Figure 3 and Figure 4 are incorrect. The X-axis label should read “Fold Change” and not “% Change”. The authors have provided corrected figures below.
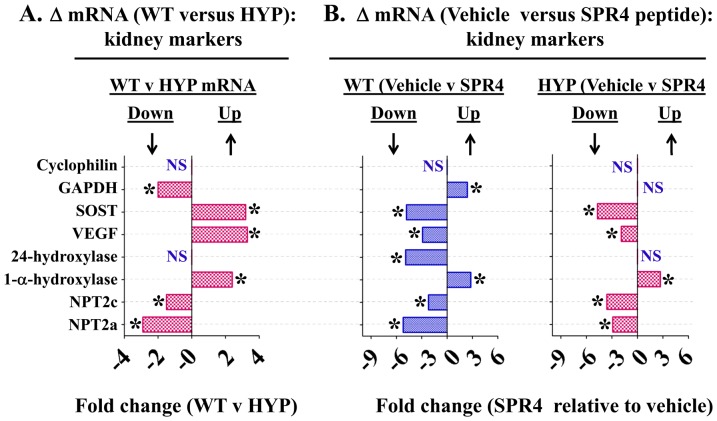
Figure 3. Whole kidney gene expression (mRNA) comparisons as measured by quantitative RT/PCR (qRT-PCR) for wild type (WT) and HYP mice infused with vehicle or SPR4 peptide for 28 days.
Column headings represent; WT = wild type mice, HYP = X-linked hypophosphatemic rickets mice, SPR4 = infused SPR4-peptide and Vehicle = Saline infused. For gene analysis mRNA was prepared from whole kidneys snap frozen in LN2 and homogenized. For qRT-PCR gene analysis fold differences in expression calculated by the Pfaffl method [163] were statistically analyzed for significance using the One Sample t-test and the Wilcoxon Signed rank-test with theoretical means set to 1. Results are significant (* = p<0.05) unless indicated by NS (see also Table 3 for detailed statistics). ND = Not done, NS = Not SignificantIndex: Cyclophilin = cyclophilin; GAPDH = Glyceraldehyde 3-phosphate dehydrogenase; SOST = Sclerostin; VEGF = Vascular Endothelial Growth factor; 24-Hydroxylase = 1,25-hydroxyvitamin D3 24-hydroxylase (CYP24A1); 1-á-Hydroxylase = 25-hydroxyvitamin D3 1-alpha-hydroxylase (CYP27B1); NPT2c = Sodium-dependent phosphate co-transporter (Slc34a3); NPT2a = Sodium-dependent phosphate co-transporter (Slc34a1); NS = not significant; * = P<0.05. Histogram bars to the left of zero on the axis indicate down regulation and to the right up regulation.
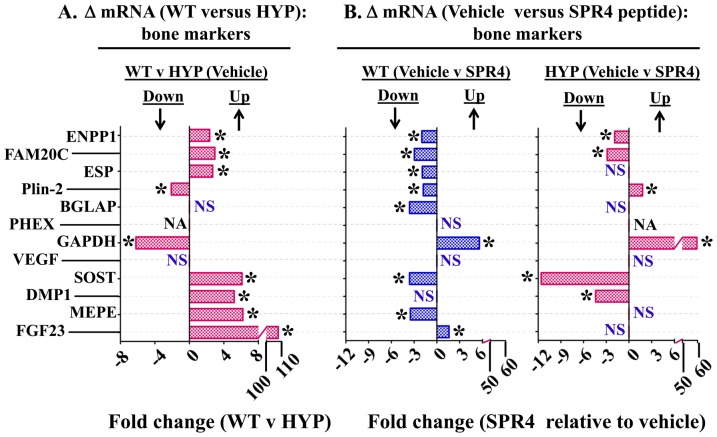
Figure 4. Bone (femur) gene expression (mRNA) comparisons as measured by quantitative RT/PCR (qRT-PCR) for wild type (WT) and HYP mice infused with vehicle or SPR4-peptide for 28 days.
Mice were sacrificed on day 28 and femurs collected for RNA purification as described in methods. Column headings represent; WT = wild type mice, HYP = X-linked hypophosphatemic rickets mice, SPR4 = infused SPR4-peptide and Vehicle = Saline infused. For gene analysis mRNA was prepared from bone marrow stromal cell “depleted” femurs as detailed in methods. For qRT-PCR gene analysis fold differences in expression calculated by the Pfaffl method [163] were statistically analyzed for significance using the One Sample t-test and the Wilcoxon Signed rank-test with theoretical means set to 1. Results are significant (* = p<0.05) unless indicated by NS (see also Table 4 for detailed statistics). Index: FAM20C = Family with sequence similarity 20, member C Kinase also known as DMP4; ENPP1 = Ectonucleotide Pyrophosphatase Phosphodiesterase 1; ESP = Osteotesticular protein tyrosine (OST-PTP); Plin-2 = Perlipin-2; phosphatase; Cyclophilin = peptidylprolyl isomerase A (cyclophilin A); BGLAP = Osteocalcin or Bone Gamma-Carboxyglutamate (gla) protein; PHEX = Phosphate-regulating gene with Homologies to Endopeptidases on the X chromosome; GAPDH = Glyceraldehyde 3-phosphate dehydrogenase; VEGF = Vascular Endothelial Growth factor; DMP1 = Dentin Matrix Protein 1; SOST = Sclerostin; MEPE = Matrix Extracellular Phosphoglycoprotein with ASARM -motif; FGF23 = Fibroblast Growth Factor 23; NS = not significant; NA = not applicable, PHEX mutated in HYP; * = P<0.05. Histogram bars to the left of zero on the axis indicate down regulation and to the right up regulation.
Reference
- 1. Zelenchuk LV, Hedge A-M, Rowe PSN (2014) PHEX Mimetic (SPR4-Peptide) Corrects and Improves HYP and Wild Type Mice Energy-Metabolism. PLoS ONE 9(5): e97326 doi:10.1371/journal.pone.0097326 [DOI] [PMC free article] [PubMed] [Google Scholar]