Abstract
Using a mutant pyrrolysyl-tRNA synthetase- pair, 3-formyl-phenylalanine is genetically incorporated into proteins at amber mutation sites in Escherichia coli. This non-canonical amino acid readily reacts with hydroxylamine dyes, leading to rapid and site-selective protein labelling.
Although the aldehyde is one of the most versatile chemical functionalities and participates in a variety of useful chemical reactions,1 it does not occur in the 20 canonical amino acids. To harness the unique reactivity of aldehydes with hydrazine and hydroxylamine dyes for protein labelling, aldehyde tags have been introduced into proteins by oxidizing an N-terminal serine or via enzymatic modifications of preinstalled substrate peptides.2 The N-terminal oxidation approach can only be applied in vitro and the enzymatic approach is generally confined for the aldehyde installation at the two protein termini. To genetically encode the keto functionality at any chosen sites in a protein, several ketone-containing non-canonical amino acids (NCAAs) have been designed and incorporated into proteins in living cells using the amber suppression approach.3 However, a method for the genetic incorporation of an aldehyde-containing NCAA has not been developed. We are interested in a genetically encoded, aldehyde-containing NCAA due to its superior reactivity with respect to ketone-containing NCAAs, and the observation that aniline can selectively enhance the reaction kinetics of the aldehyde-hydroxylamine/hydrazine condensation.4
In our previous studies, we revealed that a mutant pyrrolysyl-tRNA synthetase with mutations as N346A/C348A (PylRS(N346A/C348A)) recognizes a number of phenylalanine derivatives including meta-acetyl-phenylalanine (2 in Figure 1A) and, in coordination with , directs their genetic incorporation at amber mutation sites in Escherichia coli and mammalian cells.5 Given the size similarity between 2 and meta-formyl-phenylalanine (1), and the ability of this mutant enzyme in accepting diverse meta-substituted phenylalanine substrates, we anticipated that this same mutant enzyme would also accept 1 as its substrate. To test this prospect, 1 was readily prepared in a high yield by a three-step synthesis (Figure 1B). Its acceptance as a substrate of PylRS(N346A/C348A) were then examined. An E. coli BL21(DE3) cell that harboured two plasmids, pEVOL-pylT-PylRS(N346A/C348A) and pET-pylT-sfGFP2TAG, was employed for the investigation. Plasmid pEVOL-pylT-PylRS(N346A/C348A) contains genes coding both PylRS(N346A/C348A) and . Plasmid pET-pylT-sfGFP2TAG carries a coding gene and a sequence-optimized superfolder green fluorescent protein (sfGFP) gene with an amber mutation at its S2 position. Growing cells in M9 minimal medium supplemented with 1% glycerol, 1 mM IPTG and 0.2% arabinose, but without 1, afforded a minimal expression level of full-length sfGFP (<1 mg/L). Providing 1 at 2 mM into the medium promoted full-length sfGFP expression (Figure 2A). The expression level is comparable to that for para-propargyloxy-phenylalanine (ProY), and the electrospray ionization mass spectrometry analysis of the purified 1-containing sfGFP displayed a molecular weight that agreed well with its theoretical value (Figure 1B). Moreover, compound 1 is not toxic to E. coli cells, as phenotypes and cell pellets are comparable to control experiments.
Figure 1.
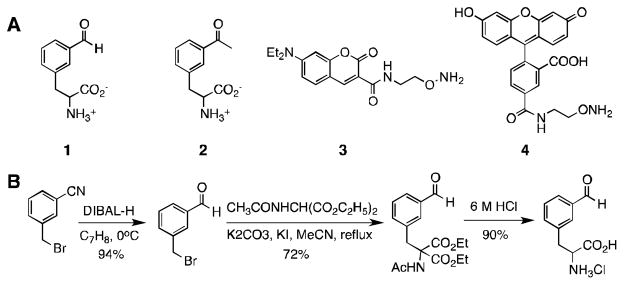
(A) Structures of 1–4. (B) The synthetic route of 1.
Figure 2.
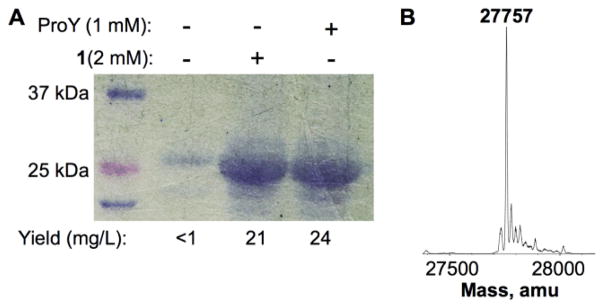
(A) The site-specific incorporation of 1 into sfGFP at its S2 position. Cells transformed with pEVOL-PylRS(N346A/C348A) and pET-pylT-sfGFP2TAG were grown in conditions with or without a NCAA supplement. (B) The deconvoluted ESI-MS spectrum of the purified 1-containing sfGFP. Its theoretical value is 27,756 Da.
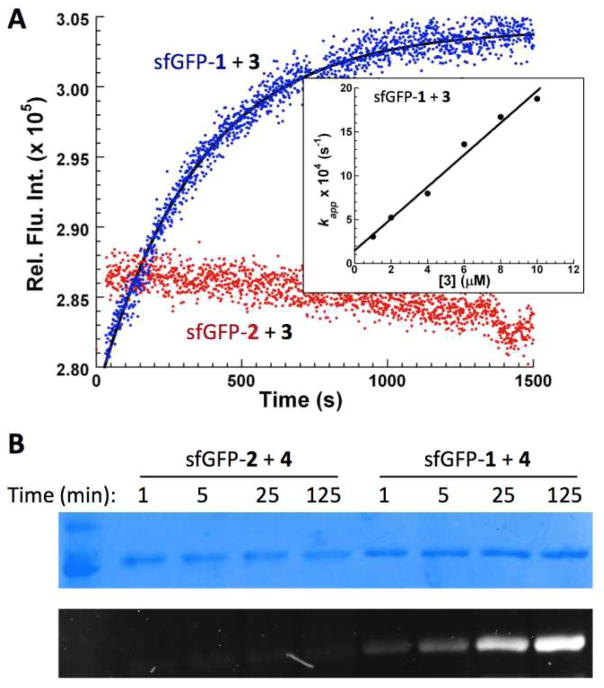
Dawson et al. previously showed that aniline could serve as an excellent catalyst for speeding the imine formation of aldehydes with hydroxylamines and hydrazines. To demonstrate this catalytically rapid reaction on proteins, sfGFP with 1 incorporated at its N149 position (sfGFP-1) was expressed and then applied to a Förster resonance energy transfer (FRET)-based kinetic investigation of its reaction with a coumarin dye 3,3d, 6 in the presence of 100 mM aniline and at pH 7 using a fluorescence spectrophotometer (Figure 1A). 3 has an excitation wavelength at 417 nm and emits at 476 nm. Its emitted light falls in the range of excitation lights of sfGFP that emits at 510 nm.7 The reaction of sfGFP-1 with 3 will place 3 in a close locality of the sfGFP fluorophore for FRET. All kinetic analyses were carried out in pseudo-first-order conditions in which 3 was at least in 5-fold excess in comparison to sfGFP-1. Data collected under these conditions were well fitted to a single exponential increase equation F = F1 − F2 × e(−k′ × t), where F was the detected fluorescent signal at a given time, F1 was the final fluorescence, F1−F2 was the background fluorescent signal, and k′ was the apparent pseudo first-order rate constant (Figure 3A). The determined k′ values were plotted against the concentrations of 3 and fitted to the equation k′ = k × [3] + C, where k was a second-order rate constant for the reaction of sfGFP-1 with 3 (inset of Figure 3A). The calculated second-order rate constant was 182±12 M−1s−1. This determined rate constant is comparable to what Dawson et al. reported for aniline-catalyzed bioconjugation, and is among one of the most rapid bioorthogonal reactions.4, 8 We also carried out the kinetic analysis in the absence of aniline. However, no obvious signal changes were observed when 0.1 μM sfGFP-1 reacted with 4 μM 3 in a period of 2 h, indicating very slow kinetics of the sfGFP-1 reaction with 3 in the absence of aniline and at pH 7. We also tried much higher concentrations of 3 to speed up the reaction rate. However, the background fluorescence from 3 in these conditions was too high to collect reliable data. As a comparison, a sfGFP variant with 2 incorporated at its N149 position (sfGFP-2) was also expressed and used for kinetic investigation of its reaction with 3 in the presence of 100 mM aniline and at pH 7. As shown in Figure 3A, at the tested conditions of 0.05 μM sfGFP-2 and 10 μM 3, no obvious signal increase due to the sfGFP-2 reaction with 3 was observed in a range of 30 min. The fluorescence decrease was caused by the bleaching effect of the excitation light.
Figure 3.
(A) Fluorescent intensities as a function of time for reactions of 0.05 μM sfGFP-1 (coloured in blue) and 0.05 μM sfGFP-2 (coloured in red) with 10 μM 3 in the presence of 100 mM aniline and at pH 7. Presented in the inset is the linear dependence of the determined pseudo first-order rate constants for the reaction of 0.1 μM sfGFP-1 with 3 on the concentrations of 3. (B) Gel imaging analysis of labelling of sfGFP-1 and sfGFP-2 with 4 with different incubation times. For both sfGFP-1 and sfGFP-2, labelling was carried out in conditions with 8 μM protein, 10 μM 4, 100 mM aniline, and pH 7. Reactions were quenched with the addition of 2 mM benzaldehyde at indicated times and analyzed by denaturing SDS-PAGE. The top panel shows proteins with Coomassie blue staining and the bottom panel presents the fluorescent imaging of the same gel before Coomassie blue staining.
The different labelling kinetics of sfGFP-1 and sfGFP-2 were also examined by fluorescent imaging of SDS-PAGE analyzed 4-labelled products. Both sfGFP-1 and sfGFP-2 at 8 μM were incubated with 10 μM 4 in the presence of 100 mM aniline and at pH 7. Reactions were quenched at different times and analyzed by denaturing SDS-PAGE to remove the intrinsic fluorescence of sfGFP. Fluorescent imaging of two proteins at different labelling times clearly showed that sfGFP-1 was efficiently labelled at 25 min but sfGFP-2 was minimally labelled even at 2 h. Labelling could also be achieved in the absence of aniline, though reaction times were significantly longer (Figure S2). Once again, sfGFP-1 showed superior labelling with respect to sfGFP-2. This result shows that, in systems where aniline may be toxic, labelling can still be achieved without it or at much lower concentrations.
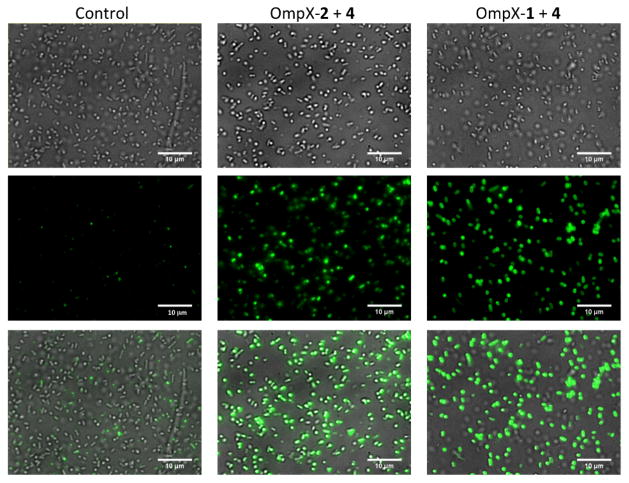
With the demonstration of fast labelling kinetics of 1 with hydroxylamine dyes, we then proceeded to test the conditions for labelling proteins that bear site-specifically incorporated 1 in living cells. Plasmid pEVOL-pylT-PylRS(N346A/C348A) and a previously constructed plasmid, pETDuet-OmpXTAG, were used to transform E. coli BL21 cells. The latter plasmid contained a gene coding for an E. coli outer membrane protein OmpX,8b with an AAAXAA (A denotes alanine and X denotes an amber mutation) insertion between two extracellular residues, 53 and 54. The transformed cells were grown in M9 minimal medium supplemented with 1% glycerol, 1 mM IPTG, 0.2% arabinose, and 2 mM 1 or 2 to express OmpX with 1 or 2 incorporated (OmpX-1 or OmpX-2). Cells were then harvested and labelled with 4 in the presence of 100 mM aniline and at pH 7 for 1 h. After labelling, cells were washed with PBS buffer for six times to remove the residual dye and then imaged by fluorescent microscopy. Cells grown in the absence of 1 or 2 were used as control. As shown in Figure 4, 4 specifically labelled cells expressing OmpX-1 and OmpX-2 were observed, but not for cells in the control experiment. Although cells expressing OmpX-2 were fluorescently labelled, their intensities are weaker than those of cells expressing OmpX-1, indicating a slower labelling kinetics of OmpX-2. However, we have found that this concentration of aniline is toxic to E. coli, which underscores the need for developing less toxic catalysts for this reaction.
Figure 4.
Selective labelling of E. coli cells expressing OmpX-1 and OmpX-2. The labelling was carried out in the presence of 100 mM aniline and at pH 7 for 1 h. The top panel shows bright field imaging of E. coli cells, the middle panel shows green fluorescent imaging of same cells, and the bottom panel shows their composite images (scale bars are 10 μm).
In summary, we reported the genetic incorporation of a readily synthesized aldehyde-containing NCAA and demonstrated its fast labelling kinetics with hydroxylamine dyes in the presence of the aniline catalyst. This rapid labelling approach was also successfully applied to label a membrane protein on the E. coli extracellular surface. Although genetically encoded ketone-containing NCAAs were reported previously, labelling of proteins with these NCAAs typically suffers low labelling efficiencies, attributing to slow labelling kinetics of ketones. This work resolves this obstacle. Given that a large variety of hydroxylamine and hydrazine dyes are commercially available, as well as the existence of numerous aldehyde-based bioconjugation strategies,9 we anticipate this approach will be quickly adopted by others for studies such as protein folding/dynamics, protein-ligand interactions, high throughput drug discovery, etc.
Supplementary Material
Acknowledgments
We thank the financial support from the National Institute of Health (grant 1R01CA161158), the National Science Foundation (grant CHE-1148684), and the Welch Foundation (grant A-1715). We also thank Dr. Yohannes H. Rezenom from the Laboratory for Biological Mass Spectrometry at Texas A&M University for characterizing our proteins with electrospray ionization mass spectrometry.
Footnotes
Electronic Supplementary Information (ESI) available: Synthesis, protein expression, protein labelling, and mass spectrometry analysis. See DOI: 10.1039/c000000x/
Notes and references
- 1.(a) McFarland JM, Francis MB. J Am Chem Soc. 2005;127:13490. doi: 10.1021/ja054686c. [DOI] [PubMed] [Google Scholar]; (b) McFarland JM, Joshi NS, Francis MB. J Am Chem Soc. 2008;130:7639. doi: 10.1021/ja710927q. [DOI] [PubMed] [Google Scholar]; (c) Han MJ, Xiong DC, Ye XS. Chem Commun (Camb) 2012;48:11079. doi: 10.1039/c2cc35738k. [DOI] [PubMed] [Google Scholar]; (d) Agarwal P, Kudirka R, Albers AE, Barfield RM, de Hart GW, Drake PM, Jones LC, Rabuka D. Bioconjug Chem. 2013;24:846. doi: 10.1021/bc400042a. [DOI] [PubMed] [Google Scholar]; (e) Agarwal P, van der Weijden J, Sletten EM, Rabuka D, Bertozzi CR. Proc Natl Acad Sci U S A. 2013;110:46. doi: 10.1073/pnas.1213186110. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Witus LS, Netirojjanakul C, Palla KS, Muehl EM, Weng CH, Iavarone AT, Francis MB. J Am Chem Soc. 2013;135:17223. doi: 10.1021/ja408868a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Geschwind II, Hao Li C. Biochimica et Biophysica Acta. 1954;15:442. doi: 10.1016/0006-3002(54)90053-0. [DOI] [PubMed] [Google Scholar]; (b) Carrico IS, Carlson BL, Bertozzi CR. Nat Chem Biol. 2007;3:321. doi: 10.1038/nchembio878. [DOI] [PubMed] [Google Scholar]; (c) Rush JS, Bertozzi CR. J Am Chem Soc. 2008;130:12240. doi: 10.1021/ja804530w. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Wu P, Shui W, Carlson BL, Hu N, Rabuka D, Lee J, Bertozzi CR. Proc Natl Acad Sci U S A. 2009;106:3000. doi: 10.1073/pnas.0807820106. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Hudak JE, Yu HH, Bertozzi CR. J Am Chem Soc. 2011;133:16127. doi: 10.1021/ja206023e. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) El-Mahdi O, Melnyk O. Bioconjug Chem. 2013;24:735. doi: 10.1021/bc300516f. [DOI] [PubMed] [Google Scholar]
- 3.(a) Wang L, Zhang Z, Brock A, Schultz PG. Proc Natl Acad Sci U S A. 2003;100:56. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhang Z, Smith BA, Wang L, Brock A, Cho C, Schultz PG. Biochemistry. 2003;42:6735. doi: 10.1021/bi0300231. [DOI] [PubMed] [Google Scholar]; (c) Huang Y, Wan W, Russell WK, Pai PJ, Wang Z, Russell DH, Liu W. Bioorg Med Chem Lett. 2010;20:878. doi: 10.1016/j.bmcl.2009.12.077. [DOI] [PubMed] [Google Scholar]; (d) Wu B, Wang Z, Huang Y, Liu WR. Chembiochem. 2012;13:1405. doi: 10.1002/cbic.201200281. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Zeng H, Xie J, Schultz PG. Bioorg Med Chem Lett. 2006;16:5356. doi: 10.1016/j.bmcl.2006.07.094. [DOI] [PubMed] [Google Scholar]
- 4.(a) Dirksen A, Hackeng TM, Dawson PE. Angew Chem Int Ed Engl. 2006;45:7581. doi: 10.1002/anie.200602877. [DOI] [PubMed] [Google Scholar]; (b) Dirksen A, Dawson PE. Bioconjug Chem. 2008;19:2543. doi: 10.1021/bc800310p. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Zeng Y, Ramya TN, Dirksen A, Dawson PE, Paulson JC. Nat Methods. 2009;6:207. doi: 10.1038/nmeth.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Wendeler M, Grinberg L, Wang X, Dawson PE, Baca M. Bioconjug Chem. 2014;25:93. doi: 10.1021/bc400380f. [DOI] [PubMed] [Google Scholar]
- 5.(a) Wang YS, Russell WK, Wang Z, Wan W, Dodd LE, Pai PJ, Russell DH, Liu WR. Mol Biosyst. 2011;7:714. doi: 10.1039/c0mb00217h. [DOI] [PubMed] [Google Scholar]; (b) Wang YS, Fang X, Wallace AL, Wu B, Liu WR. J Am Chem Soc. 2012;134:2950. doi: 10.1021/ja211972x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Wang YS, Fang X, Chen HY, Wu B, Wang ZU, Hilty C, Liu WR. ACS Chem Biol. 2013;8:405. doi: 10.1021/cb300512r. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Tharp JM, Wang YS, Lee YJ, Yang Y, Liu WR. ACS Chem Biol. 2014 doi: 10.1021/cb400917a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Tuley A, Wang YS, Fang X, Kurra Y, Rezenom YH, Liu WR. Chem Commun (Camb) 2014;50:2673. doi: 10.1039/c3cc49068h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddington SC, Tippmann EM, Jones DD. Chem Commun (Camb) 2012;48:8419. doi: 10.1039/c2cc31887c. [DOI] [PubMed] [Google Scholar]
- 7.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Nat Biotechnol. 2006;24:79. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 8.(a) Wang XS, Lee YJ, Liu WR. Chem Commun (Camb) 2014;50:3176. doi: 10.1039/c3cc48682f. [DOI] [PubMed] [Google Scholar]; (b) Lee YJ, Wu B, Raymond JE, Zeng Y, Fang X, Wooley KL, Liu WR. ACS Chem Biol. 2013;8:1664. doi: 10.1021/cb400267m. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Lang K, Davis L, Wallace S, Mahesh M, Cox DJ, Blackman ML, Fox JM, Chin JW. J Am Chem Soc. 2012;134:10317. doi: 10.1021/ja302832g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Borrmann A, Milles S, Plass T, Dommerholt J, Verkade JM, Wiessler M, Schultz C, van Hest JC, van Delft FL, Lemke EA. Chembiochem. 2012;13:2094. doi: 10.1002/cbic.201200407. [DOI] [PubMed] [Google Scholar]; (e) Plass T, Milles S, Koehler C, Szymanski J, Mueller R, Wiessler M, Schultz C, Lemke EA. Angew Chem Int Ed Engl. 2012;51:4166. doi: 10.1002/anie.201108231. [DOI] [PubMed] [Google Scholar]; (f) Blackman ML, Royzen M, Fox JM. J Am Chem Soc. 2008;130:13518. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Lorello GR, Legault MC, Rakic B, Bisgaard K, Pezacki JP. Bioorg Chem. 2008;36:105. doi: 10.1016/j.bioorg.2007.12.006. [DOI] [PubMed] [Google Scholar]; (b) Byeon JY, Limpoco FT, Bailey RC. Langmuir. 2010;26:15430. doi: 10.1021/la1021824. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ossipov DA, Yang X, Varghese O, Kootala S, Hilborn J. Chem Commun (Camb) 2010;46:8368. doi: 10.1039/c0cc03055d. [DOI] [PubMed] [Google Scholar]; (d) Raindlova V, Pohl R, Sanda M, Hocek M. Angew Chem Int Ed Engl. 2010;49:1064. doi: 10.1002/anie.200905556. [DOI] [PubMed] [Google Scholar]; (e) Rotstein BH, Rai V, Hili R, Yudin AK. Nat Protoc. 2010;5:1813. doi: 10.1038/nprot.2010.127. [DOI] [PubMed] [Google Scholar]; (f) Iyer G, Pinaud F, Xu J, Ebenstein Y, Li J, Chang J, Dahan M, Weiss S. Bioconjug Chem. 2011;22:1006. doi: 10.1021/bc100593m. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Cohen JD, Zou P, Ting AY. Chembiochem. 2012;13:888. doi: 10.1002/cbic.201100764. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Crisalli P, Hernandez AR, Kool ET. Bioconjug Chem. 2012;23:1969. doi: 10.1021/bc300344b. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Raindlova V, Pohl R, Hocek M. Chemistry. 2012;18:4080. doi: 10.1002/chem.201103270. [DOI] [PubMed] [Google Scholar]; (j) Dhal PK, Polomoscanik SC, Gianolio DA, Starremans PG, Busch M, Alving K, Chen B, Miller RJ. Bioconjug Chem. 2013;24:865. doi: 10.1021/bc300472e. [DOI] [PubMed] [Google Scholar]; (k) Ossipov D, Kootala S, Yi Z, Yang X, Hilborn J. Macromolecules. 2013;46:4105. [Google Scholar]; (l) Tanaka K, Nakamoto Y, Siwu ER, Pradipta AR, Morimoto K, Fujiwara T, Yoshida S, Hosoya T, Tamura Y, Hirai G, Sodeoka M, Fukase K. Org Biomol Chem. 2013;11:7326. doi: 10.1039/c3ob41507d. [DOI] [PubMed] [Google Scholar]; (m) Wang S, Oommen OP, Yan H, Varghese OP. Biomacromolecules. 2013;14:2427. doi: 10.1021/bm400612h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.