Abstract
Effect alleles (alleles with a polymorphism that is associated with the effect being measured) in a small number of single-nucleotide polymorphisms (SNPs) are known to influence the dietary requirement for choline. In this study, we examined a much larger number of SNPs (n=200) in 10 genes related to choline metabolism for associations with development of organ dysfunction (liver or muscle) when 79 humans were fed a low-choline diet. We confirmed that effect alleles in SNPs such as the C allele of PEMT rs12325817 increase the risk of developing organ dysfunction in women when they consume a diet low in choline, and we identified novel effect alleles, such as the C allele of CHKA SNP rs7928739, that alter dietary choline requirements. When fed a low-choline diet, some people presented with muscle damage rather than liver damage; several effect alleles in SLC44A1 (rs7873937, G allele; rs2771040, G; rs6479313, G; rs16924529, A; and rs3199966, C) and one in CHKB (rs1557502, A) were more common in these individuals. This suggests that pathways related to choline metabolism are more important for normal muscle function than previously thought. In European, Mexican, and Asian Americans, and in individuals of African descent, we examined the prevalence of the effect alleles in SNPs that alter choline requirement and found that they are differentially distributed among people of different ethnic and racial backgrounds. Overall, our study has identified novel genetic variants that modulate choline requirements and suggests that the dietary requirement for choline may be different across racial and ethnic groups.—Da Costa, K.-A., Corbin, K. D., Niculescu, M. D., Galanko, J. A., Zeisel, S. H. Identification of new genetic polymorphisms that alter the dietary requirement for choline and vary in their distribution across ethnic and racial groups.
Keywords: SNP, creatine kinase, fatty liver, muscle
Choline is a dietary component that is essential for the normal function of all cells (1). It is used as a constituent of cell membranes and the neurotransmitter acetylcholine (1), and it participates in lipid and bile transport (2). Also, it is a major source of methyl groups in the diet via the formation of betaine, which methylates homocysteine to form methionine (3). The adequate intake (AI) recommended by the Institute of Medicine for choline is 550 and 425 mg/d for men and women, respectively (4), but many Americans are not consuming the AI for choline (5–9). Diets low in choline result in fatty liver, liver damage, and muscle damage (10–12). Moreover, pregnant women eating diets low in choline have a 4-fold increased risk of having a baby with a birth defect (8, 13).
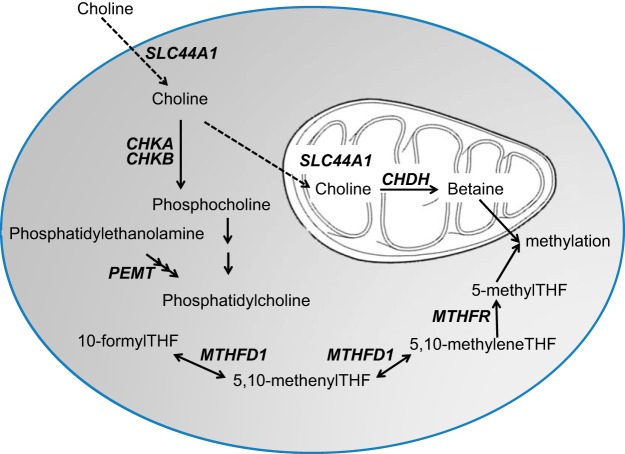
Single-nucleotide polymorphisms (SNPs) in several genes related to choline metabolism alter the dietary requirement for this nutrient (refs. 14, 15 and Fig. 1). For example, phosphatidylethanolamine N-methyltransferase (PEMT) catalyzes the formation of choline moiety as part of phosphatidylcholine (PtdCho) (16) and women possessing the C allele of PEMT rs12325817 were more susceptible to organ dysfunction when consuming a low-choline diet (15, 17). This SNP abrogates estrogen-induced expression of PEMT (18). Oxidation of choline via choline dehydrogenase (CHDH) converts choline to betaine, an important methyl donor. Effect alleles (alleles with a polymorphism that is associated with the effect being measured) in 2 SNPs in CHDH alter the dietary choline requirement (15). Effect alleles in SNPs in the genes for the enzymes involved in folate metabolism also influence the dietary requirement for choline; individuals who were carriers of the very common A allele of 5,10-methylenetetrahydrofolate dehydrogenase (MTHFD1) rs2236225 were more likely than noncarriers to develop signs of choline deficiency on a low-choline diet unless they were also treated with a folic acid supplement (14); individuals who were homozygous carriers of the T allele of methylene tetrahydrofolate reductase (MTHFR) rs1801133 had altered dietary choline requirements (19, 20). It is likely that SNPs in other genes related to the metabolism and functions of choline could also modify the dietary requirement for this nutrient.
Figure 1.
Genes with SNPs that influence dietary choline requirements. Effect alleles in SNPs in MTHFD1, MTHFR, CHDH, PEMT, and CHKA modify the dietary choline requirement in people, and SNPs in CHKB and SLC44A1 alter the phenotype for clinical presentation of choline deficiency (muscle vs. liver damage). THF, tetrahydrofolate.
In this study, we expanded on the limited SNP analyses reported previously and examined a total of 200 SNPs occurring in 10 genes [PtdCho flippase (ABCB4), betaine-homocysteine methyltransferase (BHMT), CHDH, choline kinase A/B (CHKA, CHKB), MTHFR, MTHFD1, PEMT, stearoyl CoA desaturase (SCD), and solute carrier 44A1 (SLC44A1)] related to choline metabolism and function (Fig. 1) for associations with organ dysfunction in humans fed a low-choline diet. In addition, we characterized the distribution patterns of these SNPs in European Americans (EAs), Mexican Americans (MAs), Asian Americans (AsAs), and African descendants (ADs), individuals of African descent, which included African Americans (AfAs), to determine whether potential functional differences in metabolism exist across different racial and ethnic groups.
MATERIALS AND METHODS
Human clinical study
Seventy-nine healthy volunteers (26 men and 53 women), ranging in age from 18 to 70 yr, were recruited for this study (as part of two studies, some results from these studies were previously reported; refs. 10, 17). Subject enrollment began on March 14, 2001 and on June 27, 2007. The ethnic heritages of the subjects were Caucasian (52, 66%), African American (18, 23%), Hispanic (4, 5%), Asian (3, 4%), and Native American (2, 2%), which reflected the local population characteristics of the Raleigh–Durham–Chapel Hill, NC, USA, area. Of the women, 26 were postmenopausal, and 27 were premenopausal. Postmenopausal status was defined as having had last spontaneous menstrual bleeding >12 mo previously and a follicle stimulating hormone (FSH) level of >30 IU/L. The Institutional Review Board at the University of North Carolina (UNC) at Chapel Hill approved the details of subject recruitment and the clinical protocol. Subjects were admitted to the UNC at Chapel Hill Clinical and Translational Research Center (CTRC), where they received specified diets (21), and remained under the constant supervision of study staff for the entire duration of the study.
Initially, all participants received a conventional diet of normal foods containing 550 mg choline/70 kg/d (the current AI level; ref. 4), 50 mg betaine/70 kg/d, and 400 dietary folate equivalents (DFE)/d (baseline). After 10 d on this initial baseline diet, the subjects entered a choline-depletion phase and were fed a low-choline diet containing <50 mg choline/70 kg/d (with 6 mg betaine/70 kg/d), as confirmed by chemical analysis of a sample of duplicate food portions (22). All diets were isocaloric with 30% of kilocalories from fat, 0.8 g high biological value protein per kilogram body weight, and the remaining kilocalories from carbohydrate. These diets also met or exceeded the estimated average requirement for methionine and cysteine, and the dietary reference intake for all vitamins with the exception of folate (some of the subjects received 100 μg DFE/d, and others received 400 μg DFE/d; folate intake did not influence choline-deficiency organ dysfunction; refs. 10, 14, 15). Periodic determinations of urinary choline and betaine concentrations were used to confirm compliance with the dietary restrictions (data not shown). Subjects remained on the depletion diet until they developed organ dysfunction associated with choline deficiency or for 42 d if they did not.
The subjects were deemed to have organ dysfunction associated with choline deficiency if they had a >5-fold increase of serum creatine phosphokinase (CPK) activity or a >1.5 fold increase in aspartate aminotransferase (AST), or if they had an increase in liver fat (measured by magnetic resonance imaging; ref. 10) of >28% while on the choline depletion diet. AST and CPK analyses were performed by the McClendon Clinical Laboratory [UNC Hospitals, Chapel Hill, NC, USA; Clinical Laboratory Improvement Act (CLIA) and College of American Pathologists (CAP) accredited] using blood drawn by venipuncture. Additional samples of blood were collected for genotyping.
Human population study
Human DNA samples were purchased from the Coriell Repositories (Newark, NJ, USA) for the EA, AsA, MA, and AfA populations. The EA group included CEPH samples (Utah residents with ancestry from northern and western Europe from the International HapMap project; HAPMAPPT01). For additional AD subjects, blood was collected from healthy women ranging in age from 18 to 40 yr, recruited by advertising at the Mona prenatal clinic near Kingston, Jamaica. Informed consent was obtained from all participants as part of a larger study on choline intake and cognition. The criteria for subject selection and all the details of the clinical protocol approved by the Institutional Review Board of UNC at Chapel Hill has been previously reported (17).
Genotyping
Peripheral lymphocytes were isolated from blood by Ficoll-Hypaque gradient using Vacutainer CPT tubes with sodium citrate (Becton Dickinson, Franklin Lakes, NJ, USA), and genomic DNA was extracted using phenol-chloroform (23). DNA concentration was measured by spectrometry (NanoDrop Technologies; Wilmington, DE, USA), and aliquots were sent to the Mammalian Genotyping Core at the UNC Lineberger Comprehensive Cancer Center (Chapel Hill, NC, USA) for genotyping using the Illumina GoldenGate oligo-specific extension–ligation assay (Illumina, Inc., San Diego, CA, USA; ref. 24). Briefly, 3 assay oligonucleotides, 2 allele-specific oligonucleotides (ASOs) and one locus-specific oligonucleotide (LSO), are designed for each SNP locus, and scored for likelihood of success. The ASOs for each allele and corresponding LSOs are hybridized to whole genomic DNA from each individual and washed to remove nonhybridized material. These ASOs and LSOs are then extended across the 1- to 20-bp gap between them and ligated to form the template for PCR with allele-specific fluorescently labeled universal primers, the product of which is hybridized to oligonucleotides complementary to the unique address of the LSO anchored to the bead substrate. The relative fluorescence of each allele is then quantified and the genotype determined. Positive CEPH controls and negative controls were included on each plate for quality control. The SNP chip used Genome build 36.2 [U.S. National Center for Biotechnology Information (NCBI), Bethesda, MD, USA). Data are reported with the Illumina Top format, except for rs6591331, rs4646365, and rs7873937, where the major allele was confirmed using the dbSNP database.
The SNP list used for the Illumina bead array was generated by identifying tag SNPs across our genes of interest using HaploView Tagger 4.2 (Broad Institute, Cambridge, MA, USA; ref. 25) and by literature review of publications on the genes to identify additional SNPs that have been associated with phenotypes of interest. The list was refined by eliminating minor alleles that were <1% frequency, and SNPs that were not within the gene of interest, or were intergenic and >5 kb outside the gene boundary. Gene boundaries were determined with the University of California–Santa Cruz (UCSC) Genome Browser (Genome Build 36.1; ref. 26). Several established SNPs (rs12676 and rs9001, CHDH; rs2236225, MTHFD1; rs3733890, BHMT; and rs4646343, PEMT) failed the design score test for the bead array, or had missing data, so they were genotyped by Sequenom matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF) primer extension assay (Sequenom, Inc., San Diego, CA, USA; ref. 27).
PEMT genotyping
For the PEMT SNP rs12325817, which failed the design score test for the bead array and could not be genotyped using Sequenom methodology because it was located in an Alu repeat-rich region, an allele-specific PCR (AS-PCR) method was used for genotype detection. Forward primers were designed for each allele using GeneFisher (Bielefeld University, Bielefeld, Germany; http://bibiserv.techfak.uni-bielefeld.de/genefisher) so that the SNP would be located at the 3′-end of the priming sequence that allows for specific PCR products to be amplified only if the primer is 100% complementary to its priming DNA sequences. For the PCR reactions (50 μl total volume), 80 ng DNA was added to a mixture containing 0.5 μM primers (forward for G allele, 5′-CCTGGACAACATGGTGACACTCC-3′ or forward for C allele, 5′CCTGGACAACATGGTGACACTCG-3′ and reverse, 5′-GTGGGCCCTGTACTTTTACATC-3′; Operon Biotechnologies, Huntsville, AL, USA) and 25 μl Takara enzyme SYBR mix (Takara Bio, Shiga, Japan). Each pair of primers (the forward primer with the common reverse primer, respectively) was run as a separate reaction. Successful amplification was performed on an Ep Realplex 1.5 Thermalcycler (Eppendorf, Westbury, NY, USA) using the following conditions: preheating at 95°C for 10 s; 30 cycles of denaturing at 94°C for 30 s, annealing at 65.5°C for 30 s, and extension at 72°C for 30 s; followed by final elongation at 72°C for 10 min and melting curve analysis. Products were visualized on 1% agarose gel with ethidium bromide. Genotype calls for this SNP were made based on the coding strand for PEMT. Genotype results obtained by RT-PCR were first confirmed in a subset of samples by agarose gel electrophoresis, sequencing, and PCR-restriction fragment length polymorphism (RFLP) using the following primers: forward, 5′-TAGATTGGTCATGGGAGGCTT-3′; and reverse, 5′-GGCTTTCTGCTACCCAGTAAT-3′ (15). The 1420-bp PCR products were digested with BsmBI (New England Biolabs Inc., Ipswich, MA, USA), thereby cutting the wild-type G allele into two products of 132 and 92 bp. Products were size fractionated on a 0.9% agarose gel and visualized with ethidium bromide. The genotypes were in agreement for all three methods, with 100% calls successful. For the RT-PCR, blanks and known genotypes were run as controls for every batch.
Statistical analysis
We used the Fisher's exact test to test for an association between SNPs and depletion status or ethnicity. The Benjamini-Hochberg method for false discovery rate (FDR) was used to adjust for multiple tests, utilizing the minimum number of independent tests approach or by adjusting for the number of variables tested (28). Spearman rank correlations were calculated for the allele burden of SNPs associated with muscle dysfunction. All analyses were performed using SAS 9.2 (SAS Institute, Cary, NC, USA). Assessment of linkage disequilibrium (LD) was conducted with Genome Variation Server 137 (University of Washington, Seattle, WA, USA; http://gvs.gs.washington.edu/GVS137/index.jsp). This was calculated with all available genotyping data across ethnic groups.
RESULTS
Novel SNPs that alter dietary choline requirements
Fifty of the people who completed the human choline study developed organ dysfunction (liver or muscle) when they were fed a low-choline diet. Of the 53 women in the study, 20 of the 26 postmenopausal (77%) and 11 of the 27 premenopausal (41%) women had clinical signs of choline deficiency. All but 2 had liver dysfunction, which resolved when choline was returned to the diet. Nineteen (73%) of the men developed liver dysfunction, muscle dysfunction or both (10). The remaining 29 subjects showed no clinical signs after consuming a low-choline diet for 42 d. Using DNA from these subjects, we identified several SNPs that were associated with the development of organ dysfunction (liver or muscle) in people fed a low-choline diet from a panel of 171 SNPs identified in ABCB4, BHMT, CHDH, CHKA, MTHFR, MTHFD1, PEMT and SCD (Table 1).
Table 1.
SNPs associated with organ dysfunction in humans fed a low-choline diet
| Group and gene | SNP |
P |
Direction of effect | |
|---|---|---|---|---|
| Raw | Adjusted | |||
| Women, n = 53 | ||||
| PEMT | rs12325817, G→C | 0.0003 | 0.0074 | Increased |
| PEMT | rs4646343, C→A | 0.0004 | 0.0074 | Increased |
| CHKA | rs7928739, A→C | 0.0005 | 0.0074 | Decreaseda |
| CHKA | rs10791957, A→C | 0.0006 | 0.0074 | Decreaseda |
| CHKA | rs2512612, A→G | 0.0009 | 0.0096 | Decreaseda |
| PEMT | rs3760188, G→A | 0.0047 | 0.0416 | Increased |
| Postmenopausal women, n = 26 | ||||
| PEMT | rs12325817, G→C | 0.0006 | 0.0312 | Increased |
| PEMT | rs3760188, G→A | 0.0019 | 0.0312 | Increased |
| PEMT | rs4646343, C→A | 0.0019 | 0.0312 | Increased |
| CHKA | rs6591331, A→T | 0.0034 | 0.0312 | Increasedb |
| PEMT | rs1531100, G→A | 0.0035 | 0.0312 | Increased |
| PEMT | rs4646365, G→A | 0.0035 | 0.0312 | Increased |
Effect alleles for SNPs listed have a raw value of P < 0.05. Statistical correction for multiple testing is described in Materials and Methods. For the SNP, the arrow points to the allele for which there is an effect (increased or decreased risk of developing organ dysfunction when fed a low choline diet).
Only homogyzous carriers of the effect allele for SNP were protected from choline deficiency.
There was only 1 TT in this group, so a larger sample size is needed to confirm this finding.
As discussed earlier, PEMT is an estrogen-responsive gene that catalyzes the formation of new choline moiety as part of PtdCho (29). In the groups women and postmenopausal women, the effect alleles of 3 PEMT SNPs (rs12325817, C; rs4646343, A; and rs3760188, A) were strongly associated with developing organ dysfunction when fed a low-choline diet (Table 1). Postmenopausal women who were carriers of the effect allele for PEMT rs1531100 (A) and rs4646365 (A) (these SNPs are in LD; r2=1.0) also had an increased risk of developing organ dysfunction when they consumed a diet containing ∼10% of the AI for choline (Table 1). After adjusting for multiple testing, there was no association with other SNPs and developing organ dysfunction when fed a low-choline diet in the groups of all subjects (n=79), men (n=26), or premenopausal women (n=27); however, we have listed those SNPs that have a raw value of P < 0.05 in Supplemental Table S1.
Phosphorylation by choline kinase (encoded by CHKA, CHKB) commits choline molecules to a pathway for PtdCho synthesis rather than to be used as a methyl group donor (16). Women carriers homozygous for the effect allele of the 3 CHKA SNPs rs10791957 (C), rs7928739 (C), and rs2512612 (G) were protected when they were fed the low-choline diet, while only postmenopausal women who were carriers of the T allele of CHKA rs6591331 were more likely to develop organ dysfunction when fed a low-choline diet (Table 1).
SNPs that alter the clinical presentation of choline deficiency
Thirteen percent of our subjects (8 men and 2 premenopausal women) fed a low-choline diet developed muscle damage (elevated CPK) that resolved when choline was returned to the diet. We hypothesized that these people presented with muscle rather than liver dysfunction because they had a polymorphism in a gene that influenced choline availability and metabolism in muscle. Thus, in a separate analysis, we examined the effects on low-choline-associated muscle dysfunction of 29 polymorphisms in SLC44A1 (needed for transport of choline into muscle mitochondria; refs. 30, 31) and CHKB (deletion of this gene in mice results in muscular dystrophy; ref. 32). We found several SLC44A1 polymorphisms and one CHKB polymorphism that were associated with low-choline-related muscle damage (Table 2). Those individuals who developed muscle damage when fed a low-choline diet were likely to have the G allele of SLC44A1 rs7873937 (P=0.002), and 9 of the 10 of these individuals also had the G allele of SLC44A1 rs2771040 and rs6479313 (Table 2). These two latter SNPs are in LD (r2=0.924). To determine whether a set of the effect alleles of the SNPs in Table 2 could identify muscle depleters, as compared to the other subjects, we looked at how well the SNPs were correlated in the two groups. We removed rs6479313 from the analysis because it is in high LD with rs2771040, so any correlation with rs2771040 would be the same for rs6479313 in all subjects. We found that 9 of 10 people who developed muscle damage when fed a low-choline diet were heterozygous or homozygous carriers of the effect alleles for SLC44A1 rs2771040 (G) and CHKB rs1557502 (A) (Spearman ρ=1, P<0.0001). These two polymorphisms were not correlated in people who did not develop muscle damage when fed a low-choline diet (ρ=0.03, P = 0.77).
Table 2.
SLC44A1 and CHKB SNPs were associated with subjects who developed muscle damage when fed a low-choline diet
| Gene | SNP |
P |
Direction of effect | |
|---|---|---|---|---|
| Raw | Adjusted | |||
| SLC44A1 | rs7873937, C→G | 0.0001 | 0.0022 | Increased |
| SLC44A1 | rs2771040, A→G | 0.0003 | 0.0024 | Increased |
| SLC44A1 | rs6479313, C→G | 0.0004 | 0.0024 | Increased |
| SLC44A1 | rs16924529, G→A | 0.0019 | 0.0100 | Increased |
| CHKB | rs1557502, G→A | 0.0112 | 0.0426 | Increased |
| SLC44A1 | rs440290, A→G | 0.0122 | 0.0426 | Increased |
| SLC44A1 | rs3199966, A→C | 0.0164 | 0.0493 | Increased |
SNPs listed have adjusted values of P <0.05. Statistical analyses are described in Materials and Methods.
Distribution of SNPs that alter dietary choline requirements across ethnic and racial groups
Our previous work suggests that the function of the 1-carbon metabolic pathway differs between Caucasians and AfAs and that this difference influences hepatic lipid deposition (27). In addition, other groups have shown differences in various metabolic pathways, including the 1-carbon pathway, in AfA individuals (33, 34). Therefore, we aimed to assess the distribution of SNPs that we identified to be associated with choline requirements across various ethnic groups.
The 3 PEMT effect alleles that were associated with liver dysfunction when women consumed a low-choline diet (rs12325817, C; rs4646343, A; and rs3760188, A) are in LD (r2=0.949 for rs3760188 and rs4646343; 0.898 for rs3760188 and rs12325817; and 0.826 for rs4646343 and rs12325817; Table 3). About 24% of the EA, 11% of the MA, and 1 and 0% of the AsA and AD groups, respectively, were homozygous for the effect allele of these SNPs (P<0.0001). The effect allele for PEMT rs1531100 (A) and rs4646365 (A) are in LD and were more prevalent in the EA and MA groups (77 and 69%, respectively, had the A allele) compared to the AsA and AD groups, which had a similar distribution of the effect allele, 42% as heterozygotes and 6% as homozygotes (P<0.0001; Table 3). PEMT rs7946 (effect allele A), which causes a Val → Met substitution in the enzyme, has a very different distribution in the population groups. In the AsA and AD groups, the A allele was present in 51 and 30% of people, respectively. However, >80% of the EA and MA individuals had the A allele, with 60 and 30%, respectively, being homozygous for this effect allele of the SNP (P<0.0001; Table 3).
Table 3.
Population distribution of SNPs associated with organ dysfunction in humans fed a low-choline diet
| Gene and SNP | Frequency (%) |
Differences between groups | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EA, n = 102–113 |
AD, n = 88–109 |
AsA, n = 97–100 |
MA, n = 99–100 |
||||||||||
| OO | OE | EE | OO | OE | EE | OO | OE | EE | OO | OE | EE | ||
| PEMT | |||||||||||||
| rs12325817, G→C | 26 | 50 | 24 | 80 | 20 | 0 | 72 | 27 | 1 | 38 | 50 | 12 | EA > MA > AsA > AD |
| rs4646343, C→A | 25 | 50 | 25 | 82 | 18 | 0 | 83 | 16 | 1 | 41 | 50 | 9 | EA > MA > AsA, AD |
| rs3760188, G→A | 26 | 50 | 24 | 81 | 19 | 0 | 71 | 28 | 1 | 37 | 51 | 12 | EA > MA > AsA > AD |
| rs1531100, G→A | 23 | 49 | 28 | 52 | 42 | 6 | 52 | 42 | 6 | 31 | 52 | 17 | EA, MA > AsA, AD |
| rs7946, G→A | 10 | 30 | 60 | 49 | 39 | 12 | 70 | 28 | 2 | 17 | 53 | 30 | EA > MA > AD > AsA |
| CHKA | |||||||||||||
| rs10791957, A→C | 31 | 45 | 24 | 19 | 42 | 39 | 34 | 47 | 19 | 51 | 45 | 4 | AD > AsA, EA > MA |
| rs2512612, A→G | 30 | 45 | 25 | 31 | 44 | 25 | 34 | 47 | 19 | 51 | 45 | 4 | AD, EA, AsA > MA |
| rs7928739, A→C | 32 | 45 | 23 | 11 | 41 | 48 | 34 | 47 | 19 | 50 | 47 | 3 | AD > AsA, EA > MA |
| rs6591331, A→T | 38 | 44 | 18 | 49 | 41 | 10 | 35 | 48 | 17 | 57 | 40 | 3 | AsA > AD, MA; EA > MA |
For each SNP, the distribution of the effect allele is significantly different between the 4 ethnic groups (P<0.0001; Fisher test), except for CHKA rs6591331 (P=0.0015). O, other allele (no effect); E, effect allele.
Women who were homozygous for the first 3 CHKA SNPs found in Table 3 were less likely to have clinical symptoms after consuming a low-choline diet. These 3 SNPs, rs10791957 (C), rs2512612 (G), and rs7928739 (C), are in LD (r2=0.839 for rs2512612 and rs7928739; 0.902 for rs2512612 and rs10791957; 0.901 for rs7928739 and rs10791957). Up to 48% of the AD group was homozygous for the effect alleles of these 3 SNPs, followed by the EA (24%), AsA (19%), and MA (4%) groups (P<0.0001; Table 4). For rs6591331, only 10% of the AD group was homozygous for the effect T allele of the SNP, compared to 18% in the EA and AsA groups, and 3% in the MA group (P<0.01; Table 3).
Table 4.
Population distribution of SNPs found to be associated with choline deficiency-induced muscle dysfunction
| Gene and SNP | Frequency (%) |
Differences between groups | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EA, n = 102–113 |
AD, n = 88–109 |
AsA, n = 97–100 |
MA, n = 99–100 |
||||||||||
| OO | OE | EE | OO | OE | EE | OO | OE | EE | OO | OE | EE | ||
| SLC44A1 | |||||||||||||
| rs16924529, G→A | 85 | 15 | 0 | 76 | 21 | 3 | 100 | 0 | 0 | 74 | 25 | 1 | AD, MA > EA > AsA |
| rs2771040, A→G | 76 | 22 | 2 | 29 | 51 | 20 | 100 | 0 | 0 | 69 | 30 | 1 | AD > EA, MA > AsA |
| rs3199966, A→C | 82 | 18 | 0 | 32 | 48 | 20 | 100 | 0 | 0 | 71 | 28 | 1 | AD > EA, MA > AsA |
| rs7873937, C→G | 79 | 19 | 2 | 76 | 13 | 11 | 100 | 0 | 0 | 73 | 27 | 0 | AD, EA, MA > AsA |
| CHKB | |||||||||||||
| rs1557502, G→A | 73 | 25 | 2 | 13 | 50 | 38 | 27 | 51 | 22 | 47 | 39 | 14 | AD > AsA > MA > EA |
For each SNP, the distribution of the effect allele is significantly different between the 4 ethnic groups (P<0.0001; Fisher test). O, other allele (no effect); E, effect allele.
The SLC44A1 rs7873937 effect allele G was present in ∼24% of the EA, MA, and AD groups, but was not present in the AsA group (P<0.0001; Table 4). In fact, none of the effect alleles in SLC44A1 SNPs associated with low-choline-related muscle damage was present in the AsA population (P<0.0001; we did not have data for the rs6479313 SNP in the population groups because it failed the bead array analysis). Seventy-one percent of the AD group had the SLC44A1 rs2771040 effect allele G compared with 24% of the EA group and 31% of the MA group (P < 0.0001; Table 4). This distribution was similar for rs3199966, as well. Eighty-eight percent of the AD group had the effect allele A of CHKB rs1557502 (38% were homozygous), as well as more than half of the AsA and MA groups, while only 27% of the EA group had the effect allele of this SNP (P<0.0001; Table 4).
We report distributions of other SNPs measured in Supplemental Tables S2 and S3. These data were compared to those frequencies in similar ethnic groups that have been analyzed by others and are available for viewing at the NCBI SNP Database (dbSNP; http://www.ncbi.nlm.nih.gov/projects/SNP/) and from the 1000 Genomes Browser website (http://browser.1000genomes.org). Most of our frequencies were similar or had a similar pattern to those found on the databases when the EA, AD, AsA and MA groups were compared to the Utah residents with northern and western European ancestry (CEU), Yoruba in Ibadan, Nigera (YRI), southern Han Chinese (CHS), and Mexican ancestry in Los Angeles, CA, USA (MXL) groups, respectively. There were 10% less carriers of the effect alleles of the CHKA SNPs in the AsA group (Table 3) than the CHS group, and the MA group had more heterozygous carriers of the effect alleles for the SLC44A1 SNPs compared to the MXL group (Table 4). There were also some notable differences in the AD group, which is a unique mix of Americans and Jamaicans of African ancestry: For the first 3 PEMT SNPs in Table 3, the YRI group has 10% heterozygotes while we and the composite African group (AFR) report 20%. In our AD group, 24% had the effect allele for SLC44A1 rs7873937 (Table 4) but the database reports 64% of the YRI group with this G allele, and 14% in the AFR group. For CHKB rs1557502, there was no difference in the total frequency of the A effect allele, but 50% of the AD group are heterozygous compared to the 50% that are homozygous for the A allele on the database (Table 4). Finally, the YRI group had 10% more carriers of the effect alleles of CHDH rs9001 and rs7634578, MTHFD1 rs1256146 and rs8003567, and BHMT rs3733890 compared to the AD group (Supplemental Table S2).
DISCUSSION
We identified several genetic polymorphisms that were associated with altered choline requirements in humans (Table 1). These polymorphisms occurred in PEMT and CHKA. Both of these genes are important for PtdCho synthesis; however, only PEMT produces new choline moiety by methylating phosphatidylethanolamine (choline kinase phosphorylates preexisting choline moiety; ref. 16 and Fig. 1). PEMT is induced by estrogen (29), and for this reason, these genetic polymorphisms are most important in determining the dietary choline requirement for women. Some subjects (notably young women) did not develop clinical symptoms when fed the low-choline diet for 6 wk, even though they had the effect allele of PEMT rs12325817. Though it is possible that these women are “facultative cholinevores,” the increased demand for choline during pregnancy may result in a dietary requirement for choline in these women (35).
Since the 3 PEMT SNPs described are in LD, it is not surprising that they are found within a 5-kb region of the first intron of the gene. From the work by Walkey et al. (36), it appears that rs12325817 is in the PEMT promoter region, an ideal location to change gene function, and we have previously shown that having the effect alleles of rs12325817 and rs4646343 decrease the gene's response to estrogen (18).
It is intriguing that carriers homozygous for the effect alleles from 3 CHKA SNPs were protected when dietary choline was limited. It is not known whether having the effect allele of the SNPs we report here increases or decreases CHKA activity; thus, we do not know whether the protective effect is caused by a more efficient enzyme resulting in increased flux of choline into the CDP-choline pathway for PtdCho biosynthesis (37), or by a less efficient enzyme that makes more choline available for betaine synthesis and eventual methyl group donation (ref. 38 and Fig. 1). Having at least one effect allele (C) of CHKA rs7928739 has been associated with a decreased risk of spina bifida (39). This would be consistent with the protection from low-choline-associated organ dysfunction identified in this study, as low dietary choline intake increases the risk for neural tube defects (8). Moreover, CHKA rs10791957 and rs6591331 are located in the first intron of the gene, in a region that may play a regulatory role as an enhancer, increasing the transcription levels of the gene (UCSC Genome Browser, H3K27Ac tracks; http://genome.ucsc.edu).
When fed a low-choline diet, most people develop liver dysfunction, but some (13%) present first with muscle dysfunction (10). We found that these individuals usually have effect alleles in SNPs in SLC44A1, the gene that encodes for the choline transporter needed for the nutrient to cross plasma and mitochondrial membranes (31). SLC44A1 is expressed ubiquitously, and is down-regulated by choline deficiency (40). Changes in its expression and activity, possibly due to polymorphisms, have implicated SLC44A1 in human diseases such as breast cancer, Alzheimer's disease and neonatal respiratory distress syndrome (30, 41). Perhaps, some people with polymorphisms in this gene have limited capacity to transport choline into muscle cells to maintain membrane integrity and fluidity, leaving those cells to leak CPK (42), and therefore, they are especially sensitive to diets low in choline.
Of the SLC44A1 SNPs in Table 2, only rs3199966 is exonic, causing an amino acid change from serine to alanine that can affect the function of the gene. Another important site for regulating gene function is the 3′ untranslated region (UTR) of the gene where SLC44A1 rs2771040 is located. The C allele of rs3199966 only occurs together with rs2771040 G allele on the same strand. This strong selection may be for viability; that is, the nonsynonymous substitution has to be paired with the variant in the 3′ UTR to make up for decreased specific activity by synthesizing more of the enzyme protein.
The effect allele (A) in CHKB rs1557502 was also associated with an increased risk of low-choline-associated muscle damage. As noted earlier, mutation of this gene in mice results in muscular dystrophy (32). Mutations in CHKB have also been reported in humans, causing loss of function of the enzyme, and resulting in megaconial congenital muscular dystrophy (43). This condition, characterized by mitochondrial enlargement at the periphery of muscle fibers, and loss of mitochondria in the center of the muscle fibers, underscores the importance of PtdCho synthesis in muscle, particularly PtdCho containing docosahexaenoic acid (43). It is interesting that, in the group susceptible to muscle damage, the effect alleles in the SLC44A1 and CHKB genes (on two different chromosomes) are linked. Perhaps there is an epistatic relationship between these two genes, where a variant at one locus manifests its effect when a variant at another locus is present. We suggest that pathways related to choline metabolism are more important for normal muscle function than previously thought, and investigators should consider the role of diet and genetic variation in the etiology of disorders that are associated with elevated release of CPK from muscle.
Consistent with our hypothesis that 1-carbon metabolism differs between ethnic and racial groups, carriers of the effect alleles of SNPs that alter dietary choline requirements are distributed differentially across ethnic groups and there were significant differences in distribution in all 28 SNPs examined in the population groups (Tables 3 and 4 and Supplemental Tables). Effect alleles for the SNPs studied were present in all populations; however, the prevalence of these SNPs varied greatly across the populations. For example, the AD group had the highest percentage of effect alleles in 11 SNPs (6 that decreased the risk of organ dysfunction and 5 that increased the risk when dietary choline was low), while the AsA group had the lowest percentage of effect alleles across 10 of the SNPs. Fourteen of the effect alleles in SNPs were associated with increasing the risk of organ dysfunction when dietary choline was low; 10 with decreasing the risk, and 4 are thought to influence the choline → betaine (rs9001) → homocysteine (rs3733890, rs1801133) flux. Since the AD group had the lowest number of carriers of the effect alleles from the 3 PEMT SNPs that increase the risk of organ dysfunction when on a low-choline diet (0% homozygous for the SNP), and the highest number of those with the protective CHKA SNPs, it is possible that women in the AD group would be less likely to develop organ dysfunction when dietary choline is limited. The AD group came primarily from West African tribes, transported to the Caribbean and North America as slaves. We previously reported that dietary choline intake is low in people currently living in West Africa (The Gambia; ref. 44). Perhaps, exposure to diet low in choline for many generations selects for genetic polymorphisms that reduce the dietary requirement for this nutrient. The EA group had the highest prevalence of 4 SNPs that increased the risk of organ dysfunction when a low-choline diet is consumed, perhaps reflecting a higher intake of choline in the European diet.
The development of dietary recommendations by nutrition scientists and governmental organizations has not often considered genetic diversity. For choline, our work suggests that, as well as gender and life stage, genetic polymorphisms in several genes, including novel genetic variants that we identify for the first time, influence the dietary requirement for choline, as well as influence the phenotype that people present with when fed a low-choline diet (liver vs. muscle damage). Because these polymorphisms are differentially distributed among different racial and ethnic populations, we suggest that dietary requirement for choline will differ for these different populations. Nevertheless, the easier and safer course may be to set the dietary recommendations at a level that is high enough to meet the needs of those with the greatest requirements for choline.
Supplementary Material
Acknowledgments
This research was supported by grants from the U.S. National Institutes of Health to S.H.Z. (DK55865), the University of North Carolina (UNC) Nutrition and Obesity Research Center (DK56350), the UNC Gastrointestinal Research Center (DK03498) and the UNC Clinical and Translational Research Center (M01RR00046 and UL1RR025747).
The clinical study is registered at ClinicalTrials.gov (NCT00065546; http://www.clinicaltrials.gov). The authors thank the Mammalian Genotyping Core at UNC Chapel Hill for their assistance on this project, and especially Amanda Beaty for her patience and generosity. The authors also thank Mihai George Mehedint, Huili Wang, Hye Mee Hwang and Robin Betsch for their technical assistance in this study, and Fred Wright for his statistical advice. Author contributions: K.-A.D. was responsible for sample collection, genotyping, data interpretation and preparation of the manuscript; K.D.C. assisted with the genotyping, SNP analysis, and preparation of the manuscript; M.D.N. was responsible for the design and setup of the PEMT rs12325817 genotyping; J.A.G. assisted with statistical analyses; S.H.Z. was responsible for overall experimental design and data interpretation and assisted with the preparation of the manuscript. The authors declare no conflicts of interest.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- ABCB4
- phosphatidylcholine flippase
- AD
- African descendant
- AfA
- African American
- AI
- adequate intake
- AsA
- Asian American
- ASO
- allele-specific oligonucleotide
- AS-PCR
- allele-specific PCR
- AST
- aspartate aminotransferase
- BHMT
- betaine-homocysteine methyltransferase
- CHDH
- choline dehydrogenase
- CHKA
- choline kinase A
- CHKB
- choline kinase B
- CPK
- creatine phosphokinase
- DFE
- dietary folate equivalent
- EA
- European American
- LD
- linkage disequilibrium
- LSO
- locus-specific oligonucleotide
- MA
- Mexican American
- MTHFD1
- 5,10-methylenetetrahydrofolate dehydrogenase
- MTHFR
- methylene tetrahydrofolate reductase
- PEMT
- phosphatidylethanolamine N-methyltransferase
- PtdCho
- phosphatidylcholine
- RFLP
- restriction fragment length polymorphism
- SCD
- stearoyl CoA desaturase
- SLC44A1
- solute carrier 44A1
- SNP
- single-nucleotide polymorphism
REFERENCES
- 1. Zeisel S. H. (2006) Choline: critical role during fetal development and dietary requirements in adults. Annu. Rev. Nutr. 26, 229–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li Z., Agellon L. B., Vance D. E. (2005) Phosphatidylcholine homeostasis and liver failure. J. Biol. Chem. 280, 37798–37802 [DOI] [PubMed] [Google Scholar]
- 3. Collinsova M., Strakova J., Jiracek J., Garrow T. A. (2006) Inhibition of betaine-homocysteine S-methyltransferase causes hyperhomocysteinemia in mice. J. Nutr. 136, 1493–1497 [DOI] [PubMed] [Google Scholar]
- 4. Institute of Medicine and U.S. National Academy of Sciences (1998) Choline. In Dietary Reference Intakes for Folate, Thiamin, Riboflavin, Niacin, Vitamin B12, Panthothenic Acid, Biotin, and Choline, Vol. 1, pp. 390–422, National Academy Press, Washington, DC: [PubMed] [Google Scholar]
- 5. Cho E., Willett W. C., Colditz G. A., Fuchs C. S., Wu K., Chan A. T., Zeisel S. H., Giovannucci E. L. (2007) Dietary choline and betaine and the risk of distal colorectal adenoma in women. J. Natl. Cancer Inst. 99, 1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee J. E., Giovannucci E., Fuchs C. S., Willett W. C., Zeisel S. H., Cho E. (2010) Choline and betaine intake and the risk of colorectal cancer in men. Cancer Epidemiol. Biomarkers Prev. 19, 884–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cho E., Zeisel S. H., Jacques P., Selhub J., Dougherty L., Colditz G. A., Willett W. C. (2006) Dietary choline and betaine assessed by food-frequency questionnaire in relation to plasma total homocysteine concentration in the Framingham Offspring Study. Am. J. Clin. Nutr. 83, 905–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shaw G. M., Carmichael S. L., Yang W., Selvin S., Schaffer D. M. (2004) Periconceptional dietary intake of choline and betaine and neural tube defects in offspring. Am. J. Epidemiol. 160, 102–109 [DOI] [PubMed] [Google Scholar]
- 9. Chester D., Goldman J., Ahuja J., Moshfegh A. (2011) Dietary intakes of choline. What we eat in America, NHANES 2007–2008. http://www.ars.usda.gov/SP2UserFiles/Place/12355000/pdf/DBrief/9_choline_intakes_0708.pdf
- 10. Fischer L. M., da Costa K. A., Kwock L., Stewart P. W., Lu T. S., Stabler S. P., Allen R. H., Zeisel S. H. (2007) Sex and menopausal status influence human dietary requirements for the nutrient choline. Am. J. Clin. Nutr. 85, 1275–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zeisel S. H., da Costa K.-A., Franklin P. D., Alexander E. A., Lamont J. T., Sheard N. F., Beiser A. (1991) Choline, an essential nutrient for humans. FASEB J. 5, 2093–2098 [PubMed] [Google Scholar]
- 12. Buchman A., Dubin M., Moukarzel A., Jenden D., Roch M., Rice K., Gornbein J., Ament M. (1995) Choline deficiency: a cause of hepatic steatosis during parenteral nutrition that can be reversed with intravenous choline supplementation. Hepatology 22, 1399–1403 [PubMed] [Google Scholar]
- 13. Shaw G. M., Carmichael S. L., Laurent C., Rasmussen S. A. (2006) Maternal nutrient intakes and risk of orofacial clefts. Epidemiology 17, 285–291 [DOI] [PubMed] [Google Scholar]
- 14. Kohlmeier M., da Costa K. A., Fischer L. M., Zeisel S. H. (2005) Genetic variation of folate-mediated one-carbon transfer pathway predicts susceptibility to choline deficiency in humans. Proc. Natl. Acad. Sci. U.S.A. 102, 16025–16030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. da Costa K. A., Kozyreva O. G., Song J., Galanko J. A., Fischer L. M., Zeisel S. H. (2006) Common genetic polymorphisms affect the human requirement for the nutrient choline. FASEB J. 20, 1336–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. DeLong C. J., Shen Y. J., Thomas M. J., Cui Z. (1999) Molecular distinction of phosphatidylcholine synthesis between the CDP-choline pathway and phosphatidylethanolamine methylation pathway. J. Biol. Chem. 274, 29683–29688 [DOI] [PubMed] [Google Scholar]
- 17. Fischer L. M., da Costa K. A., Kwock L., Galanko J., Zeisel S. H. (2010) Dietary choline requirements of women: effects of estrogen and genetic variation. Am. J. Clin. Nutr. 92, 1113–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Resseguie M. E., da Costa K. A., Galanko J. A., Patel M., Davis I. J., Zeisel S. H. (2011) Aberrant estrogen regulation of PEMT results in choline deficiency-associated liver dysfunction. J. Biol. Chem. 286, 1649–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shin W., Yan J., Abratte C. M., Vermeylen F., Caudill M. A. (2010) Choline intake exceeding current dietary recommendations preserves markers of cellular methylation in a genetic subgroup of folate-compromised men. J. Nutr. 140, 975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yan J., Wang W., Gregory J. F., 3rd, Malysheva O., Brenna J. T., Stabler S. P., Allen R. H., Caudill M. A. (2011) MTHFR C677T genotype influences the isotopic enrichment of one-carbon metabolites in folate-compromised men consuming d9-choline. Am. J. Clin. Nutr. 93, 348–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Busby M. G., Fischer L., Da Costa K. A., Thompson D., Mar M. H., Zeisel S. H. (2004) Choline- and betaine-defined diets for use in clinical research and for the management of trimethylaminuria. J. Am. Diet. Assoc. 104, 1836–1845 [DOI] [PubMed] [Google Scholar]
- 22. Zeisel S. H., Mar M. H., Howe J. C., Holden J. M. (2003) Concentrations of choline-containing compounds and betaine in common foods. J. Nutr. 133, 1302–1307 [DOI] [PubMed] [Google Scholar]
- 23. Sambrook J., Fritsch E., Maniatis T. (2001) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA [Google Scholar]
- 24. Shen R., Fan J. B., Campbell D., Chang W., Chen J., Doucet D., Yeakley J., Bibikova M., Wickham Garcia E., McBride C., Steemers F., Garcia F., Kermani B. G., Gunderson K., Oliphant A. (2005) High-throughput SNP genotyping on universal bead arrays. Mutat. Res. 573, 70–82 [DOI] [PubMed] [Google Scholar]
- 25. Barrett J. C., Fry B., Maller J., Daly M. J. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 [DOI] [PubMed] [Google Scholar]
- 26. Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., Haussler D. (2002) The human genome browser at UCSC. Genome Res. 12, 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Corbin K. D., Abdelmalek M. F., Spencer M. D., da Costa K. A., Galanko J. A., Sha W., Suzuki A., Guy C. D., Cardona D. M., Torquati A., Diehl A. M., Zeisel S. H. (2013) Genetic signatures in choline and 1-carbon metabolism are associated with the severity of hepatic steatosis. FASEB J. 27, 1674–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gao X., Starmer J., Martin E. R. (2008) A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet. Epidemiol. 32, 361–369 [DOI] [PubMed] [Google Scholar]
- 29. Resseguie M., Song J., Niculescu M. D., da Costa K. A., Randall T. A., Zeisel S. H. (2007) Phosphatidylethanolamine N-methyltransferase (PEMT) gene expression is induced by estrogen in human and mouse primary hepatocytes. FASEB J. 21, 2622–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Michel V., Bakovic M. (2012) The ubiquitous choline transporter SLC44A1. Cent. Nerv. Syst. Agents Med. Chem. 12, 70–81 [DOI] [PubMed] [Google Scholar]
- 31. Michel V., Bakovic M. (2009) The solute carrier 44A1 is a mitochondrial protein and mediates choline transport. FASEB J. 23, 2749–2758 [DOI] [PubMed] [Google Scholar]
- 32. Wu G., Vance D. E. (2010) Choline kinase and its function. Biochem. Cell Biol. 88, 559–564 [DOI] [PubMed] [Google Scholar]
- 33. Kimm S. Y., Glynn N. W., Aston C. E., Damcott C. M., Poehlman E. T., Daniels S. R., Ferrell R. E. (2002) Racial differences in the relation between uncoupling protein genes and resting energy expenditure. Am. J. Clin. Nutr. 75, 714–719 [DOI] [PubMed] [Google Scholar]
- 34. Hung J., Abratte C. M., Wang W., Li R., Moriarty D. J., Caudill M. A. (2008) Ethnicity and folate influence choline status in young women consuming controlled nutrient intakes. J. Am. Coll. Nutr. 27, 253–259 [DOI] [PubMed] [Google Scholar]
- 35. Zeisel S. H., Mar M.-H., Zhou Z.-W., da Costa K.-A. (1995) Pregnancy and lactation are associated with diminished concentrations of choline and its metabolites in rat liver. J. Nutr. 125, 3049–3054 [DOI] [PubMed] [Google Scholar]
- 36. Walkey C. J., Shields D. J., Vance D. E. (1999) Identification of three novel cDNAs for human phosphatidylethanolamine N-methyltransferase and localization of the human gene on chromosome 17p11.2. Biochim. Biophys. Acta 1436, 405–412 [DOI] [PubMed] [Google Scholar]
- 37. Kennedy E. P., Weiss S. B. (1956) The function of cytidine coenzymes in the biosynthesis of phospholipids. J. Biol. Chem. 222, 193–214 [PubMed] [Google Scholar]
- 38. Teng Y. W., Mehedint M. G., Garrow T. A., Zeisel S. H. (2011) Deletion of betaine-homocysteine S-methyltransferase in mice perturbs choline and 1-carbon metabolism, resulting in fatty liver and hepatocellular carcinomas. J. Biol. Chem. 286, 36258–36267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Enaw J. O., Zhu H., Yang W., Lu W., Shaw G. M., Lammer E. J., Finnell R. H. (2006) CHKA and PCYT1A gene polymorphisms, choline intake and spina bifida risk in a California population. BMC Med. 4, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Michel V., Singh R. K., Bakovic M. (2011) The impact of choline availability on muscle lipid metabolism. Food Funct. 2, 53–62 [DOI] [PubMed] [Google Scholar]
- 41. Traiffort E., O'Regan S., Ruat M. (2013) The choline transporter-like family SLC44: properties and roles in human diseases. Mol. Aspects. Med. 34, 646–654 [DOI] [PubMed] [Google Scholar]
- 42. da Costa K. A., Badea M., Fischer L. M., Zeisel S. H. (2004) Elevated serum creatine phosphokinase in choline-deficient humans: mechanistic studies in C2C12 mouse myoblasts. Am. J. Clin. Nutr. 80, 163–170 [DOI] [PubMed] [Google Scholar]
- 43. Mitsuhashi S., Ohkuma A., Talim B., Karahashi M., Koumura T., Aoyama C., Kurihara M., Quinlivan R., Sewry C., Mitsuhashi H., Goto K., Koksal B., Kale G., Ikeda K., Taguchi R., Noguchi S., Hayashi Y. K., Nonaka I., Sher R. B., Sugimoto H., Nakagawa Y., Cox G. A., Topaloglu H., Nishino I. (2011) A congenital muscular dystrophy with mitochondrial structural abnormalities caused by defective de novo phosphatidylcholine biosynthesis. Am. J. Hum. Genet. 88, 845–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dominguez-Salas P., Moore S. E., Cole D., da Costa K. A., Cox S. E., Dyer R. A., Fulford A. J., Innis S. M., Waterland R. A., Zeisel S. H., Prentice A. M., Hennig B. J. (2013) DNA methylation potential: dietary intake and blood concentrations of one-carbon metabolites and cofactors in rural African women. Am. J. Clin. Nutr. 97, 1217–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.