Abstract
Ensifer sp. TW10 is a novel N2-fixing bacterium isolated from a root nodule of the perennial legume Tephrosia wallichii Graham (known locally as Biyani) found in the Great Indian (or Thar) desert, a large arid region in the northwestern part of the Indian subcontinent. Strain TW10 is a Gram-negative, rod shaped, aerobic, motile, non-spore forming, species of root nodule bacteria (RNB) that promiscuously nodulates legumes in Thar Desert alkaline soil. It is fast growing, acid-producing, and tolerates up to 2% NaCl and capable of growth at 40oC. In this report we describe for the first time the primary features of this Thar Desert soil saprophyte together with genome sequence information and annotation. The 6,802,256 bp genome has a GC content of 62% and is arranged into 57 scaffolds containing 6,470 protein-coding genes, 73 RNA genes and a single rRNA operon. This genome is one of 100 RNB genomes sequenced as part of the DOE Joint Genome Institute 2010 Genomic Encyclopedia for Bacteria and Archaea-Root Nodule Bacteria (GEBA-RNB) project.
Keywords: root-nodule bacteria, nitrogen fixation, rhizobia, Alphaproteobacteria
Introduction
The Great Indian (or Thar) Desert is a large, hot, arid region in the northwestern part of the Indian subcontinent. It is the 18th largest desert in the world covering 200,000 square km with 61% of its landmass occupying Western Rajasthan. The landscape occurs at low altitude (<1500 m above sea level) and extends from India into the neighboring country of Pakistan [1]. The Thar Desert region is characterized by low annual precipitation (50 to 300 mm), high thermal load and alkaline soils that are poor in texture and fertility [2]. Despite these harsh conditions, the Thar Desert has very rich plant diversity in comparison to other desert landscapes [3]. Approximately a quarter of the plants in the Thar Desert are used to provide animal fodder or food, fuel, medicine or shelter for local inhabitants [4].
The Indian Thar desert harbors several native and exotic plants of the Leguminoseae family [2] including native legume members of the sub-families Caesalpinioideae, Mimosoideae and Papilionoideae that have adapted to the harsh Thar desert environment [5]. The Papilionoid genus Tephrosia can be found throughout this semi-arid to arid environment and these plants are among the first to grow after monsoonal rains. The generic name is derived from the Greek word “tephros” meaning “ash-gray” since dense trichomes on the leaves provide a greyish tint to the plant. Many species within this genus produce the potent toxin rotenone, which historically has been used to poison fish. It is a perennial shrub that has adapted to the harsh desert conditions by producing a long tap root system and dormant auxillary shoot buds.
Recently, the root nodule bacteria (RNB) microsymbionts capable of fixing nitrogen in symbiotic associations with Tephrosia have been characterized [5]. Both Bradyrhizobium and Ensifer were present within nodules, but a particularly high incidence of Ensifer was noted [5]. Ensifer was found to occupy the nodules of all four species of Tephrosia examined [5]. Here we present a preliminary description of the general features of the T. wallichii (Biyani) microsymbiont Ensifer sp. TW10 together with its genome sequence and annotation.
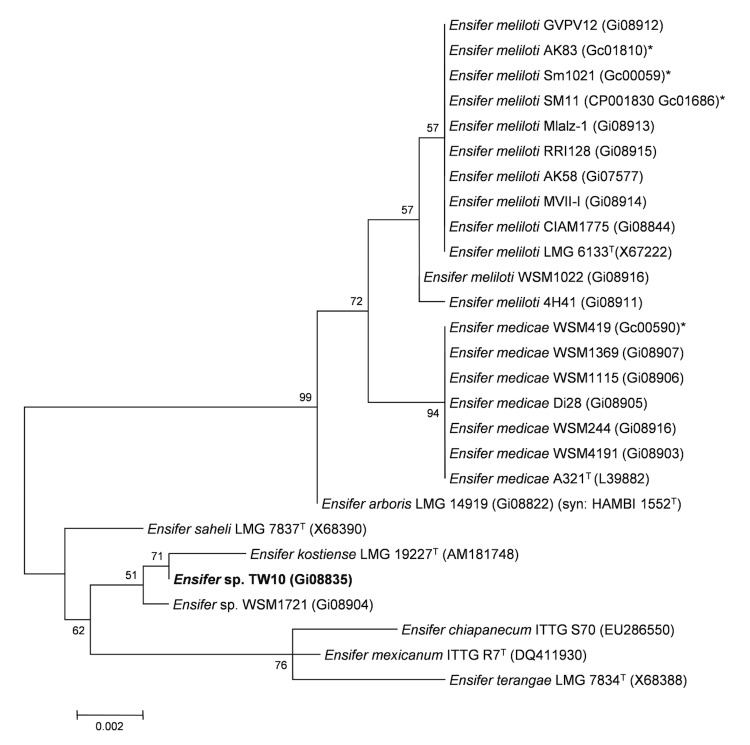
Minimum Information about the Genome Sequence (MIGS) is provided in Table 1. Figure 1 shows the phylogenetic neighborhood of Ensifer sp. strain TW10 in a 16S rRNA sequence based tree. This strain has 99% sequence identity at the 16S rRNA sequence level to E. kostiense LMG 19227 and 100% 16S rRNA sequence identity to other Indian Thar Desert Ensifer species (JNVU IC18 from a nodule of Indigofera and JNVU TF7, JNVU TP6 and TW8 from nodules of Tephrosia).
Table 1. Classification and general features of Ensifer sp. TW10 according to the MIGS recommendations [6].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [7] | |
| Phylum Proteobacteria | TAS [8] | ||
| Class Alphaproteobacteria | TAS [9,10] | ||
| Order Rhizobiales | TAS [10,11] | ||
| Family Rhizobiaceae | TAS [12,13] | ||
| Genus Ensifer | TAS [14-16] | ||
| Species Ensifer sp. | IDA | ||
| Gram stain | Negative | IDA | |
| Cell shape | Rod | IDA | |
| Motility | Motile | IDA | |
| Sporulation | Non-sporulating | NAS | |
| Temperature range | Mesophile | NAS | |
| Optimum temperature | 28°C | NAS | |
| Salinity | Non-halophile | NAS | |
| MIGS-22 | Oxygen requirement | Aerobic | TAS [5] |
| Carbon source | Varied | NAS | |
| Energy source | Chemoorganotroph | NAS | |
| MIGS-6 | Habitat | Soil, root nodule, on host | TAS [5] |
| MIGS-15 | Biotic relationship | Free living, symbiotic | TAS [5] |
| MIGS-14 | Pathogenicity | Non-pathogenic | NAS |
| Biosafety level | 1 | TAS [17] | |
| Isolation | Root nodule of Tephrosia wallichii | TAS [5] | |
| MIGS-4 | Geographic location | Jodhpur, Indian Thar Desert | TAS [5] |
| MIGS-5 | Soil collection date | Oct, 2009 | IDA |
| MIGS-4.1 | Longitude | 73.021177 | IDA |
| MIGS-4.2 | Latitude | 26.27061 | IDA |
| MIGS-4.3 | Depth | 15cm | |
| MIGS-4.4 | Altitude | Not recorded |
Evidence codes – IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [18].
Figure 1.
Phylogenetic tree showing the relationship of Ensifer sp. TW10 (shown in bold print) to other Ensifer spp. in the order Rhizobiales based on aligned sequences of the 16S rRNA gene (1,290 bp internal region). All sites were informative and there were no gap-containing sites. Phylogenetic analyses were performed using MEGA, version 5 [19]. The tree was built using the Maximum-Likelihood method with the General Time Reversible model [20]. Bootstrap analysis [21] with 500 replicates was performed to assess the support of the clusters. Type strains are indicated with a superscript T. Brackets after the strain name contain a DNA database accession number and/or a GOLD ID (beginning with the prefix G) for a sequencing project registered in GOLD [22]. Published genomes are indicated with an asterisk.
Classification and general features
Ensifer sp. strain TW10 is a Gram-negative rod (Figure 2, and Figure 3) in the order Rhizobiales of the class Alphaproteobacteria. It is fast growing, forming white-opaque, slightly domed and moderately mucoid colonies with smooth margins within 3-4 days at 28°C when grown on YMA [23].
Figure 2.
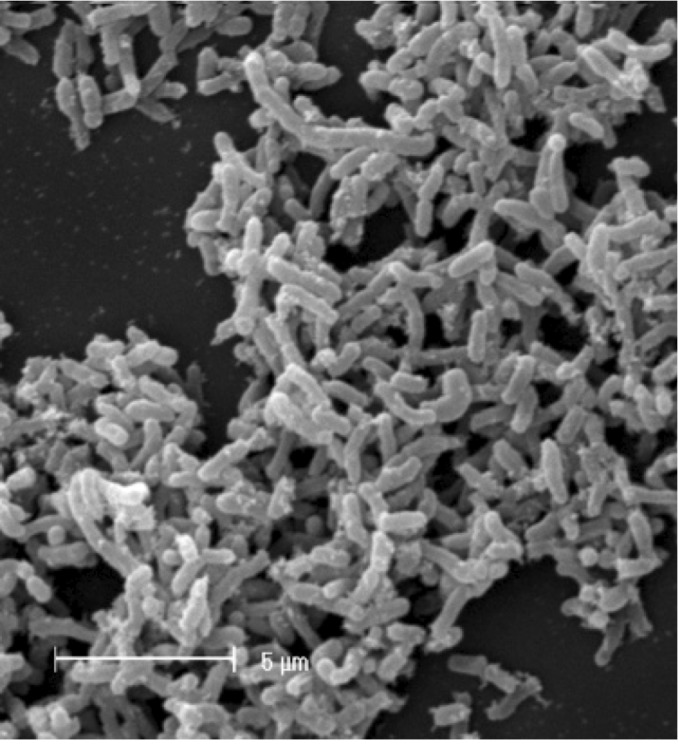
Image of Ensifer sp. TW10 using scanning electron microscopy.
Figure 3.

Image of Ensifer sp. TW10 using transmission electron microscopy.
Symbiotaxonomy
Ensifer sp. TW10 has the ability to nodulate (Nod+) and fix nitrogen (Fix+) effectively with a wide range of perennial native (wild) legumes of Thar Desert origin and with species of crop legumes (Table 2). Ensifer sp. TW10 is symbiotically competent with these species when grown in alkaline soils. TW10 can nodulate the wild tree legume Prosopis cineraria of the Mimosoideae subfamily. However, it does not form nodules on the Mimosoid hosts Mimosa hamata and M. himalayana even though these hosts are known to be nodulated by Ensifer species [5,24]. TW10 was not compatible with the host Phaseolus vulgaris, a legume of the Phaseolae tribe.
Table 2. Compatibility of Ensifer sp. TW10 with different wild and cultivated legume species.
| Species Name | Family | Wild/ Cultivar | Common Name | Habit/ Growth Type | Nod | Fix |
|---|---|---|---|---|---|---|
| Tephrosia falciformis Ramaswami | Papilionoideae | Wild | Rati biyani | Under-shrub Perennial | + | + |
|
Tephrosia purpurea (L.) Pers. sub sp. leptostachya DC. |
Papilionoideae | Wild | - | Herb Annual/ Perennial | + | + |
|
Tephrosia purpurea (L.) Pers. sub sp. purpurea (L.) Pers |
Papilionoideae | Wild | Biyani, Sarphanko | Herb Annual/ Perennial | + | + |
|
Tephrosia villosa (Linn.) Pres. |
Papilionoideae | Wild | Ruvali-biyani | Herb Annual/ Perennial | + | + |
|
Prosopis cineraria (Linn.) Druce. |
Mimosoideae | Wild/ Cultivar |
Khejari | Tree Perennial | + | + |
| Mimosa hamata Willd. | Mimosoideae | Wild | Jinjani, Jinjanio | Shrub Perennial | - | - |
| M. himalayana Gamble | Mimosoideae | Wild | Hajeru | Shrub Perennial | - | - |
|
Vigna radiata (L.) Wilczek |
Papilionoideae | Cultivar | Moong bean | Annual | + | + |
|
Vigna aconitifolia (Jacq.) Marechal |
Papilionoideae | Cultivar | Moth bean | Annual | + | + |
|
Vigna unguiculata (L.) Walp. |
Papilionoideae | Cultivar | Cowpea | Annual | + | + |
|
Macroptilium atropurpureum (DC.) Urb. |
Papilionoideae | Cultivar | Siratro | Annual | + | + |
|
Phaseolus vulgaris L. |
Papilionoideae | Cultivar | Common bean | Annual | - | - |
Nod: “+” means nodulation observed, “-” means no nodulation
Fix: “+” means fixation observed, “-” means no fixation
Genome sequencing and annotation
Genome project history
This organism was selected for sequencing on the basis of its environmental and agricultural relevance to issues in global carbon cycling, alternative energy production, and biogeochemical importance, and is part of the Community Sequencing Program at the U.S. Department of Energy, Joint Genome Institute (JGI) for projects of relevance to agency missions. The genome project is deposited in the Genomes OnLine Database [22] and standard draft genome sequence in IMG. Sequencing, finishing and annotation were performed by the JGI. A summary of the project information is shown in Table 3.
Table 3. Genome sequencing project information for Ensifer sp. strain TW10.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Standard draft |
| MIGS-28 | Libraries used | 1× Illumina library |
| MIGS-29 | Sequencing platforms | Illumina HiSeq2000 |
| MIGS-31.2 | Sequencing coverage | 330× Illumina |
| MIGS-30 | Assemblers | Allpaths, LG version r42328, Velvet 1.1.04 |
| MIGS-32 | Gene calling methods | Prodigal 1.4, |
| GenBank Genbank Date of Release GOLD ID |
pending pending Gi08835 |
|
| NCBI project ID | 210334 | |
| Database: IMG | 2509276019 | |
| Project relevance | Symbiotic N2 fixation, agriculture |
Growth conditions and DNA isolation
Ensifer sp. TW10 was cultured to mid logarithmic phase in 60 ml of TY rich medium [25] on a gyratory shaker at 28°C. DNA was isolated from the cells using a CTAB (Cetyl trimethyl ammonium bromide) bacterial genomic DNA isolation method [26].
Genome sequencing and assembly
The genome of Ensifer sp. TW10 was generated at the Joint Genome Institute (JGI) using Illumina [27] technology. An Illumina std shotgun library was constructed and sequenced using the Illumina HiSeq 2000 platform which generated 14,938,244 reads totaling 2,241 Mbp.
All general aspects of library construction and sequencing performed at the JGI can be found at the JGI website [26]. All raw Illumina sequence data was passed through DUK, a filtering program developed at JGI, which removes known Illumina sequencing and library preparation artifacts (Mingkun L, Copeland, A, and Han, J, unpublished).
The following steps were then performed for assembly: (1) filtered Illumina reads were assembled using Velvet [28] (version 1.1.04), (2) 1–3 kb simulated paired end reads were created from Velvet contigs using wgsim (https://github.com/lh3/wgsim), and (3) Illumina reads were assembled with simulated read pairs using Allpaths–LG (version r42328) [29]. Parameters for assembly steps were: 1) Velvet (velveth: 63 –shortPaired and velvetg: –veryclean yes –exportFiltered yes –mincontiglgth 500 –scaffolding no–covcutoff 10) 2) wgsim (–e 0 –1 100 –2 100 –r 0 –R 0 –X 0) 3) Allpaths–LG (PrepareAllpathsInputs:PHRED64=1 PLOIDY=1 FRAGCOVERAGE=125 JUMPCOVERAGE=25 LONGJUMPCOV=50, RunAllpath-sLG: THREADS=8 RUN=stdshredpairs TARGETS=standard VAPIWARNONLY=True OVERWRITE=True). The final draft assembly contained 57 contigs in 57 scaffolds. The total size of the genome is 6.8 Mbp and the final assembly is based on 2241Mbp of Illumina data, which provides an average 330× coverage of the genome.
Genome annotation
Genes were identified using Prodigal [30] as part of the DOE-JGI annotation pipeline [31]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) non-redundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. The tRNAScanSE tool [7] was used to find tRNA genes, whereas ribosomal RNA genes were found by searches against models of the ribosomal RNA genes built from SILVA [32]. Other non–coding RNAs such as the RNA components of the protein secretion complex and the RNase P were identified by searching the genome for the corresponding Rfam profiles using INFERNAL [33]. Additional gene prediction analysis and manual functional annotation was performed within the Integrated Microbial Genomes (IMG) platform) [34,35].
Genome properties
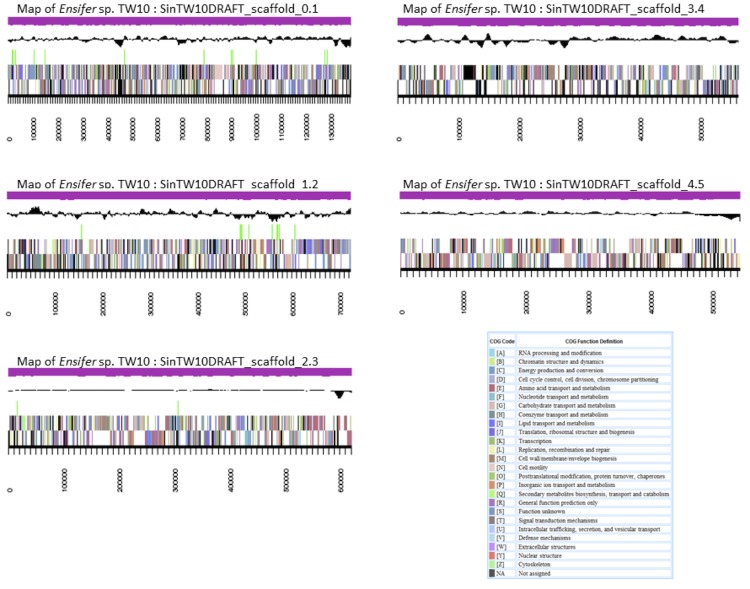
The genome is 6,802,256 nucleotides with 61.56% GC content (Table 4) and comprised of 57 scaffolds (Figure 4) of 57 contigs. From a total of 6,546 genes, 6,473 were protein encoding and 73 RNA only encoding genes. The majority of genes (77.44%) were assigned a putative function while the remaining genes were annotated as hypothetical. The distribution of genes into COGs functional categories is presented in Table 5.
Table 4. Genome Statistics for Ensifer sp. TW10.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 6,802,256 | 100.00 |
| DNA coding region (bp) | 5,800,968 | 85.28 |
| DNA G+C content (bp) | 4,187,461 | 61.56 |
| Number of scaffolds | 57 | |
| Number of contigs | 57 | |
| Total gene | 6,546 | 100.00 |
| RNA genes | 73 | 1.12 |
| rRNA operons | 1 | |
| Protein-coding genes | 6,473 | 98.88 |
| Genes with function prediction | 5,069 | 77.44 |
| Genes assigned to COGs | 5,069 | 77.44 |
| Genes assigned Pfam domains | 5,282 | 80.69 |
| Genes with signal peptides | 539 | 8.23 |
| Genes with transmembrane helices | 1,419 | 21.68 |
Figure 4.
Graphical map of five of the largest scaffolds from the genome of Ensifer sp. TW10. From bottom to the top of each scaffold: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, sRNAs red, other RNAs black), GC content, GC skew.
Table 5. Number of protein coding genes of Ensifer sp. TW10 associated with the general COG functional categories.
| Code | Value | %age | Description |
|---|---|---|---|
| J | 198 | 3.55 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0.00 | RNA processing and modification |
| K | 481 | 8.61 | Transcription |
| L | 237 | 4.24 | Replication, recombination and repair |
| B | 3 | 0.05 | Chromatin structure and dynamics |
| D | 37 | 0.66 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0.00 | Nuclear structure |
| V | 66 | 1.18 | Defense mechanisms |
| T | 262 | 4.69 | Signal transduction mechanisms |
| M | 298 | 5.34 | Cell wall/membrane biogenesis |
| N | 77 | 1.38 | Cell motility |
| Z | 0 | 0.00 | Cytoskeleton |
| W | 1 | 0.02 | Extracellular structures |
| U | 132 | 2.36 | Intracellular trafficking and secretion |
| O | 192 | 3.44 | Posttranslational modification, protein turnover, chaperones |
| C | 322 | 5.77 | Energy production conversion |
| G | 538 | 9.63 | Carbohydrate transport and metabolism |
| E | 606 | 10.85 | Amino acid transport metabolism |
| F | 96 | 1.72 | Nucleotide transport and metabolism |
| H | 194 | 3.47 | Coenzyme transport and metabolism |
| I | 199 | 3.56 | Lipid transport and metabolism |
| P | 251 | 4.49 | Inorganic ion transport and metabolism |
| Q | 139 | 2.49 | Secondary metabolite biosynthesis, transport and catabolism |
| R | 678 | 12.14 | General function prediction only |
| S | 578 | 10.35 | Function unknown |
| - | 1,477 | 22.56 | Not in COGS |
Acknowledgements
This work was performed under the auspices of the US Department of Energy’s Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396. We gratefully acknowledge funding received from the Murdoch University Strategic Research Fund through the Crop and Plant Research Institute (CaPRI), the GRDC National Rhizobium Program (UMU00032), the Council of Scientific and Industrial Research (CSIR) for a fellowship for Nisha Tak, the Department of Biotechnology (India) for a research grant (BT/PR11461/AGR/21/270/2008) and the Commonwealth of Australia for an Australia India Senior Visiting Fellowship for Ravi Tiwari.
References
- 1.Sprent JI, Gehlot HS. Nodulated legumes in arid and semi-arid environments: are they important? Plant Ecol Divers 2010; 3:211-219 10.1080/17550874.2010.538740 [DOI] [Google Scholar]
- 2.Bhandari MM. Flora of the Indian desert. Jodhpur: MPS Repros; 1990. 435 p. [Google Scholar]
- 3.Mohammed S, Kasera PK, Shukla JK. Unexploited plants of potential medicinal value from the Indian Thar Desert. Natural Product Radiance 2004; 3:69-74 [Google Scholar]
- 4.Sen DN. Non-conventional food and some medicinal plant resources of Indian Desert. In: Purkayashtha RP, editor. Economic plants and microbes: Today and Tomorrow's Printers and Publishers, New Delhi; 1991. p 67-76. [Google Scholar]
- 5.Gehlot HS, Panwar D, Tak N, Tak A, Sankhla IS, Poonar N, Parihar R, Shekhawat NS, Kuma M, Tiwari R, et al. Nodulation of legumes from the Thar Desert of India and molecular characterization of their rhizobia. Plant Soil 2012; 357:227-243 10.1007/s11104-012-1143-5 [DOI] [Google Scholar]
- 6.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen M, Angiuoli SV, et al. Towards a richer description of our complete collection of genomes and metagenomes "Minimum Information about a Genome Sequence " (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1. [Google Scholar]
- 9.Garrity GM, Bell JA, Lilburn T. Class I. Alphaproteobacteria class. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part C, Springer, New York, 2005, p. 1. [Google Scholar]
- 10.Validation List No 107. List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol 2006; 56:1-6 10.1099/ijs.0.64188-0 [DOI] [PubMed] [Google Scholar]
- 11.Kuykendall LD. Order VI. Rhizobiales ord. nov. In: Garrity GM, Brenner DJ, Kreig NR, Staley JT, editors. Bergey's Manual of Systematic Bacteriology. Second ed: New York: Springer - Verlag; 2005. p 324. [Google Scholar]
- 12.Skerman VBD, McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Bacteriol 1980; 30:225-420 10.1099/00207713-30-1-225 [DOI] [PubMed] [Google Scholar]
- 13.Conn HJ. Taxonomic relationships of certain non-sporeforming rods in soil. J Bacteriol 1938; 36:320-321 [Google Scholar]
- 14.Casida LE. Ensifer adhaerens gen. nov., sp. nov.: a bacterial predator of bacteria in soil. Int J Syst Bacteriol 1982; 32:339-345 10.1099/00207713-32-3-339 [DOI] [Google Scholar]
- 15.Young JM. The genus name Ensifer Casida 1982 takes priority over Sinorhizobium Chen et al. 1988, and Sinorhizobium morelense Wang et al. 2002 is a later synonym of Ensifer adhaerens Casida 1982. Is the combination Sinorhizobium adhaerens (Casida 1982) Willems et al. 2003 legitimate? Request for an Opinion. Int J Syst Evol Microbiol 2003; 53:2107-2110 10.1099/ijs.0.02665-0 [DOI] [PubMed] [Google Scholar]
- 16.Judicial Commission of the International Committee on Systematics of Prokaryotes The genus name Sinorhizobium Chen et al. 1988 is a later synonym of Ensifer Casida 1982 and is not conserved over the latter genus name, and the species name ' Sinorhizobium adhaerens' is not validly published. Opinion 84. Int J Syst Evol Microbiol 2008; 58:1973 10.1099/ijs.0.2008/005991-0 [DOI] [PubMed] [Google Scholar]
- 17.Agents B. Technical rules for biological agents. TRBA (http://www.baua.de) :466.
- 18.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol 2011; 28:2731-2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford University Press; 2000. [Google Scholar]
- 21.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985; 39:783-791 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- 22.Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2008; 36:D475-D479 10.1093/nar/gkm884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vincent JM. A manual for the practical study of the root-nodule bacteria. International Biological Programme. UK: Blackwell Scientific Publications, Oxford; 1970. [Google Scholar]
- 24.Gehlot HS, Tak N, Kaushik M, Mitra S, Chen WM, Poweleit N, Panwar D, Poonar N, Parihar R, Tak A, et al. An invasive Mimosa in India does not adopt the symbionts of its native relatives. Ann Bot (Lond) 2013; 112:179-196 10.1093/aob/mct112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reeve WG, Tiwari RP, Worsley PS, Dilworth MJ, Glenn AR, Howieson JG. Constructs for insertional mutagenesis, transcriptional signal localization and gene regulation studies in root nodule and other bacteria. Microbiology 1999; 145:1307-1316 10.1099/13500872-145-6-1307 [DOI] [PubMed] [Google Scholar]
- 26.DOE Joint Genome Institute user home.http://my.jgi.doe.gov/general/index.html
- 27.Bennett S. Solexa Ltd. Pharmacogenomics 2004; 5:433-438 10.1517/14622416.5.4.433 [DOI] [PubMed] [Google Scholar]
- 28.Zerbino DR. Using the Velvet de novo assembler for short-read sequencing technologies. Current Protocols in Bioinformatics 2010;Chapter 11:Unit 11 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gnerre S, MacCallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, Sharpe T, Hall G, Shea TP, Sykes S, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci USA 2011; 108:1513-1518 10.1073/pnas.1017351108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010; 11:119 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mavromatis K, Ivanova NN, Chen IM, Szeto E, Markowitz VM, Kyrpides NC. The DOE-JGI Standard operating procedure for the annotations of microbial genomes. Stand Genomic Sci 2009; 1:63-67 10.4056/sigs.632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pruesse E, Quast C, Knittel K. Fuchs BdM, Ludwig W, Peplies J, Glöckner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 2007; 35:7188-7196 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.INFERNAL http://infernal.janelia.org
- 34.Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271-2278 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]
- 35.DOE Joint Genome Institute (http://img.jgi.doe.gov/er)