Abstract
Studies of bipolar disorder (BD) suggest a genetic basis of the illness that alters brain function and morphology. In recent years, a number of genetic variants associated with BD have been identified. However, little is known about the associated genes, or brain circuits that rely upon their function. Using an anatomically comprehensive survey of the human transcriptome (The Allen Brain Atlas), we mapped the expression of 58 genes with suspected involvement in BD based upon their relationship to SNPs identified in genome wide association studies (GWAS). We then conducted a meta-analysis of structural MRI studies to identify brain regions that are abnormal in BD. Of 58 BD associated genes, 22 had anatomically distinct expression patterns that could be categorized into one of three clusters (C1–C3). Brain regions with the highest and lowest expression of these genes did not overlap strongly with anatomical sites identified as abnormal by structural MRI except in the parahippocampal gyrus, the inferior/superior temporal gyrus and the cerebellar vermis, regions where overlap was significant. Using the 22 genes in C1–C3 as reference points, additional genes with correlated expression patterns were identified and organized into sets based on similarity. Further analysis revealed that five of these gene sets were significantly associated with BD, suggesting that anatomical expression profile is correlated with genetic susceptibility to BD, particularly for genes in C2. Our data suggest that expression profiles of BD-associated genes do not explain the majority of structural abnormalities observed in BD, but may be useful in identifying new candidate genes. Our results highlight the complex neuroanatomical basis of BD, and reinforce illness models that emphasize impaired brain connectivity.
Introduction
Bipolar disorder (BD) is a severe mental illness typified by depression, mania, psychosis and neurocognitive deficits. Based upon family and twin studies, a genetic basis of the illness is strongly suspected [1], but the genes responsible for BD remain largely unknown. Large meta-analyses of genome-wide association studies (GWAS) of BD have been conducted, identifying risk alleles at ODZ4, ANK3, CACNA1C, and ITIH3 and others with weaker, but suggestive associations [2]. The evidence collected from these and other studies indicate that BD is genetically complex, with no single genetic locus explaining more than a small portion of the variance in BD. Nonetheless, approximately 25% of the genetic variance underlying BD can be explained by common gene variation, using polygenic models [3].
The underlying assumption guiding psychiatric genetics is that variation in genes ultimately affects neural pathways in the brain to cause illness. Attempts to organize genes into pathways, and deduce their function often focus on strategies based upon gene ontology (GO). However, these methods often say little about neuroanatomical context, or the tissue specific functions of a particular gene, making GO potentially susceptible to artifacts, and misleading conclusions. For instance, two calcium channel genes may cluster based upon overlap in their predicted functions, but one could be expressed only in the heart, while the other is restricted to neurons, thereby undermining the physiological significance of their association. For this reason, while some success has been reported in the use of pathways as an organizational framework for BD [4], little is known about candidate genes in BD in terms of their actual distribution in the nervous system and the capacity for biologically meaningful interaction among risk-associated genes.
From the structural perspective, brain imaging studies in BD have described changes in cortical thickness [5], and cortical volume [6], [7], but have not yet been able to formulate these findings in terms of a comprehensive understanding of the illness based on specific genetic or molecular mechanisms. While a number of studies have examined the genetic basis of brain morphology in more detail [8], [9], or conducted SNP associations with anatomical features of interest [10]–[13], the majority of these studies have either examined a small number of brain areas and/or candidate genes, or have not been conducted in clinical samples. Due to these shortcomings, it is unclear how genetic factors affect brain structure, and which brain regions are most important in BD.
We have collected information from multiple sources, including a large meta-analysis of GWAS in BD [2], 18 structural MRI studies of BD [13]–[29], and a whole genome/whole brain gene expression map [30] to investigate the relationship between genes and anatomy in BD. Using the GWAS results to prioritize a set of genes with strong evidence for BD risk-association, expression patterns were examined across ∼900 brain structures, identifying three major expression clusters. Superimposing the BD-associated gene clusters onto a structural imaging map of neuroanatomical variation in BD, we tested two hypotheses. First, that BD-risk associated gene expression will be enriched in brain regions with structural alterations in BD, and second, that genes with anatomical expression patterns similar to BD-associated genes, will themselves harbor BD risk variants. Our results reveal partial support for each hypothesis, and reinforce models of BD in which genetic risk is widely, but not uniformly spread across multiple brain regions, some of which may be nodes that are susceptible to genetic variation and structure change.
Methods
BD-associated Gene Selection
The top 100 BD-associated SNPs from the 2011 PGC-BD meta-analysis were examined, yielding a list of SNPs with genome wide association p-values ranging from 4.7×10−5 to 5.5×10−10, corresponding to the top ∼10% of genetic variants identified in the meta-analysis of >11,000 BD cases and >51,000 controls [2]. Each SNP was examined to identify those that were associated (within 100 kb) of a single gene. Intergenic variants or SNPs in close proximity to multiple genes were excluded from further analyses. In this manner, 58 genes were selected for brain mapping (Table 1).
Table 1. Genes in proximity to BD-associated SNPs.
| Gene Name | |||
| ADCY2 | FAM155A | MSI2 | SIPA1L2 |
| AKAP13 | FLJ16124 | NFIX | SNX8 |
| ANK3 | FSTL5 | NGF | SPERT |
| ANKS1A | GATA5 | NPAS3 | STK39 |
| ATP6V1G3 | GNA14 | ODZ4 | SYNE1 |
| ATXN1 | GPR81 | PAPOLG | THSD7A |
| C11orf80 | HHAT | PAX1 | TNR |
| C15orf53 | IFI44 | PBRM1 | TRANK1 |
| CACNA1C | ITIH3 | PTPRE | TRIM9 |
| CACNA1D | KDM5B | PTPRT | UBE2E3 |
| CACNB3 | KIF1A | RASIP1 | UBR1 |
| CROT | LOC150197 | RIMBP2 | ZMIZ1 |
| DLG2 | MAD1L1 | RXRG | ZNF274 |
| DNAJB4 | MAPK10 | SGCG | |
| DUSP22 | MCM9 | SH3PXD2A | |
Whole Brain, Genome Wide Expression Analysis
At the time the data were accessed (October, 2011), the Allen Brain Institute (ABI, website: http://www.brain-map.org) had complete microarray gene expression results available from two, previously healthy human male brains (age 24 and 39 yr). From these data, expression values for 58 BD-associated genes in ∼900 brain regions were selected. Overlap in the anatomical regions sampled between the brains was excellent, but for a small number of regions, only one brain sample was available. Since we first accessed the data, additional brains have become available but some of these are incomplete. For this reason, these later additions were not included in the analysis. Comprehensive brain and RNA quality control was conducted by ABI. The details of this process are available on their website. In brief, post mortem interval was 25 hr and 30 hr respectively. RNA was examined to exclude degradation, and RNA integrity values (RIN) ranged from 5.4–7.1. Brain pH >6.0 was required of all samples. Additional details of brain preparation and microarray study design were published in detail previously [30]. In all cases, at least one transcript corresponding to the gene of interest was identified, and in most cases two or more probes for the same gene were available. A total of 105 transcripts were identified. For each transcript, expression level was recorded for each anatomical region from both brains.
To identify genes with similar expression profiles, expression of each gene was analyzed by hierarchical clustering across ∼900 neuroanatomical regions, using a nearest neighbor similarity measure (StatisticXL). Data for each gene were normalized to Z-scores across brain regions in order to identify anatomical regions that are enriched or depleted in the expression of an individual gene. Average Z-scores were then determined for each cluster (C1–C3) at each anatomical site. Each of the ∼900 brain regions in the Allen Brain Atlas is associated with three dimensional X, Y, Z coordinates, which correspond to the anatomical site of origin. For improved visualization, right and left hemisphere values were averaged and regional expression blurred (FWHM = 12 mm) before being mapped to an idealized brain using Analysis of Function Images (AFNI) [31].
Structural MRI Meta-analysis
Studies were identified with an online citation indexing service (Medline) using “brain”, “volume”, and “bipolar disorder” as search terms. This search yielded 275 articles published between 1985 and 2011. These search results were filtered to include only voxel based morphometry (VBM) studies that published volume/density results as 3D coordinates (X, Y, Z) in stereotactic space, compared the target population (BD type 1) to controls, included greater than six subjects, and did not use data already published in similar analyses. Filtering the results yielded 18 articles. The data from these were then entered into the Ginger ALE program (www.brainmap.org) to identify regions of statistical significance. Subsequently, coordinates were transformed to Talairach space for studies that had published coordinates in the Montreal Neurological Institute (MNI) space according to the nonlinear Brett transformation [32] included in the BrainMap environment, to allow analysis relative to a single template. Within GingerALE study specific Gaussian blurring was applied ranging from ∼9 to ∼8.5 mm depending on the study sample size.
Two activation likelihood estimation (ALE) maps were created as described [33]. One map displays regions with increased greater gray matter volume/density in BD compared to controls, the second displays regions with greater gray matter density/volume in controls compared to BD subjects. The data consider 1,069 BD and control subjects in total. Statistical significance was determined using a permutation test of randomly generated foci. No assumptions were made concerning the distribution or spatial separation of these random foci. Five thousand permutations were computed using the same FWHM value and the same number of foci used in computing the ALE values. The test was corrected for multiple comparisons using the false discovery rate (FDR) method [34], [35]. The minimum cluster volume required was 224 mm3. All data processing was carried out using a Java version of Ginger ALE (www.brainmap.org). Whole-brain maps of the ALE values were imported into MRIcroN [36] for data visualization and overlaid onto an anatomical template generated by spatially normalizing the International Consortium for Brain Mapping (ICBM) template to Talairach space [37].
Determination of Overlap between Structural Changes and Gene Expression Clusters
The distances between foci from gene expression cluster peaks and structural abnormalities identified in the meta analysis were calculated using the following:  . To determine a distance of statistically meaningful proximity, 200,000 random brain voxel pairs were created and a cut point for an alpha of.05,.01, and.005 were determined empirically for distances (23 mm, 13 mm, and 10 mm; respectively). While this does not necessarily imply anatomical overlap, it does suggest proximity that would be unlikely to occur by chance.
. To determine a distance of statistically meaningful proximity, 200,000 random brain voxel pairs were created and a cut point for an alpha of.05,.01, and.005 were determined empirically for distances (23 mm, 13 mm, and 10 mm; respectively). While this does not necessarily imply anatomical overlap, it does suggest proximity that would be unlikely to occur by chance.
Set-based Test of Gene Expression Profiles
Each of the clustered, BD-associated genes (index genes) was analyzed using the Allen Brain Atlas Neuroblast function [38] to generate a rank ordered list of genes that correlate in expression to the index gene. In this way, genes whose expression is highly correlated with the index can be readily identified and arranged in descending order by correlation coefficient. For each index gene, the top 10 and top 25 most similarly expressed genes were identified, and defined as a set. Data were filtered so that redundant probes or probes that did not correspond to a named human transcript were discarded. No correlation coefficient threshold was employed for set inclusion, but in all cases correlation with the index gene was high (mean r = 0.68, range 0.50–0.94). A set based genetic association analysis was conducted in PLINK, using genome-wide SNP data from 2200 Caucasian BD cases and 1436 controls. BD subjects were ascertained and genotyped as two independent cohorts for GWAS studies of BD that have been described in detail previously [39], [40]. Control subjects have also been described in detail previously [41]. For each gene, all SNPs for which genotype data exist were included in the set-based analysis. SNPs were then filtered using PLINK to consider only informative markers (based on linkage disequilibrium, defined as r2≥0.50). No maximum number of SNPs was specified. After SNPs in were removed, variants in genes sets were examined to determine their association with BD. In these analyses, each set was analyzed using the genes with the top 10, and 25 (including index) most similar expression profiles. In addition, to avoid circular arguments in which index genes conferred association to the expression-based set, each analysis was repeated with the index gene removed (top 24) to ensure association of the set independently of the index gene. Each set and level of inclusion was run over 10,000 permutations. In all, 22 sets were run, each with three levels. Statistical significance was defined when p<0.05, and trend level significance was defined when p<0.10.
Results
Unsupervised Gene Clustering
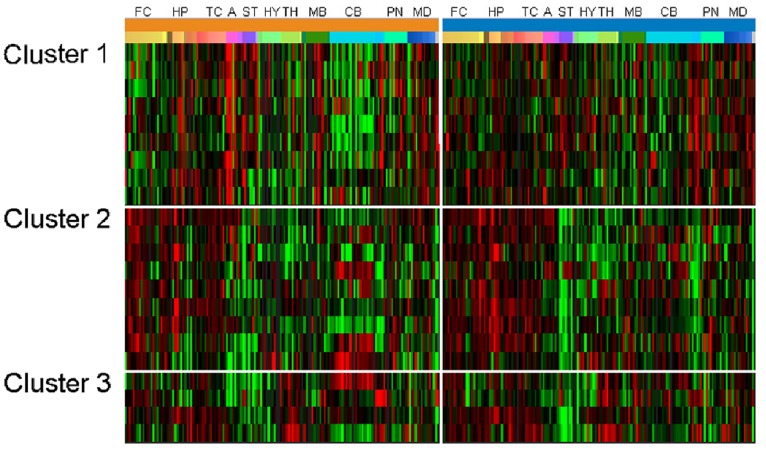
If BD preferentially alters function in specific brain structures, some BD-associated genes may be similarly expressed in patterns that include the affected regions. To determine if this was the case, we first examined the anatomical expression profile of all 58 BD-associated genes to identify common patterns of high/low expression across the brain (Table 1). Three reproducible gene expression clusters were identified, accounting for 22 genes. A fourth set of six genes reliably failed to cluster. The former sets were termed clusters 1, 2 and 3 (C1, C2, C3) and used for subsequent analysis (Figure 1). The latter set was not considered further. Within a cluster, gene expression values had pair wise correlation coefficients of 0.4–0.8, indicating moderate-high correlation across brain regions. In some cases, correlation across clusters was noted, particularly between C2 and C3. This correlation was typically less than 0.2 and always less than the correlation within the cluster. Gene cluster assignments are indicated in Table 2.
Figure 1. Whole brain expression patterns of BD-associated genes.
Expression of 22 BD-associated genes is organized by cluster for each of two brains (indicated by the thick orange and blue line). Each row represents a single gene, and each column represents one of ∼900 brain regions. Brain regions are organized anatomically, by broadly defined regions where FC: frontal cortex, HP: hippocampus, TC: temporal cortex, A: amygdala ST: striatum, HY: hypothalamus, TH: thalamus, MB: midbrain, CB: cerebellum, PN: pontine nuclei, MD: medulla.
Table 2. Bipolar disorder associated gene expression clusters.
| Cluster 1 | Cluster 2 | Cluster 3 |
| ATP6V1G3 | ADCY2 | ANK3 |
| GATA5 | CACNB3 | STK39 |
| GPR81 | DLG2 | FAM155A |
| NGF | KDM5B | SIPA1L2 |
| NPAS3 | KIF1A | |
| PAX1 | MAPK10 | |
| SPERT | RIMBP2 | |
| TNR | UBE2E3 | |
| ZMIZ1 | UBR1 |
Brain Mapping of Gene Clusters
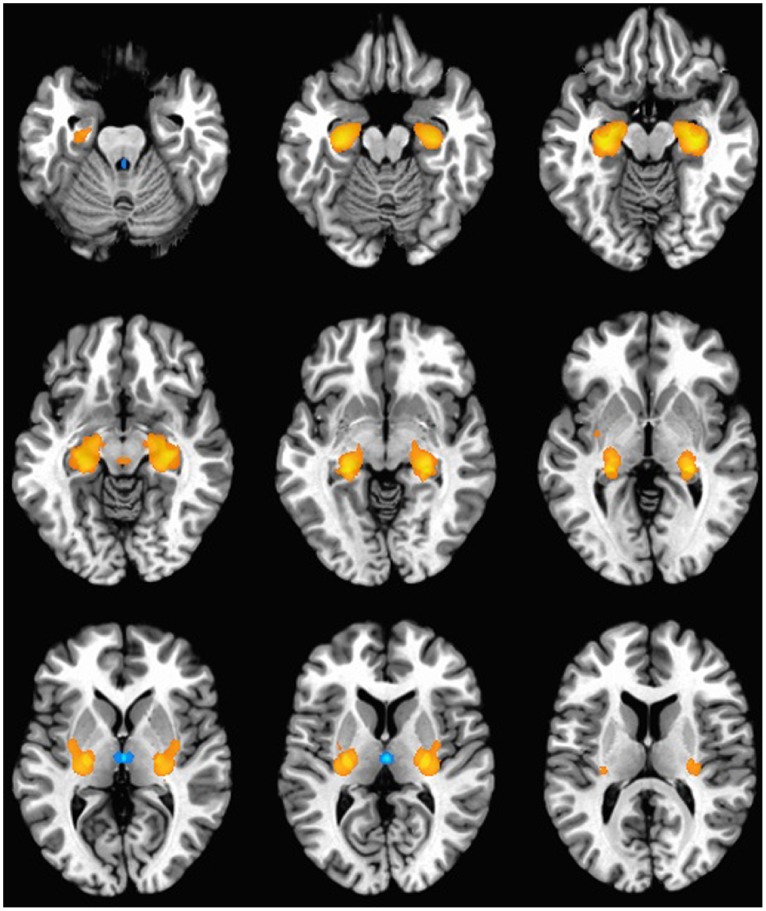
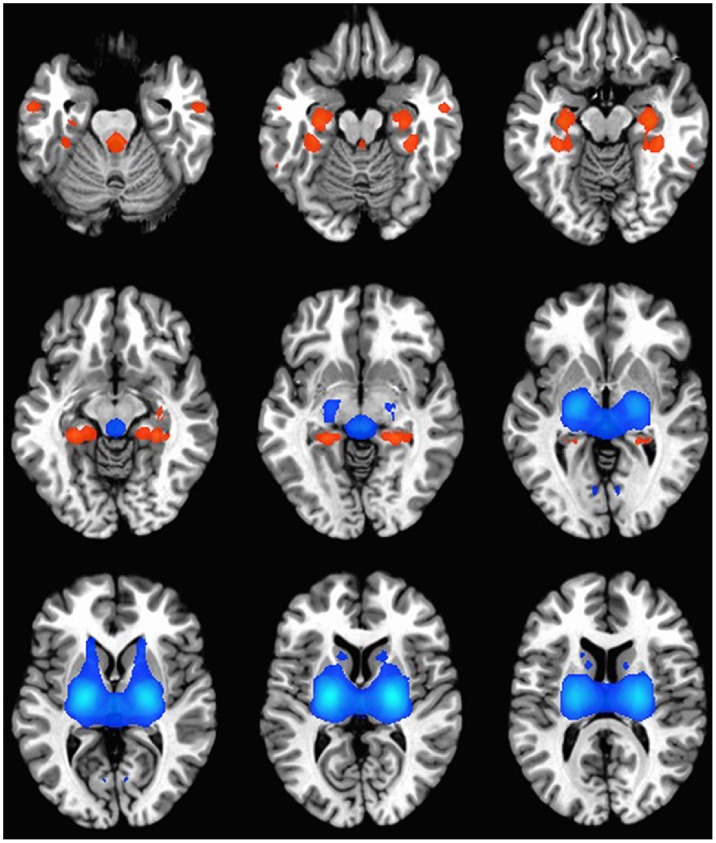
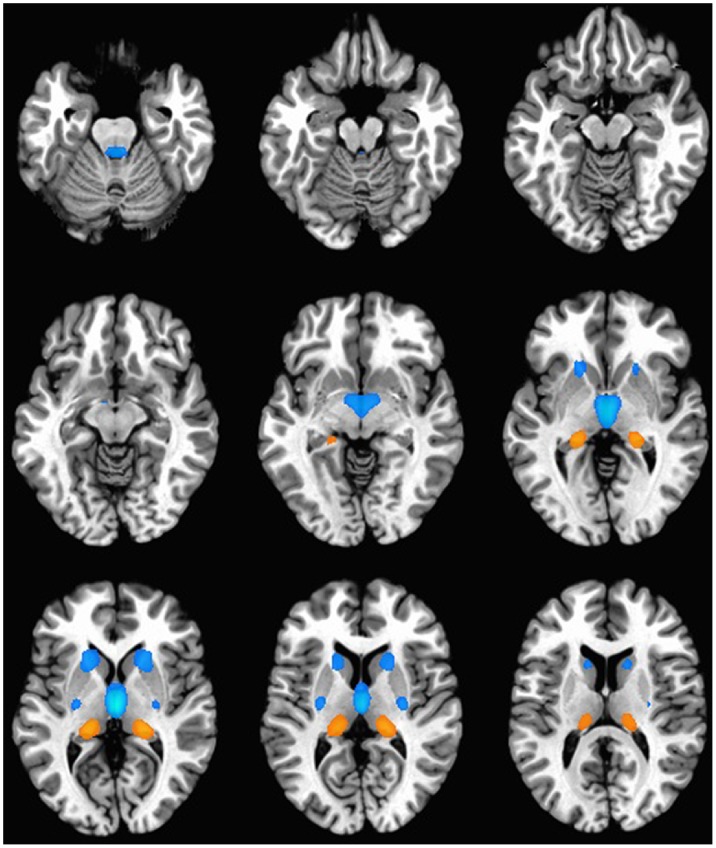
In order to map common anatomical patterns of gene expression in three dimensions, and identify areas of relative enrichment or depletion, the average expression data for regions of minimum and maximum expression for each cluster were projected onto an idealized brain. All three clusters were expressed widely across multiple brain regions, but few regions met statistical cut-offs for inclusion in mapping. Visualization of the expression patterns of the three clusters revealed that C1 was defined by high expression in the parahippocampal gyrus, hippocampus and posterior thalamus (Figure 2). C2 was expressed highly in portions of the hippocampus, temporal cortex, and midbrain, and expressed at very low levels in the striatum, and posterior thalamus (Figure 3). C3 was expressed highly in the posterior thalamus and was expressed at very low levels in the caudate, and putamen (Figure 4).
Figure 2. Cluster 1 gene expression pattern.
An idealized human brain shown in horizontal cross section indicates the anatomical regions enriched (orange and red) and depleted (blue) in gene expression associated with C1. For improved visualization, gene expression in corresponding regions of the right and left hemispheres has been consolidated and shown in mirror image bilaterally. C1 is defined by high expression in the parahippocampal gyrus, hippocampus and posterior thalamus.
Figure 3. Cluster 2 gene expression pattern.
An idealized human brain shown in horizontal cross section indicates the anatomical regions enriched (orange and red) and depleted (blue) in gene expression associated with C2. For improved visualization, gene expression in corresponding regions of the right and left hemispheres has been consolidated and shown in mirror image bilaterally. C2 is defined by high expression in portions of the hippocampus, temporal cortex, and midbrain, and very low expression in the striatum, and posterior thalamus.
Figure 4. Cluster 3 gene expression pattern.
An idealized human brain shown in horizontal cross section indicates the anatomical regions enriched (orange and red) and depleted (blue) in gene expression associated with C3. For improved visualization, gene expression in corresponding regions of the right and left hemispheres has been consolidated and shown in mirror image bilaterally. C3 is defined by high expression in the posterior thalamus and very low expression in the striatum (caudate and putamen).
Structural Differences in Bipolar vs. Control Brains
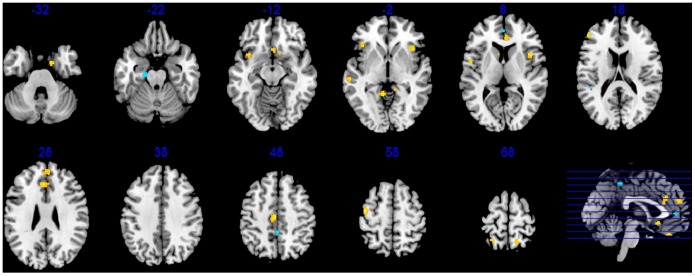
A meta-analysis of structural MRI studies was conducted to identify the most reliably affected brain regions in BD. Twenty three regions were identified that differed in density/volume between BD cases and healthy controls (Figure 5, Table 3). Twenty of these regions indicated greater gray matter density/volume in controls compared to BD subjects. Thirteen of the regions identified were in the frontal/cingulate/insula cortex, and six were located in the temporal and cortical regions. The remainder was distributed in the cerebellum and parietal cortex. Three left sided regions (cingulate gyrus, parahippocampal gyrus, paracentral lobule) were larger in BD subjects compared to controls.
Figure 5. Brain regions identified by volumetric meta analysis.
An idealized human brain shown in horizontal cross section indicates the anatomical regions that are on average, larger in volume in controls compared to BD (yellow) or smaller in controls compared to BD (blue).
Table 3. Meta analysis results for anatomical regions with differential volume.
| CON>BD | |||||
| Volume (mm3) | x | y | z | Location | ALE Value |
| 1064 | −3 | 31 | 27 | Left cingulate gyrus | 0.019 |
| 776 | −5 | −19 | 49 | Left medial frontal gyrus | 0.020 |
| 696 | 3 | 52 | 25 | Right medial frontal gyrus | 0.019 |
| 664 | 53 | −31 | 3 | Right superior temporal gyrus | 0.020 |
| 528 | −43 | −9 | 56 | Left precentral gyrus | 0.020 |
| 448 | −19 | −1 | −37 | Left uncus | 0.018 |
| 448 | −37 | 11 | −13 | Left inferior frontal gyrus | 0.018 |
| 448 | −59 | −23 | −3 | Left middle temporal gyrus | 0.018 |
| 448 | 5 | 37 | 11 | Right anterior cingulate | 0.018 |
| 432 | 13 | −55 | 71 | Right postcentral gyrus | 0.018 |
| 408 | 39 | 21 | 0 | Right insula | 0.018 |
| 400 | 2 | 19 | −9 | Right subcallosal gyrus | 0.018 |
| 392 | 43 | 11 | 9 | Right insula | 0.017 |
| 384 | 17 | −1 | −30 | Right uncus | 0.018 |
| 384 | 11 | −41 | −4 | Right cerebellar vermis | 0.018 |
| 384 | −6 | −45 | −3 | Left cerebellar vermis | 0.018 |
| 384 | −50 | 1 | 5 | Left superior temporal gyrus | 0.018 |
| 384 | −26 | −54 | 66 | Left superior parietal lobule | 0.019 |
| 360 | −40 | 26 | −5 | Left inferior frontal gyrus | 0.017 |
| 224 | −49 | 40 | 19 | Left middle frontal gyrus | 0.018 |
| BD>CON | |||||
| Volume (mm3) | x | y | z | Location | ALE Value |
| 304 | −4 | −33 | 33 | Left cingulate gyrus | 0.018 |
| 296 | −19 | −18 | −21 | Left parahippocampal gyrus | 0.016 |
| 256 | 0 | −39 | 50 | Left paracentral lobule | 0.017 |
Anatomical Comparison of Gene Expression Clusters to Structural Imaging Results
The reason for regional brain volume alteration in BD is unknown, possibly reflecting genetic predisposition, developmental abnormality and/or pathological features of illness progression. Using the results of our meta-analysis to define the anatomically affected brain areas, we tested the hypothesis that brain volume alterations in BD are primarily genetic, looking for overlap across anatomical regions that are enriched or depleted in BD-associated gene expression and those that differ in structural morphology in BD compared to controls. On the whole the overlap was modest. In general, gene expression peaks were identified primarily in sub-cortical structures whereas neuroanatomical differences were localized in the frontal and temporal cortexes. However, there were exceptions. The parahippocampal gyrus, was enriched in both C1 and C2 expression and showed evidence of increased volume among BD patients; a region spanning the inferior/superior temporal gyrus with volume differences in BD overlapped with C2 expression; and a region of the cerebellar vermis was in close proximity to expression peak of both C2 and C3 (Table 4).
Table 4. Brain region overlap between gene expression clustering and differential volume.
| MRI Region | Gene Expression Peak | Significance Level | Cluster |
| parahippocampal gyrus | parahippocampal gyrus | p<0.005 | 1 |
| parahippocampal gyrus | parahippocampal gyrus | p<0.01 | 2 |
| cerebellar vermis | cerebellar vermis | p<0.05 | 2 |
| superior temporal gyrus | inferior temporal gyrus | p<0.05 | 2 |
| cerebellar vermis | cerebellar vermis | p<0.05 | 3 |
Gene Expression Pattern Predicts Genetic Risk-association for Bipolar Disorder
It has been reported that genes expressed with similar anatomical patterns share functional similarity [30]. In our study, we hypothesized that genes with similar expression patterns share BD risk susceptibility by affecting overlapping brain circuits. We tested this idea by looking for evidence of genetic association with BD among genes that are expressed in patterns similar to genes that were identified previously as harboring risk associated alleles. Each of the 22 BD-associated genes in C1–C3 was designated as an “index”, and used for a reference against which genes with highly similar expression patterns were identified using NeuroBlast [38]. Genes expressed similarly to the index were termed “sets”. The complete list of genes in each set is shown in Table S1. Of 22 sets used for set-based genetic analysis, five showed significant evidence of BD risk-association (sets based on ATP6V1G3, ADCY2, CACNB3, RIMBP2, and UBR1 as the indexes, see example Figure 6). Two other sets demonstrated trend-level evidence of association (sets based on NGF, ZMIZ1). In two cases (RIMBP2, ATP6V1G3), removal of the index gene substantially reduced the strength of the association, suggesting the index gene was largely responsible for conferring the BD risk associated with the set (compare top 24 vs. top 25, Table 5). However, in the remaining cases, gene set associations remained significant after removing the index gene, indicating that significant genetic risk is conferred independently by the remaining genes in the set. An example is shown in Figure 6.
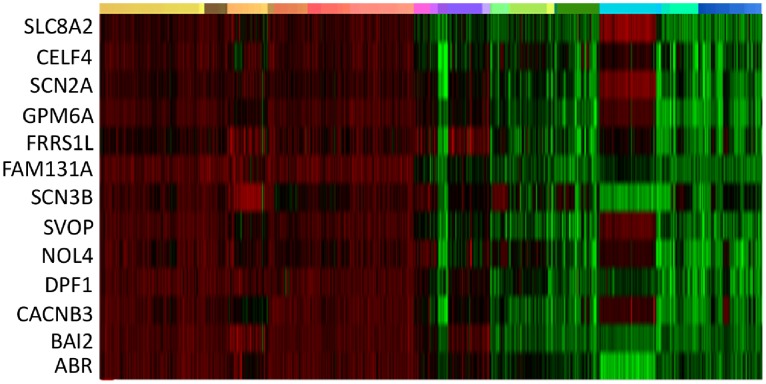
Figure 6. Gene expression pattern of CACNB3 is correlated with BD risk associated genes identified by set analysis.
Genes that are expressed in anatomical patterns similar to BD-associated genes are more likely to contain risk-associated SNPs in set based genetic analyses. The C2 gene, CACNB3 is shown as an example, in which twelve additional genes with expression patterns highly correlated with CACNB3 are shown. The same genes are in close proximity to, or contain SNPs that contributed signal to the set-based association with BD, even though they were not previously identified as having strong associations in GWAS. Brain regions are organized anatomically, FC: frontal cortex, IN: insula, CN: cingulate, HP: hippocampus, L: lingual gyrus, TC: temporal cortex, A: amygdale, ST: striatum, HY: hypothalamus, TH: thalamus, MB: midbrain, CB: cerebellum, PN: pontine nuclei, MD: medulla.
Table 5. Set-based analyses of genes sharing expression profile with C1–C3.
| Cluster | Index Gene | Top 10 | Top 24 | Top 25 | Mean Correlation |
| 1 | ATP6V1G3 * | 0.02 | 0.98 | 0.10 | 0.76 |
| 1 | GATA5 | 0.14 | 0.74 | 0.21 | 0.66 |
| 1 | GPR81 | 0.92 | 0.78 | 0.83 | 0.79 |
| 1 | NGF ** | 0.53 | 0.10 | 0.09 | 0.49 |
| 1 | NPAS3 | 0.21 | 0.16 | 0.22 | 0.91 |
| 1 | PAX1 | 0.17 | 0.67 | 0.53 | 0.48 |
| 1 | SPERT | 0.34 | 0.91 | 0.79 | 0.81 |
| 1 | TNR | 0.57 | 0.55 | 0.41 | 0.40 |
| 1 | ZMIZ1 ** | 0.29 | 0.08 | 0.10 | 0.51 |
| 2 | ADCY2 * | 0.02 | 0.02 | 0.12 | 0.80 |
| 2 | CACNB3 * | 0.18 | 0.04 | 0.12 | 0.75 |
| 2 | DLG2 | 0.14 | 0.48 | 0.26 | 0.58 |
| 2 | KDM5B | 0.88 | 0.96 | 0.96 | 0.57 |
| 2 | KIF1A | 0.29 | 0.24 | 0.16 | 0.80 |
| 2 | MAPK10 | 0.81 | 0.58 | 0.69 | 0.80 |
| 2 | RIMBP2 * | 0.07 | 0.21 | 0.04 | 0.76 |
| 2 | UBE2E3 | 0.85 | 0.61 | 0.79 | 0.73 |
| 2 | UBR1 * | 0.03 | 0.05 | 0.08 | 0.75 |
| 3 | ANK3 | 0.69 | 0.63 | 0.72 | 0.64 |
| 3 | FAM155A | 0.67 | 0.69 | 0.59 | 0.70 |
| 3 | SIPA1L2 | 0.76 | 0.49 | 0.65 | 0.63 |
| 3 | STK39 | 0.43 | 0.36 | 0.29 | 0.68 |
*significant set-based association.
**trend towards set-based association.
There was no relationship between the strength of the SNP’s BD-association in the index gene, and the subsequent set-based association. That is to say, SNPs strongly associated with BD did not necessarily make better index genes, and did not predict the BD association of expression based sets.
However, index gene clusters were not represented evenly among the enriched sets. Four out of five sets came from genes in C2, and none came from C3. Since cluster designation is organized around similarity in gene expression patterns, it is possible that overlap exists among the BD-associated gene sets (e.g. genes expressed similarly to ADCY2, may also be expressed like CACNB3). To examine whether shared genes across multiple sets contributed to any of the set-based BD-association, we examined the composition of each of the five significantly associated sets. In C1, in the set defined by ATP6V1G3-like expression, there was extensive overlap (11/25 genes) among sets that were not associated with BD, but no overlap with NGF-like or ZMIZ-like genes, the other C1 sets that did show suggestive BD association. Similarly, there was no overlap between the NGF-like and ZMIZ-like sets. This suggests that the genes conferring BD-risk association in C1 sets were uniquely associated with the index gene. In C2, the ADCY2-like and UBR1-like sets did not overlap with any of the other BD-associated sets. In contrast, the sets defined by CACNB3-like, and RIMBP2-like expression overlapped considerably with each other (4/25 genes), sharing the genes ENC1, DPF1, SCN3B, and SVOP in common, suggesting that the BD-risk association with these two sets may be attributable to common genetic factors.
Discussion
Using a comprehensive, whole brain map of gene expression, we report preliminary progress in filling in the gap that exists between our understanding of the genetic and neuroanatomical bases of BD. Our results show that gene expression reproducibly classifies 38% (22/58) of the most strongly associated genes into three clusters, indicating that many BD susceptibility genes share similar expression patterns across the brain. We made two major assumptions in conducting our work. First, we assume that BD-associated variants preferentially affect genes, a position supported by data showing that BD associated SNPs are enriched in regulatory regions likely to affect gene expression and/or alter mRNA transcripts [40], [42]. A second assumption is that brain regions with the highest/lowest levels of gene expression are the most vulnerable to perturbed expression, and that under pathological conditions, these regions may have particularly severe consequences for the progression of BD. Our data suggest that at least in terms of volumetric change, this assertion may not be true. Functional differences that do not affect gray matter volume may still follow the pattern we expected, but were not assessed by our study.
Study Limitations and Potential Confounds
Our analysis is limited by the small number of brains available for study. Furthermore, because of this small sample, it is possible that our classification scheme is influenced by artifacts from post mortem brain processing and/or RNA degradation. Because the ABI conducted careful quality control for the samples, and we selected only genes that were reliably expressed in both brain samples, we consider this risk to be small. However, replication in larger brain samples will be required to validate our expression based clustering strategies.
Instead, our meta-analysis of the structural imaging literature and other reports [6], [7], [9] support the conclusion that widespread cortical abnormalities are a key feature of BD. We have extended this finding to show that in most cases, structural brain abnormalities in BD do not overlap with regions enriched or depleted in the expression of risk-associated genes. The overlap in expression among C1/C2 genes and structural abnormalities in the hippocampus may be an important exception. Our results suggest that anatomical expression profile may be useful in identifying novel BD susceptibility genes by revealing anatomical overlap with previously detected genes associated with BD.
Comparison with Alternate Approaches
Authors from the ABI, using the same two brains used the entire human genome to perform weighted gene co-expression network analysis (WGCNA) [30]. With this much larger number of genes, 13 gene expression modules were identified (M1-13). We compared our cluster assignments to the WGCNA-modules, and found that for the 58 bipolar-associated genes we categorized, the WGCNA module system gave similar results, but with some notable differences. For C1, 3/9 genes (33%) were mapped to M4, two were mapped to M3 (22%), one was mapped to M2 (11%), while the C1 gene GPR81 was not assigned to any module. For C2, 9/9 genes (100%) we assigned to M2. C3 corresponded to M1, but FAM155A, STK39 also showed correlation with M2, and SIPA1L2 was not categorized by WGCNA. We also checked the 58 BD associated genes for omissions from clusters that would be suggested using the WGCNA modules. In this way, C15orf53 (M4) could be categorized in C1; and CACNA1C, FSTL5, MAD1L1, RASIP1, SNX8 (all M2) could be categorized in C2. Interestingly, a new cluster would be needed to include the genes CACNA1D, ODZ4, PAPOLG, PTPRE and SYNE1 that did not reliably cluster in our experiment but were all categorized as M7 using the alternative method. In general, genes that were discordant between classification methods were found to have significant cross-correlation to two or more modules, suggesting their assignment to clusters in our study did not reflect methodological error, but instead reflects genuine ambiguity in the profiles. The differences in statistical methods between studies leading to different cut-offs and subsequent group assignments may reflect differences in sample size and statistical power, but may also be to some extent arbitrary.
Structural Brain Differences in BD
The majority of studies examining gray matter in BD individuals in relation to control subjects report widespread reductions in gray matter density and/or volume in cortical regions. Consistent with this, our meta analyses identified 20 regions of reduced gray matter volume, distributed most heavily throughout frontal regions, and only three areas of increased gray matter volume in BD individuals. This empirical summary suggests that BD individuals appear to be at greatest risk for gray matter reductions in prefrontal regions.
Gene Expression Clusters Rarely Overlap with Structurally Abnormal Regions
We found little support for the hypothesis that brain regions enriched in illness-associated genes were the most implicated in structural imaging experiments. This may be true for at least two reasons. First, gene expression was mapped in cross-section in adult brains carefully selected for the absence of psychiatric illness. Hence, it may be that the intensity and/or distribution of gene expression is perturbed in the brains of BD subjects, but not controls; or that at earlier stages of development, gene expression transiently and strongly overlaps with the brain regions in question, but this overlap diminishes during maturation. Second, the majority of the structural differences were detected in cortical regions, while the majority of high/low gene-expressing regions were sub-cortical. Gene expression occurs in the cell nucleus, but the protein products of genes often function elsewhere the cell, including at axon terminals that terminate in other brain regions. Therefore, the observed mismatch of gene expression and structural change could reflect properties of BD that affect brain circuits, whereby aberrant gene function in sub-cortical regions affects the morphology in cortical structures indirectly, perhaps through synaptic processes or distal axonal projections, causing the anatomical changes in the illness to appear away from the primary site of gene action. Recent work indeed affirms this notion, identifying loss of white matter integrity in BD, and genetic liability for BD affecting association with matter connectivity [43], [44] supporting the idea that white matter, in addition to cortical and subcortical gray matter volume is an important substrate for genetic variation in BD.
Gene Expression Profile Predicts Association with Bipolar Disorder
Perhaps the most important result of our present work is that anatomical expression pattern is shared across BD-risk associated genes, and can be used to identify novel BD-associated variants. The number of BD-associated index genes we studied was small, and therefore it is likely that our index genes account for only a small portion of the total genetic risk for BD. Nonetheless, we conclude that there is information pertinent to understanding the nature of the illness in this subset of genes. This was particularly true of C2 genes (ADCY2, CACNB3, RIMBP2, UBR1) which were enriched in correlated gene sets associated with BD. C2 corresponds to the M2 module described previously, and gene ontology (GO) analysis of this module was reported to be enriched in genes associated with neocortex, neurons, and energy metabolism [30]. C2 genes are especially highly expressed in the hippocampus, and expressed very weakly in the striatum. While our focus on the extreme highs/lows diminishes emphasis on brain regions with moderate levels of expression, C2 genes are also widely expressed across cortical regions, including those associated with illness by our meta-analysis of MRI results in BD. Our set-based analyses of C1 demonstrated weaker BD association, showing trend level associations with NGF-like and ZMIZ-like genes, and association with ATP6V1G3-like genes that disappeared when the index gene was removed. C1 corresponds to M4 [30], in which GO analysis revealed strong association with signal transduction, transmembrane receptors, and G-protein coupled signaling. As discussed previously, ZMIZ expression was also correlated with C2, suggesting that the BD-association of ZMIZ-like genes may further support the association of C2. C3-like genes did not strongly associate with BD. These genes (corresponding to M1) were found previously to be enriched in parvalbumin sensory neurons, somatosensory cortex and sensory thalamic nuclei [30]. The C3 cluster contains ANK3, one of the most robustly associated BD-susceptibility genes. While its inclusion in C3 suggests a sensory processing role, it is unclear if this putative function is involved in the pathophysiology of BD, or if it has pleiotropic functions in regulating mood.
Functions of Highlighted Genes
C2 was highly expressed in the hippocampus, the only region to show strong evidence of genetic and structural association with BD. Sets of three C2-like genes (ADCY2, CACNB3, and UBR1) were associated with BD, even after the index gene was removed from the analysis, offering support for the functional significance of these genes in BD. ADCY2 encodes the enzyme adenylyl cyclase 2 (AC2) that is involved in signal transduction, generating cyclic adenosine monophasphate (cAMP) in response to the stimulation of GS-coupled receptors (e.g. dopamine D1 receptor). Like all C2 genes, AC2 is found widely throughout the brain, with enrichment in the cortex and hippocampus [45]. Of particular interest, AC2 has been implicated in synaptic plasticity and cellular differentiation [45] and was shown to be inhibited by lithium, a drug commonly used to treat BD [46]. CACNB3 encodes the β3 subunit found in ∼60% of N-type calcium channels. Animal models with the corresponding β3 subunit gene knocked-out show a number of behavioral phenotypes, including some similar to symptoms of BD: circadian increases in activity, more impulsivity-like behaviors, increased response to novelty, aggression, and memory impairments [47]–[48]. UBR1 encodes an N-terminal E3-ubiquitin ligase, an enzyme involved in protein degradation [49]. UBR1 has been implicated in a wide variety of processes in the nervous system including G-protein coupled signaling [50], apoptosis [51], neurogenesis [52], and memory [53].
Future Directions
Definitive characterization of BD-associated gene expression will require mapping studies conducted across ages, in illness affected brains, and examining both gray and white matter structures. We anticipate that brain mapping projects similar to those undertaken by the Allen Institute will be conducted in additional brains, including those from psychiatrically abnormal populations like BD. In the meantime, our work suggests a theoretical framework that could be employed in more targeted gene expression studies, suggesting that specific brain regions may be better suited to certain kinds of analyses. Multiple whole genome, microarray gene expression analyses have been conducted in brain tissues from BD patients [54], but with a limited number of exceptions, the majority of these have employed prefrontal cortex as the sample of choice. While an emphasis on pre-frontal cortex may be well suited to studies relevant to the morphological changes in brain (perhaps a late consequence of BD), subcortical structures including the hippocampus and striatum may be better suited to studies focused on genetic mechanisms involved in early stages of the illness. Better understanding of these concepts could sharpen the focus of future analyses, permitting researchers to concentrate on data most pertinent to relevant phenotypes.
Supporting Information
BD associated index genes and corresponding gene sets.
(XLS)
Acknowledgments
Thanks to the PGC-BD for providing access to GWAS data and the Allen Brain Institute for providing gene expression data and analysis resources to the research community.
Funding Statement
This research was support by grants from the Department of Veterans Affairs Office of research and Development providing support to M.J.M. (1IK2BX001275), A.D.S. (IK2CX000864), J.R.K. (I01CX000363), and A.N.S. (1I01CX000715), as well funds from the VA Center of Excellence for Stress and Mental Health. J.R.K. has additional support from NIMH and the Pharmacogenetic Research Network (MH094483 and MH092758). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. McGuffin P, Rijsdijk F, Andrew M, Sham P, Katz R, et al. (2003) The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch Gen Psychiatry 60: 497–502. [DOI] [PubMed] [Google Scholar]
- 2. PGCBD Consortium (2011) Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 43: 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, et al. (2009) Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460: 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Holmans P, Green EK, Pahwa JS, Ferreira MA, Purcell SM, et al. (2009) Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am J Hum Genet 85: 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rimol LM, Hartberg CB, Nesvag R, Fennema-Notestine C, Hagler DJ Jr, et al. (2010) Cortical thickness and subcortical volumes in schizophrenia and bipolar disorder. Biol Psychiatry 68: 41–50. [DOI] [PubMed] [Google Scholar]
- 6. Hallahan B, Newell J, Soares JC, Brambilla P, Strakowski SM, et al. (2011) Structural magnetic resonance imaging in bipolar disorder: an international collaborative mega-analysis of individual adult patient data. Biol Psychiatry 69: 326–335. [DOI] [PubMed] [Google Scholar]
- 7. Selvaraj S, Arnone D, Job D, Stanfield A, Farrow TF, et al. (2012) Grey matter differences in bipolar disorder: a meta-analysis of voxel-based morphometry studies. Bipolar Disord 14: 135–145. [DOI] [PubMed] [Google Scholar]
- 8. Panizzon MS, Fennema-Notestine C, Eyler LT, Jernigan TL, Prom-Wormley E, et al. (2009) Distinct genetic influences on cortical surface area and cortical thickness. Cereb Cortex 19: 2728–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rimol LM, Panizzon MS, Fennema-Notestine C, Eyler LT, Fischl B, et al. (2010) Cortical thickness is influenced by regionally specific genetic factors. Biol Psychiatry 67: 493–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Linke J, Witt SH, King AV, Nieratschker V, Poupon C, et al. (2012) Genome-wide supported risk variant for bipolar disorder alters anatomical connectivity in the human brain. Neuroimage 59: 3288–3296. [DOI] [PubMed] [Google Scholar]
- 11. Inkster B, Nichols TE, Saemann PG, Auer DP, Holsboer F, et al. (2010) Pathway-based approaches to imaging genetics association studies: Wnt signaling, GSK3beta substrates and major depression. Neuroimage 53: 908–917. [DOI] [PubMed] [Google Scholar]
- 12. Sprooten E, McIntosh AM, Lawrie SM, Hall J, Sussmann JE, et al. (2012) An investigation of a genome wide supported psychosis variant in ZNF804A and white matter integrity in the human brain. Magn Reson Imaging 30: 1373–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kempton MJ, Ruberto G, Vassos E, Tatarelli R, Girardi P, et al. (2009) Effects of the CACNA1C risk allele for bipolar disorder on cerebral gray matter volume in healthy individuals. Am J Psychiatry 166: 1413–1414. [DOI] [PubMed] [Google Scholar]
- 14. Adler CM, Levine AD, Delbello MP, Strakowski SM (2005) Changes in gray matter volume in patients with bipolar disorder. Biol Psychiatry 58: 151–157. [DOI] [PubMed] [Google Scholar]
- 15. Adler CM, Delbello MP, Jarvis K, Levine A, Adams J, et al. (2007) Voxel-based study of structural changes in first-episode patients with bipolar disorder. Biol Psychiatry 61: 776–781. [DOI] [PubMed] [Google Scholar]
- 16. Almeida JR, Akkal D, Hassel S, Travis MJ, Banihashemi L, et al. (2009) Reduced gray matter volume in ventral prefrontal cortex but not amygdala in bipolar disorder: significant effects of gender and trait anxiety. Psychiatry Res 171: 54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen X, Wen W, Malhi GS, Ivanovski B, Sachdev PS (2007) Regional gray matter changes in bipolar disorder: a voxel-based morphometric study. Aust N Z J Psychiatry 41: 327–336. [DOI] [PubMed] [Google Scholar]
- 18. Dickstein DP, Milham MP, Nugent AC, Drevets WC, Charney DS, et al. (2005) Frontotemporal alterations in pediatric bipolar disorder: results of a voxel-based morphometry study. Arch Gen Psychiatry 62: 734–741. [DOI] [PubMed] [Google Scholar]
- 19. Doris A, Belton E, Ebmeier KP, Glabus MF, Marshall I (2004) Reduction of cingulate gray matter density in poor outcome bipolar illness. Psychiatry Res 130: 153–159. [DOI] [PubMed] [Google Scholar]
- 20. Ha TH, Ha K, Kim JH, Choi JE (2009) Regional brain gray matter abnormalities in patients with bipolar II disorder: a comparison study with bipolar I patients and healthy controls. Neurosci Lett 456: 44–48. [DOI] [PubMed] [Google Scholar]
- 21. Janssen J, Reig S, Parellada M, Moreno D, Graell M, et al. (2008) Regional gray matter volume deficits in adolescents with first-episode psychosis. J Am Acad Child Adolesc Psychiatry 47: 1311–1320. [DOI] [PubMed] [Google Scholar]
- 22. Li M, Cui L, Deng W, Ma X, Huang C, et al. (2011) Voxel-based morphometric analysis on the volume of gray matter in bipolar I disorder. Psychiatry Res 191: 92–97. [DOI] [PubMed] [Google Scholar]
- 23. Lochhead RA, Parsey RV, Oquendo MA, Mann JJ (2004) Regional brain gray matter volume differences in patients with bipolar disorder as assessed by optimized voxel-based morphometry. Biol Psychiatry 55: 1154–1162. [DOI] [PubMed] [Google Scholar]
- 24. Lyoo IK, Kim MJ, Stoll AL, Demopulos CM, Parow AM, et al. (2004) Frontal lobe gray matter density decreases in bipolar I disorder. Biol Psychiatry 55: 648–651. [DOI] [PubMed] [Google Scholar]
- 25. McIntosh AM, Job DE, Moorhead TW, Harrison LK, Forrester K, et al. (2004) Voxel-based morphometry of patients with schizophrenia or bipolar disorder and their unaffected relatives. Biol Psychiatry 56: 544–552. [DOI] [PubMed] [Google Scholar]
- 26. Nugent AC, Milham MP, Bain EE, Mah L, Cannon DM, et al. (2006) Cortical abnormalities in bipolar disorder investigated with MRI and voxel-based morphometry. Neuroimage 30: 485–497. [DOI] [PubMed] [Google Scholar]
- 27. Stanfield AC, Moorhead TW, Job DE, McKirdy J, Sussmann JE, et al. (2009) Structural abnormalities of ventrolateral and orbitofrontal cortex in patients with familial bipolar disorder. Bipolar Disord 11: 135–144. [DOI] [PubMed] [Google Scholar]
- 28. Tost H, Ruf M, Schmal C, Schulze TG, Knorr C, et al. (2010) Prefrontal-temporal gray matter deficits in bipolar disorder patients with persecutory delusions. J Affect Disord 120: 54–61. [DOI] [PubMed] [Google Scholar]
- 29. Yatham LN, Lyoo IK, Liddle P, Renshaw PF, Wan D, et al. (2007) A magnetic resonance imaging study of mood stabilizer- and neuroleptic-naive first-episode mania. Bipolar Disord 9: 693–697. [DOI] [PubMed] [Google Scholar]
- 30. Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, et al. (2012) An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489: 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cox RW (1996) AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res 29: 162–173. [DOI] [PubMed] [Google Scholar]
- 32. Brett M, Christoff K, Cusack R, Lancaster J (2001) Using the Talairach atlas with the MNI template. Neuroimage 13: 85–85. [Google Scholar]
- 33. Turkeltaub PE, Eden GF, Jones KM, Zeffiro TA (2002) Meta-analysis of the functional neuroanatomy of single-word reading: method and validation. Neuroimage 16: 765–780. [DOI] [PubMed] [Google Scholar]
- 34. Genovese CR, Lazar NA, Nichols T (2002) Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage 15: 870–878. [DOI] [PubMed] [Google Scholar]
- 35. Laird AR, Fox PM, Price CJ, Glahn DC, Uecker AM, et al. (2005) ALE meta-analysis: controlling the false discovery rate and performing statistical contrasts. Hum Brain Mapp 25: 155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kochunov P, Lancaster J, Thompson P, Toga AW, Brewer P (2002) An optimized individual target brain in the Talairach coordinate system. Neuroimage 17: 922–927. [PubMed] [Google Scholar]
- 37. Rorden C, Brett M (2000) Stereotaxic display of brain lesions. Behav Neurol 12: 191–200. [DOI] [PubMed] [Google Scholar]
- 38. Hawrylycz M, Ng L, Page D, Morris J, Lau C, et al. (2011) Multi-scale correlation structure of gene expression in the brain. Neural Netw 24: 933–942. [DOI] [PubMed] [Google Scholar]
- 39. Smith EN, Bloss CS, Badner JA, Barrett T, Belmonte PL, et al. (2009) Genome-wide association study of bipolar disorder in European American and African American individuals. Mol Psychiatry 14: 755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Smith EN, Koller DL, Panganiban C, Szelinger S, Zhang P, et al. (2011) Genome-wide association of bipolar disorder suggests an enrichment of replicable associations in regions near genes. PLoS Genet 7: e1002134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. O’Donovan MC, Craddock N, Norton N, Williams H, Peirce T, et al. (2008) Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet 40: 1053–1055. [DOI] [PubMed] [Google Scholar]
- 42. Schork AJ, Thompson WK, Pham P, Torkamani A, Roddey JC, et al. (2013) All SNPs are not created equal: genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLoS Genet 9: e1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sprooten E, Fleming KM, Thomson PA, Bastin ME, Whalley HC, et al. (2013) White matter integrity as an intermediate phenotype: exploratory genome-wide association analysis in individuals at high risk of bipolar disorder. Psychiatry Res. 206: 223–31. [DOI] [PubMed] [Google Scholar]
- 44.Sarrazin S, Poupon C, Linke J, Wessa M, Phillips M, et al. (2014) A Multicenter Tractography Study of Deep White Matter Tracts in Bipolar I Disorder: Psychotic Features and Interhemispheric Disconnectivity. JAMA Psychiatry. 2014. [DOI] [PubMed]
- 45. Hanoune J, Defer N (2001) Regulation and role of adenylyl cyclase isoforms. Annu Rev Pharmacol Toxicol 41: 145–174. [DOI] [PubMed] [Google Scholar]
- 46. Mann L, Heldman E, Shaltiel G, Belmaker RH, Agam G (2008) Lithium preferentially inhibits adenylyl cyclase V and VII isoforms. Int J Neuropsychopharmacol 11: 533–539. [DOI] [PubMed] [Google Scholar]
- 47. Beuckmann CT, Sinton CM, Miyamoto N, Ino M, Yanagisawa M (2003) N-type calcium channel alpha1B subunit (Cav2.2) knock-out mice display hyperactivity and vigilance state differences. J Neurosci 23: 6793–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Murakami M, Ohba T, Xu F, Satoh E, Miyoshi I, et al. (2008) Modified sympathetic nerve system activity with overexpression of the voltage-dependent calcium channel beta3 subunit. J Biol Chem 283: 24554–24560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hwang CS, Shemorry A, Auerbach D, Varshavsky A (2010) The N-end rule pathway is mediated by a complex of the RING-type Ubr1 and HECT-type Ufd4 ubiquitin ligases. Nat Cell Biol 12: 1177–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee MJ, Tasaki T, Moroi K, An JY, Kimura S, et al. (2005) RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc Natl Acad Sci U S A 102: 15030–15035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Piatkov KI, Brower CS, Varshavsky A (2012) The N-end rule pathway counteracts cell death by destroying proapoptotic protein fragments. Proc Natl Acad Sci U S A 109: E1839–E1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. An JY, Seo JW, Tasaki T, Lee MJ, Varshavsky A, et al. (2006) Impaired neurogenesis and cardiovascular development in mice lacking the E3 ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Proc Natl Acad Sci U S A 103: 6212–6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Balogh SA, McDowell CS, Denenberg VH (2002) Behavioral characterization of mice lacking the ubiquitin ligase UBR1 of the N-end rule pathway. Genes Brain Behav 1: 223–229. [DOI] [PubMed] [Google Scholar]
- 54. Seifuddin F, Pirooznia M, Judy JT, Goes FS, Potash JB, et al. (2013) Systematic review of genome-wide gene expression studies of bipolar disorder. BMC Psychiatry 13: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BD associated index genes and corresponding gene sets.
(XLS)