Abstract
Purpose.
While some evidence suggests an essential role for the chaperonin containing t-complex protein 1 (CCT) in ciliogenesis, this function remains poorly understood mechanistically. We used transgenic mice, previously generated in our lab, and characterized by a genetically-induced suppression of CCT in rod photoreceptors as well as a malformation of the rod sensory cilia, the outer segments, to gain new insights into this underlying molecular mechanism.
Methods.
The CCT activity in rod photoreceptors of mice was suppressed by overexpressing the chaperonin inhibitor, phosducin-like protein short, and the ensuing changes of cellular morphology were analyzed by light and electron microscopy. Protein expression levels were studied by fluorescent microscopy and Western blotting.
Results.
Suppressing the chaperonin made the photoreceptors incompetent to build their outer segments. Specifically, the CCT-deficient rods appeared unable to expand the outer segment plasma membrane, and accommodate growth of this compartment. Seeking the molecular mechanisms underlying such a shortcoming, we found that the affected rods could not express normal levels of Bardet-Biedl Syndrome (BBS) proteins 2, 5, and 7 and, owing to that deficiency, were unable to assemble the BBSome, a multisubunit complex responsible for ciliary trafficking. A similar effect in response to the chaperonin suppression was also observed in cultured ciliated cells.
Conclusions.
Our data provide new evidence indicating the essential role of the chaperonin CCT in the biogenesis of vertebrate photoreceptor sensory cilia, and suggest that it may be due to the direct participation of the chaperonin in the posttranslational processing of selected BBS proteins and assembly of the BBSome.
Keywords: rods, chaperonin CCT, outer segments, Bardet-Biedl Syndrome, BBSome
We found that functional chaperonin containing t-complex protein 1 (CCT) is necessary for the development of rod outer segments in mice. Our data suggest that CCT is engaged in the folding of several Bardet-Biedl Syndrome (BBS) proteins required for the assembly of the ciliary trafficking complex known as the BBSome.
Introduction
The chaperonin containing t-complex protein 1, CCT (also known as TRiC), is a large ATPase complex found in all eukaryotic cells, which mediates the folding of newly translated proteins in its inner cavity.1,2 However, the list of identified chaperonin substrates is incomplete, and all known CCT clients are soluble proteins of a wide range of structures and functions.3–6 In the cell, the most abundant clients of CCT are represented by cytoskeletal proteins from the actin and tubulin families.7–11 Therefore, several studies using the ciliate Tetrahymena, which pioneered the notion of the essential role of the chaperonin in cilium biogenesis, attributed its role primarily to cytoskeleton maintenance in this organelle.12–16 However, in vivo roles of CCT in vertebrates, particularly at the organism level, are not well understood. When its function was revisited in a vertebrate zebrafish model, it was demonstrated that knockdown of the chaperonin subunits disrupts trafficking through the cilium.17 The same study also revealed a functional connection between CCT and the BBSome, a complex thought to control trafficking of molecules into the cilium.18–20 In humans, mutations in certain genes give rise to an autosomal recessive ciliopathy, known as Bardet-Biedl syndrome (BBS), characterized, among many other abnormalities, by the development of rod–cone dystrophy.21,22 Seven of those genes (BBS 1, 2, 4, 5, 7, 8, and 9) encode BBSome subunits,18,23 while the other three (BBS 6, 10, and 12) are chaperonin homologues mediating BBSome assembly.17,24–26 Thus, the rod–cone dystrophy in Bardet-Biedl syndrome patients appears to be a consequence of defective trafficking through the sensory cilia of the photoreceptor cells with the affected BBSome, a common culprit of various types of retinal neurodegeneration.27 To further explore the role of CCT in the assembly of the BBSome and ciliogenesis in vertebrate photoreceptors, we inhibited the activity of CCT in mouse rods in vivo. This manipulation completely disrupted the formation of the rod outer segments. This defect was preceded by a profound reduction in the levels of BBS2, BBS5, and BBS7 proteins that interfered with the normal assembly of the BBSome in these cells. Failure of BBS assembly as a result of suppression of the chaperonin was then demonstrated in cell culture. These data provide new evidence of the role of the chaperonin CCT in the biogenesis of the BBSome, and emphasize the importance of this complex in the development and maintenance of the rod outer segment.
Materials and Methods
Animal Model
All experiments involving Δ1-83PhLP-FLAG transgenic mice28 were performed according to procedures approved by the Animal Care and Use Committee of West Virginia University and in adherence with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Quantification of Proteins by Western Blotting
To detect Δ1-83PhLP-FLAG, two retinas were homogenized in 0.2 mL of RIPA buffer (R0278; Sigma, St. Louis, MO, USA), cellular insoluble parts were cleared by centrifugation, and each supernatant was incubated with 20 μL of anti-FLAG M2-Agarose beads (A2220, Sigma) for 1 hour at room temperature. Beads were washed four times for 3 minutes with 1.0 mL of buffer, and the captured Δ1-83PhLP-FLAG was eluted with 30 μL of urea sample buffer containing 125 mM Tris/HCl, pH 6.8, 4% SDS, 6 M urea, and bromophenol blue tracking dye. The levels of BBS proteins were compared in the retinas of Δ1-83PhLP-FLAG−/− and Δ1-83PhLP-FLAG± littermates at postnatal day 10 (P10) by quantitative Western blotting as previously described.28 Quantification of the specific bands was performed on an Odyssey Infrared Imaging System (LICOR Biosciences, Lincoln, NE, USA).
Sedimentation Analysis of BBSome Complexes
BBSome complexes were detected by Western blotting following fractionation of tissue extracts on a 10% to 40% sucrose gradient by ultracentrifugation as previously described.29 The analyses were performed using whole eyes from Δ1-83PhLP-FLAG−/− and Δ1-83PhLP-FLAG± littermates harvested at P10.
Microscopy Procedures
For light microscopy, sectioning and hematoxylin-eosin staining of ocular tissue was performed by Excaliber Pathology, Inc. (Oklahoma City, OK, USA). Four-micron thick retinal cross-sections were viewed using an Olympus AZ70 microscope (Olympus, Center Valley, PA, USA) equipped with a digital camera. For immunofluorescence microscopy, eyes were first enucleated, and fixed for 10 minutes with Oculo Fix (BBC Biochemical, Stanwood, Washington, USA). The anterior portions of the fixed eyes were then removed, and the eyecups were further fixed with Oculo Fix for 3 hours. Fixed eyecups were then cryoprotected with 30% sucrose in 1X PBS overnight. The eye cups were then transferred to a 1:1 Tissue Freezing Medium (TFM; TBS Triangle Biomedical Sciences, Inc., Durham, NC, USA) −30% sucrose solution for 2 hours, and then embedded in TFM, and frozen. The frozen blocks were then cut radially in 16-μm sections on a Leica CM1850 Cryostat (Buffalo Grove, IL, USA), and the sections stored at −20°C. Prior to immunostaining, frozen sections were thawed at room temperature for 10 minutes, hydrated in 1× PBS for 5 minutes, and blocked with goat-serum containing 5% Triton X-100 for 30 minutes. Sections were first probed with primary antibody against rhodopsin and peripherin/rds at a 1:1000 dilution overnight, and then probed with Alexa Fluor 488 or 568 conjugated secondary antibody (Invitrogen/Life Technologies, Grand Island, NY, USA) for 1 hour. The tissue sections were also counter stained with 100ng/ml DAPI (4′,6-Diamidine-2′-phenylindol dihydrochloride; Roche, Indianapolis, IN, USA) for rod nuclear staining. Images were acquired on a Zeiss LSM 510 Meta confocal microscope after adjusting the detector gain for all the fluorescent channels and the amplifier offset for background corrections, and while keeping the amplifier gain of the lasers to a minimum. Identical parameters were used when comparing samples. For electron microscopy, whole eyes were cleaned of extraneous tissue, and fixed overnight in Karnovsky's fixative containing 2% paraformaldehyde, 2.5% glutaraldehyde, and 0.1 M phosphate buffer (pH 7.4; 15720; Electron Microscopy Sciences, Hatfield, PA, USA). Whole eyes were rinsed with 0.1 M phosphate buffer, and post-fixed in 2% osmium tetroxide in distilled water for 1.5 hours. They were then gradually dehydrated in increasing concentrations of ethanol (25%–100%), transitioned in propylene oxide, and embedded in tEpon (Tousimis, Rockville, MD, USA). Sections were cut at 1 μm on a Leica UCT, examined, and the area for ultrastructural analysis chosen. Thin sections were cut at 70 nm on the same ultramicrotome. Photoreceptor images were acquired using a Philips CM-10 (Philips, Andover, MA, USA) electron microscope equipped with an SIA digital camera (SIA Scientific Instruments and Applications, Duluth, GA, USA).
Electroretinography
Mice were dark-adapted for 2 hours, and anesthetized with a 2% isoflurane/50% oxygen mixture. The animal's pupils were dilated with a mixture of 1.25% phenylephrine hydrochloride and 0.5% tropicamide ophthalmic solutions for 20 minutes. A disposable reference needle electrode (LKC Technologies, Gaithersburg, MD, USA) was inserted under the loose skin between the ears, and then custom-made silver wire electrodes were positioned on the corneas. Ganzfeld flash ERG recording was carried out in UTAS-E4000 Visual Electrodiagnostic Test System using EMWIN 8.1.1 software (LKC Technologies) according to the manufacturer's protocols.
Tissue Culture
The RPE1-hTERT cell line, expressing the green fluorescent protein (GFP)-BBS4 fusion construct (localization and purification [LAP]-BBS4), was previously described.30 The cells were maintained in Dulbecco's Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F-12 from ATCC) complete medium at 37°C, and supplemented with 10% (vol/vol) fetal bovine serum (FBS), 100 U ml−1 penicillin, 100 μg ml−1 streptomycin, 2.5 mM L-glutamine, and 15 mM HEPES. For the transient expression of Δ1-83PhLP-FLAG or phosducin,31 cells were plated in 6-well plates, and grown to at least 50% confluency. The following day, cells were transfected with 2 μg of total plasmid DNA using FuGENE 6 Transfection reagent (Roche and Promega, Madison, WI, USA) at a 6:1 FuGENE 6: plasmid DNA ratio. Empty pTriEx 4 vector was used as control. Cells were collected after 48 hours of transfection, dissolved in urea sample buffer, and the protein expression was analyzed by Western blotting using antibodies listed below. For the immunofluorescence microscopy, we used the same procedure with the exception that the cells were grown on coverslips. To distinguish transfected and nontransfected cells, pCITF2 vector expressing tdTomato, a gift from E. Tucker, West Virginia University, was made from pCIG2 vector32 by replacing the coding sequence of GFP with tdTomato (gift from R. Tsien, University of California, San Diego, USA) and also removing a stray Notl site from the tdTomato cassette. This vector was added to each plasmid vector at a 1:1 ratio. Following transfection, cells were fixed in 4% paraformaldehyde for 10 minutes, followed by chilled methanol for 5 minutes at −20°C, and blocked in 5.5% BSA in PBS. The LAP-BBS4 signal was enhanced with anti-GFP antibody. Cell nuclei were stained DAPI. Direct fluorescence of tdTomato was detected at 568 nm. Images were collected using an Olympus fluorescence microscope or a Zeiss LSM 510 Meta confocal microscope.
Antibodies
Proteins were detected using antibodies against FLAG (600-401-383; Rockland, Gilbertsville, PA, USA), BBS1 (sc-134455; Santa Cruz Biotechnology, Dallas, TX, USA), BBS2 (11188-2-AP; Proteintech, Chicago, IL, USA), BBS4 (sc-67201; Santa Cruz Biotechnology), BBS5 (14569-1-AP; Proteintech), BBS7 (18961-1-AP; Proteintech), BBS8 (HPA003310; Sigma), BBS9 (14460-1-AP; Proteintech), rhodopsin (sc-57432; Santa Cruz Biotechnology), β-tubulin (T0198; Sigma), GFP (A11122; Invitrogen), and PDE 6 α (PA1-720; Thermo Scientific, Rockford, IL, USA). Antibody against phosducin was previously described.33 Antibodies against peripherin/rds were gifts from Kathleen Boesze-Battaglia, University of Pennsylvania (Philadelphia, PA, USA) and Andrew Goldberg, Oakland University (Rochester, MI, USA).
Results
Transgenic Expression of a Short Splice Isoform of Phosducin-Like Protein in Mouse Photoreceptors
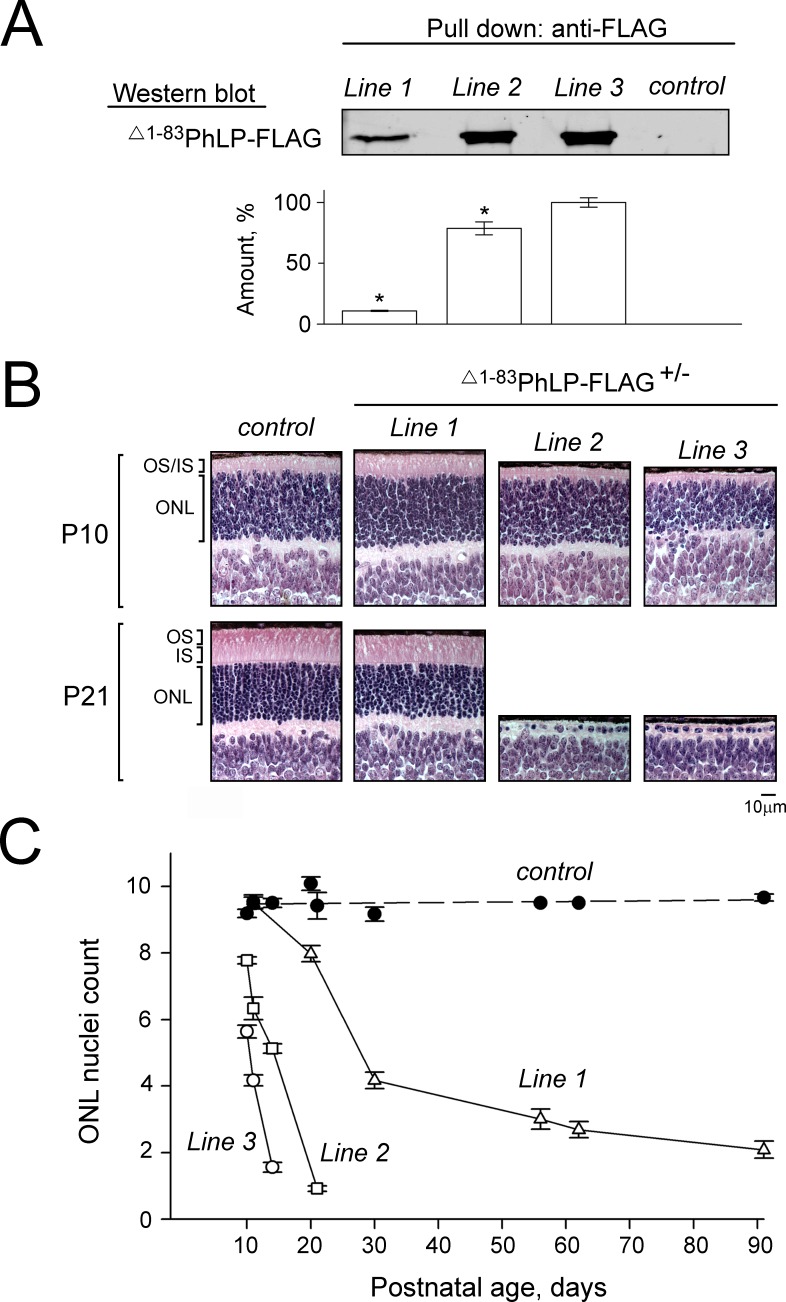
Phosducin-like protein (PhLP) is a cofactor of the chaperonin CCT that mediates the folding of heterotrimeric G-protein β subunits.34 Alternative splicing of the phosducin-like protein gene (PDCL) gives rise to an isoform designated PhLPs (for “short”), which forms a high-affinity complex with CCT, leading to its inactivation.31 We used PhLPs as a tool to inhibit the CCT activity in mice, which normally do not express this splice isoform.31 Phosducin-like proteins were expressed in retinal photoreceptors under the control of a rhodopsin promoter from a transgene designated as Δ1–83PhLP-FLAG, containing the same N-terminal deletion as PhLPs as well as a C-terminal epitope tag. Three transgenic lines (1–3) expressed different levels of Δ1–83PhLP-FLAG, detectable already at P08 (Fig. 1A). All transgenic mice developed retinal degeneration, which was particularly rapid and severe in lines 2 and 3, where all photoreceptors in the retina were gone by P21 (Fig. 1B). In general, the rate at which photoreceptor cells died directly correlated with the amount of expressed Δ1–83PhLP-FLAG (Fig. 1C). Such dose-dependent cytotoxic effect of Δ1–83PhLP-FLAG signifies the essence of the chaperonin activity for the viability of rod photoreceptors. This animal model was used to determine the specific effects of the CCT suppression on rod morphology.
Figure 1.
Suppressing the CCT activity in mouse photoreceptors by phosducin-like protein short. (A) Whole-retina extract from 8-day-old mice were analyzed by pull down with anti-FLAG agarose. A representative Western blot shows the amounts of captured Δ1-83PhLP-FLAG, as visualized with antibody against FLAG, in transgene-positive (Δ1-83PhLP-FLAG±) and control (Δ1-83PhLP-FLAG−/−) mice from the lines 1 to 3. Graph: protein bands in each experiment were quantified and their values expressed as percent of the highest value found in line 3. Bars are SEM with n = 3, and P < 0.05 as determined by paired t-test. (B) Paraffin-embedded retina cross-sections were stained with hematoxylin and eosin to visualize its cellular composition at the age P10 and P21. (C) Nuclei count across the outer nuclear layer (ONL) as a function of mouse age. Bars are SEM, n = 6. OS, outer segments; IS, inner segments.
Suppression of CCT Disrupts Formation of Rod Outer Segments
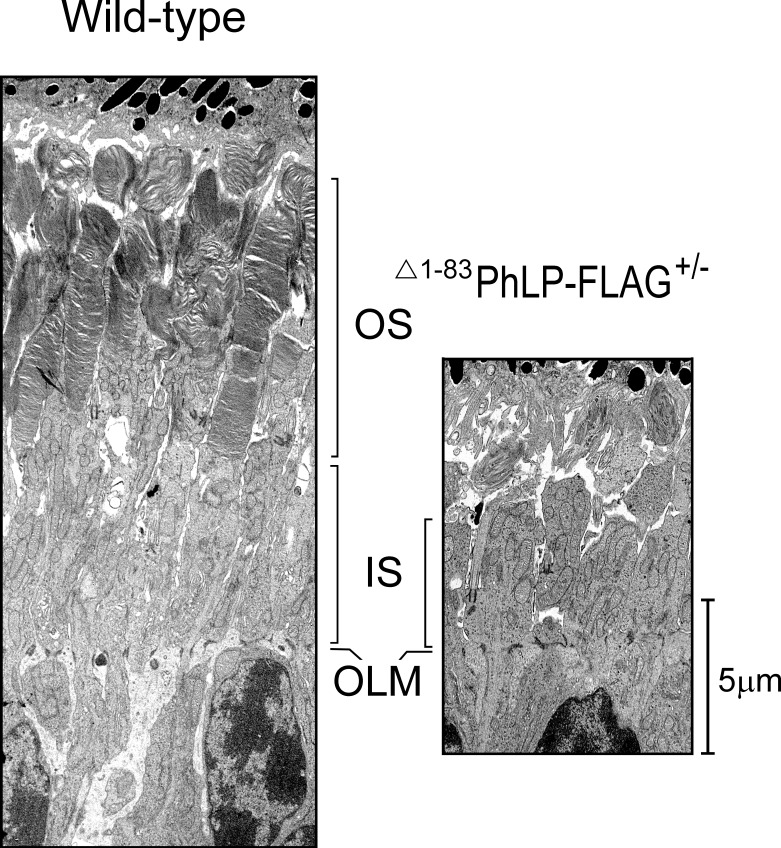
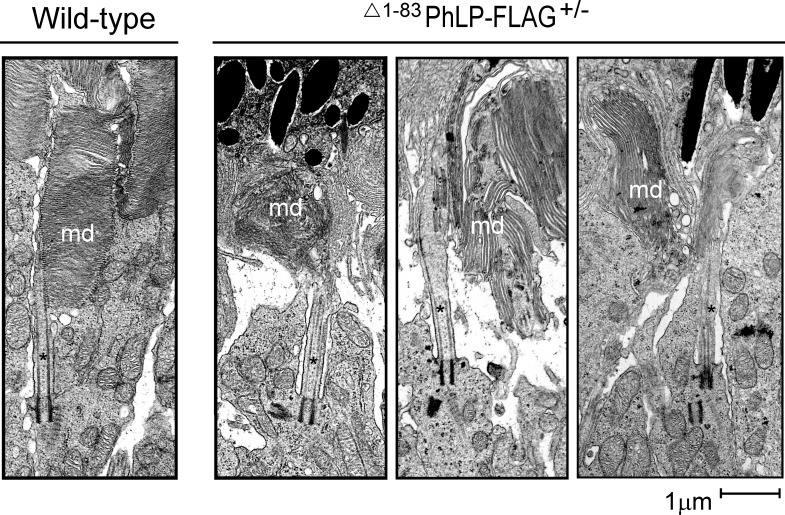
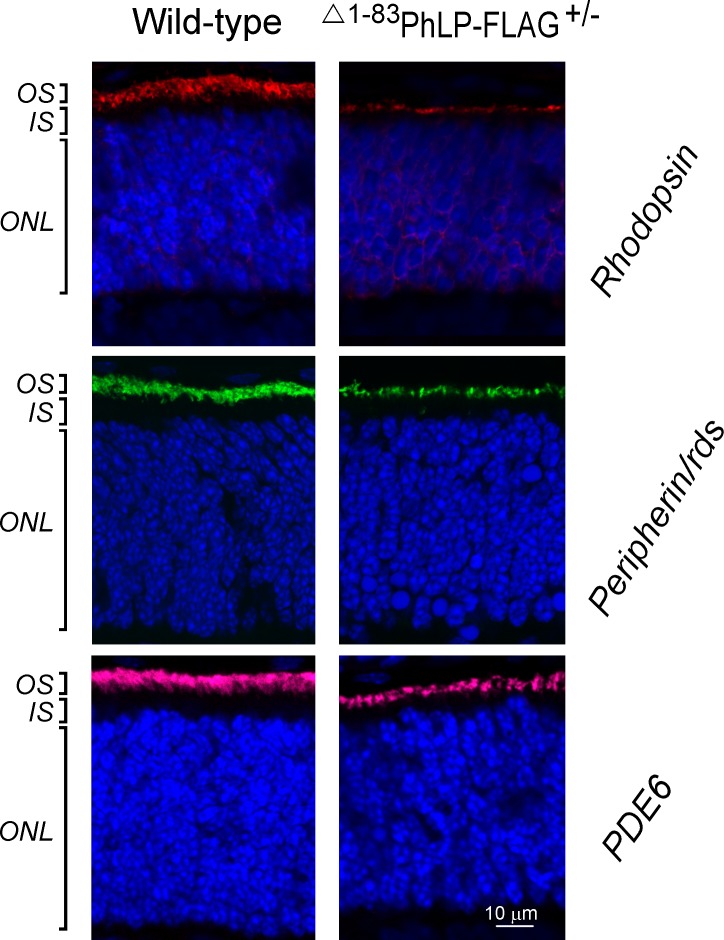
In lines 2 and 3, characterized by a higher degree of CCT suppression due to a higher level of Δ1-83PhLP-FLAG expression, the shortening of rod outer segments was noticeable already at P10 (Fig. 1B). A thorough examination of this compartment's ultrastructure at this age revealed that, in contrast to normal rods, the CCT-deficient rods had no outer segments (Fig. 2). Instead, in immediate vicinity to the connecting cilia, whose overall morphology appeared to be normal, we observed aggregates of membranes strongly resembling outer segment disks with their very characteristic flat shape (Fig. 3). Furthermore, the subcellular localization of two integral membrane proteins, rhodopsin and peripherin/rds, and a peripheral membrane protein, phosphodiesterase (PDE6), which are all normally targeted to disks, was consistent with these proteins being accumulated in these membrane aggregates (Fig. 4). In our previous studies of lines 2 and 3 we determined that the affected rods expressed 3- to 10-fold less of each of these proteins.28 Based on these observations we concluded that these membranes likely represent disks formed in the rods under conditions when these cells were unable to extend their outer segment compartments. As a result, these disks formed what appear to be extracellular aggregates, engulfed by retinal pigment epithelium, and actively destroyed by phagocytosis. The inner segments of the affected rods were also visibly reduced in length (Fig. 2). In line 1, expressing less Δ1-83PhLP-FLAG, the rod outer segments developed, however, they were shorter in length at P21 (Fig. 1B). Thus, suppressing the chaperonin activity in rod photoreceptors crucially affected their ability to build their outer segment compartments.
Figure 2.
Suppressing the CCT activity disrupts development of rod outer segments. Retinas from wild-type (left panel) and Δ1-83PhLP-FLAG± (right panel) littermates were analyzed by TEM at P10. OLM, outer limiting membrane.
Figure 3.
Formation of membrane disks in healthy and affected rods. Ultrastructure of rod photoreceptor connecting cilium (*) and OS membrane disks (md) in wild-type and Δ1-83PhLP-FLAG± littermates at P10.
Figure 4.
Targeting of proteins to the OS disks. Subcellular localization of rhodopsin (red), peripherin/rds (green), and PDE6-α (magenta) as determined by immunofluorescence microscopy in rod photoreceptors at P10. Cell nuclei are visualized with DAPI.
The Role of CCT in Posttranslational Processing of Bardet-Biedl Syndrome Proteins
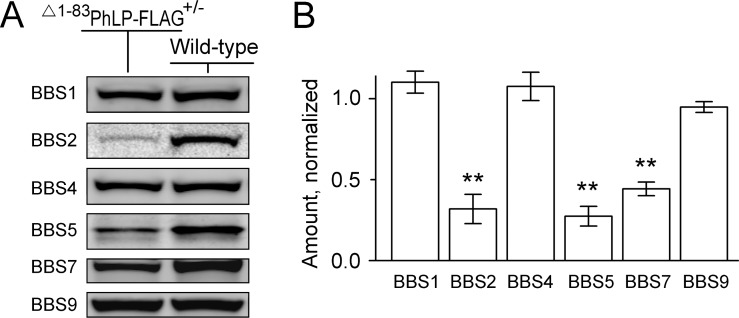
Having observed the disruption of the photoreceptor's sensory cilia, we used our animal model to test whether suppressing CCT affected the BBSome complex implicated in cilia trafficking. For that, we compared the levels of six Bardet-Biedl Syndrome proteins (BBS1, -2, -4, -5, -7, -9) that form the BBSome, in the retinas of CCT-deficient and healthy 10-day-old littermates (Fig. 5). Three of them, including BBS2, BBS5, and BBS7, were significantly reduced in CCT-deficient retinas, while the levels of BBS1, BBS4, and BBS9 remained healthy. Because these BBS proteins are expressed in all retinal cells, and photoreceptor cells comprise only approximately one-half of the retina, the actual extent of this reduction would be underestimated. Noteworthy, the reduction of these proteins likely occurred on a posttranslational level, as we found no evidence of a downregulation of their transcripts in the previous study.31
Figure 5.
Reduced levels of BBS proteins in the CCT-deficient rods. (A) A representative Western blot showing the levels of indicated BBS proteins in the whole-retina extracts of wild-type and Δ1–83PhLP-FLAG± littermates at P10. (B) The fluorescence value of each specific band of a Δ1–83PhLP-FLAG± mouse was normalized to that in a wild-type mouse, SEM, n = 4, P < 0.01 as determined by paired t-test (**).
CCT is Required for the Assembly of the BBSome in Rod Photoreceptors
To monitor the assembly of the BBSome in ocular tissues, whole-eye homogenates were analyzed by 10% to 40% sucrose density gradient ultracentrifugation coupled to Western blot analysis. In a good agreement with previous studies using the same technique,29,30 the BBSome migrated as an approximately 14S complex, composed of BBS1, BBS2, BBS4, BBS5, BBS7, BBS8, and BBS9 subunits, which predominantly appeared in fraction #11 (Fig. 6). As expected, the 14S complex was present in both, Δ1-83PhLP-FLAG−/− and Δ1-83PhLP-FLAG± mice. Importantly, an additional approximately 9S complex containing BBS1, BBS4, BBS8, and BBS9 subunits could always be detected in transgene-positive mice from lines 2 and 3 (Fig. 6). Because the 9S complex specifically lacked BBS2, BBS5, and BBS7 proteins, which were missing in the CCT-deficient rods (Fig. 5), it likely represented a partial BBSome complex assembled in these cells. Detection of the 9S complex provides the first demonstration of the assembly of an incomplete and presumably inactive BBSome in rod photoreceptors with suppressed chaperonin CCT activity in vivo.
Figure 6.
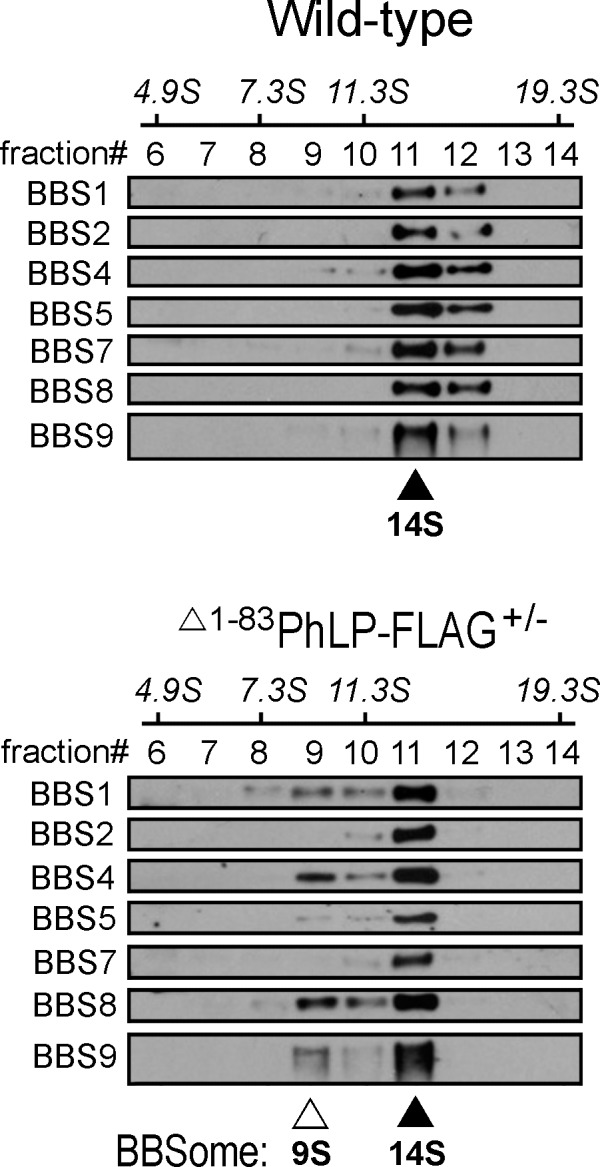
An incomplete BBSome complex is assembled in the CCT-deficient rods. Whole-eye extracts from Δ1–83PhLP-FLAG−/− and Δ1–83PhLP-FLAG± littermates at P10 were fractionated by ultracentrifugation on a 10% to 40% sucrose gradient calibrated using sedimentation (S) coefficient standards. Collected fractions were analyzed by Western blotting using specific antibodies against the indicated BBS proteins. A mature BBSome contains BBS1, -2, -4, -5, -7, -8, and -9 and migrates as a 14S complex (▴); an incomplete BBSome contains BBS1, -4, -8, -9, and migrates as a 9S complex (Δ).
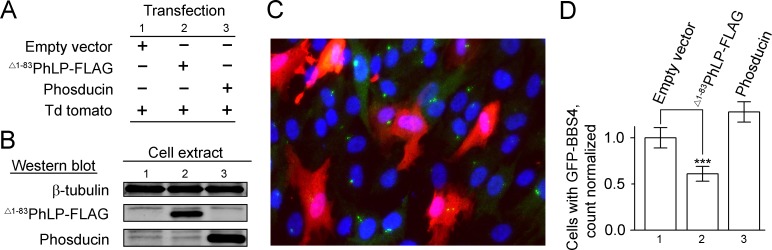
To test this conclusion by a complementary technique, we attempted to monitor the BBSome directly by confocal microscopy. Unfortunately, this approach was unsuccessful when applied for mouse retinas. This prompted us to exploit an alternative cell culture model, human hTERT RPE1 cells stably expressing GFP-labeled BBS4 subunits of the BBSome. The CCT complex in human cell lines could be efficiently sequestered by overexpressing mouse Δ1–83PhLP-FLAG.31 Phosducin, a close homolog of phosducin-like protein that doesn't interact with CCT, was used as a negative control, and tdTomato was used as a red fluorescent marker to identify the transfected cells (Figs. 7A, 7B). After immuno-enhancement with anti-GFP antibodies, the subcellular localization of GFP-BBS4 corresponding to the centrosome and centriolar satellites of the primary cilia18,23,35 was clearly visible in the cells (Fig. 7C). In response to the suppression of CCT by Δ1–83PhLP-FLAG, the number of cells with the specific GFP-BBS4 signal became significantly reduced compared to the control cells transfected with empty expression vector or expressing phosducin (Fig. 7D). This effect is consistent with GFP-BBS4 changing its subcellular localization in response to the suppression of CCT. A plausible explanation for such an effect would be underlying changes with the BBSome. Combined these data provide strong experimental evidence that the chaperonin CCT plays an essential role in the expression and assembly of the BBSome.
Figure 7.
Bardet-Biedl syndrome protein 4 becomes mislocalized in ciliated cells in response to the CCT suppression. (A) Transfection protocol for transient expression of Δ1–83PhLP-FLAG and phosducin in RPE1-hTERT cells. (B) Representative Western blotting showing the expression of the indicated proteins. (C) RPE1-hTERT cells stably expressing GFP-BBS4 fusion protein were cotransfected with empty- and tdTomato vectors. GFP-BBS4 was immuno-enhanced with antibody against GFP (green), cell nuclei were visualized with DAPI (blue), transfected cells are distinguished from nontransfected cells by the presence of tdTomato fluorescence (red). (D) A number of cells with GFP-BBS4 was determined and expressed as a fraction of all transfected (red) cells. Such fraction in the cells expressing Δ1–83PhLP-FLAG and phosducin was normalized to that found in the control cells treated with empty vector. Error bars are SEM, n = 700, P = 0.008, as determined by paired t-test (***).
Analysis of Rod Outer Segment Visual Function in Transgenic Line 1
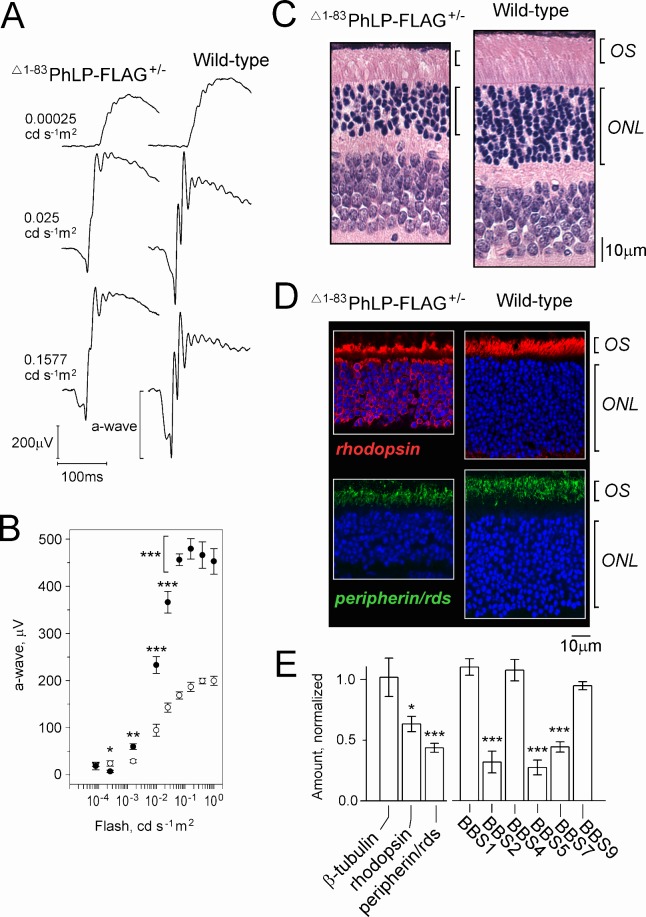
Transgenic mice from line 1, which express lower levels of Δ1–83PhLP-FLAG, and therefore display less severe retinal degeneration (Fig. 1), were suitable for the assessment of rod visual function by ERG. Soon after weaning, the rod ERG responses of these mice showed markedly reduced a-wave amplitude (Figs. 8A, 8B). At this age, the count of rod nuclei in the outer nuclear layer of the retina was reduced by 30% to 40%, and surviving rod cells had approximately 50% shorter outer segments than wild-type (Fig. 8C). The analysis of the subcellular distribution of two outer segment markers, rhodopsin and peripherin/rds, by immunofluorescence microscopy revealed a profound mislocalization of rhodopsin, but not peripherin/rds (Fig. 8D). The overall level of rhodopsin in the Δ1–83PhLP-FLAG-expressing retinas was reduced by 37 ± 3% (SEM, n = 3), which is generally consistent with the observed extent of photoreceptor loss at this age (Fig. 8C), and suggests that the level of rhodopsin in the affected rods remained normal (Fig. 8E). This was further supported by the statistically significant (P < 0.02 compared with rhodopsin), and stronger reduction of peripherin/rds by 56 ± 4% (SEM, n = 6), which was observed in the same retinal preparations (Fig. 8E). Combined, these data support the notion that the mislocalization of rhodopsin in the Δ1–83PhLP-FLAG-expressing rods was secondary to the shortening of their outer segments, which were no longer capable of accommodating this protein. That, however, was not the case for peripherin/rds, whose expression was downregulated more proportionately to the outer segment reduction. Most intriguingly, we found the levels of BBS2, BBS5, and BBS7 proteins in the retinas of adult mice from line 1 to be reduced essentially the same as in the 10-day-old mice from lines 2 and 3 (compare Figs. 8E and 5). This important observation signifies a common mechanism, whereby a short splice isoform of PhLP targets BBSome-assisted ciliary trafficking in our transgenic models and perhaps in vivo.
Figure 8.
Characterization of visual function, photoreceptor morphology, and protein expression in 21-day-old mice from transgenic line 1. (A) Representative ERG responses to 0.00025, 0.025, and 0.1577 cd s−1 m2 flashes (−40, −20, and −12 dB). (B) The a-wave amplitude as a function of flash intensity in Δ1–83PhLP-FLAG± (white circles) and wild-type (black circles); SEM, n = 6, P < 0.05 (*), P < 0.01 (**), P < 0.001 (***) as determined by t-test. (C) Paraffin-embedded retina cross-sections were stained with hematoxylin and eosin to visualize their cellular composition. (D) Subcellular localization of rhodopsin (red) and peripherin/rds (green) as determined by immunofluorescence microscopy. Cell nuclei are stained with DAPI. (E) Levels of the indicated proteins determined by Western blotting of whole retina extracts of Δ1–83PhLP-FLAG± mice were normalized to the corresponding values of wild-type mice. Error bars represent SEM, n = 3 (rhodopsin), n = 6 (peripherin/rds), n = 4 (BBS), P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***) as determined by paired t-test.
Discussion
This study demonstrates the role of the cytosolic chaperonin CCT in the biogenesis of the BBSome in vivo. Using mouse rod photoreceptors as a model, we found that suppressing the chaperonin activity in these cells by overexpressing epitope-tagged phosducin-like protein short (Δ1–83PhLP-FLAG) leads to a significant reduction in the levels of three subunits of the BBSome, including BBS2, -5, and -7. The structural and physical features of these soluble proteins, with the predicted β-propeller (BBS2 and BBS7) and plekstrin homology (BBS5) domains,20 are common among the bona fide CCT substrates, all of which are soluble proteins, often containing complex domain topology and WD-repeat domains that fold into a β-propeller structure.3–6,36 It is plausible that BBS2, BBS5, and BBS7 are substrate proteins of CCT, since CCT suppression results in posttranslational destabilization of these proteins. Because BBS2 and BBS7 are the components of the core complex (BBS2-BBS7-BBS9) initiating the assembly of the BBSome,23 their simultaneous decrease in rods would be expected to have a strong impact on the BBSome. This was recently demonstrated using BBS7-null mice, which were found to be incapable of completing BBSome assembly in testis and kidney.37 The same study also revealed a rather specific interdependence of BBS2 and BBS7 for posttranslational stability, indicating that BBS7-null mice also have reduced levels of BBS2 and vice versa. Therefore, destabilization of only one of these two proteins in the photoreceptors would already be sufficient to cause the observed phenotype. It would be of interest to test whether the partial 9S BBSome complex observed in our mouse model could also be detected in various tissues of BBS7-null mice.
The important question remains whether a concurrent decline of BBS2, BBS5, and BBS7 proteins, and arguably inactivation of the BBSome, could alone account for the severe defects of the rod outer segment observed in our mouse model. In support of this notion, BBS7-null mice exhibit a degeneration of the outer and inner segments of the photoreceptors, and have abnormal cilia and flagella.37 While the precise functions of the BBSome are not completely understood, the current understanding is that this complex is engaged in trafficking on the ciliary membrane.18–20 Thus, inactivation of the BBSome in rod photoreceptors could potentially halt movement of the membrane precursors along the connecting cilium. Consistent with such a mechanism, the affected rods in our animal model were unable to envelop their growing outer segment with plasma membrane. Surprisingly, though, these cells retained the ability to form the outer segment membrane disks. This observation further supports the notion that the formation of disks in the rod outer segment occurs via a mechanism that may be independent of ciliary plasma membrane trafficking and therefore insensitive to disruption of the BBSome. Such a mechanism may involve, for example, a direct fusion of precursor membrane carriers, already preassembled with the specific proteins, into a growing nascent disk.38,39
Recently, it was demonstrated that PhLP-null photoreceptors retain their outer segments for a significant period of time.40 This supports a notion that none of the CCT substrates folded with the direct assistance of PhLP, whose levels become significantly reduced in the PhLP-null photoreceptors, are essential for outer segment biogenesis. All of these proteins were also found to be significantly reduced in the photoreceptors of the PhLPs-expressing transgenic mice used in the present study.28,31 However, due to the broader nature of the CCT suppression by the dominant-negative PhLPs, additional proteins become affected in this animal model, including those essential for outer segment biogenesis. It should be mentioned that in this, as well as our previous studies,28,31 the suppression CCT by PhLPs didn't seem to have a significant impact on cytoskeletal proteins, actin and tubulin, known to be processed by CCT. This rather surprising observation thus makes it unlikely that destabilization of the cytoskeleton underlies the photoreceptor outer segments collapse. It is also consistent with the notion that CCT particles engaged in the folding of actin and tubulin are not available to interact with PhLPs, which predominantly targets CCT preloaded with nascent G protein β subunits.31 Therefore, the high levels of actin and tubulin expression in the cell may keep this biosynthetic pathway protected from suppression by PhLPs.
Our preliminary studies of transgenic line 1 suggest that even low amounts of PhLPs exhort a significant biological effect on the rod sensory cilium. Owing to the fact that rod photoreceptors in line 1 express approximately 10-fold less PhLPs than in lines 2 and 3, these cells don't display the same signs of acute CCT-deficiency, and many of them survive well into adulthood. Because these rods retain their outer segments, while being BBS2, -5, and -7 deficient, the severe phenotypes observed in the other two lines could not be solely attributed to deregulation of the BBSome, and was likely exacerbated by other contributing factors. Nevertheless, the physiological assessment of rods' visual function in line 1 reveals profound changes in the ERG a-wave that could not be fully reconciled in light of the observed anatomic alteration of the retina, but do indicate a significant deregulation of the phototransduction cascade. Importantly, using this line, we further confirmed that one of the specific effects of PhLPs is directed against the cilia gating complex, the BBSome, which thus makes it plausible that PhLPs interferes with the trafficking of phototransduction proteins through the cilium. The underlying molecular mechanisms remain the topic of future studies.
Acknowledgments
The authors thank Kathleen Boesze-Battaglia and Andrew Goldberg for their generous gifts of antibodies.
This work was supported by National Institutes of Health (NIH; Bethesda, MD, USA) Grants EY019665 (MS) and EY022616 (SS), and an unrestricted Research to Prevent Blindness (New York, NY, USA) grant awarded to the West Virginia University Eye Institute. The Transgenic Animal Core Facility at WVU was supported by CoBRE (NIH, Bethesda, MD, USA) Grants RR031155 and RR016440. The Microscope Imaging Facility at WVU was supported by the Mary Babb Randolph Cancer Center (Morgantown, WV, USA) and NIH Grants P20 RR016440, P30 RR032138/GM103488, and P20 RR016477.
Disclosure: S. Sinha, None; M. Belcastro, None; P. Datta, None; S. Seo, None; M. Sokolov, None
References
- 1. Munoz IG, Yebenes H, Zhou M, et al. Crystal structure of the open conformation of the mammalian chaperonin CCT in complex with tubulin. Nat Struct Mol Biol. 2011; 18: 14–19 [DOI] [PubMed] [Google Scholar]
- 2. Dekker C, Roe SM, McCormack EA, et al. The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins. Embo J. 2011; 30: 3078–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Camasses A, Bogdanova A, Shevchenko A, Zachariae W. The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol Cell. 2003; 12: 87–100 [DOI] [PubMed] [Google Scholar]
- 4. Spiess C, Meyer AS, Reissmann S, Frydman J. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 2004; 14: 598–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kubota S, Kubota H, Nagata K. Cytosolic chaperonin protects folding intermediates of Gbeta from aggregation by recognizing hydrophobic beta-strands. Proc Natl Acad Sci U S A. 2006; 103: 8360–8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yam AY, Xia Y, Lin HT, et al. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat Struct Mol Biol. 2008; 15: 1255–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Frydman J, Nimmesgern E, Erdjument-Bromage H, et al. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. Embo J. 1992; 11: 4767–4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gao Y, Thomas JO, Chow RL, et al. A cytoplasmic chaperonin that catalyzes beta-actin folding. Cell. 1992; 69: 1043–1050 [DOI] [PubMed] [Google Scholar]
- 9. Chen X, Sullivan DS, Huffaker TC. Two yeast genes with similarity to TCP-1 are required for microtubule and actin function in vivo. Proc Natl Acad Sci U S A. 1994; 91: 9111–9115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vinh DB, Drubin DG. A yeast TCP-1-like protein is required for actin function in vivo. Proc Natl Acad Sci U S A. 1994; 91: 9116–9120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stemp MJ, Guha S, Hartl FU, Barral JM. Efficient production of native actin upon translation in a bacterial lysate supplemented with the eukaryotic chaperonin TRiC. Biol Chem. 2005; 386: 753–757 [DOI] [PubMed] [Google Scholar]
- 12. Soares H, Penque D, Mouta C, Rodrigues-Pousada CA. Tetrahymena orthologue of the mouse chaperonin subunit CCT gamma and its coexpression with tubulin during cilia recovery. J Biol Chem. 1994; 269: 29299–29307 [PubMed] [Google Scholar]
- 13. Cyrne L, Guerreiro P, Cardoso AC, et al. The Tetrahymena chaperonin subunit CCT eta gene is coexpressed with CCT gamma gene during cilia biogenesis and cell sexual reproduction. FEBS Lett. 1996; 383: 277–283 [DOI] [PubMed] [Google Scholar]
- 14. Seixas C, Casalou C, Melo LV, et al. Subunits of the chaperonin CCT are associated with Tetrahymena microtubule structures and are involved in cilia biogenesis. Exp Cell Res. 2003; 290: 303–321 [DOI] [PubMed] [Google Scholar]
- 15. Seixas C, Cruto T, Tavares A, et al. CCTalpha and CCTdelta chaperonin subunits are essential and required for cilia assembly and maintenance in Tetrahymena. PLoS One. 2010; 5: e10704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bregier C, Krzemien-Ojak L, Wloga D, et al. PHLP2 is essential and plays a role in ciliogenesis and microtubule assembly in Tetrahymena thermophila. J Cell Physiol. 2013; 228: 2175–2189 [DOI] [PubMed] [Google Scholar]
- 17. Seo S, Baye LM, Schulz NP, et al. BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc Natl Acad Sci U S A. 2010; 107: 1488–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nachury MV, Loktev AV, Zhang Q, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007; 129: 1201–1213 [DOI] [PubMed] [Google Scholar]
- 19. Loktev AV, Zhang Q, Beck JS, et al. A BBSome subunit links ciliogenesis, microtubule stability, and acetylation. Dev Cell. 2008; 15: 854–865 [DOI] [PubMed] [Google Scholar]
- 20. Jin H, White SR, Shida T, et al. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010; 141: 1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Forsythe E, Beales PL. Bardet-Biedl syndrome. Eur J Hum Genet. 2013; 21: 8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Billingsley G, Bin J, Fieggen KJ, et al. Mutations in chaperonin-like BBS genes are a major contributor to disease development in a multiethnic Bardet-Biedl syndrome patient population. J Med Genet. 2010; 47: 453–463 [DOI] [PubMed] [Google Scholar]
- 23. Zhang Q, Yu D, Seo S, et al. Intrinsic protein-protein interaction-mediated and chaperonin-assisted sequential assembly of stable Bardet-Biedl syndrome protein complex, the BBSome. J Biol Chem. 2012; 287: 20625–20635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Katsanis N, Beales PL, Woods MO, et al. Mutations in MKKS cause obesity, retinal dystrophy and renal malformations associated with Bardet-Biedl syndrome. Nat Genet. 2000; 26: 67–70 [DOI] [PubMed] [Google Scholar]
- 25. Stoetzel C, Laurier V, Davis EE, et al. BBS10 encodes a vertebrate-specific chaperonin-like protein and is a major BBS locus. Nat Genet. 2006; 38: 521–524 [DOI] [PubMed] [Google Scholar]
- 26. Stoetzel C, Muller J, Laurier V, et al. Identification of a novel BBS gene (BBS12) highlights the major role of a vertebrate-specific branch of chaperonin-related proteins in Bardet-Biedl syndrome. Am J Hum Genet. 2007; 80: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gilliam JC, Chang JT, Sandoval IM, et al. Three-dimensional architecture of the rod sensory cilium and its disruption in retinal neurodegeneration. Cell. 2012; 151: 1029–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Posokhova E, Song H, Belcastro M, et al. Disruption of the chaperonin containing TCP-1 function affects protein networks essential for rod outer segment morphogenesis and survival. Mol Cell Proteomics. 2011; 10: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Seo S, Mullins RF, Dumitrescu AV, et al. Subretinal gene therapy of mice with Bardet-Biedl syndrome type 1. Invest Ophthalmol Vis Sci. 2013; 54: 6118–6132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seo S, Zhang Q, Bugge K, et al. A novel protein LZTFL1 regulates ciliary trafficking of the BBSome and Smoothened. PLoS Genet. 2011; 7: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gao X, Sinha S, Belcastro M, et al. Splice isoforms of phosducin-like protein control the expression of heterotrimeric G proteins. J Biol Chem. 2013; 288: 25760–25768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tucker ES, Lehtinen MK, Maynard T, et al. Proliferative and transcriptional identity of distinct classes of neural precursors in the mammalian olfactory epithelium. Development. 2010; 137: 2471–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sokolov M, Strissel KJ, Leskov IB, et al. Phosducin facilitates light-driven transducin translocation in rod photoreceptors. Evidence from the phosducin knockout mouse. J Biol Chem. 2004; 279: 19149–19156 [DOI] [PubMed] [Google Scholar]
- 34. Willardson BM, Howlett AC. Function of phosducin-like proteins in G protein signaling and chaperone-assisted protein folding. Cell Signal. 2007; 19: 2417–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim JC, Badano JL, Sibold S, et al. The Bardet-Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat Genet. 2004; 36: 462–470 [DOI] [PubMed] [Google Scholar]
- 36. Russmann F, Stemp MJ, Monkemeyer L, et al. Folding of large multidomain proteins by partial encapsulation in the chaperonin TRiC/CCT. Proc Natl Acad Sci U S A. 2012; 109: 21208–21215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Q, Nishimura D, Vogel T, et al. BBS7 is required for BBSome formation and its absence in mice results in Bardet-Biedl syndrome phenotypes and selective abnormalities in membrane protein trafficking. J Cell Sci. 2013; 126: 2372–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chuang JZ, Zhao Y, Sung CH. SARA-regulated vesicular targeting underlies formation of the light-sensing organelle in mammalian rods. Cell. 2007; 130: 535–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sung CH, Chuang JZ. The cell biology of vision. J Cell Biol. 2010; 190: 953–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lai CW, Kolesnikov AV, Frederick JM, et al. Phosducin-like protein 1 is essential for G-protein assembly and signaling in retinal rod photoreceptors. J Neurosci. 2013; 33: 7941–7951 [DOI] [PMC free article] [PubMed] [Google Scholar]