Cancer Res. 2005;65(22):10159–10163. doi: 10.1158/0008-5472.CAN-05-2015. [DOI] [PubMed] [Google Scholar]
- 53.Fachin AL, Mello SS, Sandrin-Garcia P, Junta CM, Donadi EA, Passos GA, Sakamoto-Hojo ET. Gene expression profiles in human lymphocytes irradiated in vitro with low doses of gamma rays. Radiat Res. 2007;168(6):650–665. doi: 10.1667/RR0487.1. [DOI] [PubMed] [Google Scholar]
- 54.Amundson SA, Lee RA, Koch-Paiz CA, Bittner ML, Meltzer P, Trent JM, Fornace AJ., Jr Differential responses of stress genes to low dose-rate gamma irradiation. Mol Cancer Res. 2003;1(6):445–452. [PubMed] [Google Scholar]
- 55.Sugihara T, Magae J, Wadhwa R, Kaul SC, Kawakami Y, Matsumoto T, Tanaka K. Dose and dose-rate effects of low-dose ionizing radiation on activation of Trp53 in immortalized murine cells. Radiat Res. 2004;162(3):296–307. doi: 10.1667/rr3223. [DOI] [PubMed] [Google Scholar]
- 56.Gridley DS, Pecaut MJ, Rizvi A, Coutrakon GB, Luo-Owen X, Makinde AY, Slater JM. Low-dose, low-dose-rate proton radiation modulates CD4(?) T cell gene expression. Int J Radiat Biol. 2009;85(3):250–261. doi: 10.1080/09553000902748609. [DOI] [PubMed] [Google Scholar]