Abstract
The Firmicutes represent a major component of the intestinal microflora. The intestinal Firmicutes are a large, diverse group of organisms, many of which are poorly characterized due to their anaerobic growth requirements. Although most Firmicutes are Gram positive, members of the class Negativicutes, including the genus Veillonella, stain Gram negative. Veillonella are among the most abundant organisms of the oral and intestinal microflora of animals and humans, in spite of being strict anaerobes. In this work, the genomes of 24 Negativicutes, including eight Veillonella spp., are compared to 20 other Firmicutes genomes; a further 101 prokaryotic genomes were included, covering 26 phyla. Thus a total of 145 prokaryotic genomes were analyzed by various methods to investigate the apparent conflict of the Veillonella Gram stain and their taxonomic position within the Firmicutes. Comparison of the genome sequences confirms that the Negativicutes are distantly related to Clostridium spp., based on 16S rRNA, complete genomic DNA sequences, and a consensus tree based on conserved proteins. The genus Veillonella is relatively homogeneous: inter-genus pair-wise comparison identifies at least 1,350 shared proteins, although less than half of these are found in any given Clostridium genome. Only 27 proteins are found conserved in all analyzed prokaryote genomes. Veillonella has distinct metabolic properties, and significant similarities to genomes of Proteobacteria are not detected, with the exception of a shared LPS biosynthesis pathway. The clade within the class Negativicutes to which the genus Veillonella belongs exhibits unique properties, most of which are in common with Gram-positives and some with Gram negatives. They are only distantly related to Clostridia, but are even less closely related to Gram-negative species. Though the Negativicutes stain Gram-negative and possess two membranes, the genome and proteome analysis presented here confirm their place within the (mainly) Gram positive phylum of the Firmicutes. Further studies are required to unveil the evolutionary history of the Veillonella and other Negativicutes.
Background
The genus Veillonella, belonging to Negativicutes, consists of anaerobic, non-fermentative, Gram-negative cocci, that are normally observed in pairs or short chains, and are non-sporulating and non-motile [1]. Veillonella spp. are abundant in the human microbiome and are found in the oral, respiratory, intestinal and genitourinary flora of humans and animals; they can make up as much as 10% of the bacterial community initially colonizing the enamel [2] and are found throughout the entire oral cavity [3], especially on the tongue dorsum and in saliva [4]. The importance of Veillonella spp. in human infections is uncertain, and they are generally considered to be of low virulence. Veillonella form biofilms, often with Streptococcus spp., and species of these genera have been found to be more abundant in the oral microflora of people with poor oral health [5]. Studies have shown that during formation of early dental plaque, the fraction of Veillonella spp. changes in mixed-microbial colonies with streptococci [6]. Thus, Veillonella spp. may play a role in caries formation as they utilize the lactic acid produced by the organisms conducive to caries [7]. Veillonella are also among the most common anaerobic species reported from pulmonary samples and are frequently recovered from cystic fibrosis cases [8]. The organisms are also abundant in the human gut flora, where their numbers were found to be higher in children with type I diabetes compared to healthy controls [9]. Currently, 12 species of Veillonella have been characterized [10,11] including V. parvula, V. atypica and V. dispar, which are found in the human oral cavity.
The Negativicutes are the only diderm (literally 'two skins') members of the phylum Firmicutes as they possess an inner and an outer membrane. Their placement within the Firmicutes has been widely accepted, and has been confirmed by 16S rRNA analysis [12]. However, their genomes have not been analyzed in detail to confirm their taxonomic position. This work presents a broad analysis of the Negativicutes with focus on the Veillonella spp. using comparative microbial genomics. A total of 24 genomes from the Negativicutes were compared to 121 genomes covering most of the taxonomic span of sequenced bacterial genomes. We investigated how the Negativicutes genomes compared to other bacterial genomes using three different and complementary approaches: 1) phylogenetic trees to visualize the relative distance of the Negativicutes genomes to other genomes; 2) amino acid composition, nucleotide tetramer frequency and metabolism analysis using 2-D clustering and heatmaps to compare genomes; and 3) proteomic comparison across the Negativicutes genomes.
Materials and Methods
Genome sequences used for analysis
The set of 145 genomes included in this study (24 Negativicutes genomes and 121 other prokaryotic genomes covering 26 phyla) are listed in Table 1.
Table 1. Genomes used in this study.
| Phylum | Name of organism and strain | Strain designation | Type strain? | NCBI Taxon ID | NCBI Project ID |
|---|---|---|---|---|---|
| Acidobacteria | Acidobacterium capsulatum | ATCC 51196 | Yes | 240015 | 28085 |
| Acidobacteria | “Korebacter versatiles” | Ellin 345 | 204669 | 15771 | |
| Acidobacteria | “Solibacter usitatus” | Ellin6076 | 234267 | 12638 | |
| Actinobacteria | Bifidobacterium bifidum | 317B | No | 1681 | 42863 |
| Actinobacteria | Catenulispora acidiphila | ID139908, DSM 44928 | Yes | 479433 | 21085 |
| Actinobacteria | Corynebacterium pseudotuberculosis | C231 | No | 681645 | 40875 |
| Actinobacteria | Segniliparus rugosus | ATCC BAA-974 | Yes | 679197 | 40685 |
| Actinobacteria | Streptomyces bingchenggensis | BCW-1 | Name not validly published | 749414 | 46847 |
| Actinobacteria | Tropheryma whipplei | Twist | Yes | 203267 | 95 |
| Aquificae | Persephonella marina | EX-H1 | Yes | 123214 | 12526 |
| Aquificae | Sulfurihydrogenibium sp. | YO3AOP1 | No type strain available | 436114 | 18889 |
| Aquificae | Thermocrinis albus | HI 11/12, DSM 14484 | Yes | 638303 | 37275 |
| Bacteroidetes | Bacteroides thetaiotaomicron | VPI-5482 | Yes | 226186 | 399 |
| Bacteroidetes | Candidatus Sulcia muelleri | DMIN | 641892 | 37785 | |
| Bacteroidetes | Chitinophaga pinensis | UQM 2034, DSM 2588 | Yes | 485918 | 27951 |
| Bacteroidetes | Paludibacter propionicigenes | WB4, DSM 17365 | Yes | 694427 | 42009 |
| Chlamydiae | Protochlamydia amoebophila | UWE25 | Yes | 264201 | 10700 |
| Chlamydiae | Chlamydia trachomatis | E/Sweden2 | No | 634464 | 43167 |
| Chlamydiae | Chlamydophila pneumoniae | AR39 | No | 115711 | 247 |
| Chlamydiae | Waddlia chondrophila | WSU 86-1044 | Yes | 716544 | 43761 |
| Chlorobi | “Chlorobium chlorochromatii” | CaD3 | Name not validly published | 340177 | 13921 |
| Chlorobi | Chlorobium tepidum | TLS | Yes | 194439 | 302 |
| Chloroflexi | Chloroflexus aggregans | DSM 9485 | Yes | 326427 | 16708 |
| Chloroflexi | Dehalococcoides sp | BAV1 | No | 216389 | 15770 |
| Chloroflexi | Herpetosiphon aurantiacus | ATCC 23779 | Yes | 316274 | 16523 |
| Chloroflexi | Roseiflexus sp. | RS-1 | No type strain available | 357808 | 16190 |
| Cyanobacteria | Anabaena variabilis 3 | ATCC 2941 | No | 240292 | 10642 |
| Cyanobacteria | Cyanothece sp. | PCC 7822 | No | 497965 | 28535 |
| Cyanobacteria | Prochlorococcus marinus | MIT9301 | No | 167546 | 15746 |
| Cyanobacteria | Synechocystis sp. | PCC6803 | No | 1148 | 60 |
| Deferribacteres | Calditerrivibrio nitroreducens | Yu37-1, DSM 19672 | Yes | 768670 | 49523 |
| Deferribacteres | Deferribacter desulfuricans | SSM1, DSM 14783 | Yes | 197162 | 37285 |
| Deferribacteres | Denitrovibrio acetiphilus | N2460, DSM 12809 | Yes | 522772 | 29431 |
| Deinococcus-Thermus | Oceanithermus profundus | 506, DSM 14977 | Yes | 670487 | 40223 |
| Deinococcus-Thermus | Thermus thermophilus | HB8 | Yes | 300852 | 13202 |
| Deinococcus-Thermus | Truepera radiovictrix | RQ-24, DSM 17093 | Yes | 649638 | 38371 |
| Dictyoglomi | Dictyoglomus turgidum | DSM 6724 | Yes | 515635 | 29175 |
| Elusimicrobia | Elusimicrobium minutum | Pei 191 | Yes | 445932 | 19701 |
| Fibrobacteres | Fibrobacter succinogenes | S85 | Yes | 59374 | 32617 |
| Firmicutes | Acetohalobium arabaticum | Z-7288, DSM 5501 | Yes | 574087 | 32769 |
| Firmicutes | Acidaminococcus fermentans | VR4, DSM 20731 | Yes | 591001 | 33685 |
| Firmicutes | Acidaminococcus sp. | D21 | No type strain available | 563191 | 34117 |
| Firmicutes | Alkaliphilus oremlandii | OhILAs | Yes | 350688 | 16083 |
| Firmicutes | Bacillus subtilis subsp. subtilis | 168 | Yes | 224308 | 76 |
| Firmicutes | Clostridium botulinum | F Langeland | No | 441772 | 19519 |
| Firmicutes | Clostridium cellulolyticum | H10 | Yes | 394503 | 17419 |
| Firmicutes | Clostridium difficile | 630 (epidemic type X) | No | 272563 | 78 |
| Firmicutes | “Desulfotomaculum reducens” | MI-1 | Name not validly published | 349161 | 13424 |
| Firmicutes | Dialister invisus | DSM 15470 | Yes | 592028 | 33143 |
| Firmicutes | Dialister micraerophilus | Oral Taxon 843 DSM 19965 | Yes | 888062 | 53029 |
| Firmicutes | Dialister micraerophilus | UPII-345-E | No | 910314 | 59521 |
| Firmicutes | Enterococcus faecalis | V583 | No | 226185 | 70 |
| Firmicutes | Eubacterium cylindroides | T2-87 | No | 717960 | 45917 |
| Firmicutes | Eubacterium rectale | A1-86, DSM 17629 | No | 39491 | 39159 |
| Firmicutes | Exiguobacterium sibiricum | 255-15 | Yes | 262543 | 10649 |
| Firmicutes | Geobacillus kaustophilus | HTA426 | Yes | 235909 | 13233 |
| Firmicutes | Lactococcus lactis | cremoris MG1363 | No | 416870 | 18797 |
| Firmicutes | Lysinibacillus sphaericus | C3-41 | No | 444177 | 19619 |
| Firmicutes | Megamonas hypermegale | ART12/1 | No | 158847 | 39163 |
| Firmicutes | Megasphaera genomo sp. | type 1 28L | No type strain available | 699218 | 42553 |
| Firmicutes | Megasphaera micronuciformis | F0359 | No | 706434 | 43125 |
| Firmicutes | Mitsuokella multacida | A 405-1, DSM 20544 | Yes | 500635 | 28653 |
| Firmicutes | Paenibacillus sp. | JDR-2 | No | 324057 | 20399 |
| Firmicutes | Phascolarctobacterium sp. | YIT 12067 | No | 626939 | 48505 |
| Firmicutes | Selenomonas artemidis | F0399 | No | 749551 | 47277 |
| Firmicutes | Selenomonas flueggei | ATCC 43531 | Yes | 638302 | 37273 |
| Firmicutes | Selenomonas noxia | ATCC 43541 | Yes | 585503 | 34641 |
| Firmicutes | Selenomonas sp. | Oral Taxon 137 F0430 | No type strain available | 879310 | 52055 |
| Firmicutes | Selenomonas sp. | Oral Taxon 149 67H29BP | No type strain available | 864563 | 50535 |
| Firmicutes | Selenomonas sputigena | DSM 20758 | Yes | 546271 | 51247 |
| Firmicutes | Staphylococcus aureus aureus | ED98 | No | 681288 | 39547 |
| Firmicutes | Streptococcus pneumoniae | TIGR4 | No | 170187 | 277 |
| Firmicutes | Thermoanaerobacter sp. | X514 | Name not validly published | 399726 | 16394 |
| Firmicutes | Thermosinus carboxydivorans | Nor1 | Yes | 401526 | 17587 |
| Firmicutes | Turicibacter sp. | PC909 702450 42765 | No | ||
| Firmicutes | Veillonella atypica | ACS-049-V-Sch6 | No | 866776 | 51075 |
| Firmicutes | Veillonella atypica | ACS-134-V-Col7a | No | 866778 | 51079 |
| Firmicutes | Veillonella dispar | ATCC 17748 | Yes | 546273 | 30491 |
| Firmicutes | Veillonella parvula | ATCC 17745 | No | 686660 | 41557 |
| Firmicutes | Veillonella parvula | Te3, DSM 2008 | Yes | 479436 | 21091 |
| Firmicutes | Veillonella sp. | 3 1 44 | Name not validly published | 457416 | 41975 |
| Firmicutes | Veillonella sp. | 6 1 27 | Name not validly published | 450749 | 41977 |
| Firmicutes | Veillonella sp. | Oral Taxon 158 F0412 | Name not validly published | 879309 | 52053 |
| Fusobacteria | Fusobacterium nucleatum nucleatum | ATCC 25586 | Yes | 190304 | 295 |
| Fusobacteria | Ilyobacter polytropus | CuHBu1, DSM 2926 | Yes | 572544 | 32577 |
| Fusobacteria | Leptotrichia buccalis | C-1013-b, DSM 1135 | Yes | 523794 | 29445 |
| Fusobacteria | Sebaldella termitidis | NCTC 11300 | Yes | 526218 | 29539 |
| Fusobacteria | Streptobacillus moniliformis | 9901, DSM 12112 | Yes | 519441 | 29309 |
| Planctomycetes | Pirellula staleyi | DSM 6068 | Yes | 530564 | 29845 |
| Planctomycetes | Planctomyces limnophilus | Mu 290, DSM 3776 | Yes | 521674 | 29411 |
| Proteobacteria | Acinetobacter baumannii | SDF | No | 509170 | 13001 |
| Proteobacteria | Alkalilimnicola ehrlichii | MLHE-1 | Yes | 187272 | 15763 |
| Proteobacteria | Arcobacter nitrofigilis | DSM 7299 | Yes | 572480 | 32593 |
| Proteobacteria | Burkholderia xenovorans | (fungorum) LB400 | Yes | 266265 | 254 |
| Proteobacteria | Campylobacter jejuni | doylei 269.97 | No | 360109 | 17163 |
| Proteobacteria | Candidatus Pelagibacter ubique | SAR11 HTCC1062 | Name not validly published | 335992 | 13989 |
| Proteobacteria | Candidatus Zinderia insecticola | CARI | Name not validly published | 871271 | 51243 |
| Proteobacteria | Cellvibrio japonicus | Ueda107 | Yes | 498211 | 28329 |
| Proteobacteria | Cupriavidus taiwanensis | LMG19424 | Yes | 164546 | 15733 |
| Proteobacteria | Escherichia coli | K-12, MG1655 | No | 511145 | 225 |
| Proteobacteria | Geobacter uraniireducens | Rf4 | Yes | 351605 | 15768 |
| Proteobacteria | Hahella chejuensis | KCTC 2396 | Yes | 349521 | 16064 |
| Proteobacteria | Haliangium ochraceum | SMP-2, DSM 14365 | Yes | 502025 | 28711 |
| Proteobacteria | Helicobacter pylori | 908 | No | 869727 | 50869 |
| Proteobacteria | Lawsonia intracellularis | PHE/MN1-00 | No | 363253 | 183 |
| Proteobacteria | Magnetococcus sp. | MC-1 | Name not validly published | 156889 | 262 |
| Proteobacteria | Methylobacterium nodulans | ORS2060 | Yes | 460265 | 20477 |
| Proteobacteria | Neisseria meningitidis | Z2491 | No | 122587 | 252 |
| Proteobacteria | Neorickettsia sennetsu | Miyayama | Yes | 222891 | 357 |
| Proteobacteria | Nitrosomonas eutropha | C91 (C71) | Yes | 335283 | 13913 |
| Proteobacteria | Photorhabdus luminescens laumondii | TT01 | Yes | 243265 | 9605 |
| Proteobacteria | Polynucleobacter necessarius | STIR1 | No | 452638 | 19991 |
| Proteobacteria | Pseudomonas aeruginosa | LESB58 | No | 557722 | 31101 |
| Proteobacteria | Pseudomonas fluorescens | SBW25 | No | 216595 | 31229 |
| Proteobacteria | Pseudomonas stutzeri | A1501 | No | 379731 | 16817 |
| Proteobacteria | Salmonella enterica enterica | PT4 P125109 | No | 550537 | 30687 |
| Proteobacteria | Shewanella oneidensis | MR-1 | Yes | 211586 | 335 |
| Proteobacteria | Sorangium cellulosum | So ce56 | No | 448385 | 28111 |
| Proteobacteria | Stigmatella aurantiaca | DW4 /3-1 | No | 378806 | 52561 |
| Proteobacteria | Sulfurospirillum deleyianum | 5175, DSM 6946 | No | 525898 | 29529 |
| Proteobacteria | Vibrio cholerae | O395 | No | 345073 | 32853 |
| Spirochaetes | Borrelia turicatae | 91E135 | Yes | 314724 | 13597 |
| Spirochaetes | Brachyspira murdochii | 56-150, DSM 12563 | Yes | 526224 | 29543 |
| Spirochaetes | Leptospira interrogans | lai 56601 | No | 189518 | 293 |
| Synergistetes | Thermanaerovibrio acidaminovorans | Su883, DSM 6589 | Yes | 525903 | 29531 |
| Tenericutes | Acholeplasma laidlawii | PG-8A | No | 441768 | 19259 |
| Tenericutes | Candidatus Phytoplasma asteris | yellows witches'-broom AY-WB 322098 | Name not validly published | 13478 | |
| Tenericutes | Candidatus Phytoplasma mali | AT | Name not validly published | 37692 | 25335 |
| Tenericutes | Mycoplasma genitalium | G37 | Yes | 243273 | 97 |
| Tenericutes | Mycoplasma pneumoniae | FH | No | 722438 | 49525 |
| Tenericutes | Ureaplasma parvum | sv 3, ATCC 27815 | No | 505682 | 19087 |
| Thermotogae | Fervidobacterium nodosum | Rt17-B1 | Yes | 381764 | 16719 |
| Thermotogae | Kosmotoga olearia | TBF 19.5.1 | Yes | 521045 | 29419 |
| Thermotogae | Petrotoga mobilis | SJ95 | Yes | 403833 | 17679 |
| Thermotogae | Thermotoga naphthophila | RKU-10 | Yes | 590168 | 33663 |
| Verrucomicrobia | Akkermansia muciniphila | ATCC BAA-835 | Yes | 349741 | 20089 |
| Verrucomicrobia | Opitutus terrae | Yes | PB90-1 | 452637 | |
| Crenarchaeota | Sulfolobus solfataricus | P2 | 273057 | 108 | |
| Crenarchaeota | Thermosphaera aggregans | M11TL, DSM 11486 | Yes | 633148 | 36571 |
| Euryarchaeota | Halogeometricum borinquense | PR3, DSM 11551 | Yes | 469382 | 20743 |
| Euryarchaeota | Methanocella sp. | RC-I | Name not validly published | 351160 | 19641 |
| Euryarchaeota | Methanothermus fervidus | V24S, DSM 2088 | Yes | 523846 | 33689 |
| Korarchaeota | Candidatus Korarchaeum cryptofilum | OPF8 | Name not validly published | 374847 | 16525 |
| Nanoarchaeota | “Nanoarchaeum equitans” | Kin4-M | Name not validly published | 228908 | 9599 |
16S rRNA tree
For this analysis, 16S rRNA sequences were predicted from the whole genome sequences of the selected organisms, using the RNAmmer algorithm [13]. These sequences were aligned using the MAFFT program, with the iterative refinement algorithm using maximum iteration (1000) and default parameters for gap penalties [14]. A distance tree was constructed using MEGA5 [15] with the Neighbor-joining algorithm [16] and 1,000 bootstrap re-samplings. The taxa in the resulting tree were collapsed to phyla, except for the Negativicutes.
Composition Vector Tree (CV)
A Composition Vector Tree was constructed based on protein sequences of the 145 selected genomes using a webserver (available at http://tlife.fudan.edu.cn/cvtree/) with the K parameter set at 6 [17]. The outcome from the program is a distance matrix based on amino acid sequence comparisons, which is then used to generate a phylogenetic tree with the neighbor-joining method. In the shown tree, the outgroup chosen was Methanothermus fervidus (an Archaea). After tree visualization with MEGA5, branches were collapsed wherever possible with the exception of the Negativicutes branch, which remained expanded.
Consensus tree of conserved genes
Using the list of universally conserved core genes, previously identified by Ciccarelli et al. [18], and an implementation of BLAST, a set of genes that was shared among all 145 genomes was identified. Proteins that had no match in at least one genome or showed poor E-value were eliminated. The 27 conserved core genes were extracted (Table 1) and a multiple alignment was produced using MUSCLE software [19]. A set of phylogenetic trees was constructed by PAUP [20] and a best-fit consensus tree was generated using Phylogeny Inference package (PHYLIP) as described elsewhere [21]. Bootstrap values were found after 27 re-samplings, which is equal to the number of gene families conserved in all the analyzed genomes.
DNA tetramer analysis and amino acid usage
A tetramer frequency heatmap was constructed from the observed ratios of tetra-nucleotide frequencies divided by estimated tetra-nucleotide frequencies for each genome [22]. The estimated tetra-nucleotides were computed from the genomes' base composition. The ratio of observed over expected frequency was used for hierarchical clustering using complete linkage and Euclidean distance, which was subsequently performed with respect to both strain and tetramer frequencies.
The amino acid heatmap is based on frequencies of deduced proteomic amino acids from each genome normalized with respect to the total number of amino acids in each genome. The amino acid frequencies for each genome were clustered using complete linkage and Euclidean distance with respect to both genomes and amino acids. The heatmap was made using the R package ggplot2 [23].
Comparison of metabolism potential
The protein sequences of Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology categories [24] were downloaded and only the Bacterial sequences were considered. The Hidden Markov model (HMM) of each ortholog was generated using HMMER version 3 [25] based on the multiple alignment of each orthologous set of KEGG proteins, using MUSCLE software [19]. The 145 proteomes were queried against the HMMs to infer their ontology. A cutoff of 1×10−30 was used for statistical significance. A heatmap of each pathway and process derived from the database KEGG was illustrated based on normalized abundance of the enzymes present in each pathway. The heatmap and hierarchical clustering were performed in the software R [23].
Construction of BLAST matrix and proteome comparison
Reciprocal BLAST was performed between each genome pair. The program blastall version 2.2.25 was used for BLAST implementation using default settings (BLASTp, E-value set to 1×10−5 for non-homologs and 1×10−8 for homologs, without filtering). A hit was considered significant at a BLAST cutoff of 95% identity and 95% coverage (of the longest gene in comparison). The number of hits was then given as a percentage of the genes in the column representing the corresponding genome. The diagonal designates internal homologs, computed by blasting each genome with itself. To avoid including identical genes, the second highest scoring hits were used. Furthermore, we also performed homology reduction of the diagonal hits, using an implementation of the Hobohm algorithm [26].
Results
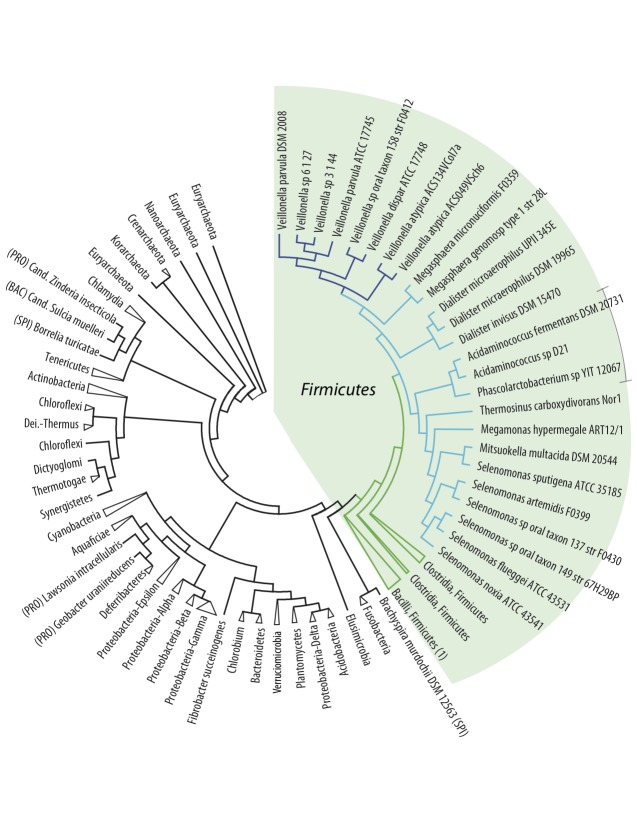
Twenty-four Negativicutes genomes were compared to 121 other prokaryotic genomes covering 22 Bacterial and 4 Archaeal phyla. When available, at least two genomes were included for every phylum. The first analysis presented here is based on 16S rRNA alignments. A single 16S rRNA gene was extracted from each of the genomes and an alignment was produced spanning the maximum length of the gene. A phylogenetic tree was constructed based on this alignment, as shown in Figure 1. With the exception of the Negativicutes, branches of the tree were collapsed in those cases where the analyzed species within a phylum clustered together. With the exception of some Firmicutes, the analyzed genomes cluster according to their phylum, although the Deferribacteres phylum is mixed with the Proteobacteria phyla, and two members of Proteobacteria are not positioned with other members of their phylum (Lawsonia intracellularis and Magnetococcus). That most phyla could be collapsed is consistent with the weight of 16S rRNA similarities in currently accepted taxonomic descriptions of prokaryotes. The Firmicutes, however, show less consistency. Although most of the analyzed Firmicutes cluster together, two species are separated from the Firmicutes branch (Eubacterium cylindroides and Thermoanaerobacter sp., both members of Clostridia). The Negativicutes are positioned within the Firmicutes cluster, and this part of the tree is expanded in the figure for clarity. As can be seen, phylogeny of the 16S rRNA gene provides good resolution between the different genera of the analyzed Negativicutes. All Veillonella spp. are clustered within one branch of the Negativicutes. The Acidaminococcaceae (to which Phascolarctobacterium spp. also belong) are placed within the cluster of the Veillonellaceae, in accordance with their current classification [27]. The Acidaminococcaceae used to be recognized as a separate family within the Negativicutes, just like the Veillonellaceae, and during preparation of this contribution these two families were presented as such in the Taxonomy database at NCBI. Of note is the relatively close relationship between Negativicutes and two Clostridium species (C. botulinum and C. cellulolyticum), which does not cluster with other members of the Clostridium genus (Figure 1). That genus displays a high degree of variation and re-classification of some of the members of this genus is in progress (see for example [27]). That two members of the Clostridia are even placed outside the Firmicutes phylum is an indication of 16S rRNA gene sequence heterogeneity within this class.
Figure 1.
Phylogenetic neighbor-joining tree based on 16S rRNA genes extracted from 145 genomes (24 Negativicutes and 121 prokaryotic genomes representing 26 phyla). Bootstrap values of 50 and higher are indicated. With the exception of the Negativicutes, branches where all organisms belong to the same phyla are collapsed and named by the phyla they represent. The green shading indicates the position of Firmicutes. The collapsed branch of the Bacilli, marked (1), contains Turicibacter sanguinis, a Firmicutes member of the Erysipelotrichales as well as Bacilli members. An uncollapsed tree is included in the supplementary material.
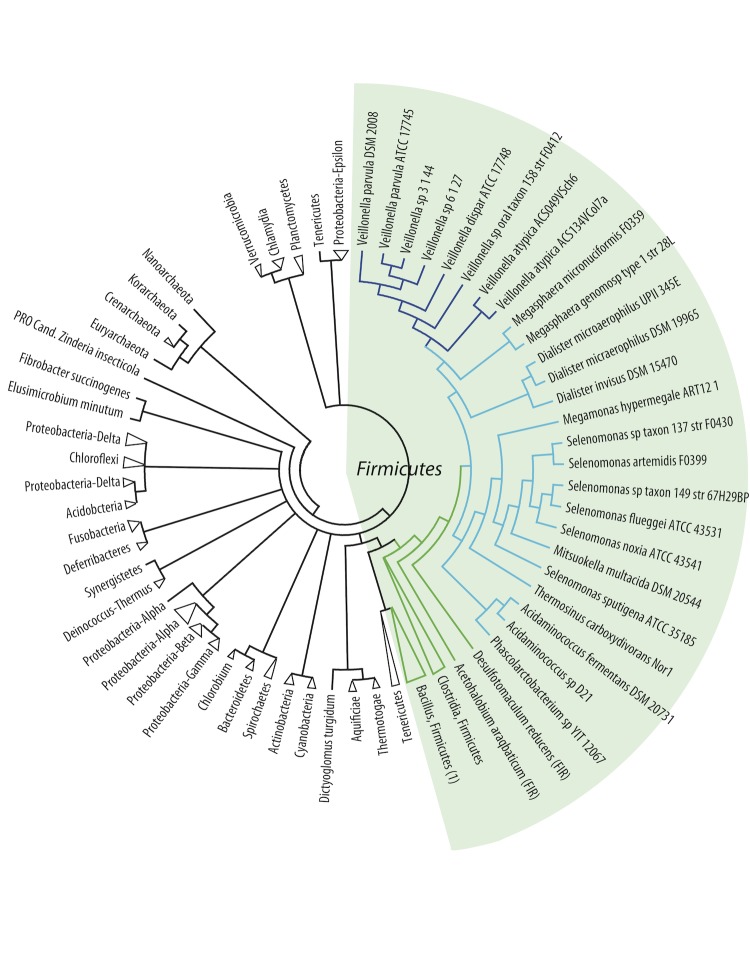
Next, all protein-coding genes of the analyzed genomes were compared and a composition vector tree (CVtree) was produced, based on amino acid sequences (Figure 2). The topology of the resulting tree is generally in accordance with the 16S rRNA tree shown in the previous figure. As indicated by the collapsed branches, the CVtree grouped most genomes according to their known taxonomic phyla, although not all Spirochaetes cluster together. In contrast to the 16S rRNA tree, in this protein tree all the Firmicutes cluster together, and are distinct from other phyla. The Negativicutes genomes, nested within the Firmicutes, again have the Acidaminococcaceae placed within the Veillonellaceae, while all Veillonella spp. are found in one cluster. All Clostridia, this time divided into two collapsed branches, are positioned as the closest relatives to Negativicutes. It is of interest that among the closest relatives to Firmicutes, based on this analysis, are the Fusobacteria and the Elusimicrobia; these are atypical diderm bacteria that produce lipopolysaccharides [28]. However, the spirochete, Brachyspira murdochii, does not possess two membranes, but is nevertheless grouped with atypical diderms. On the other hand while the Synergistetes are atypical diderm bacteria, they are placed elsewhere in the tree (Figure 2).
Figure 2.
Phylogenetic tree based on composition vector analysis (CVtree) of all protein coding genes (amino acid sequences) derived from the analyzed genomes. Note that the branch lengths in this plot are artificial. The coloring is the same as in Figure 1 and branches have been collapsed. The Firmicutes branch Bacilli, marked (1), contains Turicibacter sanguinis. An uncollapsed tree is included in the supplementary material.
A third analysis was based on a subset of proteins found conserved amongst all analyzed genomes. These conserved proteins were selected based on a protein BLAST (a cutoff of 50% identity and 50% coverage of the query length was used) and single linkage clustering. The analysis identified 29 genes that are shared among all 145 genomes [Table 2]. A consensus tree was constructed based on these 29 conserved proteins (Figure 3). The results confirm the global observations of the other two phylogenetic analyses: the Negativicutes cluster together and are most closely related to Clostridia (in this case the most closely related species are Desulfotomaculum reducens and Acetohalobium arabaticum). As before, the Acidaminococcaceae cluster together but within the Veillonellaceae. The position of Turicibacter sanguinis within the Bacilli group of Firmicutes is consistent with the other two trees but contrasts with its taxonomic description at NCBI as a member of the Erysipelotrichia.
Table 2. Universally conserved COGs.
| Group | Average length (aa) | Annotation |
|---|---|---|
| COG0012 | 380 | Predicted GTPase, probable translation factor |
| COG0016 | 423 | Phenylalanine-tRNA synthethase alpha subunit |
| COG0048 | 137 | Ribosomal protein S12 |
| COG0049 | 182 | Ribosomal protein S7 |
| COG0052 | 240 | Ribosomal protein S2 |
| COG0080 | 154 | Ribosomal protein L11 |
| COG0081 | 230 | Ribosomal protein L1 |
| COG0087 | 288 | Ribosomal protein L3 |
| COG0091 | 157 | Ribosomal protein L22 |
| COG0092 | 240 | Ribosomal protein S3 |
| COG0093 | 130 | Ribosomal protein L14 |
| COG0094 | 182 | Ribosomal protein L5 |
| COG0096 | 131 | Ribosomal protein S8 |
| COG0097 | 177 | Ribosomal protein L6P/L9E |
| COG0098 | 220 | Ribosomal protein S5 |
| COG0100 | 145 | Ribosomal protein S11 |
| COG0102 | 167 | Ribosomal protein L13 |
| COG0103 | 172 | Ribosomal protein S9 |
| COG0172 | 442 | Seryl-tRNA synthetase |
| COG0184 | 154 | Ribosomal protein S15P/S13E |
| COG0186 | 122 | Ribosomal protein S17 |
| COG0197 | 175 | Ribosomal protein L16/L10E |
| COG0200 | 166 | Ribosomal protein L15 |
| COG0201 | 445 | Preprotein translocase subunit SecY |
| COG0202 | 323 | DNA-directed RNA polymerase, alpha subunit |
| COG0256 | 178 | Ribosomal protein L18 |
| COG0495 | 854 | Leucyl-tRNA synthetase |
| COG0522 | 199 | Ribosomal protein S4 and related proteins |
| COG0533 | 375 | Metal-dependent proteases with chaperone activity |
Figure 3.
Consensus tree based on the phylogenetic trees of 27 genes conserved in all 145 genomes. The collapsed branch of the Bacilli, marked (1), contains Turicibacter sanguinis. An uncollapsed tree is available as a supplemental figure.
In conclusion, based on three independent phylogenetic analyses, the closest relatives to the Negativicutes seem to be the Clostridiaceae. The observed clustering of species within the Negativicutes is consistent with their assigned taxonomy. Furthermore, these analyses show that Veillonella spp. form a distinct branch, most different from the other Negativicutes, while the recent change of status of the Acidaminococcaceae (they are no longer a separate family) is confirmed by these analyses.
Apart from comparing proteins and genes, genomes can also be compared based on nucleotide composition irrespective of their coding capacity. For instance, the frequency of nucleotide combinations can reveal similarities between genomes that are independent of protein-coding information. We compared the frequency of tetranucleotides for all 145 genomes. The observed frequency of all 64 tetranucleotide combinations was extracted for each genome and these frequencies were divided by the theoretically calculated, expected frequencies (corrected for differences in base composition). This ratio, which could be interpreted as a genomic signature, was expected to reflect taxonomic divisions [29]. However, although the analysis identified a high similarity in tetranucleotide frequency for all of the analyzed Veillonella genomes, most of the clustering observed was not in accordance with known taxonomic relationships. Not only were Negativicutes other than Veillonella separated from each other and strewn across the phyla, but also several other Firmicutes were distributed over various branches (data shown as supplementary material). In fact, for most of the analyzed genomes, members of identical phyla did not cluster together and even the Archaea were mixed with Bacteria, although some closely related species were indeed clustered. This may explain why all Veillonella genomes grouped together. Several organisms with similar tetranucleotide frequencies did not share a common ecological niche, in contrast to previously reported observations (reviewed in [30]). Neither was the obtained clustering dictated by GC-content. The conclusion from this analysis was that tetranucleotide analysis is only taxonomically informative for closely related genomes.
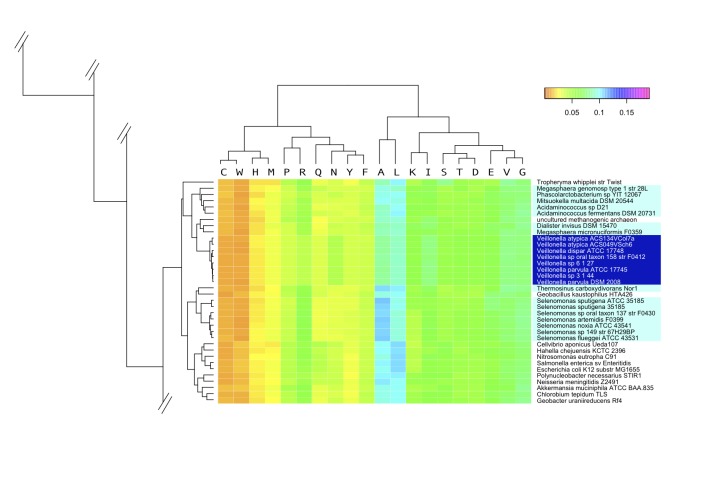
We also compared whole-genome amino acid frequencies in each of the deduced proteomes. Although the results are slightly more in agreement with known taxonomy as compared with the genomic signatures discussed above, this analysis does not cluster organisms according to their phyla, and again some Archaea are mixed with Bacteria. The relevant part of the heatmap based on amino acid frequency is shown in Figure 4. All Veillonella genomes cluster together within the Negativicutes, with the exception of two of the three Dialister genomes, which are found most closely related to Clostridium species (See supplemental information for a version of this figure showing all the genomes). The major Negativicutes cluster also contains a Geobacillus (which is a Gram-positive Firmicutes) and a methanogenic Archaean. Interestingly, the closest relatives to this cluster are not Clostridia, as the previous phylogenetic trees suggest, but a number of Proteobacteria. It is striking that the amino acid frequency analysis detects similarities to Proteobacteria, with which the Negativicutes have their two membranes in common.
Figure 4.
A zoomed heatmap of the amino acid frequency found in the deduced proteomes of all 145 genomes. A fragment of the heatmap is shown, presenting the cluster in which all but two Negativicutes are found. The remaining two, both Dialister microaerophilus genomes, are positioned elsewhere in the tree, closest to Clostridium cellulolyticum (not shown in this zoom). The color scale indicates highly underrepresented (orange) to highly overrepresented amino acid frequency (magentum). The full figure is available as supplementary information.
The metabolic properties encoded by the genomes were analyzed next, based on KEGG comparisons [24]. The results are again visualized in a heatmap (Figure 5). We hypothesized that this analysis could identify similarities based on niche adaptation. For simplicity, only a selected number of phyla are shown: apart from the Firmicutes, genomes are included that represent Bacteroidetes and Proteobacteria (both of which contain members frequently found in the oral or gut microbiome), while Cyanobacteria are included as representatives of a phylum that occupy an environmental niche. Since the genomes are compared based on predicted proteomes, their annotation was standardized in order to reduce artificial variation caused by gene annotation differences. As can be seen in Figure 5, the Veillonella genomes all cluster together at the right-hand side of the plot, within a larger cluster containing most of the other Negativicutes and some Firmicutes. The three Dialister species are placed outside the Negativicutes cluster. The other Firmicutes that are found combined with the Negativicutes, based on their metabolic potential, are Clostridium cellulolyticum, Eubacterium rectale, Lactococcus lactis, Streptococcus pneumoniae and Turicibacter sanguinis. These are all common members of the oral or intestine microbiome. As expected, the metabolic pathway for lipopolysaccharide biosynthesis is shared between the Negativicutes and other Gram-negative species, as indicated by the arrows in Figure 5. Interestingly, the Cyanobacteria form a small cluster within, not outside the tree, together with a Haliangium and a Sorangium species as their closest neighbors (both are social Myxococcales belonging to the Deltaproteobacteria). The exclusive ability of carbon fixation by Cyanobacteria is apparent from the dark red square in the block 'energy'. The lanes of Veillonella in Figure 5 are dominated by light colors, indicative of medium metabolic potential; that is, in contrast to some genomes where most of the pathways are present (dark red for Proteobacteria for example) or missing (dark green for other Negativicutes), the Veillonella genomes have partial pathways (based on knowledge primarily from aerobic genomes). There is no reason to believe that the Veillonella genomes should have less metabolic potential than other Negativicutes. Indeed, it is likely that the differences in metabolic potential of Veillonella are truly reflective of alternative capabilities for these bacteria.
Figure 5.
Heatmap of metabolism potential, based on Kyoto Encyclopedia of Genes and Genomes ontology (KEGG). The green color in the heatmap indicates weak metabolic potential, while red signals strong potential. The arrows to the right indicate the scores for lipopolysaccharide biosynthesis. A version summarizing the metabolism pathways and showing the species legend is available as supplementary material.
It was further investigated how conserved the predicted proteomes are within the Negativicutes. As a quantitative measure for homology, shared protein-coding genes were identified by pairwise BLASTP comparison and expressed as a percentage of the combined proteomes. The results are shown in a matrix (Figure 6). In addition to the proteomes of the 24 Negativicutes, the comparison includes Clostridium botulinum, Cl. cellulolyticum and Desulfotomaculum reducens, as these Firmicutes were shown to share characteristics with Negativicutes in previous analyses (cf. Figures 1 and 3). The proteome of E. coli K12 is included as an example of a Gram-negative intestinal bacterium. The BLAST matrix was constructed using reciprocal best BLAST hits to determine the presence of shared protein family between two genomes. Inspection of Figure 6 shows that the genus Veillonella is relatively homogeneous; any two members of this genus share between 67% and 90% homology (1,357 to 1,682 protein families), irrespective of the species. The genus Selenomonas is more heterogeneous, with pairwise homology varying from 42% to 82% between any two species (980 to 1659 protein families). The three proteomes of Dialister spp., covering two species, share between 40% and 84% homology. The highest homologous fraction identified between two members of different genera within the Negativicutes is 43% (Mitsuokella multacida compared to Selenomonas sputigena, whereas the lowest homology is 15% (Dialister spp. compared to Thermosinus carboxydivorans). Negativicutes share between 9% and 33% homology with the analyzed Firmicutes, whereas slightly lower homology is detected with E. coli (between 7% and 24%).
Figure 6.
Proteome comparison represented by a BLAST matrix, based on 24 Negativicutes genomes with reciprocal best hits. The genomes of Clostridium botulinum, Cl. cellulolyticum, Desulfotomaculum reducens and E. coli are added for comparison. Inter-genus comparisons are indicated by black squares. A version reporting the numerical values of homology percentages is available as supplementary information.
Finally, we assessed the gene pool conserved within all analyzed Negativicutes. Using the same cutoff for protein BLAST comparison as before, a core-genome is identified that contains about 300 conserved protein families (data not shown). This is a relatively low number of conserved proteins, reflective of the extensive genetic heterogeneity within this bacterial class.
Discussion
The availability of complete sequences for a large and diverse set of Bacterial genomes has helped in exploring the conundrum of the genus Veillonella, a genus within the Negativicutes class, all of which are Gram negative Firmicutes. The 16S rRNA tree shown as Figure 1 illustrates how “close” the Negativicutes are to other Firmicutes. The closest Gram positive Clostridium species are actually quite distant to Veillonella and other Negativicutes genomes, as can be seen in the low fraction of shared protein families in Figure 6. The Gram-negative Firmicutes are even more distant to other Gram negatives, such as Proteobacteria (e.g., E. coli). It should be noted that the family Clostridiaceae is a largely diverse group with many members being re-classified [27]. It is therefore possible that the taxonomic description of some Clostridium genomes may change in future. However, our analyses did not identify one single Gram-positive Firmicutes (Clostrida or others) that consistently was identified as most closely related to Veillonella. As seen from three types of phylogenetic analysis, the Negativicutes class genomes form a distinct cluster within the Firmicutes, and the Veillonella genus forms a relatively homogeneous group of species within the Negativicutes, with relatively conserved metabolic properties (Figure 5). In comparison, the Selenomonas genus is more heterogeneous, at least based on their total gene comparison, as illustrated in Figure 6.
In contrast to expectations, relatively little homology between Negativicutes and other Gram-negative genomes was detected in our analyses. Neither gene-dependent phylogenetic analysis, nor gene-independent DNA tetramer analysis identified a significant commonness between Negativicutes and, say, Proteobacteria. Only whole-genome frequency analysis of amino acid usage identified some similarity to a few Proteobacteria, and this might be more reflective of environment the organism is adapted to, and not phylogeny. Using KEGG pathways for metabolic comparison of the proteomes we found few pathways in common, with the exception of a shared lipopolysaccharide biosynthesis pathway. From all analyses combined, it is clear that the taxonomic placement of Negativicutes within the Firmicutes reflects their genetic and genomic characteristics, although the proteins encoded by the Negativicutes genomes are quite distinct from their Gram-positive cousins. It could be speculated that the double membrane of the Negativicutes evolved in a lineage that used to be a single-membrane (Gram-positive) Firmicute. Whether this event co-evolved independently of the formation of other Gram-negative phyla, or was the result of lateral gene transfer, cannot be stated for certain at present; estimations of horizontally transferred regions in Veillonella parvula DSM 2008, the only fully assembled Veillonella genome available, using the least conservative method on the Islandviewer web-site [31], revealed that only 2% of the genome is of foreign origin. In comparison, 9% of the E. coli K-12 subsp. MG1655 genome was predicted as horizontally transferred. Further analyses are therefore needed to assess this in more detail.
Author’s contributions
Tammi Vesth was a main contributor to the writing of the manuscript and to the organization of the work. Trudy Wassenaar helped considerably in editing and improving the manuscript. Individual contributions: Asli Ozen (16s rRNA and CV tree), Oksana Lukjancenko (consensus tree), Sandra Andersen (initial investigations and background research, early version of the manuscript), Rolf Sommer Kaas (BLAST matrix), Jon Bohlin (tetramer and amino acid usage heatmaps), Intawat Nookaew (metabolism heatmaps). David Ussery provided the original idea for this manuscript, suggested the figures, helped in early drafts of the manuscript, and supervised the project.
Acknowledgements
This research was supported by grants from the Danish Research Council, and in part by a grant 09-067103/DSF from the Danish Council for Strategic Research.
References
- 1.Delwiche EA, Pestka JJ, Tortorello ML. The veillonellae: gram-negative cocci with a unique physiology. Annu Rev Microbiol 1985; 39:175-193 10.1146/annurev.mi.39.100185.001135 [DOI] [PubMed] [Google Scholar]
- 2.Diaz PI, Chalmers NI, Rickard AH, Kong C, Milburn CL, Palmer RJ, Kolenbrander PE. Molecular Characterization of Subject-Specific Oral Microflora during Initial Colonization of Enamel. Appl Environ Microbiol 2006; 72:2837-2848 10.1128/AEM.72.4.2837-2848.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the Normal Bacterial Flora of the Oral Cavity. J Clin Microbiol 2005; 43:5721-5732 10.1128/JCM.43.11.5721-5732.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol 2003; 30:644-654 10.1034/j.1600-051X.2003.00376.x [DOI] [PubMed] [Google Scholar]
- 5.Olson JC, Cuff CF, Lukomski S, Lukomska E, Canizales Y, Wu B, Crout RJ, Thomas JG, McNeil DW, Weyant RJ, et al. Use of 16S ribosomal RNA gene analyses to characterize the bacterial signature associated with poor oral health in West Virginia. BMC Oral Health 2011; 11:7 10.1186/1472-6831-11-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalmers NI, Palmer RJ, Cisar JO, Kolenbrander PE. Characterization of a Streptococcus sp.-Veillonella sp. Community Micromanipulated from Dental Plaque. J Bacteriol 2008; 190:8145-8154 10.1128/JB.00983-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leuckfeld I, Paster BJ, Kristoffersen AK, Olsen I. Diversity of Veillonella spp. from subgingival plaque by polyphasic approach. APMIS 2010; 118:230-242 10.1111/j.1600-0463.2009.02584.x [DOI] [PubMed] [Google Scholar]
- 8.Tunney MM, Field TR, Moriarty TF, Patrick S, Doering G, Muhlebach MS, Wolfgang MC, Boucher R, Gilpin DF, McDowell A, Elborn JS. Detection of anaerobic bacteria in high numbers in sputum from patients with cystic fibrosis. Am J Respir Crit Care Med 2008; 177:995-1001 10.1164/rccm.200708-1151OC [DOI] [PubMed] [Google Scholar]
- 9.Murri M, Leiva I, Gomez-Zumaquero JM, Tinahones FJ, Cardona F, Soriguer F, Quepo-Ortuño MI. Gut microbiota in children with type I diabetes differs from that in healthy children: a case-control study. BMC Med 2013; 11:46 10.1186/1741-7015-11-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolenbrander PE, Moore LVH. The genus Veillonella in:H.G. Balows, M. Trüper, W. Dworkin, W. Harder, K.H. Schleifer (Eds.), The prokaryotes (2nd ed.), Springer, New York (1992), pp. 2034–2047. [Google Scholar]
- 11.Mashima I, Nakazawa F. Identification of Veillonella tobetsuensis in tongue biofilm by using a species-specific primer pair. Anaerobe 2013; 22:77_81. [DOI] [PubMed]
- 12.De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB. Volume 3: The Firmicutes In Bergey’s Manual of Systematic Bacteriology, Springer 2009. [Google Scholar]
- 13.Lagesen K, Hallin P, Rødland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 2007; 35:3100-3108 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katoh K, Toh H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 2010; 26:1899-1900 10.1093/bioinformatics/btq224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol 2011; 28:2731-2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987; 4:406-425 [DOI] [PubMed] [Google Scholar]
- 17.Xu Z, Hao B. CVTree update: a newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic Acids Res 2009; 37(suppl 2):W174-W178 10.1093/nar/gkp278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science 2006; 311:1283-1287 10.1126/science.1123061 [DOI] [PubMed] [Google Scholar]
- 19.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004; 32:1792-1797 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fink WL. Microcomputers and phylogenetic analysis. Science 1986; 234:1135-1139 10.1126/science.234.4780.1135 [DOI] [PubMed] [Google Scholar]
- 21.Retief JD. Phylogenetic analysis using PHYLIP. Methods Mol Biol 2000; 132:243-258 [DOI] [PubMed] [Google Scholar]
- 22.Karlin S, Burge C. Dinucleotide relative abundance extremes: a genomic signature. Trends Genet 1995; 11:283-290 10.1016/S0168-9525(00)89076-9 [DOI] [PubMed] [Google Scholar]
- 23.Wickham H: ggplot2: Elegant Graphics for Data Analysis (Use R!). Springer New York, 2009. ISBN-10: 0387981403 | ISBN-13: 978-0387981406
- 24.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res 2004; 32:277-280 10.1093/nar/gkh063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eddy SR. BIOINFORMATICS REVIEW Profile hidden Markov models. [PubMed]. Bioinformatics 1998; 14:755-763 10.1093/bioinformatics/14.9.755 [DOI] [PubMed] [Google Scholar]
- 26.Hobohm U, Sander C. Enlarged representative set of protein structures. Protein Sci 1994; 3:522-524 10.1002/pro.5560030317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludwig W, Schleifer KH, Whitman W. Revised road map to the phylum Firmicutes. In Bergey’s Manual of Systematic Bacteriology, Springer 2009:1-13. [Google Scholar]
- 28.Gupta RS. Origin of diderm (Gram-negative) bacteria: antibiotic selection pressure rather than endosymbiosis likely led to the evolution of bacterial cells with two membranes. Antonie van Leeuwenhoek 2011; 100:171-182 10.1007/s10482-011-9616-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pride DT, Meinersmann RJ, Wassenaar TM, Blaser MJ. Evolutionary implications of microbial genome tetranucleotide frequency biases. Genome Res 2003; 13:145-158 10.1101/gr.335003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dutta C, Paul S. Microbial lifestyle and genome signatures. Curr Genomics 2012; 13:153-162 10.2174/138920212799860698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langille MG, Brinkman FS. IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics 2009; 25:664-665 10.1093/bioinformatics/btp030 [DOI] [PMC free article] [PubMed] [Google Scholar]