Abstract
It has become apparent that plants have extensively diversified their arsenal of labdane-related diterpenoids (LRDs), in part via gene duplication and neo-functionalization of the ancestral ent-kaurene synthase (KS) required for gibberellin metabolism. For example, castor bean (Ricinus communis) was previously shown to produce an interesting set of biosynthetically related diterpenes, specifically ent-sandracopimaradiene, ent-beyerene, and ent-trachylobane, in addition to ent-kaurene, using four separate diterpene synthases, albeit these remain unidentified. Notably, despite mechanistic similarity of the underlying reaction to that catalyzed by KSs, ent-beyerene and ent-trachylobane synthases have not yet been identified. Given our interest in LRD biosynthesis, and the recent availability of the castor bean genome sequence, we applied a synthetic biology approach to biochemically characterize the four KS(-like) enzymes [KS(L)s] found in Ricinus communis [i.e., the RcKS(L)s]. In particular, using bacteria engineered to produce the relevant ent-copalyl diphosphate precursor and synthetic genes based on the predicted RcKS(L)s, although this ultimately required correction of a “splicing” error in one of the predicted genes, highlighting the dependence of such a synthetic biology approach on accurate gene sequences. Nevertheless, we can assign each of the four RcKS(L)s to one of the previously observed diterpene synthase activities, providing access to functionally novel enzymes. Intriguingly, the product distribution of the RcKS(L)s seems to support the distinct diterpene synthase reaction mechanism proposed by quantum chemical calculations, rather than the classically proposed pathway.
Keywords: natural products biosynthesis, diterpenoids, terpene synthases
1. Introduction
Plants are particularly prolific producers of terpenoids, of which there are more than 55,000 known (Köksal et al., 2011). Prominent among these are the labdane-related diterpenoids (LRDs), with ~7,000 known, and whose biosynthesis in plants can be traced back to the requisite production of gibberellin phytohormones (Peters, 2010). These natural products are characterized by their derivation from a sequential pair of terpene synthase (TPS) catalyzed reactions. First, (bi)cyclization of the general diterpenoid precursor (E,E,E)-geranylgeranyl diphosphate (GGPP), generally to the eponymous labdadienyl/copalyl diphosphate (CPP) intermediate, mediated by a class II diterpene cyclase. This is followed by the action of a more typical class I (di)terpene synthase. In the case of gibberellins, this pair of reactions yields ent-kaurene (1) via ent-CPP, as catalyzed by a CPP synthase (CPS) and subsequently acting ent-kaurene synthase (KS). Given their homology to the ancestral KS, those class I diterpene synthases acting on CPP are often termed KS-like (KSL), and these fall into what has been designated the TPS-e sub-family (Chen et al., 2011). This sub-family is further distinguished by the almost universal presence of an additional/insertional γ-domain relative to other plant class I TPS.
It is now evident that monocots, particularly cereal crop plants, have significantly expanded their arsenal of LRD natural products (Schmelz et al., 2014). In part, this wide range of LRDs evolved via gene duplication of the ancestral KS and neo-functionalization of the resulting KSLs, which produce the multicyclic hydrocarbon backbone structures that characterize the resulting various families of LRDs (Peters, 2010). The evolutionary radiation of KSLs in monocots was first discovered in rice (Peters, 2006), but more recent work has demonstrated that this applies at least to other cereal crop plants such as wheat (Zhou et al., 2012), as well as maize (Schmelz et al., 2014). In particular, the identified monocot KSLs cluster with the monocot KSs, while the dicot KSs form a separate group. Nevertheless, such derivation of KSLs from KS appears to have occurred early in at least the Poaceae (grass) plant family, as suggested by clustering of KSLs from all three investigated species, which diverged early in evolution of the grasses (Schmelz et al., 2014), separate from the monocot KSs. The continuing nature of this gene family radiation is further indicated by the presence of KSLs in the KS containing cluster, as well as the number of KSLs in each species (≥ 4). Indeed, comparison of functionally distinct alleles of a rice KSL led to identification of single residue “switch” for product outcome that applies in KSs as well (Xu et al., 2007).
By contrast, little is known about the production of LRDs in dicots (Zi et al., 2014), although a number are known to produce more specialized LRDs (i.e., other than gibberellins). While various species from the Lamiaceae plant family produce LRDs, the relevant KSLs identified to date generally do not contain the otherwise typical γ-domain (Caniard et al., 2012; Gao et al., 2009; Schalk et al., 2012), highlighting their unusual evolutionary origin (Hillwig et al., 2011), and complicating comparison of these to KSs – e.g., for analysis of catalytic mechanism (Zi et al., 2014). Although full-length KSLs have been identified from dicots (Sallaud et al., 2012; Zerbe et al., 2013), these are only distantly related to each other and have not yet led to any mechanistic insights.
Castor bean has been reported to produce an interesting set of four biosynthetically related diterpenes derived from ent-CPP. Specifically, ent-beyerene (2), ent-sandaracopimaradiene (3) and ent-trachylobane (4), as well ent-kaurene (Robinson and West, 1970a). Critically, this has been shown to result from the activity of four distinct enzymes rather than being produced by a smaller number of promiscuous cyclases (Robinson and West, 1970b; Spickett et al., 1994). Notably, no KSL specifically producing either ent-beyerene (2) or ent-trachylobane (4), which can be envisioned as arising from deprotonation of plausible intermediates en route to ent-kaurene (Figure 1), has yet been reported. With the recent report of the castor bean genome sequence (Chan et al., 2010), it seemed possible to functionally identify the relevant RcKSLs via a synthetic biology approach. Here we expressed the potentially relevant enzymes from the predicted transcriptome using synthetic gene constructs, in Escherichia coli engineered to produce the necessary ent-CPP precursor via a previously described modular metabolic engineering system (Cyr et al., 2007). These studies not only led to functional identification of novel enzymes, but also provided insights into diterpenoid evolution and the catalytic mechanism of diterpene synthases.
Figure 1.
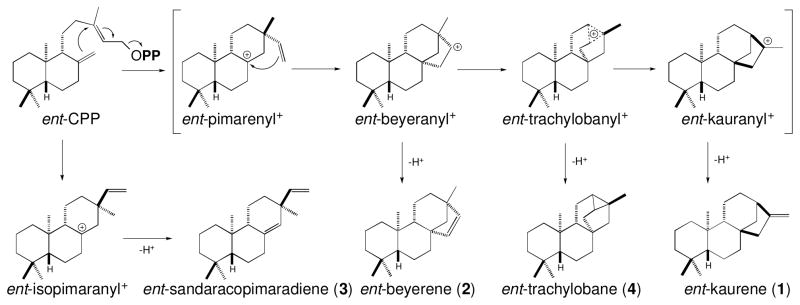
Cyclization mechanisms for the diterpene synthase activities previously identified in castor bean by assays with cell-free extracts (Robinson and West, 1970b; Spickett et al., 1994), and their relationship to the classical mechanism for production of ent-kaurene (1). This classic mechanism proceeds via ionization of the allylic diphosphate ester bond in ent-CPP to trigger initial cyclization to the depicted ent-pimarenyl+ intermediate, followed by secondary cyclization to the depicted ent-beyeranyl+ intermediate that rearranges via the depicted ent-trachylobanyl+ intermediate en route to the ent-kauranyl+ intermediate that is quenched by deprotonation to yield ent-kaurene. As shown, the production of ent-beyerene (4) and ent-trachylobane (2) similarly arises from deprotonation of the corresponding carbocations.
2. Results
2.1 Initial identification of KS(L)s from castor bean
To begin investigating the interesting set of castor bean diterpene synthases, BLAST searches of the available Ricinus communis sequence information were carried out using the KS from Arabidopsis thaliana (AtKS) as a probe for full-length (i.e., γ-domain containing) KS(L)s, as well as the miltiradiene synthase from Salvia miltiorrhiza (SmMS) as a probe for shorter (i.e., non γ-domain containing) KSLs. From this bioinformatic search four full-length (but no shorter) KS(L)s were found among the predicted genes from the reported genome sequence (Chan et al., 2010), termed here RcKS(L)1-4 in the order in which they were listed in the BLAST results (i.e., similarity to AtKS). Initial molecular phylogenetic analysis indicated that the top hit was significantly more closely related to dicot KSs and was termed RcKS(L)1, while the other three clustered separately and were termed RcKSL2-4. RcKSL2-4 were found in close proximity to each other, within a region of 65 kb, with RcKSL2 and 4 occurring as a tandem gene pair. Rather than attempting to clone full-length cDNA for each of these predicted genes, synthetic open reading frames, codon optimized for expression in E. coli, were obtained.
2.2 Biochemical analysis of the RcKS(L)s
We have previously developed a modular metabolic engineering system that enables facile production of terpenoids in E. coli (Cyr et al., 2007). Of particular interest here, this system enables co-expression of a GGPP synthase (GGPS) with potential diterpene synthases, including both a CPS and KS(L). Accordingly, the synthetic RcKS(L) were truncated to remove the N-terminal plastid-directing transit peptide sequences, individually sub-cloned into compatible expression vectors and each co-expressed with either just the GGPS, or the GGPS along with a CPS. This enabled analysis of the ability of the RcKS(L)s to react with either GGPP or any of the three known stereoisomers of CPP (normal, ent or syn). In particular, by extraction of the relevant recombinant culture, which yields any products (i.e., olefins or alcohols) resulting from the removal of the allylic diphosphate group from any of the potential substrates (i.e., GGPP or various stereoisomers of CPP), which were then analyzed by GC-MS (Figure 2). Consistent with its closer relationship to other dicot KSs, RcKS(L)1 was found to only react with ent-CPP, producing small amounts of ent-kaurene (1), and we hereafter refer to this as RcKS1. Both RcKSL2 and RcKSL3 also were found to selectively react with ent-CPP, with RcKSL2 producing primarily ent-trachylobane (2, ~70%) as well as smaller amounts of ent-kaurene (1, ~30%), while RcKSL3 produces ent-sandaracopimaradiene (3, ~94%) along with small amounts of ent-labdatriene (~3%) and ent-pimaradiene (~3%). However, RcKSL4 seemed to be inactive, with no products evident from any substrate.
Figure 2.
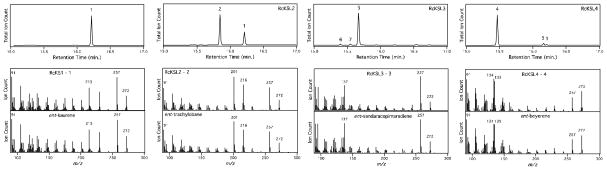
Identification of products formed by the RcKS(L)s by comparison to authentic standards by GC-MS. Chromatographs of the products formed from ent-CPP by corrected RcKS1, RcKSL2, RcKSL3 and corrected RcKSL4, as indicated (with corrections as described in the text). Peaks are labeled with numbering corresponding to that used in the text (with structures shown in Figures 1 and/or 5), along with ent-labdatriene (6) and ent-pimaradiene (7). Also shown are mass spectra of the major enzymatic products, their retention times (RT) were 16. 22 min. for RcKSL1; 15.84 min. for RcKSL2; 15.67 min. for RcKSL3; and 15.46 min. for RcKSL4, along with those from the corresponding authentic standards [RT = 16.23 min. for ent-kaurene (1); 15.85 min. for ent-trachylobane (2); 15.67 min. for ent-sandaracopimaradiene (3); and 15.47 min. for ent-beyerene (4)].
2.3 Correction of RcKSL4
Given that an ent-beyerene synthase was not yet evident, and our interest in such novel catalytic activity, we further investigated the possibility that RcKSL4 might encode this expected enzyme. In particular, we carefully inspected the amino acid (aa) sequence predicted for RcKSL4 and found that this contained a small region that was quite divergent from that present in other KS(L)s (Figure S1). Moreover, the GenBank entry for this predicted gene notes that it spans a gap in the sequencing data and might be missing an exon. Hypothesizing that the observed divergent region might represent incorrect “splicing” by the automated gene prediction algorithm, we cloned a fragment of the RcKSL4 cDNA covering this region, demonstrating that this portion of the predicted RcKSL4 was in fact incorrect. Upon synthesis of a codon-optimized gene for the correct aa sequence and functional analysis as described above, RcKSL4 was demonstrated to selectively react with ent-CPP and produce largely ent-beyerene (4, ~95%) along with very small amounts of ent-atiserene (5, see Figure 5, ~4%) and ent-kaurene (1, ~1%)(Figure 2). The correct exon is not evident in the underlying genome sequence, and presumably lies in the internal gap in the currently available genome sequence for RcKSL4. During this inspection of gene structure, we also noted that the predicted RcKSL4 contains a small insertion in its N-terminal transit peptide that is derived from a small, 39 nt exon, which is not found in the other RcKSLs. We speculate that this exon is incorrectly included, and present the corresponding shorter aa sequence for RcKSL4 here (Figure 3).
Figure 5.
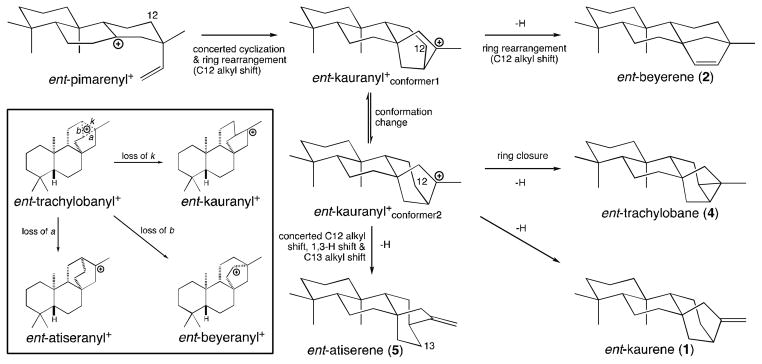
Cyclization mechanism derived from quantum chemical calculation (QCC) for the formation of ent-kaurene (1) and related diterpenes arising from secondary cyclization of the ent-pimarenyl+ intermediate formed by initial cyclization of ent-CPP. The QCC mechanism postulates concerted formation of ent-kaurenyl+, which then serves as a central intermediate en route to all other tetracyclic diterpenes, as well as the pentacyclic ent-trachylobane (2), with the corresponding ent-trachylobanyl+ intermediate not serving as a transition state (i.e., as has been suggested in the classic mechanism – see insert), but rather as arising independently.
Figure 3.
Amino acid sequence alignment of the corrected RcKS1, RcKSL2, RcKSL3 and corrected RcKSL4 (corrections as described in the text). The primary DDxxD and secondary (N/D)Dxx(S/T)xxxE divalent magnesium ion binding motifs are underlined, with an asterisk (*) underneath the previously identified single residue switch position controlling progression to the secondary cyclization step of the KS reaction mechanism.
2.4 Suggested corrections to other RcKS(L)s
Examination of the GenBank entries for all the RcKS(L)s revealed that the entry for RcKSL3 also has been annotated as spanning a gap in the underlying genome sequence data. Nevertheless, our results demonstrate that the predicted RcKSL3 encodes an active enzyme. However, the predicted RcKSL3 aa sequence does appear to have a significantly longer N-terminal transit peptide than the other RcKSL. Examination of the predicted gene structure revealed that this was due to the inclusion of a short 38 nt exon at the 5′ end of the predicted gene that is over 900 nt upstream of the rest of the gene, and an analogous exon is not found in any of the other RcKSLs. We suggest that inclusion of this exon may be incorrect, and present the corresponding shorter open reading frame for RcKSL3 here (i.e., in Figure 3), and the corresponding aa residue numbering also is used here.
Although it was possible to recombinantly express the predicted RcKS1 and observe some production of ent-kaurene, this aa sequence is completely missing the N-terminal transit peptide sequence, and appears to have an extended C-terminus relative to other KS(L)s, which is derived from the last/3′-most exon. We speculate that both the missing and extended sequences are the result of incorrect “splicing” of the predicted RcKS1. Specifically, the absence of two exons at the 5′ end and addition of an “extra” exon on the 3′ end. For example, simply extending the penultimate exon of the predicted gene by 4 nt would incorporate an in-frame stop codon and the resulting protein would closely match the RcKSLs in length. Consistent with this suggestion, simply truncating RcKS1 at this point leads to the production of significantly more (~4-fold) ent-kaurene (1), as an exclusive product, upon incorporation into our metabolic engineering system (Figure 2), and we present the corresponding shorter aa sequence for RcKS1 here (Figure 3).
2.5 Verification and mutation of the secondary Mg2+ binding motif
In our inspection of aa sequence alignment of the RcKS(L)s (Figure 3), we noted that RcKSL2 contained a significant deviation from the usually well-conserved (N/D)Dxx(S/T)xxxE sequence that serves as a secondary magnesium (Mg2+) binding motif, specifically the residues in bold (Christianson, 2006). Previous work has demonstrated that the middle Ser/Thr residue is less well-conserved, with the presence of Gly at this position not infrequently observed, and such substitution is not particularly deleterious, although mutation to Ala was found to severely reduce catalytic activity (Zhou and Peters, 2009). Accordingly, we were surprised to observe that the predicted RcKSL2 aa sequence contains an Ala residue at this position (Ala676). We hypothesized that this might be incorrect and cloned a fragment of the RcKSL2 cDNA covering this region. However, RcKSL2 was found to in fact encode for Ala at this position, and RcKSL2 is active. To investigate the functional consequences of this natural substitution, we changed this Ala to the presumably ancestral Thr. Strikingly, incorporation of the resulting RcKSL2:A676T mutant into our metabolic engineering system led to significantly more product (~60-fold increase), composed of essentially the same mix of ent-trachylobane (4) and ent-kaurene (1) observed with the wild type enzyme. It is unclear if the attenuated activity of the native RcKSL2 provides some unknown selective advantage or if RcKSL2 is undergoing gene decay. Although RcKSL3 contains a Gln at this position, Gln at this position is observed in other class I TPS, and mutation of this to Thr only slightly decreases the amount of ent-sandaracopimaradiene (3) produced upon incorporation of this RcKSL3:Q674T mutant into our metabolic engineering system (~2-fold).
2.7 Molecular phylogenetic analysis
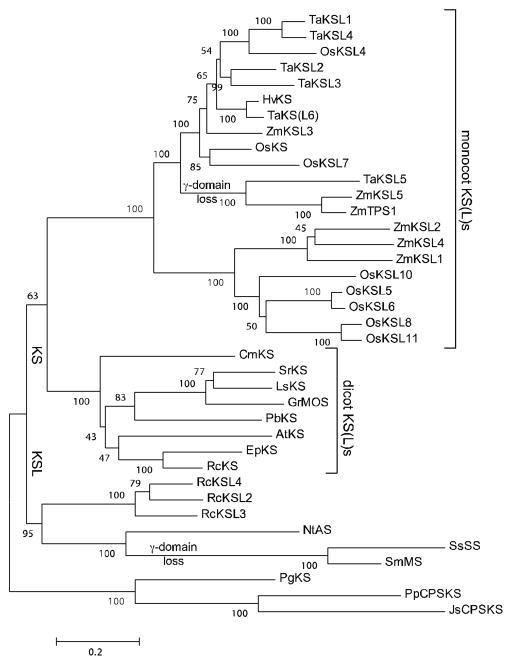
To investigate the origin of the functionally divergent RcKS(L)s identified here, we carried out molecular phylogenetic analysis using the native open reading frame nucleotide sequences of not only these, but also other previously characterized KS(L)s (i.e., TPS-e sub-family members). To enable inference of gene evolutionary history, the resulting phylogenetic tree (Figure 4) has been rooted with the only known gymnosperm (Picea glauca) KS (Keeling et al., 2010), as well as the bifunctional CPS/KS from basal land plants – i.e., the bryophyte Physcomitrella patens (Hayashi et al., 2006) and lycophyte Jungermannia subulata (Kawaide et al., 2011). Notably, the RcKSLs cluster with other functionally distinct TPS-e sub-family members (i.e., none of these are KSs). While such functional diversification might underlie the observed phylogenetic separation (i.e., reflecting positive selection for evolution of novel function), these enzymes actually seem to be under strong purifying selection instead. In particular, the number of synonymous changes per site (dS) exceeds that of non-synonymous changes per site (dN) by > 5 in pairwise comparison of these functionally divergent KSLs with any other TPS-e sub-family member, leading to p-values < 10−6 for purifying selection (i.e., dS/dN is significantly > 1). This may reflect the conservation of more general catalytic activity, as these all still ionize the allylic diphosphate ester of CPP, albeit of various stereochemistry and with differing product outcome. In any case, the separate phylogenetic clustering of these TPS-e sub-family members then is not a result of their neo-functionalization and does indicate homologous origin. Accordingly, despite the lack of any monocot family members in the RcKSL containing cluster, the relevant KS gene duplication event may predate the separation of monocots and dicots, as the TPS-e sub-family otherwise is subsequently split into dicot and monocot clusters.
Figure 4.
Phylogenetic tree for the biochemically characterized members of the TPS-e family. Constructed using MEGA5 (Tamura et al., 2011), specifically the Maximum-Likelihood method from codon-based alignment of the relevant open reading frames as described in the Experimental section. The gymnosperm derived PgKS and non-vascular plant derived PpCPSKS and JsCPSKS serve as the outgroup rooting the tree.
3. Discussion
The TPS-e sub-family of terpene synthases was originally defined as containing only the KSs required for gibberellin biosynthesis (Bohlmann et al., 1998). However, over the last decade it has become evident that this family contains a number of functionally diverse KSLs, which react with one of the various stereoisomers of CPP and catalyze a range of product outcomes, leading to the production of more specialized LRDs (Zi et al., 2014). KSs catalyze a complex carbocation cascade reaction, which can be envisioned as proceeding by initial cyclization of ent-CPP to a pimaradienyl+ intermediate, followed by a secondary cyclization to an ent-beyeranyl+ intermediate that then undergoes ring rearrangement, potentially through an ent-trachylobanyl+ intermediate, to form the final ent-kauranyl+ intermediate that is quenched by specific deprotonation at the neighboring C16 methyl group (Figure 1). While some insight has been reported into enzymatic determinants controlling progression to secondary cyclization in KS(L)s (Morrone et al., 2008; Xu et al., 2007; Zhou and Peters, 2011), little else is known of the structure-function relationships underlying the later steps in production of ent-kaurene (1).
Previous evidence demonstrated that castor bean contains diterpene synthases that react with ent-CPP to produce ent-trachylobane (2) or ent-beyerene (4), as well as ent-sandaracopimaradiene (3) and the requisite ent-kaurene (1)(Robinson and West, 1970b; Spickett et al., 1994). No enzymes catalyzing the production of ent-trachylobane (2) or ent-beyerene (4) had been previously identified. Thus, the recent report of the castor bean genome sequence offered potential access to these functionally novel diterpene synthases (Chan et al., 2010). Here, by incorporating synthetic genes into our modular metabolic engineering system, it was possible to assign each of the four RcKS(L)s found in the castor bean genome to the four expected catalytic activities (Figure 2). Consistent with recent discussion (Keasling et al., 2012), our results support the utility of such a synthetic biology approach to elucidation of natural products biosynthesis. However, our results also highlight the difficulties in applying such an approach on the basis of plant genome sequences alone, as these are most often drafts littered with gaps that impede accurate gene prediction, particularly in the absence of extensive transcriptome sequence information (e.g., from RNA-Seq studies), yet accurate open reading frames are obviously required for biochemical analysis of the encoded enzymes. Here we simply carried out sufficient sequencing to enable such biochemical characterization.
The phylogenetic distance between RcKS1 and the RcKSLs clearly indicates that the RcKSLs are derived from an ancient KS gene duplication and neo-functionalization event (Figure 4). In addition, there is substantial sequence divergence between the RcKSLs themselves, as well as even the most closely related pair RcKSL2 and 4 share only ~75% aa sequence identity. Accordingly, comparison of their sequences, or even molecular models, does not reveal any readily evident determinants for the ability of RcKSL2 and 4 to catalyze similar reactions that nevertheless lead to distinct products. Although RcKSL4 contains a Val in place of the otherwise conserved Ile in the previously identified single residue switch position controlling progression to secondary (tetra)cyclization (Figure 3), the effect of this residue depends on its ability to electrostatically stabilize the initially formed pimarenyl+ (Zhou and Peters, 2011), and such conservative aliphatic substitution does not alter product outcome in KSs – i.e., block secondary cyclization (Wilderman and Peters, 2007). Accordingly, this Val is not expected to have a key role in the production of tetracyclic ent-beyerene (4) by RcKSL4.
Quantum chemical calculation (QCC) analysis of diterpene cyclization indicates that secondary cyclization of the ent-pimarenyl tertiary carbocation intermediate proceeds via concerted, albeit asynchronous cyclization and ring arrangement (Figure 5), in which ent-beyeranyl + and ent-trachylobanyl+ do not serve as a transition state intermediates, leading to direct formation of an ent-kauranyl+ intermediate (Hong and Tantillo, 2010). Recently, experimental support for such counter-intuitive cyclization mechanisms proposed by QCC has been reported (Zu et al., 2012). However, aza analogs of ent-beyeranyl+ and ent-trachylobanyl+ readily bind and inhibit KSs, with synergistic effects from inorganic pyrophosphate (mimicking the diphosphate co-product in the catalyzed reaction) increasing affinity beyond that implied for the substrate by the pseudo-binding constant KM, indicating that the enzyme can stabilize these potential high-energy intermediates (Roy et al., 2007). Thus, the KS catalyzed reaction might differ from that implied by QCC.
Intriguingly, the somewhat counterintuitive mechanism suggested by QCC is consistent with the observed specificity of the characterized KSs, all of which exclusively produce ent-kaurene (1). This includes not only plant KSs such as the RcKS1 characterized here (Figure 2), but also the phylogenetically unrelated KSs from fungi and bacteria (Hershey et al., 2014; Kawaide et al., 1997; Morrone et al., 2009; Toyomasu et al., 2000). More speculatively, such facile and selective formation might underlie the original selection of ent-kaurene (1) for elaboration to a phytohormone (i.e., the gibberellins).
In any case, the previously reported QCC analysis further indicated that the production of ent-trachylobane (2) or ent-beyerene (4) also proceeds via initial formation of an ent-kauranyl+ intermediate, rather than reflecting deprotonation of the corresponding carbocationic intermediates en route to ent-kauranyl+ (Hong and Tantillo, 2010). Given that the minor products from TPS catalyzed reactions must represent accessible carbocation intermediates, particularly including intermediates on the major reaction pathway, the production of small amounts of ent-kaurene (1) by the ent-trachylobane and ent-beyerene synthases RcKSL2 and 4, respectively (Figure 2), is consistent with an intermediary role for ent-kauranyl+ in the catalyzed reactions. Perhaps even more informative is the absence of ent-trachylobane (2) production by RcKSL4, despite its production of ent-atiserene (5) and ent-kaurene (1) as well as its major ent-beyerene (4) product. In particular, the production of all three of these can be envisioned as proceeding through ent-trachylobanyl+ (Figure 5, inset), but the observed lack of ent-trachylobane (2) production is consistent with the previously reported QCC analysis (Hong and Tantillo, 2010), which indicated that this distributed carbonium cation is not an intermediate en route to any of the observed products [i.e., ent-beyerene (4), ent-atiserene (5) and ent-kaurene (1)]. Similarly, the production of only ent-kaurene (1), and not ent-beyerene (4) or ent-atiserene (7), as side-products by the ent-trachylobane synthase RcKSL2 also argues against a central role for the ent-trachylobanyl+ intermediate. Accordingly, the product distribution observed here (Figure 3) seems to be consistent with the mechanism derived by QCC analysis, with facile rearrangement to a central ent-kauranyl+ intermediate, rather than the series of carbocations classically assigned to its formation (c.f., Figures 1 and 5).
4. Conclusions
In summary, we report here the use of a synthetic biology approach to identify novel diterpene synthases from castor bean, RcKS(L)s. While this approach offers the ability to quickly functionally characterize such biosynthetic enzymes, such rapid progress was impeded here by inaccurate gene predictions, which were a result of the draft nature of the available castor bean genome and lack of in-depth transcriptome sequence information. Nevertheless, it was possible to biochemically characterize all four of the RcKS(L)s from the castor bean genome, which matched the ent-kaurene (RcKS1), ent-trachylobane (RcKSL2), ent-sandaracopimaradiene (RcKSL3), and ent-beyerene (RcKSL4) synthases expected from previous cell-free extract assay based studies. Interestingly, the observed product distributions of these diterpene synthases are consistent with the concerted formation of a central ent-kauranyl+ intermediate indicated by quantum chemical calculations, rather than the preceding formation of ent-beyeranyl+ and ent-trachylobanyl+ intermediates classically invoked in formation of ent-kaurene. Looking forward, while there is substantial phylogenetic distance between the RcKS(L)s, these nevertheless provide a set of related KS(L)s for further mechanistic investigation.
5. Experimental
5.1 General
Unless otherwise noted, all chemical reagents were purchased from Fisher Scientific (Loughborough, Leicestershire, UK), and molecular biology reagents from Invitrogen (Carlsbad, CA, USA). All bacterial (E. coli) growth was carried out using TB media, using the TOP10 strain for molecular biology manipulations (e.g., sub-cloning) and the OverExpress C41 strain (Lucigen, Middleton, WI, USA) for recombinant expression. Gas chromatography with mass spectrometric detection (GC-MS) analyses were performed using a Varian (Palo Alto, CA, USA) 3900 instrument with HP-5ms narrowbore (30 m long/0.25 mm diameter/0.25 μm film) column and Saturn 2100 ion trap mass spectrometer in electron ionization (70 eV) mode. Samples (1 μL) were injected in splitless mode and an injector temperature of 250 °C, with a 1.2 mL/min flow rate of helium and an oven temperature of 50 °C, after a 3 min. hold, the temperature raised at 14 °C/min. to 300 °C, where it was held for 3 min. MS data was collected from m/z from 90 to 600, starting 12 min. after the injection until the end of the run. Sequence alignments were generally carried out using the CLC Main Workbench software package (version 6.9.1) – i.e., other than for phylogenetic analysis, which is described in more detail below.
5.2 Identification, synthesis and cloning of the RcKS(L)s
BLAST searches were carried out using the aa sequence of AtKS (AAC39443) and, separately, SmMS (ABV08817), to probe the sequences available for castor bean in GenBank (TaxID 3988). The identified RcKS(L)1-4 correspond to accessions XM_002533648, XM_002525795, XM_002525790 and XM_002525796, respectively. These further correspond to GenBank GeneIDs RCOM_0306870, RCOM_0823630, RCOM_0823080 and RCOM_0823640, respectively, with the proximity of RcKSL2-4 implied by the numbering of their GeneIDs verified by visual inspection of their genomic context using the GenBank genome viewer (none of the other genes in this region appear to have a plausible role in diterpenoid biosynthesis). The corresponding genomic sequence and flanking regions for gene structure analysis were obtained from Phytozome (www.phytozome.net). Synthetic genes corresponding to the aa sequences predicted for RcKS(L)1-4 were obtained from GenScript (Piscataway, NJ, USA). The full-length RcKSLs (see supporting information for synthetic genes) were truncated to remove the N-terminal plastid targeting peptide sequence (corresponding to the first 49 aa residues shown in Figure 3) by PCR amplification and sub-cloned into pENTR/SD/D-TOPO. These clones were subsequently transferred via directional recombination to the T7-based N-terminal GST fusion expression vector pDEST15.
5.3 Verification and correction of the RcKS(L)
In order to investigate the lack of activity with the predicted RcKSL4, a suspect exon was investigated by partial cloning of a fragment of the RcKSL4 mRNA that covered this region (see Figure S1). For this purpose, castor bean plants were grown and a cDNA library prepared from leaf material using SuperScript III reverse transcriptase. A fragment of RcKSL4 was amplified by PCR using primers RcKSL4-F1 and RcKSL4-R1 (Table S1), cloned into pZeroBluntII and sequenced. The sequence of this fragment demonstrated that the predicted RcKSL4 was in fact incorrect. A synthetic gene corresponding to the correct sequence was then obtained, again from GenScript. The secondary Mg2+ binding motif of RcKSL2 was verified in much the same fashion – i.e., amplification of a covering fragment of the RcKSL2 mRNA by PCR using the same cDNA library as template, and primers RcKSL2-F1 and RcKSL2-R1 (Table S1). As suggested by aa sequence alignment and comparison of the underlying gene structure to that of the RcKSLs, the predicted RcKS1 was truncated to remove the last exon. This version of RcKS1 corresponds to the aa sequence shown in Figure 3. Expression constructs (pDEST15 based) for these corrected versions of RcKSL4 and RcKS1 were generated by the same cloning procedure described above.
5.4 Mutagenesis
Site directed mutants were constructed by PCR amplification of the relevant truncated pENTR constructs using the primers described in Table S1 and Pfx DNA polymerase. The resulting constructs were verified by complete gene sequencing before being transferred via directional recombination to the T7-based N-terminal GST fusion expression vector pDEST15.
5.5 Biochemical characterization via metabolic engineering
The RcKS(L)s were biochemically characterized by use of our previously described modular metabolic engineering system (Cyr et al., 2007). Briefly, class I labdane-related diterpene synthases such as these RcKS(L)s are co-expressed, from pDEST expression vectors, with pACYC-Duet (Novagen/EMD) derived plasmids that carry a GGPP synthase and CPS (pGGxC). These pGGxC plasmids are compatible with pET based vectors such as the pDEST15 constructs described above for the RcKS(L)s, and can be induced to produce any one of the three most common stereoisomers of CPP, specifically pGGnC leads to production of normal CPP, pGGeC leads to production of ent-CPP, and pGGsC leads to production of syn-CPP. Accordingly, the RcKS(L)s were separately co-expressed with each of the three pGGxC vectors (i.e., in all possible pairings), along with the additionally co-compatible pIRS, which enables induction of key steps of the endogenous isoprenoid precursor pathway (Morrone et al., 2010), increasing flux to (di)terpenoid production. The resulting recombinant bacteria were then analyzed as previously described (Morrone et al., 2009). Briefly, 50 mL TB liquid media cultures were grown with shaking at 200 rpm to A600 ~ 0.8 at 37 °C, the temperature reduced to 16 °C for 1 hr prior to induction with IPTG (added to a final concentration of 0.5 mM), along with 10 mM pyruvate and 5 mM MgCl2, followed by continued fermentation at 16 °C for an additional ~72 hr. These cultures were then extracted with an equal volume of hexanes (overlaid, swirled gently for 30 sec. and left to settle overnight at 4 °C prior to separation), dried under N2, and resuspended in 1 mL fresh hexanes for GC-MS analysis, with product identification accomplished by comparison to authentic standards.
5.6 Molecular phylogenetic analysis
All phylogenetic analysis was carried out using the MEGA 5.2.2 software package for Mac (Tamura et al., 2011). These analyses were based on the corrected open reading frame sequences for the RcKS(L)s described here, along with those of other biochemically characterized KS(L)s (i.e., TPS-e sub-family members) listed here: AtKS (AF034774); Cucurbita maxima KS (CmKS, U43904); Lactuca sativa KS (LsKS, AB031205); Euphorbia peplus KS (EpKS, KC702395); Plectranthus barbatus KS (PbKS, KC702394); Stevia rebaudiana KS (SrKS, AF097311); Grindelia hirsutula KSL/manoyl oxide synthase (GrMOS, KC702399); Nicotiana tabacum KSL/Z-abienol synthase (NtAS, HE588140); SmMS, (EF635966); Salvia sclarea KSL/sclareol synthase (SsSS, JN133922); rice (Oryza sativa) KS(L)s (OsKS, AB126933; OsKSL4, AY616862; OsKSL5, DQ823352; OsKSL6, DQ823353; OsKSL7, DQ823354; OsKSL8, AB118056; OsKSL10, DQ823355; OsKSL11, DQ100373); wheat (Triticum aestivum) KS(L)s (TaKS(L6), AB597962; TaKSL1, AB597957; TaKSL2, AB597958; TaKSL3, AB597959; TaKSL4, AB597960; TaKSL5, AB597961); maize (Zea mays) KS(L)s (ZmKSL1, AFW61735; ZmKSL2, DAA54948; ZmKSL3, DAA36069; ZmKSL4, DAA49845; ZmKSL5, NM_001148416; ZmTPS1, NM_001111627) barley (Hordeum vulgare) KS (HvKS, AY551436); Picea glauca KS (PgKS, GU045756); P. patens CPS/KS (PpCPSKS, AB302933); and J. subulata CPS/KS (JsCPSKS, AB563712). These nucleotide sequences were aligned by codons using the MUSCLE algorithm with default settings (e.g., gap opening penalty of −12, gap extension penalty of −1, and UPGMB clustering). This alignment was used to calculate phylogenetic trees by both the Neighbor-Joining and Maximum-Likelihood methods, with the reliability in each case tested by bootstrapping with 100 replicates. The Maximum-Likelihood tree is shown here (Figure 4), as this directly yielded the expected PgKS, PpCPSKS and JsCPSKS containing outgroup, although both yielded essentially equivalent trees. This Maximum-Likelihood tree was generated using the calculated optimal parameters, namely the General Time Reversible (GTR) model with discrete Gamma distribution to model evolutionary rate differences between sites (+G), allowing some sites to be evolutionarily invariable (+I). All possible codon positions were included, although any position with less than 95% coverage was excluded, leaving a total of 2163 positions in the final dataset. Evolutionary pressure was probed by codon-based Z-tests of selection using the Nei-Gojobori method for analysis with variance calculated by bootstrapping with 100 replicates. All ambiguous positions were removed for each sequence pair, leaving a total of 958 positions in the final dataset. Such analysis averaged over all the sequence pairs demonstrated that the TPS-e family is under strong purifying selection, with dS – dN > 32 and a p-value of < 10−10. Such analysis between all pairs of sequences further demonstrated that even the functionally divergent cluster that the RcKSLs are found in also is under strong purifying selection, with any pairwise comparison exhibiting dS – dN > 5 and p-values < 10−6.
Supplementary Material
Highlights.
Functional genomics + synthetic biology approach to castor bean diterpene synthases
These catalyze mechanistically related cyclization reaction
Product outcome supports quantum chemical calculations of diterpene cyclization
Acknowledgments
We thank Dr. Robert M. Coates (Univ. of Illinois) for graciously providing authentic samples of ent-trachylobane (2) and ent-beyerene (4). This study was supported in part by a grant from the NIH to R.J.P. (GM076324).
Abbreviations
- aa
amino acid
- KS
ent-kaurene synthase
- KSL
KS-like
- RcKS(L)
castor bean (Ricinius communis) KS(L)
- AtKS
Arabidopsis thaliana KS
- CPP
copalyl diphosphate
- CPS
CPP synthase
- GC-MS
gas chromatography with mass spectrometric detection
- GGPP
(E,E,E)-geranygeranyl diphosphate
- nt
nucleotide
- TPS
terpene synthase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bohlmann J, Meyer-Gauen G, Croteau R. Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA. 1998;95:4126–4133. doi: 10.1073/pnas.95.8.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caniard A, Zerbe P, Legrand S, Cohade A, Valot N, Magnard JL, Bohlmann J, Legendre L. Discovery and functional characterization of two diterpene synthases for sclareol biosynthesis in Salvia sclarea (L.) and their relevance for perfume manufacture. BMC Plant Biol. 2012;12:119. doi: 10.1186/1471-2229-12-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AP, Crabtree J, Zhao Q, Lorenzi H, Orvis J, Puiu D, Melake-Berhan A, Jones KM, Redman J, Chen G, Cahoon EB, Gedil M, Stanke M, Haas BJ, Wortman JR, Fraser-Liggett CM, Ravel J, Rabinowicz PD. Draft genome sequence of the oilseed species Ricinus communis. Nat Biotechnol. 2010;28:951–956. doi: 10.1038/nbt.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tholl D, Bohlmann J, Pichersky E. The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011;66:212–229. doi: 10.1111/j.1365-313X.2011.04520.x. [DOI] [PubMed] [Google Scholar]
- Christianson DW. Structural biology and chemistry of the terpenoid cyclases. Chem Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- Cyr A, Wilderman PR, Determan M, Peters RJ. A Modular Approach for Facile Biosynthesis of Labdane-Related Diterpenes. J Am Chem Soc. 2007;129:6684–6685. doi: 10.1021/ja071158n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Hillwig ML, Huang L, Cui G, Wang X, Kong J, Yang B, Peters RJ. A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org Lett. 2009;11:5170–5173. doi: 10.1021/ol902051v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Kawaide H, Notomi M, Sakigi Y, Matsuo A, Nozaki H. Identification and functional analysis of bifunctional ent-kaurene synthase from the moss Physcomitrella patens. FEBS Lett. 2006;580:6175–6181. doi: 10.1016/j.febslet.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Hershey DM, Lu X, Zi J, Peters RJ. Functional conservation of the capacity for ent-kaurene biosynthesis and an associated operon in certain rhizobia. J Bact. 2014;196:100–106. doi: 10.1128/JB.01031-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillwig ML, Xu M, Toyomasu T, Tiernan MS, Gao W, Cui G, Huang L, Peters RJ. Domain loss has independently occurred multiple times in plant terpene synthase evolution. Plant J. 2011;68:1051–1060. doi: 10.1111/j.1365-313X.2011.04756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YJ, Tantillo DJ. Formation of beyerene, kaurene, trachylobane, and atiserene diterpenes by rearrangements that avoid secondary carbocations. J Am Chem Soc. 2010;132:5375–5386. doi: 10.1021/ja9084786. [DOI] [PubMed] [Google Scholar]
- Kawaide H, Hayashi K, Kawanabe R, Sakigi Y, Matsuo A, Natsume M, Nozaki H. Identification of the single amino acid involved in quenching the ent-kauranyl cation by a water molecule in ent-kaurene synthase of Physcomitrella patens. FEBS J. 2011;278:123–133. doi: 10.1111/j.1742-4658.2010.07938.x. [DOI] [PubMed] [Google Scholar]
- Kawaide H, Imai R, Sassa T, Kamiya Y. Ent-kaurene synthase from the fungus Phaeosphaeria sp L487 cDNA isolation, characterization, and bacterial expression of a bifunctional diterpene cyclase in fungal gibberellin biosynthesis. J Biol Chem. 1997;272:21706–21712. doi: 10.1074/jbc.272.35.21706. [DOI] [PubMed] [Google Scholar]
- Keasling JD, Mendoza A, Baran PS. Synthesis: A constructive debate. Nature. 2012;492:188–189. doi: 10.1038/492188a. [DOI] [PubMed] [Google Scholar]
- Keeling CI, Dullat HK, Yuen M, Ralph SG, Jancsik S, Bohlmann J. Identification and functional characterization of monofunctional ent-copalyl diphosphate and ent-kaurene synthases in white spruce reveal different patterns for diterpene synthase evolution for primary and secondary metabolism in gymnosperms. Plant Physiol. 2010;152:1197–1208. doi: 10.1104/pp.109.151456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köksal M, Jin Y, Coates RM, Croteau R, Christianson DW. Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis. Nature. 2011;469:116–120. doi: 10.1038/nature09628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone D, Chambers J, Lowry L, Kim G, Anterola A, Bender K, Peters RJ. Gibberellin biosynthesis in bacteria: Separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum. FEBS Lett. 2009;583:475–480. doi: 10.1016/j.febslet.2008.12.052. [DOI] [PubMed] [Google Scholar]
- Morrone D, Lowry L, Determan MK, Hershey DM, Xu M, Peters RJ. Increasing diterpene yield with a modular metabolic engineering system in E. coli: comparison of MEV and MEP isoprenoid precursor pathway engineering. Appl Microbiol Biotechnol. 2010;85:1893–1906. doi: 10.1007/s00253-009-2219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone D, Xu M, Fulton DB, Determan MK, Peters RJ. Increasing complexity of a diterpene synthase reaction with a single residue switch. J Am Chem Soc. 2008;130:5400–5401. doi: 10.1021/ja710524w. [DOI] [PubMed] [Google Scholar]
- Peters RJ. Uncovering the complex metabolic network underlying diterpenoid phytoalexin biosynthesis in rice and other cereal crop plants. Phytochemistry. 2006;67:2307–2317. doi: 10.1016/j.phytochem.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Peters RJ. Two rings in them all: The labdane-related diterpenoids. Nat Prod Rep. 2010;27:1521–1530. doi: 10.1039/c0np00019a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DR, West CA. Biosynthesis of cyclic diterpenes in extracts from seedlings of Ricinus communis L. I. Identification of diterpene hydrocarbons formed from mevalonate. Biochemistry. 1970a;9:70–79. doi: 10.1021/bi00803a010. [DOI] [PubMed] [Google Scholar]
- Robinson DR, West CA. Biosynthesis of cyclic diterpenes in extracts from seedlings of Ricinus communis L. II. Conversion of geranylgeranyl pyrophosphate into diterpene hydrocarbons and partial purification of the cyclization enzymes. Biochemistry. 1970b;9:80–89. doi: 10.1021/bi00803a011. [DOI] [PubMed] [Google Scholar]
- Roy A, Roberts FG, Wilderman PR, Zhou K, Peters RJ, Coates RM. 16-Aza-ent-beyerane and 16-Aza-ent-trachylobane: Potent Mechanism Based Inhibitors of Recombinant ent-Kaurene Synthase from Arabidopsis thaliana. J Am Chem Soc. 2007;129:12453–12460. doi: 10.1021/ja072447e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallaud C, Giacalone C, Topfer R, Goepfert S, Bakaher N, Rosti S, Tissier A. Characterization of two genes for the biosynthesis of the labdane diterpene Z-abienol in tobacco (Nicotiana tabacum) glandular trichomes. Plant J. 2012;72:1–17. doi: 10.1111/j.1365-313X.2012.05068.x. [DOI] [PubMed] [Google Scholar]
- Schalk M, Pastore L, Mirata MA, Khim S, Schouwey M, Deguerry F, Pineda V, Rocci L, Daviet L. Towards a Biosynthetic Route to Sclareol and Amber Odorants. J Am Chem Soc. 2012;134:18900–18903. doi: 10.1021/ja307404u. [DOI] [PubMed] [Google Scholar]
- Schmelz EA, Huffaker A, Sims J, Christensen S, Lu X, Okada K, Peters RJ. Biosynthesis, regulation and roles of monocot terpenoid phytoalexins. Plant J. 2014 doi: 10.1111/tpj.12436. in press. [DOI] [PubMed] [Google Scholar]
- Spickett CM, Ponnamperuma K, Abell C. The resolution of diterpene cyclase activities from Ricinus communis. Phytochemistry. 1994;37:971–973. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomasu T, Kawaide H, Ishizaki A, Shinoda S, Otsuka M, Mitsuhashi W, Sassa T. Cloning of a full-length cDNA encoding ent-kaurene synthase from Gibberella fujikuroi: functional analysis of a bifunctional diterpene cyclase. Biosci Biotechnol Biochem. 2000;64:660–664. doi: 10.1271/bbb.64.660. [DOI] [PubMed] [Google Scholar]
- Wilderman PR, Peters RJ. A single residue switch converts abietadiene synthase into a pimaradiene specific cyclase. J Am Chem Soc. 2007;129:15736–15737. doi: 10.1021/ja074977g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Wilderman PR, Peters RJ. Following evolution’s lead to a single residue switch for diterpene synthase product outcome. Proc Natl Acad Sci USA. 2007;104:7397–7401. doi: 10.1073/pnas.0611454104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbe P, Hamberger B, Yuen MM, Chiang A, Sandhu HK, Madilao LL, Nguyen A, Hamberger B, Bach SS, Bohlmann J. Gene discovery of modular diterpene metabolism in nonmodel systems. Plant Physiol. 2013;162:1073–1091. doi: 10.1104/pp.113.218347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Peters RJ. Investigating the conservation pattern of a putative second terpene synthase divalent metal binding motif in plants. Phytochemistry. 2009;70:366–369. doi: 10.1016/j.phytochem.2008.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Peters RJ. Electrostatic effects on (di)terpene synthase product outcome. ChemComm. 2011;47:4074–4080. doi: 10.1039/c0cc02960b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Xu M, Tiernan MS, Xie Q, Toyomasu T, Sugawara C, Oku M, Usui M, Mitsuhashi W, Chono M, Chandler PM, Peters RJ. Functional characterization of wheat ent-kaurene(-like) synthases indicates continuing evolution of labdane-related diterpenoid metabolism in the cereals. Phytochemistry. 2012;84:47–55. doi: 10.1016/j.phytochem.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi J, Mafu S, Peters RJ. To gibberellins and beyond! Surveying the evolution of diterpenoid metabolism. Annu Rev Plant Biol. 2014 doi: 10.1146/annurev-arplant-050213-035705. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu L, Xu M, Lodewyk MW, Cane DE, Peters RJ, Tantillo DJ. Effect of isotopically sensitive branching on product distribution for pentalenene synthase: support for a mechanism predicted by quantum chemistry. Journal of the American Chemical Society. 2012;134:11369–11371. doi: 10.1021/ja3043245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.