Summary
Studies over the past several years have revealed that steps in gene expression are extensively coupled to one another both physically and functionally. Recently, in vitro systems were developed for understanding the mechanisms involved in coupling transcription by RNA polymerase II to RNA processing. Here we describe an efficient two-way system for coupling transcription to splicing and a robust three-way system for coupling transcription, splicing, and polyadenylation. In these systems a CMV-DNA construct is incubated in HeLa cell nuclear extracts in the presence of 32P-UTP to generate the nascent transcript. Transcription is then stopped by addition of μ-amanitin followed by continued incubation to allow RNA processing.
Keywords: RNAP II
1. Introduction
During gene expression, pre-mRNAs are synthesized in the nucleus by RNAP II and then undergo several processing steps, including capping, splicing, and polyadenylation. These steps are extensively coupled to one another via an extensive network of interactions (1-3). A number of systems have been developed to investigate the mechanisms for coupling transcription to splicing (4-9), coupling transcription to polyadenylation (10), and coupling transcription to both polyadeylation and splicing (11). Here we describe methods for two systems that we developed, one for coupling transcription to splicing and one for coupling transcription, splicing and polyadenylation. In these systems, pre-mRNAs are synthesized by RNAP II in HeLa cell nuclear extracts followed by RNA processing. The method employs nuclear extracts similar to those that were originally optimized for splicing 32P-labeled pre-mRNA synthesized with bacteriophage RNA polymerases (12). These nuclear extracts are typically prepared in bulk from 10 to 50 liters of cells grown in suspension (13), but for small-scale applications, can also be prepared using a few 150 mm plates of HeLa cells grown as adherent monolayers (14). Preparation of the nuclear extracts was optimized for use in the coupled systems (5). The DNA template used in the coupled systems is a PCR product containing the CMV promoter fused to a DNA template encoding a splicing substrate. The bovine growth hormone (BGH) polyA signal is also present in the DNA template for the system using polyadenylation.
2. Materials
All solutions are prepared using analytical grade reagents and ultrapure water (Milli-Q water – purified deionized water at a sensitivity of 18 MΩ cm at 25°C). Storage temperature of each reagent is listed below.
2.1. Materials for preparation of CMV-DNA constructs
Plasmid encoding CMV-Ftz DoF construct containing or lacking BGH polyA signal or encoding constructs of interest. Plasmids should be stored at −20°C in 1× TE buffer (Tris-HCl, pH 8.0, 1 mM EDTA) at a concentration of 5 ng/μL.
Primers for coupled transcription/splicing: Make 500 μL aliquots of Forward primer (5′ tgg agg tcg ctg agt agt gc 3′) and Reverse primer (5′ tag aag gca cag tcg agg 3′) at a final concentration of 1.6 μM. Store at −20°C.
Primers for coupled transcription/splicing/polyadenylation: Make 500 μL aliquots of Forward primer (5′ tgg agg tcg ctg agt agt gc 3′) and Reverse primer (5′ cca cac cct aac tga gac 3′) at a final concentration of 1.6 μM. Store at −20°C.
10 mM dNTPs (GenScript, Cat#: C01582). Store at −20°C.
50 mM MgSO4. Store at −20°C.
Platinum Taq HiFi and 10× HiFi Buffer provided by supplier (Invitrogen, Cat#: 11304-029). Store at −20°C.
10× TBE: Combine 432 g Tris-Base, 220 g Boric acid and 37.2 g EDTA. Add water to a final volume of 4 L. Store at room temperature.
10 mg/mL Ethidium bromide (Ethidium bromide solution BioReagent, for molecular biology, 10 mg/mL in H2O, Sigma, Cat# E1510-10 ML)
Agarose HS standard/High Melt (Denville, Cat#: CA3510-8). Store at room temperature.
3 M Sodium Acetate. Store at room temperature.
100% and 70 % Ethanol diluted from 200 Proof pure ethanol. Store at room temperature.
Phenol/Chloroform, pH 7.9 (Ambion, Cat#: AM9732). Store at 4°C.
1 kb DNA Ladder (NEB, Cat#: N3232S). Store at −20°C.
2.2. Components of Coupled Transcription/splicing Reaction
12.5 mM ATP. Filter, make 100 μL aliquots and store at −20°C.
0.5 M Creatine Phosphate di-Tris salt (CrPh): (Sigma, Cat#P1937). Filter, make 100 μL aliquots and store at −20°C.
80 mM MgCl2: Filter, make 100 μL aliquots and store at −20°C.
CMV-DNA template. Make 50 μL aliquots at 200 ng/μL (see Note 1). Store at - 20°C.
[α-32P]-UTP (EasyTide, 800 Ci/mmol, 250 μCi, PerkinElmer, Cat#: BLU507X250UC). Store at 4°C.
HeLa cell nuclear extract (see Note 2). Store at −80°C
α-amanitin: dilute to 10 ng/μL with water from 1 mg/mL stock (Sigma, Cat#: A2263). Store at −20°C.
2× Proteinase K buffer (PK buffer): Mix 20 mL 1M Tris pH 8.0, 5 mL 0.5M EDTA, 6 mL 5M NaCl, 10 mL 20% Sodium Dodecyl Sulfate. Add water up to 100 mL. Filter and store at room temperature.
Proteinase K (PK). Add water to PK powder to prepare a 10 mg/mL stock (Roche, Cat#: 03115879001). Make 100 μL aliquots. Store at −20°C.
Glycogen, 20 mg/mL (Roche, Cat#901393). Store at −20°C.
Formamide Gel Loading Dye: add 16 mL Formamide (Formamide DI™ deionized, American Bioanalytical, Cat#: AB00600-00500), 0.4 mL 0.5 M EDTA, 0.8 mL 2.5% Xylene Cyanol and 0.8 mL 2.5% Bromophenol Blue. Mix well, and make 1 mL aliquots. Store at −20°C.
Phenol:Chloroform:Isoamyl Alcohol, pH 6.6 (Ambion Cat#: AM9732). Store at 4°C.
2.3. Components for Coupled Transcription/splicing/polyadenylation Reaction
12.5 mM ATP. Filter, make 100 μL aliquots and store at −20°C.
0.5 M Creatine Phosphate di-Tris salt (CrPh): (Sigma, Cat#P1937). Filter, make 100 μL aliquots and store at −20°C.
160 mM MgCl2: Filter, make 100 μL aliquots and store at −20°C.
CMV-DNA template with BGH polyA signal. Make 50 μL aliquoted at 200 ng/μL. Store at −20°C.
15% (w/v) polyvinyl alcohol (PVA) dissolved in water. Autoclave to solubilize the PVA. Use low molecular weight PVA (Sigma, Cat#: P-8136). Make 1 mL aliquots and store at −20°C.
50 μM GTP, CTP, UTP [G, C, U]: Mix 5μL of each NTP from 10 mM stock solutions (Promega, Cat#: P1221) with 85 μL water and store at −20°C.
10 mM UTP (Promega, Cat#: P1221). Store at −20°C.
2.4. Denaturing Polyacrylamide Gel Materials
5% Denaturing Gel Solution: Mix 215 g Urea with 50 mL of 10X TBE and 62.5 mL 40% Acrylamide-Bisacrylamide (Acrylamide-Bisacrylamide, Solution, OmniPur*. 40% (w/v, 29:1), EMD Millipore, Cat#: EM-1700). Bring volume up to 500 mL with water. Filter and store at 4°C.
N,N,N’,N’-Tetramethylethylenediamine (TEMED) (Sigma, Cat#: T9281-50ML). Store at 4°C.
10% Ammonium Persulfate (APS). Store at 4°C.
Model V16 Polyacrylamide Gel Electrophoresis System (Labrepco, Cat#: E21070010), PROTEAN II xi Spacers 0.5mm (Bio-Rad, Cat#: 165-1841), PROTEAN II xi Comb (Bio-Rad, Cat#: #165-1870).
Gel Loading Tips: Flat Orifice, 83 mm × 0.33 mm diameter (Denville, Cat#: P3083).
Whatman Paper 3 MM Chr (Whatman, Cat#: 3030 917).
Bio-Rad Model 583 Gel Dryer (Bio-Rad, Cat#: 165-1745).
Bio-Rad HydroTech Vacuum Pump (Bio-Rad, Cat#: 165-1781)
Phosphoimager cassette: Mounted General Purpose, 20 × 25 cm, screen and cassette (Ge HealthCare. Cat#: 63-0034-89)
Phosphoimager: Personal Molecular Imager™ (PMI) System (Bio-Rad, Cat#: 170-9400)
3. Methods
Carry out all procedures on ice unless otherwise specified.
3.1. Preparation of the CMV-DNA template
PCR reaction: Mix 2 μL of the CMV-DNA plasmid (CMV-DoF) (see Note 3), 2 μL 10 mM dNTPs, 2 μL 50 mM MgSO4, 5 μL 10× HiFi Buffer, 12.5 μL of each primer, 13.6 μL of Milli-Q water and 0.4 μL Platinum Taq HiFi. Start the PCR reaction at 94°C for 5 minutes followed by 32 cycles at 94°C for 30 seconds, 55°C for 30 seconds and 68°C for 2 minutes. The final cycle at 72°C for 10 minutes and store the PCR reaction at 15°C.
After the PCR reaction, bring the volume up to 150 μL with water and run a small aliquot (2 μL) on a mini-agarose gel. The PCR product for CMV-DoF should be ~1.5 kb.
Purify the DNA template by extracting the PCR reaction with an equal volume of Phenol/Chloroform (pH 7.9). Transfer the supernatant (aqueous phase) to a new eppendorf tube.
Add 2 μL of glycogen, mix well. Add 1/10 volume 3 M sodium acetate, mix well. Add 3 volumes of 100% Ethanol. Centrifuge at 16,000 × g for 15 minutes to pellet the DNA. Remove the supernatant, without disturbing the pellet. The precipitated DNA should form a translucent pellet at the bottom of the tube. Wash once with 1 mL 70 % Ethanol. Air-dry the pellet and dissolve it in 100 μL of water. Estimate the concentration of DNA by running 2 μL on an agarose gel and comparing the intensity of the band to the known concentrations of bands in the 1 kb DNA ladder (see Notes 4 and 5).
3.2. Coupled Transcription and Splicing Protocol
Preheat two water baths to 30°C and 37°C.
Prepare a master mix for the total number of reactions you plan to perform. A 1× reaction mixture contains 1 μL CMV-DNA template, 1 μL 12.5 mM ATP, 1 μL 0.5 M CrPh, 1 μL 80 mM MgCl2, 1 μL α-32P-UTP and 5 μL autoclaved Milli-Q water.
Aliquot 10 μL of the master mix per tube.
Add 15 μL of nuclear extract and pipet up and down gently to mix (see Note 6).
Incubate the reaction mixtures at 30°C for 8 minutes to allow RNAP II transcription (see Note 7).
Add 1 μL of α-amanitin (10 ng/μL) per 25-μL reaction mixture, and pipet up and down gently to mix (see Note 8).
Remove a 4 μL aliquot of the reaction at the 8-minute time point and transfer it to a microfuge tube containing 100 μL of 2× PK Buffer and 91 μL autoclaved Milli-Q water (see Note 9).
Repeat step #7 for all subsequent time points.
Add 5 μL of PK to each sample, mix well, and incubate at 37°C for 15 minutes.
Add 200 μL of Phenol/Chloroform pH 6.6 to each sample and mix well by pipetting up and down. Centrifuge for 10 minutes at 16,000 × g.
Transfer 175 μL of the aqueous phase to a new tube containing 2 μL of Glycogen. Mix by pipetting up and down.
Add 500 μL of 100% Ethanol and mix well by pipetting up and down.
Spin at 16,000 × g for 15 minutes. The RNA pellet should look white/translucent.
Carefully remove the supernatant using a pipet.
Quick spin at 16,000 × g.
Remove the rest of the supernatant using a P200 pipetman (see Note 10).
Resuspend the pellet in 15 μL Formamide Dye and mix carefully by pipetting.
Prepare a gel by combining 15 mL of 5% Denaturing Gel Solution with 15 μL TEMED and 150 μL 10% APS. Wait 10 minutes until the gel is polymerized.
Place samples in hot water (75 to 90°C) for 10 minutes.
Pre-run the denaturing gel at 20 mAmps for 10 minutes.
Briefly centrifuge the samples and load 7.5 μL of each on the pre-run denaturing gel. Run the gel at 20 mAmps, constant current for 30 to 45 minutes (see Note 11).
Transfer the gel to Whatman paper and dry the gel on a gel dryer for 30 minutes at 80°C
Place the gel in a phosphoimager cassette, expose 1-12 hrs, and scan the gel using a phosphoimager.
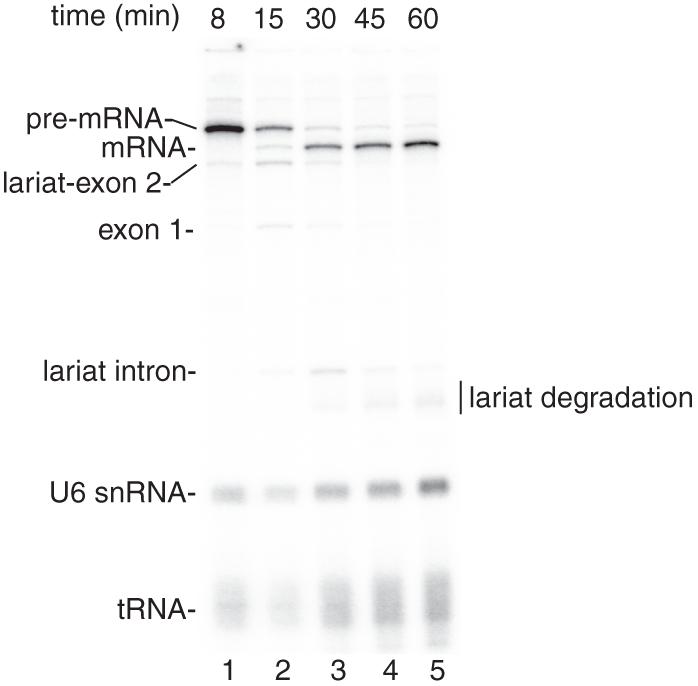
See representative results in Figure 1.
Figure 1.
Coupled RNAP II transcription and pre-mRNA splicing in vitro. (A) Structure of the DoF CMV-DNA template. The sizes of the exons and intron are indicated. B. 32P-UTP and the CMV-DoF DNA template were incubated under transcription/splicing conditions for 8 minutes. α-amanitin was added after the 8-minute time point and incubation was continued for the indicated times. Pre-mRNA and the splicing intermediates are indicated.The endogenous U6 snRNA and tRNA in the extract are labeled by the 32P-UTP (see Reddy et al. [1987].
3.3. Coupled Transcription/Splicing/Polyadenylation Protocol
Prepare the CMV-DNA template containing the BGH polyA signal by following the steps outlined in 3.1. Use the Reverse primer specifically designed for the coupled Transcription/Splicing/Polyadenylation system.
Prepare a master mix for the total number of reactions you plan to perform. A 1× reaction mixture contains 1 μL CMV-DNA template, 0.5 μL 160 mM MgCl2 and 4.1 μL 15% PVA. Add 1 μL autoclaved Milli-Q water.
Aliquot 10 μL of master mix per tube.
Add 15 μL of nuclear extract to each reaction mixture and pipet up and down gently to mix (see Note 6).
Incubate the tubes at 30°C for 20 min (see Note 12).
Add 1 μL 0.5 M CrPh, 2 μL 12.5 mM ATP, 2 μL α-32P-UTP, and 0.5 μL 50 μM [G, C, U]. Mix well.
Incubate the tubes at 30°C for 2 to 5 min (see Note 13).
Add cold UTP to each sample to a final concentration of 2 mM (see Note 14).
Incubate again at 30°C for 5 to 8 minutes.
Add 1 μL of α-amanitin (10 ng/μL) per 25-μL reaction mixture, pipet up and down to mix (see Note 8).
Follow all of the steps from 3.2.7 to 3.2.24.
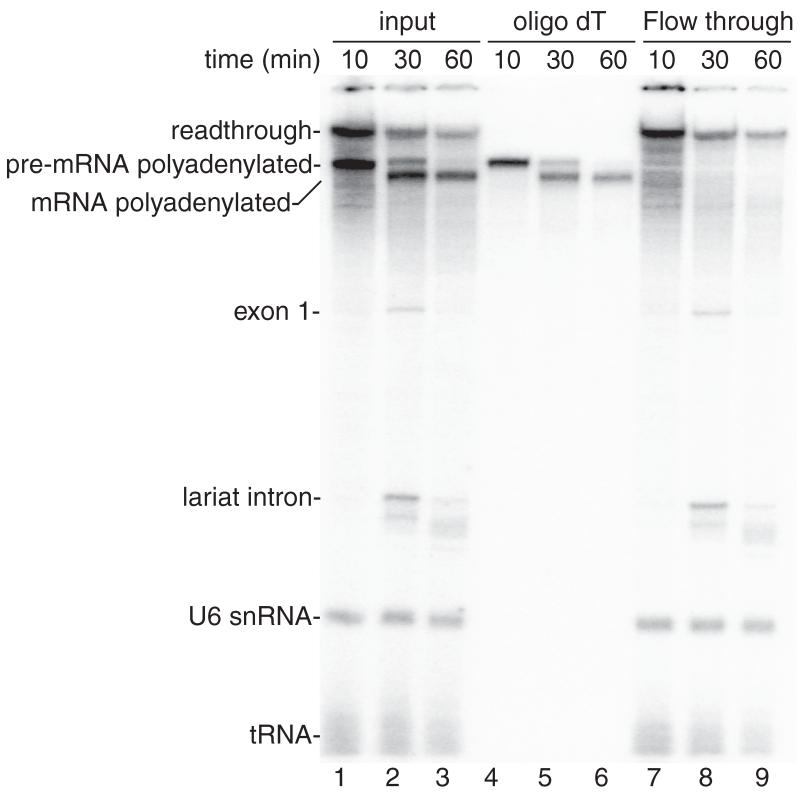
See Figure 2 for representative results.
Figure 2.
Coupled RNAP II transcription/splicing/polyadenylation in vitro. (A) Structure of the DoF CMV-DNA template showing the pA signal. The sizes of the exons and intron are indicated. B. 32P-UTP and the CMV-DoF/BGH DNA template were incubated for 20 minutes to assemble a pre-initiation complex followed by addition of 32P-UTP, ATP, and CrPhos and continued incubation for 10 minutes. The readthrough transcript and polyadenylated pre-mRNA are generated by this time point (lane 1). Spliced polyadenylated mRNA is generated at the subsequent time points (lanes 2, 3). Total RNA from aliquots of the reactions shown in lanes 1-3 were isolated and passed through an oligo dT column. The polyadenylated pre-mRNA and mRNA bound to the oligo dT (lanes 4-6) whereas the readthrough transcript, exon 1, the lariat intron, U6 snRNA and tRNA are detected in the flow through from the oligo dT column (lanes 7-9).
Acknowledgement
This work was supported by NIH grant GM043375.
Footnotes
The amount of CMV-DNA template should be titrated to obtain optimal RNAP II transcription efficiency. For our 1.5 kb CMV-DoF DNA, 200 ng/25 μL coupled reaction is optimal.
The main difference between the nuclear extract used for the coupled systems versus uncoupled systems is the omission of the spin after the dialysis at the end of the standard Dignam protocol.
In some preparations of nuclear extract, RNAP II transcribes end-to-end in a promoter-independent manner. If you are using your own DNA template, use the smallest possible PCR fragment that contains your sequence of interest to avoid large end-to-end transcription products.
RNAP II can initiate at nicks in DNA. To avoid nicked DNA, store aliquots at – 20°C in 1× TE, and dilute to 200 ng/μL before use.
PCR products should not be purified using mini-columns because these templates are not transcribed well.
Time points are taken in 4 μL aliquots from 25 μL reaction mixtures. Typically, 3-5 time points are taken. If more time points are needed, it is best to set up one larger reaction mixture and divide it into 25 μL aliquots for incubation rather than incubating a large-volume reaction.
For our CMV-DoF DNA template, time points ranging from 5 to 60 minutes are used and should be optimized for different DNA templates.
α-amanitin is used to block transcription, and the time of addition should be optimized for different DNA templates and nuclear extracts.
Samples in PK buffer are stable at room temperature and can be stored until all of the time points have been collected. At this step, samples can also be stored at - 20°C overnight and processed further later.
Ensure that the pellets are dry before adding the formamide loading dye because any remaining ethanol will add to the volume of the sample as well as distort the migration of the bands on the gel.
To resolve splicing products generated from CMV-DoF DNA template, the gel should be run until the bromophenol blue is at the bottom of the gel.
The nuclear extract and CMV-DNA template are incubated with MgCl2 and PVA to assemble a Pre-Initiation Complex (PIC). The PIC is necessary for efficient polyadenylation.
An incubation of 2 minutes is usually sufficient for the first incubation after PIC formation using CMV-DoF. However, this step should be optimized for each preparation of nuclear extract and DNA template.
Because the radioactive UTP is usually limiting, addition of cold UTP is used as a chase to generate full-length transcripts.
References
- 1.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 2.Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 3.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Hicks MJ, Yang CR, Kotlajich MV, Hertel KJ. Linking splicing to Pol II transcription stabilizes pre-mRNAs and influences splicing patterns. PLoS Biol. 2006;4:e147. doi: 10.1371/journal.pbio.0040147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das R, Dufu K, Romney B, Feldt M, Elenko M, Reed R. Functional coupling of RNAP II transcription to spliceosome assembly. Genes Dev. 2006;20:1100–1109. doi: 10.1101/gad.1397406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 7.Yu Y, Das R, Folco EG, Reed R. A model in vitro system for co-transcriptional splicing. Nucleic Acids Res. 2010;38:7570–7578. doi: 10.1093/nar/gkq620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh S, Garcia-Blanco MA. Coupled in vitro synthesis and splicing of RNA polymerase II transcripts. Rna. 2000;6:1325–1334. doi: 10.1017/s1355838200992537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Natalizio BJ, Garcia-Blanco MA. In vitro coupled transcription splicing. Methods. 2005;37:314–322. doi: 10.1016/j.ymeth.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 10.Rigo F, Kazerouninia A, Nag A, Martinson HG. The RNA tether from the poly(A) signal to the polymerase mediates coupling of transcription to cleavage and polyadenylation. Mol Cell. 2005;20:733–745. doi: 10.1016/j.molcel.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 11.Rigo F, Martinson HG. Functional coupling of last-intron splicing and 3′-end processing to transcription in vitro: the poly(A) signal couples to splicing before committing to cleavage. Mol Cell Biol. 2008;28:849–862. doi: 10.1128/MCB.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krainer AR, Maniatis T, Ruskin B, Green MR. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- 13.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Folco EG, Lei H, Hsu JL, Reed R. Small-scale nuclear extracts for functional assays of gene expression machineries. J. Vis. Exp. 2012;(64):e4140. doi: 10.3791/4140. DOI: 10.3791/4140. [DOI] [PMC free article] [PubMed] [Google Scholar]