Abstract
Background
The shape of grass leaves possesses great value in both agronomy and developmental biology research. Leaf rolling is one of the important traits in rice (Oryza sativa L.) breeding. MYB transcription factors are one of the largest gene families and have important roles in plant development, metabolism and stress responses. However, little is known about their functions in rice.
Results
In this study, we report the functional characterization of a rice gene, OsMYB103L, which encodes an R2R3-MYB transcription factor. OsMYB103L was localized in the nucleus with transactivation activity. Overexpression of OsMYB103L in rice resulted in a rolled leaf phenotype. Further analyses showed that expression levels of several cellulose synthase genes (CESAs) were significantly increased, as was the cellulose content in OsMYB103L overexpressing lines. Knockdown of OsMYB103L by RNA interference led to a decreased level of cellulose content and reduced mechanical strength in leaves. Meanwhile, the expression levels of several CESA genes were decreased in these knockdown lines.
Conclusions
These findings suggest that OsMYB103L may target CESA genes for regulation of cellulose synthesis and could potentially be engineered for desirable leaf shape and mechanical strength in rice.
Keywords: OsMYB103L, Leaf rolling, MYB transcription factor, Cellulose
Background
Leaves are the main organs of photosynthesis and play a crucial role in plant development. Leaf rolling is a frequently observed phenotype in fields and rice mutant populations [1-3]. The appropriate leaf rolling has been regarded as one of the most important elements in rice ideotype breeding as it improves photosynthetic efficiency, accelerates dry-matter accumulation, reduces solar radiation on leaves, decreases leaf transpiration under drought stress and raises grain yield [2,4-9]. However, severe leaf rolling could lead to growth retardation, developmental defects and crop yield reduction.
To date, many studies have been performed to characterize the genes controlling leaf rolling by analyses of the rolled leaf mutants in rice. Twelve rolled leaf (rl) mutants in rice are reported and the corresponding genes are mapped genetically, of which, six rl genes are mapped on rice chromosomes by morphological markers and the rest are directly mapped in rice genome by molecular markers [10-14]. Among these 12 rl mutants, rl9 is the first to be cloned and analyzed in detail. ROLLED LEAF9 (RL9)/SHALLOT-LIKE1 (SLL1) encodes a KANADI transcription factor, and rl9/sll1 mutants display extremely incurved leaves due to the defective development of sclerenchymatous cells on the abaxial side [1,15]. Some other genes are also found to be related to leaf rolling in rice. Loss-of-function of OsCSLD4 gene, which encodes a cellulose synthase-like protein, results in phenotypes of reduced leaf width and semi-rolled leaves, possibly due to the significantly smaller bulliform cells in mutants [16-19]. Loss-of-function of CONSTITUTIVELY WILTED1 (OsCOW1)/NARROW LEAF7 (NAL7), encoding a member of the YUCCA protein, causes rice leaves to roll upward [20,21]. Loss-of-function of CURLY FLAG LEAF (CFL1), encoding a WW domain protein, induces rolled leaves which have a wrinkled epidermis and contain papillae with reduced wax content [22]. ROLLED LEAF 14 (RL14), encoding a 2OG-Fe (II) oxygenase family protein, is reported to modulate rice leaf rolling by affecting secondary cell wall formation and bulliform cell development [2]. Mutation of SEMI-ROLLED LEAF 1 (SRL1), encoding a putative GPI-anchored protein, results in leaves rolled adaxially in rice due to the increased number of bulliform cells at the adaxial cell layers [23]. In addition, recent studies show that overexpression of rice gene OsAGO7, which encodes an Argonaute (AGO) family member, results in the leaf blades curling upward [3]. OsHB1 is a member of the Class III homeodomain leucine zipper family of genes, overexpression of its microRNA166-resistant version could result in adaxially rolled leaves due to the reduced sclerenchyma and formation of bulliform cells on the abaxial side [24]. Taken together, the major factors possibly affecting leaf rolling include the alteration of osmotic pressure and/or turgidity in bulliform cells, the impairment of cell polarity establishment and cell differentiation. However, the mechanisms underlying leaf rolling in rice and, particularly, the involved transcription factors remain largely unknown [1].
MYB proteins belong to a super-family of transcription factors, which prevail in all eukaryotes and share a highly conserved DNA-binding domain, MYB [25-27]. Depending on the number of adjacent repeats in the MYB domain, MYB proteins can be classified into four subfamilies, including 1R-MYB, R2R3-MYB, 3R-MYB, and 4R-MYB [26,28]. Among them, R2R3-MYB subfamily is a typical representative one, whose members contain two MYB repeats in their DNA-binding domains [29,30]. MYB proteins in plants are involved in a variety of plant-specific processes, including primary and secondary metabolism, cell fate and identity, developmental processes, and responses to various stresses [25-27,31-40]. Genome-wide analysis shows that there are approximately 183 MYB genes in rice, of which 109 encode R2R3-MYB proteins [41,42]. So far, only a few number of MYB proteins, including OsGAMYB, OsMYB2, OsMyb4, OsMYB3R-2, CSA, MYBS3 and OsMYB2P-1 [43-49], have been functionally characterized. The functions of most of MYB proteins are still unknown in rice.
To characterize functions of MYB transcription factors in rice, we overexpressed several MYB genes in Kasalath, an indica cultivar, with the transgene constructs containing the full-length cDNAs of rice MYB genes, driven by maize ubiquitin promoter. Of the transgenic lines, one line overexpressing the full-length cDNA of Os08g05520, designated as OsMYB103L hereafter, displays a rolled leaf phenotype. OsMYB103L encodes an R2R3-MYB transcription factor. Our study shows that it localizes in the nucleus and possesses transcriptional activity. We detail the phenotypes of OsMYB103L overexpressing (OE) and RNA interference (RNAi) knockdown plants, including the altered leaf shape, the changed cellulose content, and the impaired mechanical strength. The roles of OsMYB103L in leaf shape formation and cellulose synthesis are discussed. We propose the potential application of OsMYB103L in molecular breeding of rice.
Results
OsMYB103L encodes an R2R3-MYB transcription factor
To discover transcription factors controlling leaf development, we screened the rice lines ectopically expressing rice MYB genes under the control of maize ubiquitin promoter. One line overexpressing LOC_Os08g05520 was selected for further study due to its particular leaf shape, such as upward curling of the leaf blade. According to the rice genome annotation database (http://rice.plantbiology.msu.edu), LOC_Os08g05520 encodes a putative R2R3-MYB family transcription factor with a length of 359 amino acids and a molecular mass of approximately 40 kD. The Pfam database (http://pfam.sanger.ac.uk/) shows that the deduced protein has two MYB DNA-binding domains (PF00249) at the N-terminus (Figure 1A). As revealed by phylogenetic analysis of the related MYB transcription factors in Arabidopsis thaliana and rice, Os08g05520 is closely related to At1g63910 (AtMYB103) [50] (Figure 1B). Protein sequence alignment showed that they are highly conserved in the predicted R2- and R3-MYB DNA-binding domains (Figure 1A). We hereby designated Os08g05520 as OsMYB103L (Oryza sativa MYB103 Like).
Figure 1.
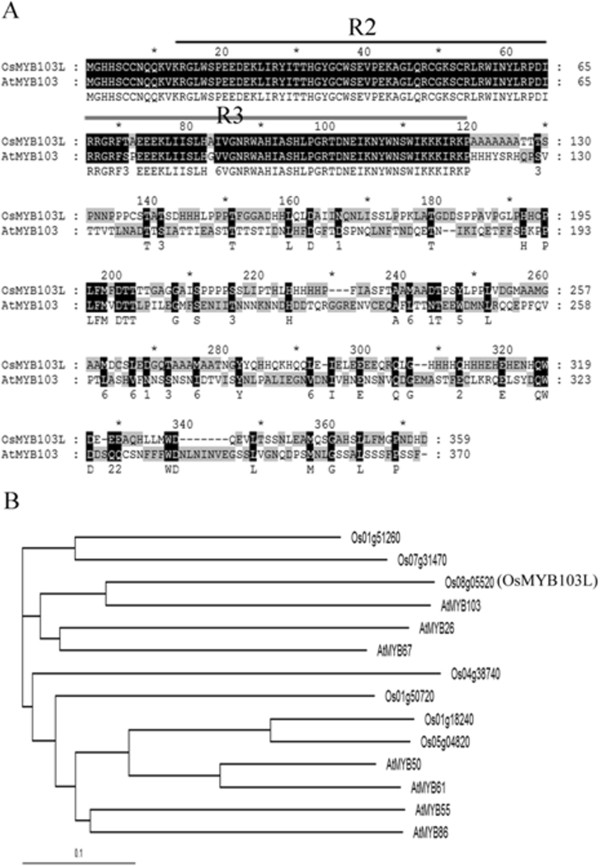
OsMYB103L (Os08g05520) is an R2R3-MYB transcription factor and has the most sequence homology to AtMYB103 (At1g63910) in Arabidopsis thaliana. (A) Protein sequences alignment of OsMYB103L and AtMYB103. OsMYB103L and AtMYB103 are highly conserved in predicted R2- (black line) and R3-MYB DNA-binding domains (grey line). (B) Phylogenetic relationships among OsMYB103L and related R2R3-MYB proteins in rice and Arabidopsis thaliana. An unrooted phylogenetic tree was generated with the full-length amino acid sequences.
Nuclear localization is one of the significant features of transcription factors. To determine its subcellular localization, we fused green fluorescent protein (GFP) to the C-terminus of OsMYB103L to produce an OsMYB103L-GFP fusion protein. We monitored the fluorescence of the transiently expressed fusion protein in rice protoplasts and onion (Allium cepa L.) epidermis cells. In both cases, the fluorescence signals of the fusion protein were observed predominantly in nuclei (Figure 2A and Additional file 1: Figure S1). While in the GFP alone control, fluorescence signals were observed in nuclei and cytoplasm (Figure 2A and Additional file 1: Figure S1). These results indicate that OsMYB103L is a nuclear-localized protein.
Figure 2.
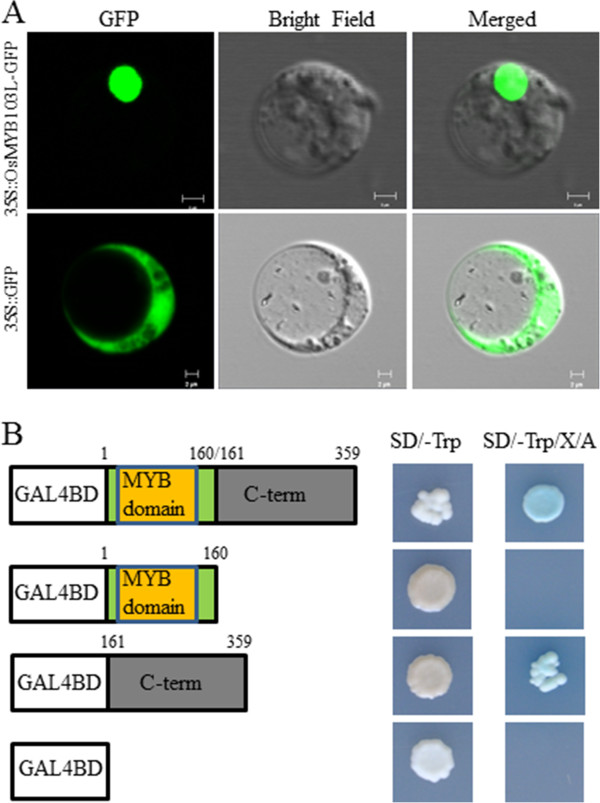
Subcellular localization and transactivation analysis of OsMYB103L. (A) Subcellular localization of OsMYB103L. GFP and OsMYB103L-GFP fusion gene under the control of the CaMV35S promoter were expressed transiently in rice protoplasts. Left to right: GFP fluorescence image, transmission image and merged image. Bar = 2 μm. (B) Transactivation analysis of different regions of OsMYB103L fused with the GAL4 DNA binding domain in yeast. The full-length, N-terminal MYB DNA-binding domain (1–160 amino acids) or the C-terminal putative activation domain (161–359 amino acids) were respectively cloned into pBD-GAL4 vector containing AUR1-C and MEL1 reporter genes and then transformed into yeast host strain Y2HGold. The pBD vector was used as a negative control. The expression of AUR1-C confers strong resistance to the highly toxic drug Aureobasidin A (Aba). The activities of α-galactosidase encoded by MEL-1 were examined by X-α-gal staining.
Transcriptional activation is a characteristic feature of many transcription factors. The full-length, N-terminal MYB DNA-binding domain and C-terminal putative activation domain were fused in-frame with the GAL4 DNA binding domain, respectively. We conducted a transcriptional activation analysis in yeast by using the fusion proteins. The analysis showed that the full-length and C-terminal region of OsMYB103L activated the expression of AUR1-C and MEL1 reporter genes in yeast (Figure 2B), indicating that OsMYB103L has the capability of transcriptional activation.
Expression of OsMYB103L varies in different organs
We determined the expression level of OsMYB103L in various rice organs, including root, culm, leave, and panicle using quantitative real-time PCR (qRT-PCR). The expression level of OsMYB103L was highest in culm, lower in root, leaf and mature panicle, and lowest in young panicle (Figure 3A).
Figure 3.
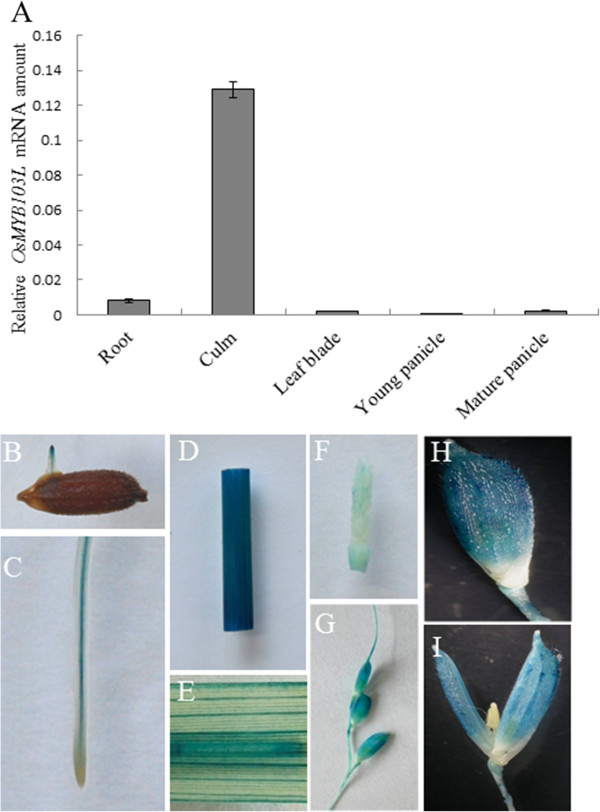
Expression analysis of OsMYB103L. (A) Expression of OsMYB103L in various organs determined by qRT-PCR analysis. Roots were harvested from 2-week-old plants. Culms, leaf blades and mature panicles were harvested from rice plants at heading stage. Young panicles were collected at the 5 cm stage. Rice ACTIN1 gene was used as internal control. Error bars represent the SD of transcript levels determined from three independent replicates. (B) to (I) Examination of GUS activity in the transgenic plants expressing OsMYB103L pro::GUS. The GUS activity is shown in coleoptiles (B), root (C), culm (D), leaf (E), young panicle (F) and mature panicle (G-I). GUS activity was also detected in pedicel, lemma and palea in mature panicle (H), while no GUS activity in sterile lemma, lodicule, pistil and stamen (I).
In addition, the expression pattern of OsMYB103L was examined by the β-glucuronidase (GUS) reporter gene driven by OsMYB103L putative promoter in rice plants. Examination of GUS activity in three independent transgenic lines revealed that the GUS signals were stronger in culm and weaker in vascular bundles of coleoptile, root and leaf (Figure 3B to 3I). GUS staining was stronger in mature panicle than young panicle (Figure 3F and 3G). Further examination showed that GUS activity was detectable in pedicel, lemma and palea while no GUS activity was detected in sterile lemma, lodicule, pistil, and stamen in mature panicle (Figure 3H and 3I). These results indicate OsMYB103L has diverse expressions in various tissues and organs in rice.
Overexpression of OsMYB103L results in a leaf rolling phenotype
Twelve independent transgenic lines overexpressing OsMYB103L (OE) were obtained. The expression level of OsMYB103L was determined by qRT-PCR (Figure 4A). Three independent transgenic lines (OE-1, OE-4 and OE-8) with the highest expression levels were selected for further study (Figure 4A).The most significant phenotype of OE lines was upward curling of the leaf blades (Figure 4B). This phenotype appeared first at the seedling stage and then maintained through the rest of the plant growth (Figure 4B). The measurement of leaf rolling index (LRI) was performed at the time of flowering. The LRIs of the flag leaf, the second leaf from top, and the third leaf from top were approximately 57%, 52%, and 50%, respectively (Figure 4C). The leaf blade cross-sections did not show significant differences between wild type (WT) and OE in terms of organization of the sclerenchyma and vascular tissues. It suggests that there is no obvious alteration of leaf polarity in transgenic plants. However, in contrast to WT, the leaves of transgenic plants displayed smaller bulliform cells in the rolled regions (Figure 4D).
Figure 4.
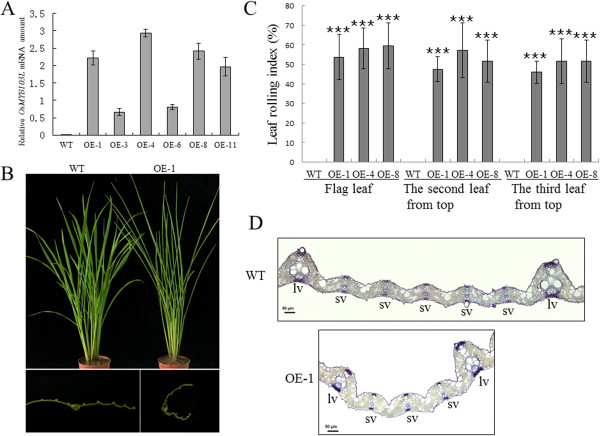
Phenotypes and molecular analysis of OsMYB103L overexpressing plants (OE). (A)OsMYB103L expression in OE as determined by qRT-PCR. Rice ACTIN1 gene was used as internal control. Each column represents an average of three replicates and bars indicate the SDs. (B) Morphology of 2-month-old WT and OE leaves. OE showed inward rolled leaves compared to WT (top panel); Cross sections of leaves of WT and OE (bottom panel). Bars = 10 cm (top panel) or 3 mm (bottom panel). (C) Statistical analysis of LRIs at the time of flowering for three OE lines of the top three leaf blades (n = 20). Asterisks (***) indicate a significant difference between transgenic plants and WT controls at P < 0.001, by Student’s t-test. (D) Transverse sections of the flag leaves in the middle part of WT and OE. Shown are regions between two large veins. lv, large vein; sv, small vein. Bars = 50 μm.
Overexpression of OsMYB103L affects the expression levels of several genes encoding cellulose synthase (CESA)
To explore the possible molecular mechanism of OsMYB103L’s effects on rice leaf development, we examined the expression profile in OE-1 and WT leaf blades using Digital Gene Expression profiling analysis (DGE). In all, 3,036 annotated genes showed expressions with more than twofold difference between OE-1 and WT, of which 2,026 genes were up-regulated and 1,010 genes were down-regulated (Additional file 1: Figure S2 and Additional file 2: Table S1). To obtain an ontological profile of the up- or down-regulated genes, ontological terms were assigned, and the enrichment significance was analyzed by AgriGO [51]. Twenty-one Gene Ontology (GO) terms were significantly enriched in the up-regulated gene set, showing that up-regulated genes were involved in multiple biological processes, such as metabolism, biogenesis, localization, and cellular processes (Figure 5A and Additional file 1: Figure S3). Meanwhile, we noticed that two GO terms, cellulose metabolic process (GO:0030243) and cellulose biosynthetic process (GO:0030244), were possibly related to the leaf rolling trait as reported in rl14 mutant [2] (Additional file 1: Figure S3). Therefore, we compared the expression levels of the rice CESA genes [52]. Our DGE data showed that OsCESA1, OsCESA4, OsCESA7, OsCESA8, OsCESA9, and OsCESA11 were up-regulated while OsCESA5 and OsCESA6 were down-regulated (Additional file 1: Table S2). We verified the expressions by qRT-PCR for these CESA genes in three independent lines (OE-1, OE-4 and OE-8). Four CESA genes, OsCESA4, OsCESA7, OsCESA8, and OsCESA9, showed significantly increased transcript levels; while one gene, OsCESA6, was down-regulated in the OE plants (Figure 5B). The qRT-PCR analysis was largely consistent with the DGE analysis except three genes, OsCESA1 and OsCESA11 of which, due to their relatively low Log2Ratio in DGE data. CESAs are proteins responsible for cellulose synthesis in plants [52-54], thus alteration of CESA expression may change the cellulose content. Therefore, we measured cellulose content in leaf blades of WT and OE plants. The results showed that OE leaf blades had 13% higher cellulose content compared to WT (Figure 5C). These results suggest that OsMYB103L regulates CESA gene expression and may consequently influence cellulose synthesis in rice.
Figure 5.
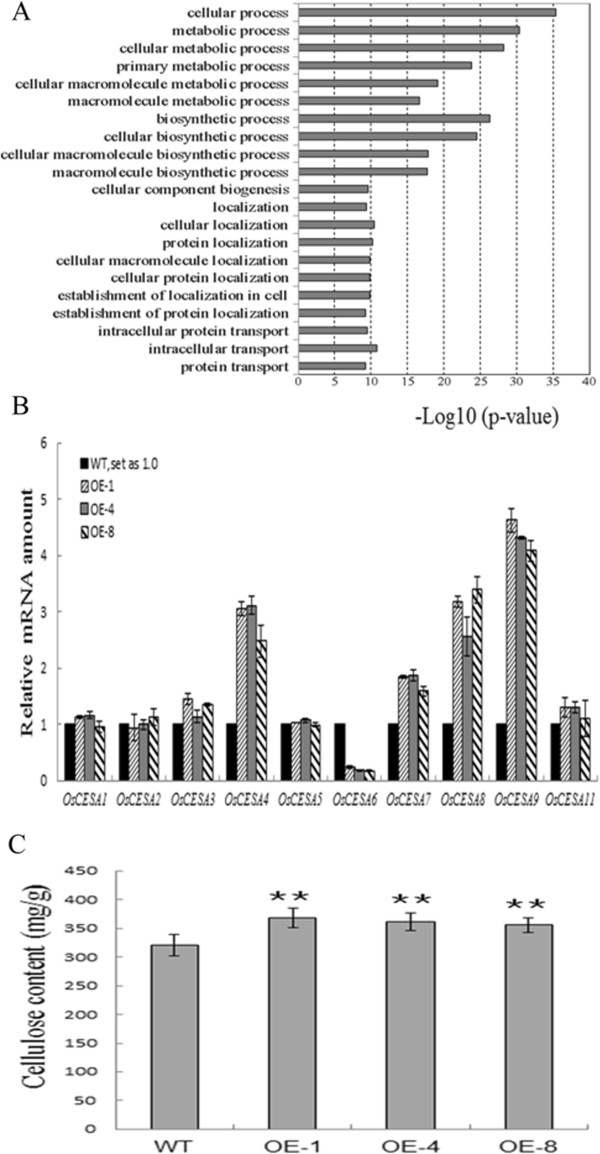
DGE, qRT-PCR and chemical analysis of wild type (WT) and OsMYB103L overexpression (OE) plants. (A) Main clusters identified by GO enrichment analysis of genes up-regulated in OE compared with WT. In each GO biological process category, the bar indicates the fold enrichment, which is defined as -Log10 (P value). The up-regulated genes are involved in multiple biological processes, such as metabolism, biogenesis, localization and transport. (B) Relative expression of rice CESA genes in WT and OE was determined by qRT-PCR analysis. The transcript levels of genes were normalized to the levels of ACTIN1 gene expression. The transcript levels of these genes in WT were arbitrarily set to 1. Values are the mean ± SD of three replicates. (C) Measurement of cellulose content in WT and OE. Cellulose content (milligrams per gram of total cell wall residues) of the flag leaf segments from WT and OE plants. The error bars were obtained from five measurements. Asterisks (**) indicate a significant difference between transgenic plants and WT controls at P < 0.01, by Student’s t-test.
Knockdown of OsMYB103L decreases the mechanical strength and cellulose content of leaves
To explore the intrinsic function of OsMYB103L, we analyzed the phenotypes of mutants with decreased expression levels. Since the loss-of-function mutants were not available, we created RNAi-knockdown plants for OsMYB103L. Six independent RNAi transgenic lines were obtained. Quantitative RT-PCR analyses showed that the expression level of OsMYB103L was significantly decreased in leaves of the RNAi lines compared with that of WT (Figure 6A). There were no obvious morphological changes in RNAi lines. However, in the heading stage of the RNAi plants, we noticed that the leaves were brittle. When the leaf blade was bent, WT leaves were flexible while the leaves of RNAi lines were broken easily, especially in the midribs (Figure 6B). The force required to break the leaf blades was 50% less for the RNAi lines than that for WT (Figure 6C). The weakened mechanical strength may relate to the altered cellulose content in leaf blades. We measured the cellulose content in leaf blades of WT and RNAi plants. Compared with WT, the levels of cellulose content in RNAi lines were significantly decreased (Figure 6D). Therefore, the decreased mechanical strength of RNAi plants might result from the decreased cellulose content. We determined the expression levels of ten CESA genes by qRT-PCR, and found that three CESA genes, OsCESA4, OsCESA7, and OsCESA9 were significantly down-regulated in the RNAi lines (Figure 6E). These results suggest that the down-regulated CESA genes in OsMYB103L RNAi plants may lead to a decreased level of cellulose content and the weakened mechanical strength of transgenic leaf blades.
Figure 6.
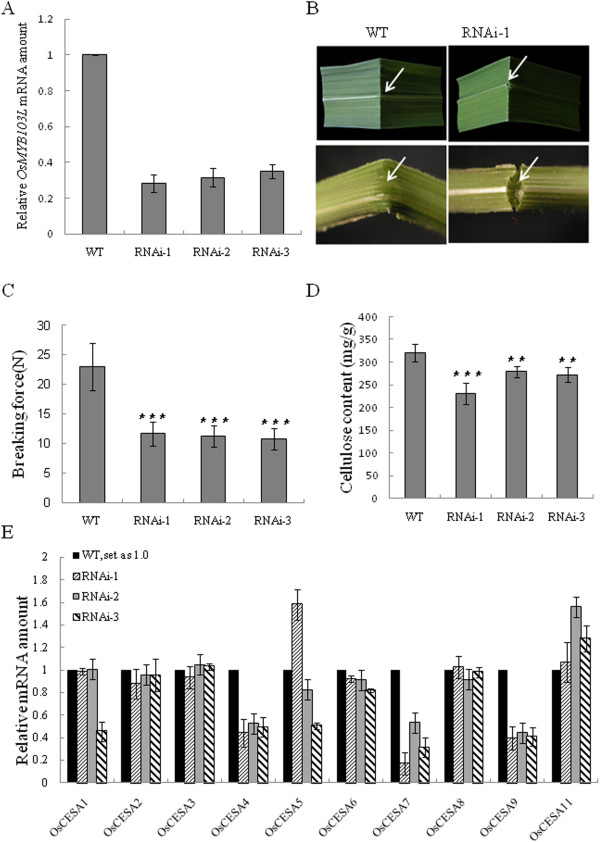
Down-regulation of OsMYB103L decreases mechanical strength and affected cellulose biosynthesis. (A) Quantitative RT-PCR analysis for RNAi transgenic plants. The transcript levels of OsMYB103L in WT were arbitrarily set to 1. Values are the mean ± SD of three replicates. (B) An easily broken OsMYB103L RNAi-1 leaves, as indicated by the arrow. (C) The force required to break WT and OsMYB103L RNAi lines flag leaves. Error bar represents SD (n =10 in each measurement). (D) Comparison of cellulose content between the wild type and OsMYB103L RNAi lines flag leaves. Error bar represents SD (n = 5 in each measurement). (E) Relative expression of rice cellulose synthase (CESA) genes in wild type (WT) and OsMYB103L RNAi plants was determined by qRT-PCR analysis. The transcript levels of genes were normalized to the levels of ACTIN1 gene expression. The transcript levels of these genes in wild type were arbitrarily set to 1. Values are the mean ± SD of three replicates. Asterisks (** or ***) indicate a significant difference between transgenic plants and WT controls at P < 0.01 or 0.001, respectively, by Student’s t-test.
Discussion
By screening phenotypes of a transgenic rice population overexpressing rice genes in MYB family, we identified a rice leaf rolling-related R2R3-MYB gene, OsMYB103L. We showed its activity of transcriptional activation and described its influence on leaf rolling. We further showed that the altered expression of OsMYB103L resulted in changes in CESA gene expression, cellulose content, and mechanical strength in rice leaf. These results suggest that OsMYB103L may have important biological functions in rice leaf development.
Leaf rolling is one of the frequently observed phenotypes in rice and other cereals. It is one of the important agronomic traits for high yield rice varieties [6-9]. Although many cultivars and mutants in rice show several types of leaf rolling phenotypes and a number of leaf rolling related genes have been identified [1-3,19,20,55], the mechanisms underlying the rolling phenomena remain to be elucidated. It is generally realized that two major factors can influence leaf rolling. The first one is the establishment of cell polarity and differentiation, and the second is physiological factors, such as osmotic pressure or turgidity in bulliform cells [1,2,56,57]. Previous studies on mutants defective in leaf development in Arabidopsis thaliana, maize, and rice have shown that leaf rolling is related to the defective development of the leaf polarity along the adaxial–abaxial axis [1,58]. However, we did not observe obvious alterations in leaf polarity in the OsMYB103L overexpressing plants by histological analysis (Figure 4D) and SEM observations (Additional file 1: Figure S4). Instead, we found that the bulliform cells in the rolled regions of OE leaves were smaller compared with that of WT leaves (Figure 4D). Several investigations have shown that bulliform cells are involved in leaf rolling [2,59]. Loss of turgor pressure in bulliform cells could lead to leaf “roll up” during water stress [56]. Here the observed abnormal bulliform cells in the leaves of OsMYB103L overexpressing plants are consistent with those described in a number of known rolled leaf mutants [2,20]. For instance, cow1 and rl14 mutants showed rolled leaves with serious shrinkage and smaller bulliform cells, but with no obvious defects in leaf polarity [2,20]. Therefore, our results suggest that the abnormal bulliform cells, rather than defects in leaf polarity, may account for the rolled leaf phenotype in OsMYB103L overexpressing plants to some extent.
Secondary cell walls constitute skeletal frameworks and furnish the plant body with mechanical strength [60,61]. Molecular and genetic studies in the model plant, Arabidopsis thaliana, revealed that a group of secondary wall-associated NAC domain (SWN) transcription factors are master switches regulating a cascade of down-stream transcription factors, such as MYB transcription factors that leads to the activation of the secondary wall biosynthetic program [62,63]. It has been reported that AtMYB103 (At1g63910) is directly activated by the master switches, SWNs [62,63]. In Arabidopsis thaliana, overexpression of R2R3-MYB gene AtMYB103 led to a noticeable increase in the secondary wall thickness, while its dominant repression caused reduction in secondary wall thickness of both xylary fibers and interfascicular fibers [50,64]. It has been proposed that AtMYB103 has the potential to induce the expression of genes involved in cellulose biosynthesis [50]. The transcriptional network regulating secondary cell wall biosynthesis may be evolutionarily conserved in plants, however, little is known about the detailed regulatory programs in grass species, such as rice [65].
Based on our analyses on the OsMYB103L and its encoded protein, we deduced that OsMYB103L might be an ortholog of AtMYB103 in rice. We wondered whether it has the conserved functions in rice, a model plant in monocotyledon, and addressed this question by reverse genetics, including overexpression and RNAi-knockdown approaches. Our study showed that overexpression of OsMYB103L resulted in rolled leaves and knockdown of OsMYB103L led to brittle leaves (Figures 4B and 6B). Further analyses disclosed that the expression of four rice CESA genes, OsCESA4, OsCESA7, OsCESA8, and OsCESA9, was increased significantly in transgenic overexpression rice plants (Figure 5B and Additional file 2: Table S1) whereas three of these CESA genes, OsCESA4, OsCESA7, and OsCESA9, were significantly down-regulated in the RNAi lines (Figure 6E). OsMYB103L is a putative R2R3 MYB transcription factor, which is expected to regulate gene expression by binding to the promoters of its downstream genes. Putative MYB binding sequences (MBSs) can be identified by using the tools described in PLACE (A Database of Plant Cis-acting Regulatory DNA Elements; http://www.dna.affrc.go.jp/PLACE/) [66]. We did find two and even more CNGTTR sites, the putative MBSs, in the respective promoter regions of these CESA genes (Additional file 1: Figure S5). Therefore, we hypothesized that these CESA genes would be the targets of OsMYB103L. Further investigation, such as chromatin immunoprecipitation (ChIP) and electrophoretic mobility shift assay (EMSA), could be carried out to prove it.
In higher plants, CESAs were characterized as catalytic subunits of cellulose synthase complexes [67,68]. In this study, our results showed that the cellulose content of mature leaves was higher in overexpression lines than that in WT (Figure 5C) while the OsMYB103L RNAi lines had decreased cellulose content in the leaf blades (Figure 6D). The results indicate that OsMYB103L may regulate the expression levels of CESA genes, which could subsequently alter the cellulose content. However, we also noticed that the contents of cellulose were not positively correlated with the expression levels of OsMYB103L in transgenic plants, especially in the overexpression lines. Plant CESA genes are members of multigene families [69]. Arabidopsis has 10 CESA isoforms which can be classified into six orthologous groups having non-redundant functions in cellulose synthesis as revealed by mutational analysis [53]. Rice also has 10 CESA isoforms, of which OsCESA4, OsCESA7 and OsCESA9 were showed non-redundant functions in cellulose synthesis specifically in secondary walls [52,70]. Based on these, we speculate that most rice CESAs might also play distinctive roles in cellulose synthesis. Some particular CESA gene expression induction may not lead to significant change of cellulose content. Therefore, changed expressions of several CESAs in OsMYB103L transgenic lines may not induce obviously changes in cellulose content of the transgenic plants.
In addition, our study demonstrated that OsMYB103L RNAi-knockdown plants possessed brittle leaves with obviously decreased mechanical strength and reduced cellulose content in the leaf blades (Figure 6B- 6D). Of note, reduced cellulose content and inferior mechanical strength are the common phenotypes of CESA mutants, such as irx1 to irx3 in Arabidopsis and brittle culm13 (bc13) in rice [70-72]. Thus, we wondered whether mechanical strength was elevated due to the increased cellulose in OsMYB103L overexpression plants. Therefore, we also measured the mechanical strength of the overexpression lines of OsMYB103L. Nevertheless, the leaves of overexpression plants did not show statistically significant changes in mechanical strength compared to wild type (Additional file 1: Figure S6). Mechanical strength is a complex trait, which can be affected by many factors [73,74]. Perhaps, in our case, the increase of cellulose contents was not enough to cause the change of mechanical strength in OsMYB103L overexpression plants.
Moderate leaf rolling has been regarded as one of the important agronomic traits in high yield rice cultivars [6-9], and selection of this trait has been applied for rice breeding directly or indirectly in China [5,6]. One of the successful examples is the breeding of super-hybrid rice, Liang-You-Pei-Jiu (LYP9) [75,76]. LYP9 has moderate xrolling leaves and is widely cultivated throughout southern China due to its high yield and seed quality. The successful cultivation of LYP9 is therefore taken as an example for improving crop yield and quality using the rolled leaf trait [77]. Therefore, identification of genes that regulate leaf rolling would be beneficial to breeding. In this study, we report that overexpression of OsMYB103L in rice results in leaf rolling, which suggests that OsMYB103L could be a potential target for genetic manipulation of leaf shape and plant architecture in rice. In addition, leaf brittleness is an important agronomic trait for rice and other crops since it affects not only grain production but also the utilization of cereal straw as animal forage [74]. Here, we found that down-regulation of the expression level of OsMYB103L decreases mechanical strength in rice plants, suggesting that OsMYB103L could be engineered for favorable leaf brittleness in rice, as well as in other cereal and forage crops.
Conclusions
In this study, an R2R3 MYB transcription factor, OsMYB103L, has been identified in rice. We show that overexpression of OsMYB103L in rice results in leaf rolling, increased cellulose content, and elevated expression of genes encoding cellulose synthase (CESA). Besides, the RNA interference (RNAi)-knockdown of OsMYB103L leads to a weakened mechanical strength and decreased cellulose content in leaves. Moreover, the expression levels of several CESA genes are up-regulated in OsMYB103L overexpression lines while down-regulated in OsMYB103L-knockdown lines. Collectively, this work demonstrates that OsMYB103L impacts leaf shape, cellulose synthesis, and mechanical strength in rice.
Methods
Plant materials and growth conditions
Rice (Oryza sativa L.) subspecies indica cultivar Kasalath and japonica cultivar Nipponbare were used for analysis. Rice seeds from control wild type and transgenic lines were immersed in water for 2 days and grown in a soil seed bed for approximately 30 days, and then the seedlings were transplanted to the field and/or greenhouse for further analysis. Plants maintained in greenhouses were grown under standard conditions at 28°C with 16 h of light. The fields were located in the Experimental Stations (Beijing and Hainan) of the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China.
Bioinformatics analysis
A search for OsMYB103L homologs was performed using the National Center for Biotechnology Information (NCBI) BLAST server (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Domain prediction was performed using the Pfam database (http://pfam.sanger.ac.uk) and the NCBI Conserved Domains database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). Alignment was performed with ClustalW using default setting (http://clustalw.ddbj.nig.ac.jp/). Unrooted neighbor-joining phylogenetic tree was constructed using the neighbor-joining algorithm with PHYLIP version 3.572c [78]. Bootstrap analysis was carried out with 1,000 replicates. TreeView was used to generate the graphical output [79].
Subcellular localization
For detection of subcellular localization of OsMYB103L, the open reading frame (ORF) of OsMYB103L was amplified from the FL-cDNA clone (AK111808) and then was cloned into vector pEZR(K)-LN to create the 35S:OsMYB103L-GFP vector. The 35S: OsMYB103-GFP and empty vector (pEZR(K)-LN) were transformed into rice protoplasts by polyethylene glycol-mediated transformation method [80] or onion epidermis by particle bombardment using a PDS-1000/He biolistic particle delivery system (Bio-Rad, California, USA), respectively. Subcellular distribution of the OsMYB103L protein was visualized by fluorescent confocal microscopy (Leica TCS SP5, California, USA).
Transactivation assay in yeast cells
The yeast strain Y2HGold, containing the AUR1-C and MEL1 reporter genes with GAL4 binding elements in the promoters, respectively, was used as an assay system (Clontech, USA). The expression of AUR1-C confers strong resistance to the highly toxic drug Aureobasidin A (Aba). The activities of α-galactosidase which MEL-1 encoded were examined by X-α-gal staining. The full-length, N-terminal MYB DNA-binding domain (1–160 Amino acids) or the C-terminal putative activation domain (161–359 Amino acids) were respectively cloned into pBD-GAL4 vector. The pBD vector was used as a negative control. All these plasmids were individually introduced into cells of yeast strain Y2HGold. The yeast transformants were grown on SD medium in the absence of tryptophan (SD/-Trp) for 2–3 days at 30°C. Transferred yeast cells were also grown on SD/-Trp in the presence of Aba and X-α-gal for 2–3 days at 30°C to monitor the generation of blue color.
Promoter-GUS analysis
The promoter of the OsMYB103L (about 1.4 kb upstream of ATG) was amplified from the rice genomic DNA (cv. Nipponbare) using the primers 5′-GGATCCACTCCTGCGAGGCTCTGATACG-3′ and 5′-CCATGGATGCACTCT-AGATATCACTG-3′. Then, the DNA fragment was cloned into the pCAMBIA1301 (CAMBIA, Australia) vector resulting in fusion of the promoter and the GUS reporter gene. The ProOsMYB103: GUS construct was transformed into wild type rice cultivar Kasalath. About 20 independent transgenic lines were obtained after screening. GUS staining was performed according to the method of Jefferson et al. [81].
Binary vectors construction and rice transformation
For the overexpression construct of OsMYB103L, gene-specific primers, 5′-GGATCCTGCTAGCAGCTAGATCAAG-3′ and 5′-ACTAGTGTCATCCTCCTG-TGTTTATT-3′ (BamH I and Spe I sites italicized) were used to amplify the coding sequence of OsMYB103L using the FL-cDNA clone (AK111808) as template. The FL-cDNA clone was from the National Institute of Agrobiological Sciences, Japan [78]. The sequencing-confirmed PCR fragment was cloned into the vector pTCK303 [82] to create the overexpression vector of OsMYB103L driven by a maize ubiquitin promoter.
For the RNA interference (RNAi) construct of OsMYB103L, a fragment of approximately 325 bp was amplified from OsMYB103L with two primers, 5′-GGTACCACTAGTCCGTGAAGCTGGCGATGAAC-3 (Kpn I and Spe I sites italicized) and 5′-GGATCCGAGCTCCACGACGAGCCCGAACAAC-3′ (BamH I and Sac I sites italicized). The sequencing-confirmed PCR fragment was cloned into the vector pTCK303 as previously described [82] to create the RNAi vector of OsMYB103L driven by a maize ubiquitin promoter.
These constructs were transferred to Agrobacterium tumefaciens strain LBA4404 by electroporation, and were transformed into an indica rice cv. Kasalath, respectively, according to the method of Hiei et al. [83].
RNA extraction and qRT-PCR analysis
Total RNA was extracted from different tissues using Trizol reagent (Invitrogen, California, USA) according to the manufacturer’s protocol. After treatment with DNase to remove genomic DNA contamination, the first strand of cDNA was synthesized by M-MLV reverse transcriptase (Promega, USA). Relative quantification of gene expression by real-time PCR was performed on a Bio-Rad Chromo 4 Real Time PCR System (Bio-Rad, USA) with TransStart SYBR Green qPCR SuperMix Kit (TransGen, Beijing, China). The housekeeping gene ACTIN1 was used as an internal control for normalization of RNA samples. Three replicates were carried out for each gene and each analysis was biologically repeated at least twice. Gene-specific primers are shown in Additional file 1: Table S3 line.
Measurement of the leaf rolling index (LRI)
To determine the leaf rolling index (LRI), two measurements were taken, Lw (Expand the leaf blade and determine the greatest width of the leaf blade) and Ln (Measure the natural distance of the leaf blade margins at the same site). LRI was calculated as LRI (%) = (Lw-Ln)/Lw × 100 [3]. Data were collected on the flag (uppermost), second and third leaves on twenty individual plants at the time of flowering.
Cellulose measurement
Mature leaves were cut into pieces and ground into powder under liquid nitrogen. The materials were then extracted twice with 70% ethanol at 70°C for 1 h. Cell wall materials were dried under vacuum for cellulose content measurement [84]. Whatman 3MM filter paper was used to establish a standard curve for quantification of cellulose. The anthrone reagent was used to determine the cellulose content.
Breaking force test
To examine the mechanical strength in leaf blade, the leaf blades of flag leaves from developmentally matched transgenic rice plants and wild type plants were collected and immediately used for measurement. The forces that break the samples at leaf blades were measured with a digital force/length tester (5848 Microtester; Instron, USA).
Scanning electron microscopy (SEM) observation
Samples were fixed in 2.5% glutaraldehyde solution. Fixed samples were dehydrated with gradual ethanol series, dried by critical-point drying method using liquid carbon dioxide (Model HCP-2, Hitachi, Tokyo), gold-coated with an Edwards E-1010 ion sputter coater (Hitachi, Tokyo), and then observed using a S-3000 N variable pressure scanning electron microscope (Hitachi, Tokyo).
Microscopy
For light microscopic analysis, the samples were fixed in 2.5% glutaraldehyde solution, and then dehydrated through a graded acetone series. The samples were embedded in Epon812 (SPI CHEM) and polymerized at 70°C. Sections were cut with a microtome (Leica RM2265, Germany) and stained with 0.25% toluidine blue, and finally observed with a microscope (OLYMPUS BX41, Japan).
Digital gene expression profiling (DGE) analysis
For digital gene expression analysis, transgenic overexpressor (OE-1) and wild type flag leaves were harvested for RNA extraction using TRIzol (Invitrogen) as described by the manufacturer. The digital expression analysis was performed by the Beijing Genomics Institute (http://www.genomics.cn) using the standardized procedure. The main reagents and supplies are Illumina Gene Expression Sample Prep Kit and Illumina Sequencing Chip (flowcell), and the main instruments are Illumina Cluster Station and Illumina HiSeqTM 2000 System. All clean tags were mapped to the reference sequences (ftp://ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_Dbs/pseudomolecules/version_6.1/all.dir/all.cDNA) and only 1 bp mismatch is considered. Clean tags mapped to reference sequences from multiple genes were filtered. Remainder clean tags were designed as unambiguous clean tags. The number of unambiguous clean tags for each gene was calculated and then normalized to TPM (number of transcripts per million clean tags).
The original data set is deposited in the National Institutes of Health Gene Expression Omnibus database under accession number GSE52394.
Accession numbers
Sequence data used in this manuscript can be found in the Arabidopsis Information Resource (TAIR, http://www.arabidopsis.org) and the rice genome annotation database (http://rice.plantbiology.msu.edu) under the following accession numbers: AtMYB103 (At1g63910), AtMYB26 (At3g13890), AtMYB50 (At1g57560), AtMYB55 (At4g01680), AtMYB61 (At1g09540), AtMYB67 (At3g12720), AtMYB86 (At5g26660), OsMYB103L (Os08g05520), Os01g51260, Os01g18240, Os05g04820, Os07g31470, Os01g50720, Os04g38740, OsActin1 (Os03g50890), OsCESA1 (Os05g08370), OsCESA2 (Os03g59340), OsCESA3 (Os07g24190), OsCESA4 (Os01g54620), OsCESA5 (Os03g62090), OsCESA6 (Os07g14850), OsCESA7 (Os10g32980), OsCESA8 (Os07g10770), OsCESA9 (Os09g25490), OsCESA11 (Os06g39970).
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YC and DL carried out the most of molecular genetic studies, data analysis and drafted the manuscript. YC, XLiu, and XLi carried out the rice transformation. CJ participated in microscopy analysis. LH participated in bioinformatics analysis. XZ participated in phenotype analysis. CC and ZC participated in the data analysis. LZ and DL conceived of the study, and participated in its design, coordination and drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
OsMYB103L-GFP is located to nucleus in onion epidermal cells. Figure S2. Number of differentially expressed genes in Digital Gene Expression profiling analysis (DGE) between wild type and OsMYB103L overexpressing plants. Figure S3. GO enrichment analysis of genes up-regulated in wild type and OsMYB103L overexpressing plants. Figure S4. Scanning electron micrographs (SEM) analysis. Figure S5. Schematic diagrams of the promoter regions of OsCESA genes. Figure S6. The force required to break flag leaves in wild type and OsMYB103L overexpression plants. Table S2. Expression levels of CESA genes in DGE analysis between wild type (WT) and OsMYB103L overexpressing plants (OE-1). Table S3. Primers of qRT-PCR used in this article.
Genes that up- or down-regulated two-fold in DGE data of OsMYB103L overexpressing plants compared to that of the wild type (Excel File).
Contributor Information
Chunhua Yang, Email: chyang@genetics.ac.cn.
Dayong Li, Email: dyli@genetics.ac.cn.
Xue Liu, Email: xliu@genetics.ac.cn.
Chengjun Ji, Email: jicj@pku.edu.cn.
Lili Hao, Email: haolili@big.ac.cn.
Xianfeng Zhao, Email: xfzhao@genetics.ac.cn.
Xiaobing Li, Email: xbli@genetics.ac.cn.
Caiyan Chen, Email: cychen@isa.ac.cn.
Zhukuan Cheng, Email: zkcheng@genetics.ac.cn.
Lihuang Zhu, Email: lhzhu@genetics.ac.cn.
Acknowledgements
This work was supported by the grants from the National Natural Science Foundation of China (31123007) and the State Key Laboratory of Plant Genomics, China (2012B0301). We thank Prof. Shouyi Chen and Dr. Hongfeng Zou (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for their kind helps on the transcriptional activation activity analysis. We thank Dr. Yihua Zhou (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for her kind helps on breaking force test. We thank Dr. Kang Chong (Institute of Botany, Chinese Academy of Sciences) for providing the vector pTCK303. We thank the National Institute of Agrobiological Sciences, Japan, for providing the rice FL-cDNA clones. We also thank Brian Fanella and Dr. Maoyin Li (Donald Danforth Plant Science Center) and Dr. Donglei Zhang (University of Chicago) for their critical reading and helpful discussion on the manuscript.
References
- Zhang GH, Xu Q, Zhu XD, Qian Q, Xue HW. SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell. 2009;21(3):719–735. doi: 10.1105/tpc.108.061457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Zhao F, Cong Y, Sang X, Du Q, Wang D, Li Y, Ling Y, Yang Z, He G. Rolling-leaf14 is a 2OG-Fe (II) oxygenase family protein that modulates rice leaf rolling by affecting secondary cell wall formation in leaves. Plant Biotechnol J. 2012;10(5):524–532. doi: 10.1111/j.1467-7652.2012.00679.x. [DOI] [PubMed] [Google Scholar]
- Shi Z, Wang J, Wan X, Shen G, Wang X, Zhang J. Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta. 2007;226(1):99–108. doi: 10.1007/s00425-006-0472-0. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, Tanaka H, Kitano H, Matsuoka M. Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol. 2006;24(1):105–109. doi: 10.1038/nbt1173. [DOI] [PubMed] [Google Scholar]
- Yuan L. Super-high yield hybrid rice breeding. Hybrid Rice. 1997;12:1–6. [Google Scholar]
- Chen Z, Pan X, Hu J. Relationship between rolling leaf and ideal plant type of rice (Oryza sativa L.) Jiangsu Agricultural Res. 2001;22:88–91. [Google Scholar]
- Zhu D, Lin X, Cao W. Comparison of leaf photosynthetic characteristics among rice hybrids with different leaf rolling index. Acta Agronomica Sin. 2001;27(1):329–333. [Google Scholar]
- Lang Y, Zhang Z, Gu X, Yang J, Zhu Q. Physiological and ecological effects of crimpy leaf character in rice (Oryza sativa L.) I. Leaf orientation, canopy structure and light distribution. Acta Agronomica Sin. 2004;30(1):739–744. [Google Scholar]
- Lang Y, Zhang Z, Gu X, Yang J, Zhu Q. Physiological and ecological effects of crimpy leaf character in rice (Oryza sativa L.) II. Photosynthetic character, dry mass production and yield forming. Acta Agronomica Sin. 2004;30(1):883–887. [Google Scholar]
- Khush GS, Kinoshita T. In: Rice Biotechnology. Khush GS, Toenniesen GH, editor. Wallingford, UK: CAB International; 1991. Rice karyotype, marker genes, and linkage group; pp. 83–107. [Google Scholar]
- Kinoshita T. In: Biology of Rice. Tsunoda S, Takahashi N, editor. Tokyo: JSSP/Elsevier; 1984. Gene analysis and linkage map. pp(1): 187–274. [Google Scholar]
- Li SG, Ma YQ, He P, Li HY, Chen Y, Zhou KD, Zhu LH. Genetics analysis and mapping the flag leaf roll in rice (Oryza sativa L.) J Sichuan Agric Uni. 1998;16(1):391–393. [Google Scholar]
- Shao YJ, Pan CH, Chen ZX, Zuo SM, Zhang YF, Pan XB. Fine mapping of an incomplete recessive gene for leaf rolling in rice (Oryza sativa L.) Chin Sci Bull (in Chinese) 2005;50(1):2466–2672. [Google Scholar]
- Luo Z, Yang Z, Zhong B, Li Y, Xie R, Zhao F, Ling Y, He G. Genetic analysis and fine mapping of a dynamic rolled leaf gene, RL10(t), in rice (Oryza sativa L.) Genome. 2007;50(9):811–817. doi: 10.1139/G07-064. [DOI] [PubMed] [Google Scholar]
- Yan S, Yan CJ, Zeng XH, Yang YC, Fang YW, Tian CY, Sun YW, Cheng ZK, Gu MH. ROLLED LEAF 9, encoding a GARP protein, regulates the leaf abaxial cell fate in rice. Plant Mol Biol. 2008;68(3):239–250. doi: 10.1007/s11103-008-9365-x. [DOI] [PubMed] [Google Scholar]
- Li M, Xiong G, Li R, Cui J, Tang D, Zhang B, Pauly M, Cheng Z, Zhou Y. Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J. 2009;60(6):1055–1069. doi: 10.1111/j.1365-313X.2009.04022.x. [DOI] [PubMed] [Google Scholar]
- Hu J, Zhu L, Zeng D, Gao Z, Guo L, Fang Y, Zhang G, Dong G, Yan M, Liu J, Qian Q. Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol Biol. 2010;73(3):283–292. doi: 10.1007/s11103-010-9614-7. [DOI] [PubMed] [Google Scholar]
- Luan W, Liu Y, Zhang F, Song Y, Wang Z, Peng Y, Sun Z. OsCD1 encodes a putative member of the cellulose synthase-like D sub-family and is essential for rice plant architecture and growth. Plant Biotechnol J. 2010;9:513–524. doi: 10.1111/j.1467-7652.2010.00570.x. [DOI] [PubMed] [Google Scholar]
- Wu C, Fu Y, Hu G, Si H, Cheng S, Liu W. Isolation and characterization of a rice mutant with narrow and rolled leaves. Planta. 2010;232(2):313–324. doi: 10.1007/s00425-010-1180-3. [DOI] [PubMed] [Google Scholar]
- Woo YM, Park HJ, Su’udi M, Yang JI, Park JJ, Back K, Park YM, An G. Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol Biol. 2007;65(1–2):125–136. doi: 10.1007/s11103-007-9203-6. [DOI] [PubMed] [Google Scholar]
- Fujino K, Matsuda Y, Ozawa K, Nishimura T, Koshiba T, Fraaije MW, Sekiguchi H. NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol Genet Genomics. 2008;279(5):499–507. doi: 10.1007/s00438-008-0328-3. [DOI] [PubMed] [Google Scholar]
- Wu R, Li S, He S, Wassmann F, Yu C, Qin G, Schreiber L, Qu LJ, Gu H. CFL1, a WW domain protein, regulates cuticle development by modulating the function of HDG1, a class IV homeodomain transcription factor, in rice and Arabidopsis. Plant Cell. 2011;23(9):3392–3411. doi: 10.1105/tpc.111.088625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang JJ, Zhang GH, Qian Q, Xue HW. Semi-rolled leaf1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol. 2012;159(4):1488–1500. doi: 10.1104/pp.112.199968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J, Hibara K, Sato Y, Nagato Y. Developmental role and auxin responsiveness of Class III homeodomain leucine zipper gene family members in rice. Plant Physiol. 2008;147(4):1960–1975. doi: 10.1104/pp.108.118679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Martin C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol. 1999;41(5):577–585. doi: 10.1023/A:1006319732410. [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15(10):573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011;66(1):94–116. doi: 10.1111/j.1365-313X.2010.04459.x. [DOI] [PubMed] [Google Scholar]
- Rosinski JA, Atchley WR. Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. J Mol Evol. 1998;46(1):74–83. doi: 10.1007/PL00006285. [DOI] [PubMed] [Google Scholar]
- Martin C, Paz-Ares J. MYB transcription factors in plants. Trends Genet. 1997;13(2):67–73. doi: 10.1016/S0168-9525(96)10049-4. [DOI] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol. 2001;4(5):447–456. doi: 10.1016/S1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell. 1991;67(3):483–493. doi: 10.1016/0092-8674(91)90523-2. [DOI] [PubMed] [Google Scholar]
- Bender J, Fink GR. A Myb homologue, ATR1, activates tryptophan gene expression in Arabidopsis. Proc Natl Acad Sci U S A. 1998;95(10):5655–5660. doi: 10.1073/pnas.95.10.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeren FU, Dolferus R, Wu Y, Peacock WJ, Dennis ES. Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. Genetics. 1998;149(2):479–490. doi: 10.1093/genetics/149.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell. 1999;99(5):473–483. doi: 10.1016/S0092-8674(00)81536-6. [DOI] [PubMed] [Google Scholar]
- Preston J, Wheeler J, Heazlewood J, Li SF, Parish RW. AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J. 2004;40(6):979–995. doi: 10.1111/j.1365-313X.2004.02280.x. [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007;50(4):660–677. doi: 10.1111/j.1365-313X.2007.03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53(5):814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- Zhou J, Lee C, Zhong R, Ye ZH. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell. 2009;21(1):248–266. doi: 10.1105/tpc.108.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Kondo M, Aya K, Miyao A, Sato Y, Antonio BA, Namiki N, Nagamura Y, Matsuoka M. Identification of transcription factors involved in rice secondary cell wall formation. Plant Cell Physiol. 2013;54(11):1791–1802. doi: 10.1093/pcp/pct122. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Sakamoto S, Kawai T, Kobayashi Y, Sato K, Ichinose Y, Yaoi K, Akiyoshi-Endo M, Sato H, Takamizo T, Ohme-Takagi M, Mitsuda N. Engineering the Oryza sativa cell wall with rice NAC transcription factors regulating secondary wall formation. Frontiers in plant science. 2013;4:383. doi: 10.3389/fpls.2013.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yang X, He K, Liu M, Li J, Gao Z, Lin Z, Zhang Y, Wang X, Qiu X, Shen Y, Zhang L, Deng X, Luo J, Deng XW, Chen Z, Gu H, LJ Q. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol. 2006;60(1):107–124. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- Katiyar A, Smita S, Lenka SK, Rajwanshi R, Chinnusamy V, Bansal KC. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics. 2012;13:544. doi: 10.1186/1471-2164-13-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MR, Yun KY, Mohanty B, Herath V, Xu F, Wijaya E, Bajic VB, Yun SJ, De Los Reyes BG. Supra-optimal expression of the cold-regulated OsMyb4 transcription factor in transgenic rice changes the complexity of transcriptional network with major effects on stress tolerance and panicle development. Plant Cell Environ. 2010;33(12):2209–2230. doi: 10.1111/j.1365-3040.2010.02221.x. [DOI] [PubMed] [Google Scholar]
- Su CF, Wang YC, Hsieh TH, Lu CA, Tseng TH, Yu SM. A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol. 2010;153(1):145–158. doi: 10.1104/pp.110.153015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Wang Y, Yang A, Zhang WH. OsMYB2P-1, an R2R3 MYB transcription factor, is involved in the regulation of phosphate-starvation responses and root architecture in rice. Plant Physiol. 2012;159(1):169–183. doi: 10.1104/pp.112.194217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Inukai Y, Ueguchi-Tanaka M, Itoh H, Izawa T, Kobayashi Y, Hattori T, Miyao A, Hirochika H, Ashikari M, Matsuoka M. Loss-of-function mutations of the rice GAMYB gene impair alpha-amylase expression in aleurone and flower development. Plant Cell. 2004;16(1):33–44. doi: 10.1105/tpc.017327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Dai X, Xu Y, Guo J, Liu Y, Chen N, Xiao J, Zhang D, Xu Z, Zhang X, Chong K. Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 2009;150(1):244–256. doi: 10.1104/pp.108.133454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Xu C, He Y, Zong J, Yang X, Si H, Sun Z, Hu J, Liang W, Zhang D. Mutation in CSA creates a new photoperiod-sensitive genic male sterile line applicable for hybrid rice seed production. Proc Natl Acad Sci U S A. 2013;110(1):76–81. doi: 10.1073/pnas.1213041110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Dai X, Zhang WH. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot. 2012;63(7):2541–2556. doi: 10.1093/jxb/err431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell. 2008;20(10):2763–2782. doi: 10.1105/tpc.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38(Web Server issue):W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Guo K, Li Y, Tu Y, Hu H, Wang B, Cui X, Peng L. Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 2010;10:282. doi: 10.1186/1471-2229-10-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C. Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol. 2006;22:53–78. doi: 10.1146/annurev.cellbio.22.022206.160206. [DOI] [PubMed] [Google Scholar]
- Richmond T. Higher plant cellulose synthases. Genome Biol. 2000;1(4):REVIEWS3001. doi: 10.1186/gb-2000-1-4-reviews3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Shi ZY, Shen GZ, Wang XQ, An LS, Zhang JL. Overexpression of ACL1 (abaxially curled leaf 1) increased Bulliform cells and induced abaxial curling of leaf blades in rice. Mol Plant. 2010;3(5):807–817. doi: 10.1093/mp/ssq022. [DOI] [PubMed] [Google Scholar]
- Price AH, Young EM, Tomos AD. Quantitative trait loci associated with stomatal conductance, leaf rolling and heading date mapped in upland rice (Oryza sativa) New Phytol. 1997;137(5):83–91. [Google Scholar]
- Hsiao TC, O’Toole JC, Yambao EB, Turner NC. Influence of osmotic adjustment on leaf rolling and tissue death in rice (Oryza sativa L.) Plant Physiol. 1984;75(2):338–341. doi: 10.1104/pp.75.2.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JM, Lane B, Freeling M. Expression of a mutant maize gene in the ventral leaf epidermis is sufficient to signal a switch of the leaf’s dorsoventral axis. Development. 2002;129(19):4581–4589. doi: 10.1242/dev.129.19.4581. [DOI] [PubMed] [Google Scholar]
- Alvarez JM, Rocha JF, Machado SR. Bulliform cells in Loudetiopsis chrysothrix (Nees) conert and Tristachya leiostachya Nees (Poaceae): structure in relation to function. Braz Arch Biol Technol. 2008;55(5):113–119. [Google Scholar]
- Roberts K. Structures at the plant cell surface. Curr Opin Cell Biol. 1990;2(5):920–928. doi: 10.1016/0955-0674(90)90093-T. [DOI] [PubMed] [Google Scholar]
- Albersheim P, An J, Freshour G, Fuller MS, Guillen R, Ham KS, Hahn MG, Huang J, O’Neill M, Whitcombe A, Williams MV, York WS, Darvill A. Structure and function studies of plant cell wall polysaccharides. Biochem Soc Trans. 1994;22(2):374–378. doi: 10.1042/bst0220374. [DOI] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Ye ZH. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell. 2007;19(9):2776–2792. doi: 10.1105/tpc.107.053678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey SG, Mizrachi E, Spokevicius AV, Bossinger G, Berger DK, Myburg AA. SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biol. 2011;11:173. doi: 10.1186/1471-2229-11-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. Regulation of cell wall biosynthesis. Curr Opin Plant Biol. 2007;10(6):564–572. doi: 10.1016/j.pbi.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, McCarthy RL, Reeves CK, Jones EG, Ye ZH. Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol. 2011;52(10):1856–1871. doi: 10.1093/pcp/pcr123. [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27(1):297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci U S A. 1996;93(22):12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Hofte H, Plazinski J, Birch R, Cork A, Glover J, Redmond J, Williamson RE. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279(5351):717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Hofte H, Gonneau M, Vernhettes S. Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2007;104(39):15572–15577. doi: 10.1073/pnas.0706569104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Murata K, Yamazaki M, Onosato K, Miyao A, Hirochika H. Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol. 2003;133(1):73–83. doi: 10.1104/pp.103.022442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SR, Somerville CR. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell. 1997;9(5):689–701. doi: 10.1105/tpc.9.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song XQ, Liu LF, Jiang YJ, Zhang BC, Gao YP, Liu XL, Lin QS, Ling HQ, Zhou YH. Disruption of secondary wall cellulose biosynthesis alters cadmium translocation and tolerance in rice plants. Mol Plant. 2013;6(3):768–780. doi: 10.1093/mp/sst025. [DOI] [PubMed] [Google Scholar]
- Ning J, Zhang B, Wang N, Zhou Y, Xiong L. Increased leaf angle1, a Raf-like MAPKKK that interacts with a nuclear protein family, regulates mechanical tissue formation in the Lamina joint of rice. Plant Cell. 2011;23(12):4334–4347. doi: 10.1105/tpc.111.093419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Qian Q, Zhou Y, Yan M, Sun L, Zhang M, Fu Z, Wang Y, Han B, Pang X, Chen M, Li J. BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell. 2003;15(9):2020–2031. doi: 10.1105/tpc.011775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Tao Y, Liu G, Chen C, Luo R, Xia H, Gan Q, Zeng H, Lu Z, Han Y, Li X, Song G, Zhai H, Peng Y, Li D, Xu H, Wei X, Cao M, Deng H, Xin Y, Fu X, Yuan L, Yu J, Zhu Z, Zhu L. A transcriptomic analysis of superhybrid rice LYP9 and its parents. Proc Natl Acad Sci U S A. 2009;106(19):7695–7701. doi: 10.1073/pnas.0902340106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L. Super Hybrid Rice Research (ed.L.Yuan) Shanghai, China: Shanghai Scientific & Technical Publishers; 2006. pp. 2–3. [Google Scholar]
- Shen NW, Qian Q, Zhang GH. Research progress on rice rolled leaf and its application in breeding program. Molecular Plant Breeding. 2009;7(5):852–860. [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, Hotta I, Kojima K, Namiki T, Ohneda E, Yahagi W, Suzuki K, Li CJ, Ohtsuki K, Shishiki T, Otomo Y, Murakami K, Iida Y, Sugano S, Fujimura T, Suzuki Y, Tsunoda Y, Kurosaki T, Kodama T, Masuda H. Foundation of Advancement of International Science Genome Sequencing Analysis Group et al. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science. 2003;301(5631):376–379. doi: 10.1126/science.1081288. [DOI] [PubMed] [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12(4):357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Bart R, Chern M, Park CJ, Bartley L, Ronald PC. A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods. 2006;2:13. doi: 10.1186/1746-4811-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6(13):3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Chen C, Xu Y, Jiang R, Han Y, Xu Z, Chong K. A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.) Plant Mol Biol. 2004;22:409–417. doi: 10.1007/BF02772683. [DOI] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6(2):271–282. doi: 10.1046/j.1365-313X.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Updegraff DM. Semimicro determination of cellulose in biological materials. Anal Biochem. 1969;32:420–424. doi: 10.1016/S0003-2697(69)80009-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
OsMYB103L-GFP is located to nucleus in onion epidermal cells. Figure S2. Number of differentially expressed genes in Digital Gene Expression profiling analysis (DGE) between wild type and OsMYB103L overexpressing plants. Figure S3. GO enrichment analysis of genes up-regulated in wild type and OsMYB103L overexpressing plants. Figure S4. Scanning electron micrographs (SEM) analysis. Figure S5. Schematic diagrams of the promoter regions of OsCESA genes. Figure S6. The force required to break flag leaves in wild type and OsMYB103L overexpression plants. Table S2. Expression levels of CESA genes in DGE analysis between wild type (WT) and OsMYB103L overexpressing plants (OE-1). Table S3. Primers of qRT-PCR used in this article.
Genes that up- or down-regulated two-fold in DGE data of OsMYB103L overexpressing plants compared to that of the wild type (Excel File).


