Abstract
HIV-1 infection disrupts the intestinal immune system, leading to microbial translocation and systemic immune activation. We investigated the impact of HIV-1 infection on the intestinal microbiome and its association with mucosal T cell and dendritic cell (DC) frequency and activation, as well as with levels of systemic T cell activation, inflammation and microbial translocation. Bacterial 16S ribosomal DNA sequencing was performed on colon biopsies and fecal samples from subjects with chronic, untreated HIV-1 infection and uninfected control subjects. Colon biopsies of HIV-1 infected subjects had increased abundances of Proteobacteria and decreased abundances of Firmicutes compared to uninfected donors. Furthermore at the genus level, a significant increase in Prevotella and decrease in Bacteroides was observed in HIV-1 infected subjects, indicating a disruption in the Bacteroidetes bacterial community structure. This HIV-1-associated increase in Prevotella abundance was associated with increased numbers of activated colonic T cells and myeloid DCs. Principal coordinates analysis demonstrated an HIV-1-related change in the microbiome that was associated with increased mucosal cellular immune activation, microbial translocation and blood T cell activation. These observations suggest that an important relationship exists between altered mucosal bacterial communities and intestinal inflammation during chronic HIV-1 infection.
INTRODUCTION
Clinical manifestations of gastro-intestinal (GI) abnormalities, such as diarrhea, weight loss, and malnutrition, have long been observed in HIV-1 infected individuals.1 Disruption of the intestinal epithelial barrier occurs early in HIV-1 infection and is characterized by epithelial cell apoptosis2 and altered expression of genes related to epithelial barrier repair and regeneration as well as up-regulation of genes associated with inflammation.3 In conjunction with this early disturbance in epithelial integrity, a significant and substantial loss of mucosal CD4 T cells occurs which persists throughout the course of HIV-1 infection.4 This loss of CD4 T cells is marked by a significant depletion of T helper (Th17) cells,5 T cells that are involved in intestinal epithelial barrier homeostasis as well as in mucosal defense6. Furthermore, alterations in intestinal homeostasis occur due to the rapid influx of cytotoxic T cells,2 and the presence of high levels of pro-inflammatory cytokines and chemokines7.
A fundamental consequence of an altered intestinal microenvironment is the increased movement of microbes and microbial products that would normally be contained within the intestinal lumen by an intact epithelium, into the lamina propria, draining lymph nodes and ultimately the systemic circulation. Initial studies by Brenchley and colleagues showed that levels of plasma lipopolysaccharide (LPS), a cell wall component of Gram-negative bacteria known as endotoxin, and bacterial 16S ribosomal DNA (16S rDNA), a gene conserved among bacteria, were higher in HIV-1 infected individuals and associated with peripheral blood T cell activation.8,9 These observations provided the first indication that a relationship likely existed between intestinal microbial translocation and systemic immune activation. Since those seminal studies, numerous groups have linked increased HIV-1-associated microbial translocation, assessed by plasma LPS or soluble CD14 (sCD14) levels, with peripheral blood T cell activation and proliferation, decreased intestinal Th17 and gut-homing T cells frequencies, overall mortality in HIV-1 disease and with neurocognitive disorders (reviewed in 10). Importantly, plasma LPS levels in the first years of chronic HIV-1 infection predicted HIV-1 disease progression.11
The intestinal microbiota is a critical factor in the development and maintenance of physiological responses within the host. Indeed, numerous diseases and metabolic disorders have been linked to alterations in the intestinal bacterial community structure (“dysbiosis”)12, including obesity,13 inflammatory bowel disease (IBD),14 Type 1 diabetes,15 periodontal disease,16 and most recently atherosclerosis and cardiovascular disease.17,18 Gori and colleagues were the first to address potential intestinal dysbiosis in early HIV-1 infection and observed a high prevalence of Pseudomonas aeruginosa and Candida albicans as well as lower abundance of Bifidobacteria and Lactobacilli in the fecal microbiota compared to historical controls.19 Positive associations between stool Bacteroidales and blood CD8 T cell activation as well as negative associations between Enterobacteriales and duodenal CD4 count were observed in treatment-naïve HIV-1 infected subjects.20 In a more recent study, meta-analysis demonstrated that the fecal microbiota of HIV-1 infected subjects resembled the Prevotella-rich bacterial communities observed in agrarian cultures.21 Higher proportions of Lactobacillales were associated with higher CD4 counts, lower viral loads and reduced microbial translocation in a cohort of untreated subjects with early HIV-1 infection, and these associations continued following antiretroviral therapy (ART).22 Finally, in a recent study investigating bacterial community structure in rectosigmoid biopsies from untreated and treated HIV-1 infected subjects, an altered mucosal community of bacteria was found to associate with a number of markers of disease progression, systemic inflammation and higher levels of tryptophan catabolism.23
In our study, we sought to understand not only how untreated, chronic HIV-1 infection altered the intestinal microbiome, but whether specific changes in mucosal bacterial communities were linked to innate and adaptive mucosal immunity as well as systemic inflammation and markers of HIV-1 pathogenesis. We utilized high-throughput bacterial 16S ribosomal DNA gene sequencing to evaluate colonic mucosal tissue microbiota and examined the relationship between HIV-1-associated altered bacterial communities and intestinal T cell and dendritic cell (DC) immunological parameters, as well as systemic indicators of microbial translocation, inflammation and epithelial barrier integrity.
RESULTS
Intestinal microbial communities were assessed in 18 untreated, chronic HIV-1 infected and 14 age and sex-matched uninfected subjects (Table 1). To evaluate the impact of HIV-1 infection on the intestinal microbiome, we first determined if there was an overall difference in the relative abundance of colonic mucosa-adherent bacteria at the phylum, family or genera taxonomic levels between uninfected and HIV-1 infected subjects. We then directly compared the relative abundances of specific bacteria between the two subject groups and evaluated whether these alterations were also observed in the stool microbiome. Finally, we evaluated associations between the sampled microbiome from colonic biopsies with diet, body mass index (BMI), and clinical and immune status of the cohort.
Table 1.
Subject Characteristics
| Uninfected subjects | HIV-1 infected subjects | |
|---|---|---|
| Number of subjects | 14 | 18 |
| Age (yrs) | 31 (23–54) | 32.5 (22–58)n/s |
| Male/Female Ratio | 9/5 | 13/5 n/s |
| CD4 count (cells/µl) | 724 (468–1071) | 425 (238–782)* |
| Plasma Viral Load (HIV-1 RNA copies/ml) | - | 51350 (2880 – 207000) |
| Years since first HIV-1 seropositive test | - | 4.75 (0.25–15) |
| Body Mass Index (kg/m2) | 25.3 (18.5–32.3)# | 25.4 (17.4–34.7) |
| Ethnicity: n/s | ||
| Non-Hispanic | 11 (78.6%) | 17 (94.4%) |
| Hispanic | 3 (21.4%) | 1 (5.6%) |
| Race: n/s | ||
| White/Caucasian | 10 (71.4%) | 12 (66.7%) |
| Black/African American | 2 (14.2%) | 5 (27.8%) |
| Asian | 2 (14.2%) | 1 (5.5%) |
Values are shown as median (range) except for Ethnicity and Race which are shown as the number and percentage of each cohort. Statistical analysis was performed using Mann-Whitney U test for comparisons between uninfected and HIV-1 infected subjects and the Fisher Exact test or Chi-square test for comparison of categorical data.
n/s: not statistically different;
p=0.0002.
One subject had no weight or height values recorded at time of study therefore n=13.
Relative abundance of phyla in colon biopsies
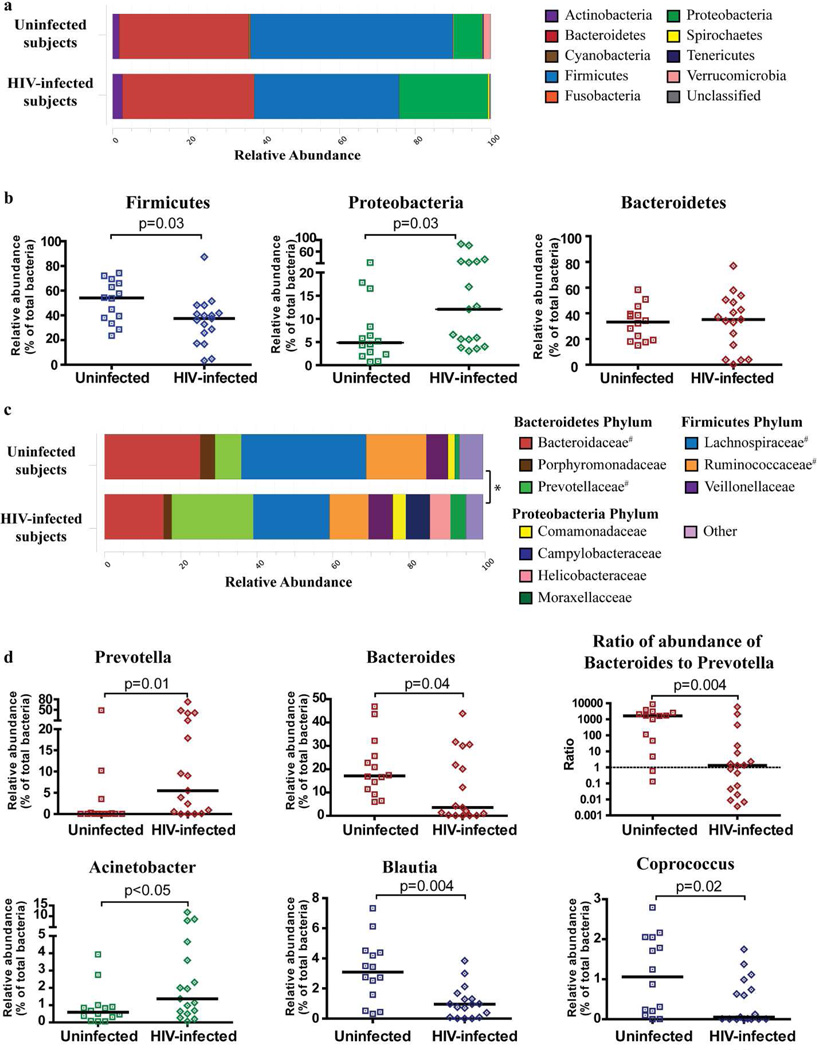
The three most dominant phyla detected within colon biopsies of both uninfected and HIV-1 infected subjects were Firmicutes, Bacteroidetes and Proteobacteria (Figure 1a). The remaining phyla each accounted for less that 3% of the overall abundance of bacteria. No overall significant difference existed at the phylum level between uninfected and HIV-1 infected subjects (p=0.11). However, when the relative abundance of the three major phyla were individually compared between uninfected and HIV-1 infected subjects, the median relative abundance of Firmicutes from HIV-1 infected subjects was significantly lower (p=0.03) whereas that of Proteobacteria was significantly higher (p=0.03) than in uninfected controls (Figure 1b). The relative abundances of Bacteroidetes were comparable between the two cohorts (Figure 1b), and there were no significant differences in the relative abundance of the remaining phyla between HIV-1 infected and uninfected subjects (data not shown).
Figure 1. Colonic mucosal dysbiosis in chronic, untreated HIV-1 infection is dominated by increased abundance of Prevotella.
Bacterial taxa were identified in colon biopsies from 17 subjects with chronic, untreated HIV-1 infection and 14 uninfected control subjects using bacterial 16S ribosomal DNA sequencing. (a) Stacked bar representing the average relative abundance of the top 9 most abundant phyla within colon biopsies of uninfected and HIV-1 infected subjects with the average percentage of the 3 most dominant phyla shown. Bacteria that were not identified as belonging to any known phylum were termed “Unclassified”. Statistical analysis was performed using the adonis function of the vegan community ecology R package and no overall significant difference existed at the phylum level between uninfected and HIV-1 infected subjects (p=0.11). (b) Comparison of the relative abundance of the Firmicutes, Proteobacteria and Bacteroidetes phyla within uninfected and HIV-1 infected subjects. Values are shown as a fraction of the total bacteria detected within each individual. Lines represent the median value. Statistical analysis was performed using the Mann Whitney U test. (c) Stacked bar plot representing the average relative abundance of the top 10 most abundant bacterial families (from 91 total families detected) within colon biopsies of uninfected and HIV-1 infected subjects with a statistical difference observed in the overall average relative abundance between these two cohorts (*: p=0.02). #Statistical difference observed when the relative abundance of families within uninfected individuals was compared to HIV-1 infected individuals using the Mann-Whitney U test. (d) Plots showing individual relative abundance of various genera that were statistically different between uninfected and HIV-1 infected subjects assessed using the Mann-Whitney U test. Values are shown as a fraction of the total bacteria detected within each individual. Lines represent the median value. The ratio of the relative abundance of Bacteroides to Prevotella was calculated for each individual and plotted as shown. Dotted line designates a ratio of 1.0 (relative abundance of Bacteroides = relative abundance of Prevotella). False discovery rate corrections were not performed and P values are uncorrected.
Relative abundance of family and genera in colon biopsies
Comparisons of bacteria at the family level demonstrated a significant difference in the overall relative abundances between uninfected and HIV-1 infected subjects (p=0.02). Specifically, evaluation of the relative abundance within individuals (Supplementary Figure S1) and the average relative abundance of the top ten most abundant families for both uninfected and HIV-1 infected subjects (Figure 1c) highlighted that the microbiota within colonic mucosal tissue was dominated by Bacteroidaceae, Lachnospiraceae, Rumminococceae and Prevotellaceae. In direct comparisons, the relative abundance of Prevotellaceae (p=0.03) was statistically higher in HIV-1 infected subjects whereas Lachnospiraceae (p=0.009), Christensenellaceae (p=0.04), Ruminococcaceae (p=0.04) and Bacteroidaceae (p=0.04) were all lower in HIV-1 infected subjects relative to uninfected individuals (Supplementary Table S1). The HIV-1-associated increase in relative abundance in the Proteobacteria phylum was attributable to increased Brucellaceae (p=0.02), Xanthomonadaceae (p=0.04) and Moraxellaceae (p=0.05) in conjunction with a decrease in relative abundance of Rhodospirillaceae (p=0.02).
Comparisons of bacterial genera demonstrated a significant difference in the overall relative abundances between uninfected and HIV-1 infected subjects (p=0.009; data not shown). Within the Bacteroidetes phylum, differences were most notable for the significant increase in Prevotella and decrease in Bacteroides in HIV-1 infected subjects (Figure 1d). A significant decrease in Bacteroides:Prevotella ratio was also observed among HIV-1 infected subjects (Figure 1d). More than 85% (12/14) of the uninfected subjects had greater abundance of Bacteroides than Prevotella, whereas in HIV-1 infected subjects, only 52% (9/17) had a Bacteroides-dominant profile. HIV-1 infected subjects also had reduced relative abundance of other genera belonging to the phylum Bacteroidetes including Alistipes (p=0.02) and Barnesiella (p=0.02).
The relative abundance of Acinetobacter of the Proteobacteria phylum was increased in HIV-1 infected subjects (Figure 1d) whereas Thalassospira was decreased (p=0.01). Within the Firmicutes phylum, the relative abundance of Blautia and Coprococcus genera were also reduced in HIV-1 infected subjects (Figure 1d). Given that it has previously been reported that bacterial communities may cluster into distinct enterotypes distinguished primarily by levels of Bacteroides, Prevotella and Ruminococcus24, and that we had observed alterations in both Bacteroides and Prevotella, we evaluated if Ruminococcus, a member of the Firmicutes phylum, was also altered in HIV-1 infection. In uninfected subjects, the relative abundance of Ruminococcus was generally low (median relative abundance: 0.69% of total bacteria, range: 0–6.62%), and although the median relative abundance was lower in HIV-1 infected subjects (0.07%, 0–2.12%), this difference did not reach statistical significance (p=0.15).
A summary flow chart illustrating the differences in the relative abundance of mucosa-adherent bacteria at each taxonomic level between uninfected and HIV-1 infected subjects is shown in Supplementary Figure S2.
Comparisons of microbial diversity in colon biopsies
A comparison of both the number of genera observed (Sobs) as well as the estimated number of genera (SChao1) showed similarity in richness values between uninfected and HIV-1 infected subjects (Table 2). No significant difference was found in the estimated diversity (H) or evenness (H/Ho) of microbiome data between donor groups, although a trend towards greater evenness in HIV-1 infected individuals was noted (p=0.08).
Table 2.
Richness and diversity within colonic mucosal tissue from uninfected and HIV-1 infected subjects.
| Uninfected | HIV-1 infected | p valuea | |
|---|---|---|---|
| subjects (n=14) | subjects (n=16) | ||
| Observed Richness (Sobs) | 56.6 (34.78–78.16) | 54.71 (33.5–76.1) | p=0.79 |
| Estimated Richness (SChao1) | 67.3 (42.4–96.1) | 67.0 (45.1–86.7) | p=0.49 |
| Shannon Diversity (H) | 3.8 (2.8–4.2) | 3.4 (1.3–4.5) | p=0.25 |
| Evenness (%) | 64.2 (49.9–73.4) | 59.5 (23.0–73.9) | p=0.08 |
Values are shown as median (range) of the mean value calculated for each individual. OUT richness and diversity values were calculated for genus-level OTUs.
Statistical tests were performed using Mann Whitney U test.
Relationships between mucosa-adherent microbiota, diet and BMI
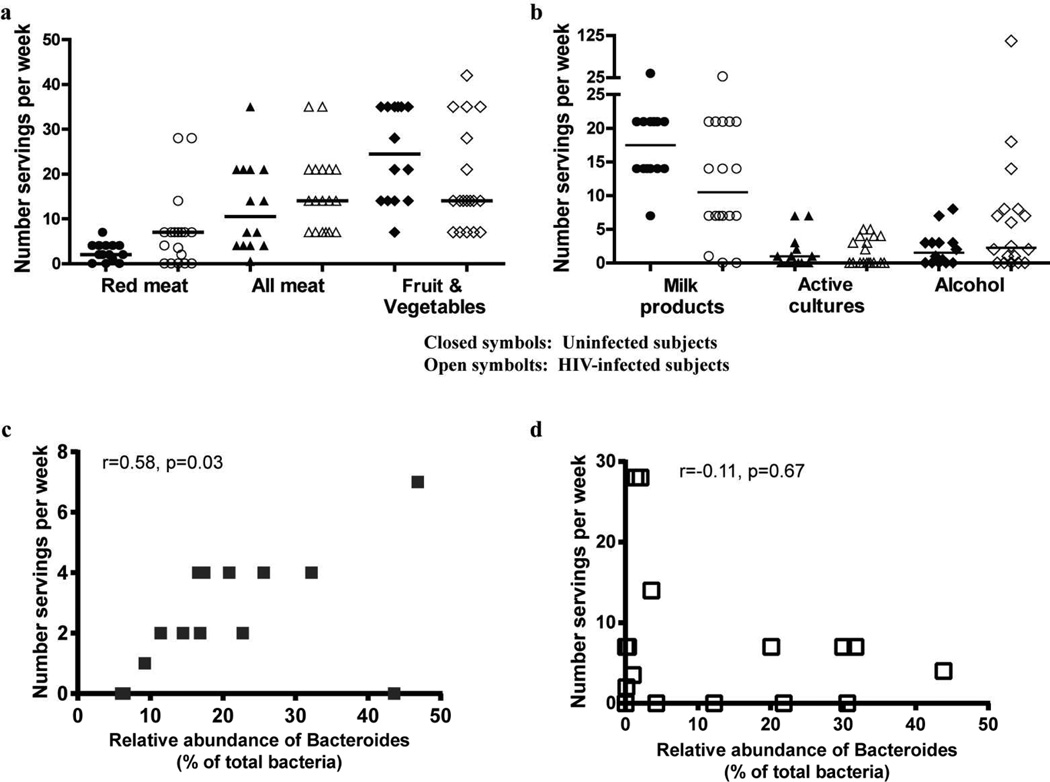
Dietary habits have been associated with alterations in the composition of the intestinal microbiome.13 As part of our Study Specific Interview, we obtained a limited diet history from each individual. No significant differences between HIV-1 infected and uninfected subjects were observed in the number of servings per week reported for meat, fruits and vegetables (Figure 2a), milk products, active cultures (e.g. yogurt) or alcohol consumption (Figure 2b), although trends towards an increase in the median number of servings of meat and decreased servings of fruits, vegetables and milk products in HIV-1 infected subjects were noted.
Figure 2. Dietary habits of uninfected and HIV-1 infected subjects.
(a,b) Uninfected and HIV-1 infected subjects completed a short questionnaire at the time of study visit detailing the average number of servings of various diet-related food groups per day or per week. Standardized values are shown as “number of servings per week” for both uninfected (closed symbols) and HIV-1 infected (open symbols) subjects. Lines represent median values. Statistical analysis was performed using the Mann Whitney U test with no comparisons reaching a statistically significant p value (p<0.05). (c,d) Association between the relative abundance of Bacteroides and number of servings of red meat per week in (c) uninfected subjects and (d) HIV-1 infected subjects. Statistical analysis was performed using the Spearman t test.
Given that abundances of Bacteroides and Prevotella were most dramatically different between uninfected and HIV-1 infected subjects, we next evaluated associations between diet and the relative abundances of these two genera. In uninfected subjects, a significant positive association was observed between the number of servings of red meat and the relative abundance of Bacteroides (Figure 3c, Table 3), with a trend towards a positive association when servings of ‘all meat’ were evaluated (Table 3). However, in HIV-1 infected subjects, such associations were not observed (Figure 3d, Table 3). Interestingly, a trend towards a negative association between the number of servings of milk products per week and the relative abundance of Prevotella was observed within HIV-1 infected subjects (p=0.06), whereas no association was noted in uninfected subjects (p=0.82) (Table 3).
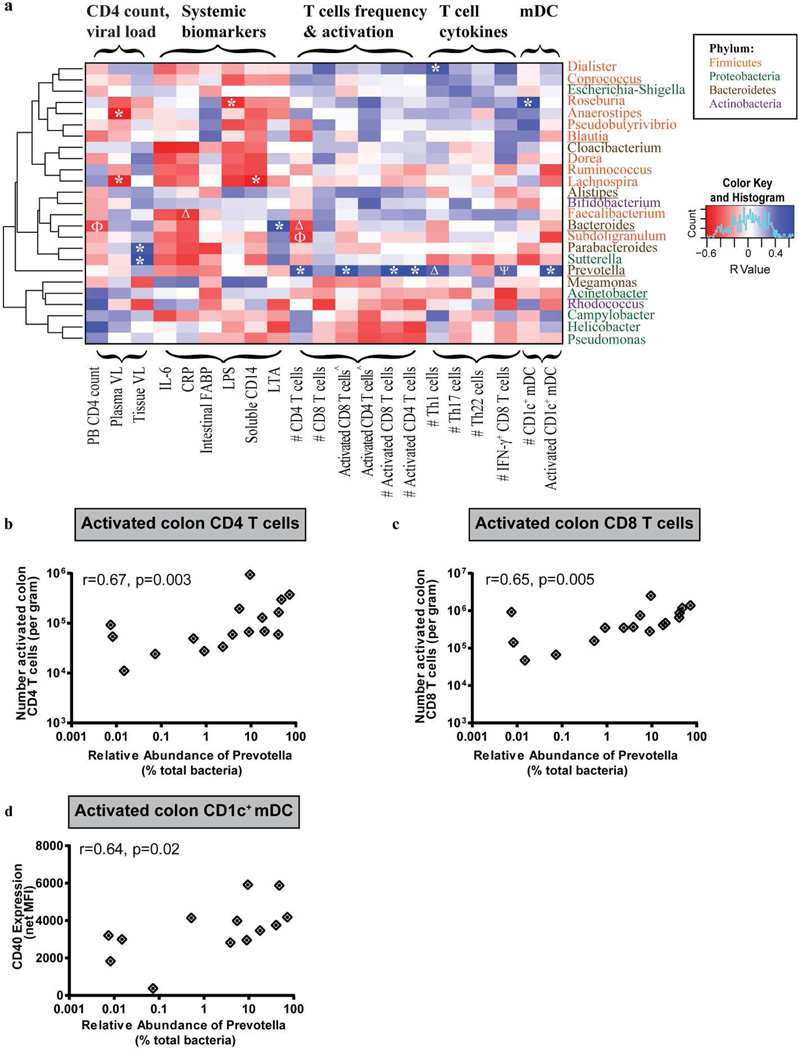
Figure 3. HIV-1 associated increase in the relative abundance of Prevotella is associated with colonic T cell and mDC activation.
Associations between the top 25 most abundant genera within HIV-1 infected subjects with various clinical and immune parameters were assessed. (a) Heatmap representing positive (blue shading) and negative (red shading) associations between CD4 count, plasma and tissue viral load, mucosal T cell frequency, activation, T helper cell (Th)1, Th17 and Th22 and IFN-γ-producing CD8 T cell frequencies as well as CD1c+ mDC frequency and activation. Clustering was performed based on genera associations with the clinical and immune parameters. Genera are color-coded based on their respective phylum. #: number per gram of mucosal tissue; ^Percent of activated CD4+ or CD8+ T cells within viable, CD45+ cells. *p<0.05, Δp<0.06, ϕp<0.07, ψp<0.08. Genera that are altered in HIV-1 infected subjects relative to uninfected subjects are underlined. (b, c and d) Associations between the relative abundance of Prevotella and numbers of activated colon CD4 (b) and CD8 (c) T cells and CD1c+ mDC CD40 expression levels (d) in HIV-1 infected subjects (n=17 for colon T cell activation; n=13 for CD1c+ mDC). Statistical analysis was performed using the Spearman t test. False discovery rate corrections were not performed and P values are uncorrected.
Table 3.
Associations between dietary habits (servings per week) of HIV-1 infected subjects and the relative abundance of Bacteroides or Prevotella in colonic mucosal tissue.
| Uninfected (n=14) | HIV-1 infected (n=17) | |
|---|---|---|
| Red Meat | ||
| Bacteroides | r= 0.58, p=0.03 | r=−0.11, p=0.67 |
| Prevotella | r=−0.13, p=0.65 | r= 0.19, p=0.45 |
| All Meat | ||
| Bacteroides | r= 0.45, p=0.10 | r= 0.16, p=0.53 |
| Prevotella | r=−0.09, p=0.77 | r=−0.09, p=0.77 |
| Fruits and Vegetables | ||
| Bacteroides | r=−0.07, p=0.81 | r=−0.17, p=0.51 |
| Prevotella | r= 0.03, p=0.91 | r= 0.03, p=0.90 |
| Milk Products | ||
| Bacteroides | r= 0.05, p=0.88 | r= 0.28, p=0.27 |
| Prevotella | r=−0.07, p=0.82 | r=−0.46, p=0.06 |
| Active Cultures | ||
| Bacteroides | r= 0.25, p=0.41 | r=−0.21, p=0.43 |
| Prevotella | r=−0.32, p=0.28 | r= 0.02, p=0.94 |
| Alcohol | ||
| Bacteroides | r= 0.17, p=0.57 | r= 0.08, p=0.75 |
| Prevotella | r=−0.44, p=0.12 | r= 0.28, p=0.27 |
Statistical analysis performed using the Spearman rank correlation test. (Bold values: significant association; bold italics: trending towards significance).
Obesity has also been linked to alterations in intestinal microbiota.25 The median BMI values for uninfected (25.3) and HIV-1 infected (25.4) subjects were not statistically different (Table 1). The similarities in BMI between the two cohorts suggests that the differences observed in the microbiota between uninfected and HIV-1 infected subjects were not likely due to dysbiosis secondary to obesity or being overweight. Indeed, we did not observe any associations between BMI and the relative abundances of either Bacteroides or Prevotella for uninfected or HIV-1 infected subjects (data not shown).
Associations between Prevotella in colon biopsies and immune parameters
Intestinal homeostasis is established and maintained, in part, through a complex relationship between the host immune system and the microbiota. Given that HIV-1 infection results in alterations in mucosal immunity as well substantial structural damage within the GI tract,26 we evaluated whether the specific microbiota of HIV-1 infected individuals was associated with various mucosal innate and adaptive immune parameters or with plasma biomarkers indicative of systemic microbial translocation, inflammation and epithelial damage.
We focused this analysis on the 25 most abundant bacterial genera that accounted for 90% of the total bacteria detected in the HIV-1 infected subjects. Figure 3a shows a heat map summarizing Spearman rank correlation coefficients between the relative abundance of the abundant genera with clinical readouts (CD4 count, plasma viral load), colon tissue viral load, plasma biomarkers indicative of inflammation (IL-6, C-Reactive Protein (CRP)), intestinal epithelial damage (intestinal Fatty Acid Binding Protein (iFABP)) and microbial translocation (LPS, sCD14, Lipoteichoic acid (LTA)), as well as mucosal T cell frequency, activation and cytokine production and CD1c+ myeloid dendritic cell (mDC) frequency and activation. It is notable that within each phylum, not all genera clustered together, indicating that associations between genera and clinical or immune parameters were not determined solely by higher taxonomic rank (Figure 3a. Supplementary Figure S3). Furthermore, Prevotella did not cluster with any other bacteria, and the relative abundance of Prevotella had significant positive associations with a number of different immune parameters, including mucosal T cell and DC activation (Figure 3a). Specifically, the number of activated (CD38+HLA-DR+) mucosal CD4 and CD8 T cells per gram of mucosal tissue strongly associated with the relative abundance of Prevotella (Figure 3b,c). The relative abundance of Prevotella also positively associated with the number of colon Th1 and IFN-γ+ CD8 T cells, although these associations did not reach statistical significance (Th1: r=0.49, p=0.06; IFN-γ+ CD8 T cells: r=0.48, p=0.08). The level of CD1c+ mDC activation based on CD40 expression correlated strongly with the relative abundance of Prevotella (Figure 3d). Conversely, no significant associations between Prevotella abundance and mucosal T cell and CD1c+ mDC activation levels were observed in uninfected subject samples (data not shown). Few significant associations were observed for the other genera that were statistically different in abundance in HIV-1 infected subjects, although Bacteroides abundance positively associated with plasma LTA (r=0.60, p=0.01) and trended towards a negative association with CD4 T cell count (r= −0.45, p=0.08).
Patterns of HIV-1-associated dysbiosis among stool and mucosal samples
In all but one of the other published studies addressing intestinal dysbiosis during HIV-1 infection, microbial communities were assessed in more easily obtainable stool samples.19–23 However, some studies have suggested that the microbial structure of human fecal samples may differ to the mucosa-adherent bacteria associated with the colonic mucosa.27 Therefore, we determined if dysbiosis in mucosa-adherent bacteria was limited to the colonic mucosal tissue or was also reflected in subject-matched fecal aspirates or stool samples (Supplementary Figure S4). The increased relative abundance of Proteobacteria found in mucosal samples from HIV-1 infected subjects was not observed in fecal aspirates or rectal swabs, suggesting that the increased abundance of Proteobacteria may be restricted to the colonic mucosal tissue bacterial communities. The HIV-1-associated decrease in the relative abundance of Firmicutes in colonic mucosal tissue was also observed in fecal aspirates, but abundances were similar in stool samples from HIV-1 infected and uninfected subjects. Similarly, the observed HIV-1-associated differences in the Proteobacteria and Firmicutes families and genera of mucosa-adherent bacteria were not consistently observed in either fecal aspirates or in stool samples. However, changes in relative abundance of families and genera belonging to the Bacteroidetes phylum appeared to be more similar across the 3 samples. Taken together, these observations suggest that fecal aspirates and stool samples generally reflected the same altered patterns of the Bacteroidetes families and genera, but HIV-1-associated changes in Proteobacteria and Firmicutes seemed to be mucosa-specific.
In additional analyses, we compared the relative abundance of Prevotella in mucosal tissue with the relative abundances observed in fecal aspirates and in rectal stool samples. A significant positive correlation was observed between the relative abundance of mucosa-adherent Prevotella to that in fecal aspirates, but not to that in stool samples (Supplementary Figure S5). Further, no significant associations were evident between the relative abundance of Prevotella in either fecal aspirates or rectal stool samples and any mucosal immune parameters in HIV-1 infected subjects (data not shown). These findings are in contrast with the strong associations observed between mucosal immune parameters and the relative abundance of mucosa-associated Prevotella (Figure 3).
We next evaluated whether significant associations observed between other mucosa-adherent bacterial abundances and the various immunological and virological parameters were also observed in fecal aspirates or in stool samples. Within the Firmicutes, only the associations between Dialister and the number of colon Th1 cells (r=0.53, p<0.05) and between Roseburia and the number of colon CD1c+ DC (r=0.67, p=0.01) were observed in fecal aspirates and only the negative correlation between Lachnospira and plasma HIV-1 viral load (r=-0.59, p=0.01) observed in rectal stool samples. Within the Bacteroidetes phylum, the relative abundance of Bacteroides in fecal aspirates positively associated with plasma levels of LTA (r=0.63, p=0.008), but no other significant associations were observed. Taken together, these observations further highlight the specificity of the associations between the mucosa-associated microbiome and the various immunological and virological parameters.
Characterization of patterns of mucosa-adherent bacterial communities with clinical and immunological parameters
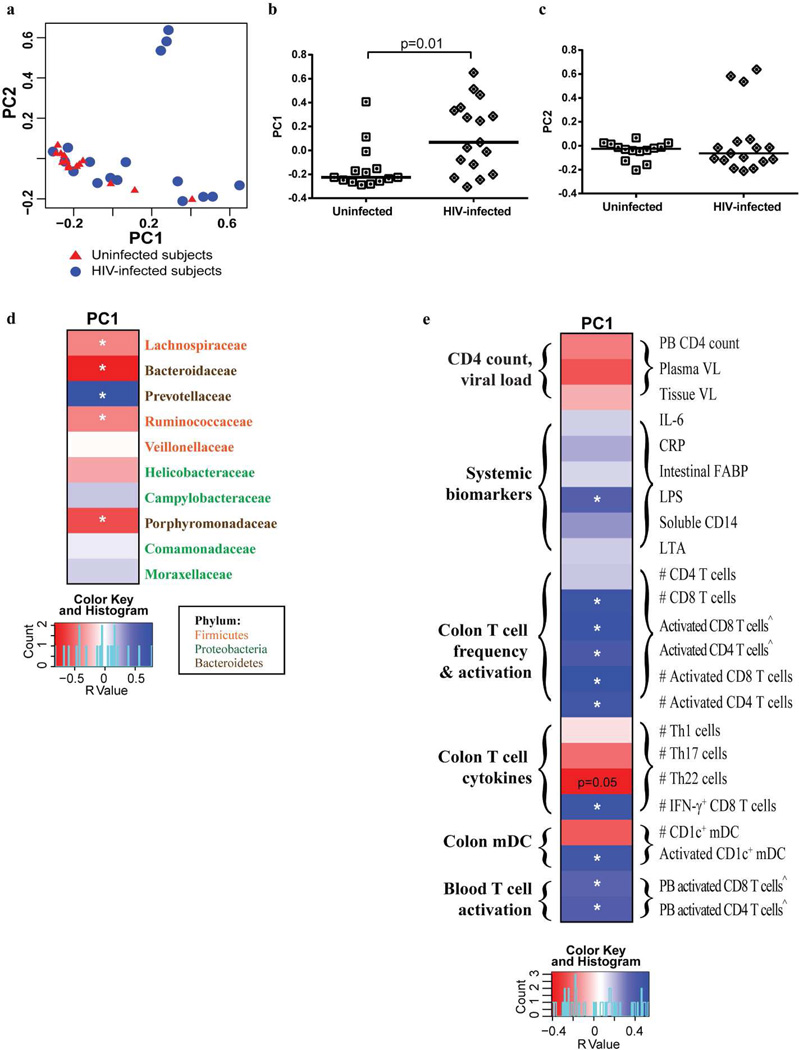
A broad approach was next taken to evaluate overall patterns in the microbiomes of all study subjects by utilizing principal coordinates analysis with distance matrices calculated using the Bray-Curtis ecological dissimilarity index. In this analysis, HIV-1 infection status was significantly associated with PC1 scores (Figure 4a, b), but not PC2 (Figure 4a, c) or other PC scores (data not shown). Further analysis of the 3 HIV-1 infected subjects with high PC2 values demonstrated that the microbiome of each of these subjects was dominated by a high relative abundance of one bacterial genus in each of the subjects (Subject 1: Helicobacter: 68.7% of total bacteria; Subject 2: Staphylococcus: 20.4%; Subject 3: Campylobacter: 81.5%). Higher PC1 values were significantly associated with greater abundance of Prevotellaceae in conjunction with lower abundance of Bacteroidaceae and Porphyromonadaceae and of bacteria within the Firmicutes phyla (Lachnospiraceae and Ruminococcaceae) (Figure 4d, Supplementary Figure S6). PC1 values were also significantly associated with levels of plasma LPS, colon T cell and DC activation levels, and with peripheral blood CD4 and CD8 T cell activation levels (Figure 4e, Supplementary Figure S7).
Figure 4. Colonic mucosal dysbiosis is associated with chronic, untreated HIV-1 infection.
(a) Principal coordinates (PC) plots demonstrating an association of PC1 values with HIV-1 infection. (b,c) Direct comparisons of PC1 (b) and PC2 (c) scores in uninfected and HIV-1 infected individuals. Lines represent the median value. Statistical analysis was performed using the Mann Whitney U test. (d,e) Heatmaps illustrating associations (positive: blue shading; negative: red shading) between PC1 and bacterial families (d) and CD4 count, plasma and tissue viral load (HIV-infected subjects only), mucosal T cell frequency, activation, T helper cell (Th)1, Th17 and Th22, IFN-γ-producing CD8 T cell frequencies, CD1c+ mDC frequency and activation and peripheral blood (PB) T cell activation (e). Families are color-coded based on their respective phylum. #Number per gram of mucosal tissue; ^Percent of activated CD4+ or CD8+ T cells within viable, CD45+ cells. Statistical analysis was performed using the Spearman t test. *p<0.05. Trending associations are noted as p=0.05. False discovery rate corrections were not performed and P values are uncorrected.
DISCUSSION
In this cross-sectional study, we compared the colonic mucosal microbiome in untreated, chronic, HIV-1 infected individuals to that of sex and age-matched uninfected control subjects. One advantage of using mucosal biopsies for microbiome analysis is that bacterial species directly adherent to the GI tract epithelium and thus more likely to translocate in the event of epithelial barrier breakdown would be better represented. We also measured colon tissue DC and T cell frequencies and function, systemic T cell activation and indicators of inflammation, microbial translocation and epithelial barrier dysfunction in order to evaluate the relationship between these parameters and HIV-1 associated alterations in intestinal bacterial communities.
Our observations of increased Proteobacteria, decreased Firmicutes including members of the Clostridia class, and significant depletion in Bacteroides and Alistipes in the colonic mucosal tissue of untreated, HIV-1 infected subjects are in agreement with a recently published study.23 In addition, HIV-associated reductions in Firmicutes including genera of the Lachnospiraceae, Ruminococcaceae and Rikenellaceae were observed in rectal microbiota samples obtained from anally-inserted sponges.28 In a third recent study in which fecal samples were evaluated, abundances of Bacteroidaceae, Rikenellaceae and Porphyromonadaceae were decreased concurrent with an increase in Prevotellaceae in HIV-infected samples, also in general agreement with our findings.21 Moreover, we identified an HIV-related intestinal mucosal microbiome profile that was significantly associated with levels of mucosal cellular immune activation, with markers of systemic microbial translocation, and with levels of blood T cell activation, thereby linking mucosal dysbiosis to a marker of HIV-1 disease progression.29 These findings support those by McCune and colleagues, who also showed an association between altered mucosa-adherent intestinal microbiota and markers of HIV-1 disease progression.23 In conjunction with these 3 recently published studies21,23,28 and earlier work,19,20,22 our observations further highlight the considerable impact of HIV-1 infection on the intestinal microbiome.
One of the most striking findings of our study was the increased relative abundance of the Gram-negative anaerobe Prevotella in conjunction with decreased Bacteroides in HIV-1 infected individuals. The HIV-1 disease-associated increase in mucosa-adherent Prevotella significantly and strongly associated with mucosal DC and T cell activation, suggesting that a link exists between the altered microbiome and local cellular immune activation and inflammation. We also observed an association between Prevotella abundance and numbers of colonic CD4 and CD8 T cells with the potential to produce the pro-inflammatory cytokine, IFN-γ. Increased mucosal IFN-γ production not only potentially contributes to local inflammation, but could also increase epithelial barrier leakiness by increasing paracellular permeability and bacterial translocation, as has been shown in experimental intestinal epithelial models.30
Alterations along a Prevotella-Bacteroides gradient were also observed by a separate group of investigators in fecal samples from untreated HIV-1 infected subjects, and these alterations were only minimally impacted by ART.21 In our study, we were able to directly compare the microbiota in the colonic mucosal tissue with subject-matched stool samples. The specific alterations in the Bacteroidetes genera were generally mirrored in colonic luminal bacteria (fecal aspirates), as well as in stool samples, which potentially reflect the entire gut microbiome. However, it is important to note that similarities in the altered mucosa adherent-bacterial communities were not observed for all bacteria at these distal sites. Furthermore, the association between the altered microbiota and mucosal cellular immune activation appeared to be specific to the mucosa-adherent bacterial communities, with similar direct associations not observed between markers of immune activation and Prevotella abundance in fecal aspirates or stool samples. Using a limited dietary questionnaire, we did not identify dietary differences between uninfected and HIV-infected cohorts that could account for HIV-1-associated microbiome differences; however, a more detailed and prospective dietary questionnaire would be required to conclusively exclude any impact of diet on our observations.
Although we cannot determine in this cross-sectional study whether the increased relative abundance of Prevotella is causative or a consequence of mucosal immune activation in HIV-1 infection, a number of studies have provided evidence that Prevotella is capable of acting as a pathobiont, a potentially ‘pathogenic symbiont’ within the microbiota. Alterations in abundance of Prevotella have been observed in a number of inflammatory disease states including periodontal disease31 and in active ulcerative colitis,32 and exacerbation of colitis in an inflammasome knockout mouse model was linked to Prevotellaceae and the induction of CCL5 by epithelial cells, with subsequent recruitment of immune cells leading to spontaneous inflammation. A recent study by Koeth et al. found that Prevotella spp. were associated with higher plasma levels of the pro-atherogenic metabolite trimethylamine-N-oxide (TMAO).17 HIV-1 infected individuals are at increased risk of coronary heart disease33 and while there are numerous factors that are known to contribute to this increased risk, it is tempting to speculate that the altered proportions of Prevotella and Bacteroides observed in our HIV-1 infected subjects may be an additional contributing factor. Finally, Prevotella may also augment disruption of mucosal barrier function via its ability to degrade mucin,34 thereby potentially further contributing to the epithelial breakdown and microbial translocation observed in HIV-1 infected individuals.
Previous studies have shown an increase in pathogenic and decrease in probiotic fecal-associated bacteria in early HIV-1 infection19, and higher proportions of probiotic Lactobacillales were associated with better immune function in this clinical setting.22 In our study, the increased abundance of Prevotella was a proportional shift, observed in combination with decreased relative abundance of other mucosa-associated bacteria. This raises the possibility that a reduced relative abundance of specific bacteria, especially those with anti-inflammatory properties35 and/or those that contribute to epithelial barrier integrity, may also contribute to increased mucosal inflammation during HIV-1 infection. For example, butyrate is a short chain fatty acid that is a major energy source for the colonic epithelium.36 Reduced abundance of butyrate-producing bacteria could perpetuate the epithelial barrier disruption observed in HIV-1 infected individuals. Indeed, in our cohort, HIV-1 infected subjects had significantly reduced relative abundances of Lachnospiraceae and Ruminococcaceae, families of bacteria in the Firmicutes phylum that contain many of the known butyrate-producing bacterial species.37
Although our study has focused on understanding the impact of HIV-1 infection on the intestinal microbiome by identifying taxonomic changes in bacterial composition, this approach does not provide information about microbial community function. In a recent study by McHardy et al., the use of bioinformatics tools to impute bacterial metagenomic functions based on microbial diversity analysis suggested that differing metabolic functions, such as amino acid metabolism and vitamin biosynthesis, would be present in HIV-infected subjects due to alterations in microbial community structure.28 In future studies, this type of comprehensive approach may identify additional key bacterial functional pathways that are altered as a result of HIV-associated dysbiosis and lead to a better understanding of the interplay between the host microbiota and immune system. Comprehensive approaches may also provide new and exciting avenues of treatment founded on an ecological-based approach to restore the ‘good’ bacteria and optimize their growth and function while limiting the ‘bad’ taxa and their potential detrimental effects on the host.38
In summary, our study demonstrates that significant dysbiosis occurs in the colonic mucosal tissue of untreated, chronic HIV-1 infected individuals. This dysbiosis was dominated by increased relative abundance of the pathobiont Prevotella, which was associated with increased mucosal DC and T cell activation, systemic T cell activation, and a marker of systemic microbial product translocation. These observations indicate a fundamental shift in host-microbiota interactions in the intestine during HIV-1 infection and suggest that an important relationship may exist between mucosal bacterial communities, intestinal inflammation and systemic immune activation during chronic, untreated HIV-1 infection.
METHODS
Study Participants and Study Design
Eighteen HIV-1 infected individuals and 14 HIV-1 seronegative (uninfected) controls who met the entry criteria were enrolled in this cross-sectional study at the University of Colorado Anschutz Medical Campus. Efforts were made to enroll HIV-1 seronegative subjects who were matched for age and sex to the HIV-1 infected subjects. HIV-1 infected subjects were ART-treatment naïve or had not been on treatment for more than 7 days in the preceding 6 months. Subject characteristics and a list of exclusion criteria are detailed in Supplementary Materials. The clinical details for study subjects are provided in Table 1. All subjects voluntarily gave written, informed consent. This study was approved by the Colorado Multiple Institutional Review Board (COMIRB) at the Anschutz Medical Campus.
All subjects underwent a study-specific interview and physical examination, phlebotomy, stool sample collection by rectal swab, and flexible sigmoidoscopy at the Clinical and Translational Research Center (CTRC) and at the University of Colorado Hospital endoscopy clinic. Collection and storage of stool samples, fecal aspirates, colon biopsies and PBMC are detailed in Supplementary Materials. As part of the study-specific interview, subjects provided clinical history and completed a dietary questionnaire that asked subjects to detail the average number of servings of 1) fruit and vegetables, 2) red meat, 3) all types of meat (all meat), 4) milk products, 5) active cultures and 6) alcohol they consumed per day. These data were then collated to reflect the average number of servings per week for each individual.
Processing of colon biopsies, plasma, serum and PBMC
Colon pinch biopsies were collagenase-digested as detailed in Supplementary Materials. Collection of plasma, serum and PBMC from whole blood was performed as previously described39,40 and is detailed in Supplementary Materials.
Mitogenic stimulation of single cell colon biopsy preparations
Isolated colon cells were resuspended at 1×106 cells/ml in RPMI + 10% human AB serum (Gemini Bioproducts, West Sacramento, CA) + 1% penicillin/streptomycin/L-glutamine (complete RPMI) and stimulated with 250ng/ml PMA and 1µg/ml ionomycin (both Sigma-Aldrich) in the presence of Brefeldin A (1µg/ml; BD Biosciences) for 16hrs at 37°C, 5% CO2.
Surface and intracellular flow cytometry staining assays, acquisition and analysis
Standard flow cytometry staining protocols to identify expression of surface markers and intracellular cytokines were followed as previously described39,41 and detailed in Supplementary Materials. All flow cytometry data was acquired on an LSRII Flow Cytometer (BD Biosciences). Routine quality control using the Cytometer Setup & Tracking feature within the BD FACSDiva software version 6.1.2 (BD Biosciences) was performed daily as previously detailed.39 Electronic compensation was performed using positive anti-mouse Igk beads (BD Bioscience) stained with the appropriate individual antibodies and mixed 1:1 with negative control beads (BD Biosciences). To compensate for the Aqua viability dye, ArC Amine Reactive Beads (Invitrogen) were stained following the manufacturer’s protocol.
Bacterial 16S ribosomal RNA gene sequencing
Bacterial profiles of study participants were generated by broad-range amplification and sequence analysis of bacterial 16S rRNA genes following our previously described methods42 and as detailed in Supplementary Materials. In brief, DNA was extracted from rectal swabs, fecal aspirates and colon biopsies using the UltraClean Fecal DNA Kit (MO BIO Laboratories Inc., Carlsbad, CA). 16S gene templates were quantified using the protocol of Nadkarni et al.43 Amplicons were generated using oligonucleotide primers that target approximately 300 b.p. of the V4 variable region of the 16S rRNA gene (primers 515F and 805R)44 and also include barcode45 and Illumina adapter sequences. Illumina paired-end sequencing was performed on the Miseq platform with version 2.0 of the Miseq Control Software, using a 500-cycle (v.2) reagent kit. Paired-end sequences were quality-filtered and demultiplexed as previously described42 and as detailed in Supplementary Materials. The software package Explicet46 (v2.8.3) was used to analyze, display, and generate figures of microbiome data.
A total of 13,683,660 high quality 16S ribosomal DNA sequences were generated (5,739,143 from aspirates, 2,640,168 from biopsies, and 5,304,349 from swabs). The median number of high-quality sequences generated per specimen was 191,724, 87,558, and 185,250.5, for aspirates, biopsies, and swabs, respectively; biopsies generated significantly fewer sequences than did aspirates or swabs (p=3.3e-06 and p=1.2e-05), respectively for aspirates and swabs, assessed by two-tailed t-test. Nonetheless, the Good’s coverage index for genus level OTUs (calculated through 1000 replicate samples with 10,000 sequences per sample) was greater than 99% for every 16S library indicating that most biodiversity was captured for each subject. Raw paired-end Illumina MiSeq reads were submitted to the NCBI Small Read Archive under BioProject accession number PRJNA227062.
Determination of colon tissue viral load
Total RNA was extracted from pre-weighed colon biopsies with the RNeasy Mini Kit (Qiagen, Valencia, CA) and HIV cDNA generated using the SuperScript III first-strand synthesis kit (Invitrogen) according to the manufacturers’ protocol. HIV RNA was quantified by real-time (RT)-PCR in an ABI 7900HT thermocycler (Applied Biosystems, Foster City, CA). A detailed protocol for determination of colon tissue viral load appears in Supplementary Materials.
Plasma Biomarker Measurement
EDTA plasma samples were assessed for LPS and IL-6 levels, heparin plasma samples for sCD14, iFABP) and CRP and serum samples for LTA. LPS levels were measured using the Limulus Amebocyte Lysate (LAL) assay (Lonza, Switzerland) with plasma samples diluted 1:10 in endotoxin free water and heat-inactivated at 80°C for 15min. Commercially available ELISAs were used to evaluate levels of IL-6, sCD14, and iFABP (all R&D Systems, Minneapolis, MN). CRP levels were determined using a high-sensitivity immunoturbidimetric assay (Beckman Coulter). Manufacturer’s protocols were followed for all assays. Serum LTA levels were assessed using a custom ELISA as previously reported.47
Statistical Analysis
Non-parametric statistics were performed with no adjustments for multiple comparisons due to the exploratory nature of this study. Statistical comparisons of bacterial communities were performed with the R software package (v 2.15.2). The overall statistical differences between the gut microbiomes of uninfected and HIV-1 infected subjects were assessed at the phylum, family and genus level using the adonis function of the vegan community ecology R package.48 Statistical differences in OTUs at each taxonomic level were analyzed using the Wilcoxon test. Subsequent analysis and graphing of those OTUs deemed significantly different using the Wilcoxon rank-sum test were performed in Explicet v. 2.8.546 or GraphPad Prism v. 6. Spearman rank correlation tests were performed to evaluate associations between two variables. Fisher exact tests and Chi-squared tests were used for comparisons of categorical data. A p value of <0.05 was considered significant. For principal coordinates analysis, distance matrices were calculated using the Bray-Curtis ecological dissimilarity index (vegdist function of vegan R package) and applied to the cmdscale function in R. False discovery rate corrections were not performed and P values are uncorrected.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to express our gratitude to all the study participants as well as the physicians and staff at the University of Colorado Infectious Disease Group Practice Clinic. We would also like to acknowledge the staff at the Clinical and Translational Research Center (CTRC) and the University Hospital endoscopy clinic for their assistance with our clinical study. We thank the staff at University of Colorado Hospital CTRC Core Lab for performing the IL-6 and hs-CRP assays and the San Diego CFAR Translational Virology Core for performing viral RNA measurements on colon tissue. We gratefully acknowledge Kevin Rufner for his contributions to clinical study development. Finally, we thank Jennifer Manuzak for assistance with DNA extraction.
This study was supported by National Institutes of Health Grants 1RO1 DK088663, AI36214 and, in part, by NIH/NCATS Colorado CTSI Grant Number UL1 TR000154.
Footnotes
DISCLOSURE: The authors declare no conflict of interest.
REFERENCES
- 1.Kotler DP, Gaetz HP, Lange M, Klein EB, Holt PR. Enteropathy associated with the acquired immunodeficiency syndrome. Annals of internal medicine. 1984;101:421–428. doi: 10.7326/0003-4819-101-4-421. [DOI] [PubMed] [Google Scholar]
- 2.Epple HJ, et al. Acute HIV infection induces mucosal infiltration with CD4+ and CD8+ T cells, epithelial apoptosis, and a mucosal barrier defect. Gastroenterology. 2010;139:1289–1300. doi: 10.1053/j.gastro.2010.06.065. [DOI] [PubMed] [Google Scholar]
- 3.Sankaran S, et al. Rapid onset of intestinal epithelial barrier dysfunction in primary human immunodeficiency virus infection is driven by an imbalance between immune response and mucosal repair and regeneration. Journal of virology. 2008;82:538–545. doi: 10.1128/JVI.01449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenchley JM, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenchley JM, et al. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood. 2008;112:2826–2835. doi: 10.1182/blood-2008-05-159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaschitz C, Raffatellu M. Th17 cytokines and the gut mucosal barrier. J Clin Immunol. 2010;30:196–203. doi: 10.1007/s10875-010-9368-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGowan I, et al. Increased HIV-1 mucosal replication is associated with generalized mucosal cytokine activation. J Acquir Immune Defic Syndr. 2004;37:1228–1236. doi: 10.1097/01.qai.0000131846.12453.29. [DOI] [PubMed] [Google Scholar]
- 8.Brenchley JM, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 9.Jiang W, et al. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated HIV infection. The Journal of infectious diseases. 2009;199:1177–1185. doi: 10.1086/597476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marchetti G, Tincati C, Silvestri G. Microbial translocation in the pathogenesis of HIV infection and AIDS. Clin Microbiol Rev. 2013;26:2–18. doi: 10.1128/CMR.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marchetti G, et al. Microbial translocation predicts disease progression of HIV-infected antiretroviral-naive patients with high CD4+ cell count. AIDS. 2011;25:1385–1394. doi: 10.1097/QAD.0b013e3283471d10. [DOI] [PubMed] [Google Scholar]
- 12.Frank DN, Zhu W, Sartor RB, Li E. Investigating the biological and clinical significance of human dysbioses. Trends in microbiology. 2011;19:427–434. doi: 10.1016/j.tim.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown K, DeCoffe D, Molcan E, Gibson DL. Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients. 2012;4:1095–1119. doi: 10.3390/nu4081095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathis D, Benoist C. The influence of the microbiota on type-1 diabetes: on the threshold of a leap forward in our understanding. Immunol Rev. 2012;245:239–249. doi: 10.1111/j.1600-065X.2011.01084.x. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, et al. Metagenomic sequencing reveals microbiota and its functional potential associated with periodontal disease. Sci Rep. 2013;3:1843. doi: 10.1038/srep01843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koeth RA, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang WH, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. The New England journal of medicine. 2013;368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gori A, et al. Early impairment of gut function and gut flora supporting a role for alteration of gastrointestinal mucosa in human immunodeficiency virus pathogenesis. Journal of clinical microbiology. 2008;46:757–758. doi: 10.1128/JCM.01729-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellis CL, et al. Molecular characterization of stool microbiota in HIV-infected subjects by panbacterial and order-level 16S ribosomal DNA (rDNA) quantification and correlations with immune activation. J Acquir Immune Defic Syndr. 2011;57:363–370. doi: 10.1097/QAI.0b013e31821a603c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lozupone CA, et al. Alterations in the Gut Microbiota Associated with HIV-1 Infection. Cell host & microbe. 2013;14:329–339. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez-Santiago J, et al. Gut lactobacillales are associated with higher CD4 and less microbial translocation during HIV infection. AIDS. 2013;27 doi: 10.1097/qad.0b013e3283611816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vujkovic-Cvijin I, et al. Dysbiosis of the Gut Microbiota Is Associated with HIV Disease Progression and Tryptophan Catabolism. Science translational medicine. 2013;5:193ra191. doi: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol. 2010;26:5–11. doi: 10.1097/MOG.0b013e328333d751. [DOI] [PubMed] [Google Scholar]
- 26.Brenchley JM, Douek DC. The mucosal barrier and immune activation in HIV pathogenesis. Curr Opin HIV AIDS. 2008;3:356–361. doi: 10.1097/COH.0b013e3282f9ae9c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zoetendal EG, et al. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Applied and environmental microbiology. 2002;68:3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McHardy IH, et al. HIV infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome. 2013;1:26. doi: 10.1186/2049-2618-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunt PW. Role of immune activation in HIV pathogenesis. Curr HIV/AIDS Rep. 2007;4:42–47. doi: 10.1007/s11904-007-0007-8. [DOI] [PubMed] [Google Scholar]
- 30.Beaurepaire C, Smyth D, McKay DM. Interferon-gamma regulation of intestinal epithelial permeability. J Interferon Cytokine Res. 2009;29:133–144. doi: 10.1089/jir.2008.0057. [DOI] [PubMed] [Google Scholar]
- 31.Kumar PS, et al. New bacterial species associated with chronic periodontitis. J Dent Res. 2003;82:338–344. doi: 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- 32.Lucke K, Miehlke S, Jacobs E, Schuppler M. Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J Med Microbiol. 2006;55:617–624. doi: 10.1099/jmm.0.46198-0. [DOI] [PubMed] [Google Scholar]
- 33.Boccara F, et al. HIV and coronary heart disease: time for a better understanding. J Am Coll Cardiol. 2013;61:511–523. doi: 10.1016/j.jacc.2012.06.063. [DOI] [PubMed] [Google Scholar]
- 34.Wright DP, Rosendale DI, Robertson AM. Prevotella enzymes involved in mucin oligosaccharide degradation and evidence for a small operon of genes expressed during growth on mucin. FEMS Microbiol Lett. 2000;190:73–79. doi: 10.1111/j.1574-6968.2000.tb09265.x. [DOI] [PubMed] [Google Scholar]
- 35.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nature reviews. Immunology. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamer HM, et al. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 37.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate- producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 38.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dillon SM, et al. HIV-1 infection of human intestinal lamina propria CD4+ T cells in vitro is enhanced by exposure to commensal Escherichia coli. J Immunol. 2012;189:885–896. doi: 10.4049/jimmunol.1200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manuzak J, Dillon S, Wilson C. Differential interleukin-10 (IL-10) and IL-23 production by human blood monocytes and dendritic cells in response to commensal enteric bacteria. Clin Vaccine Immunol. 2012;19:1207–1217. doi: 10.1128/CVI.00282-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dillon SM, et al. Human intestinal lamina propria CD1c+ dendritic cells display an activated phenotype at steady state and produce IL-23 in response to TLR7/8 stimulation. J Immunol. 2010;184:6612–6621. doi: 10.4049/jimmunol.1000041. [DOI] [PubMed] [Google Scholar]
- 42.Markle JG, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 43.Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148:257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- 44.Lane DJ. 16S23S rRNA sequencing. Wiley; 1991. pp. 115–175. [Google Scholar]
- 45.Frank DN. BARCRAWL and BARTAB: software tools for the design and implementation of barcoded primers for highly multiplexed DNA sequencing. BMC bioinformatics. 2009;10:362. doi: 10.1186/1471-2105-10-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robertson CE, et al. Explicet: Graphical user interface software for metadata-driven management, analysis, and visualization of microbiome data. Bioinformatics. 2013 doi: 10.1093/bioinformatics/btt526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siewe B, Keshavarzian A, French A, Demarais P, Landay A. A Role for TLR Signaling During B-Cell Activation in Antiretroviral (ART) Treated HIV Individuals. AIDS Res Hum Retroviruses. 2013 doi: 10.1089/aid.2013.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oksanen J, et al. The Vegan Package: Community Ecology Package Version 1.15-1. 2008 < http://cran.r-project.org/, http://vegan.r-forge.r-project.org.>.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.