Abstract
Background
Crop plants are highly sensitive to ambient temperature, with a 1 ºC difference in temperature sufficient to affect development and yield. Monocot crop plants are particularly vulnerable to higher temperatures during the reproductive and grain-filling phases. The molecular mechanisms by which temperature influences grain development are, however, unknown. In Arabidopsis thaliana, H2A.Z-nucleosomes coordinate transcriptional responses to higher temperature. We therefore investigated whether the effects of high temperature on grain development are mediated by H2A.Z-nucleosomes.
Results
We have analyzed the thermal responses of the Pooid grass, Brachypodium distachyon, a model system for crops. We find that H2A.Z-nucleosome occupancy is more responsive to increases in ambient temperature in the reproductive tissue of developing grains compared withvegetative seedlings. This difference correlates with strong phenotypic responses of developing grain to increased temperature, including early maturity and reduced yield. Conversely, temperature has limited impact on the timing of transition from the vegetative to generative stage, with increased temperature unable to substitute for long photoperiod induction of flowering. RNAi silencing of components necessary for H2A.Z-nucleosome deposition is sufficient to phenocopythe effects of warmer temperature on grain development.
Conclusions
H2A.Z-nucleosomes are important in coordinating the sensitivity of temperate grasses to increased temperature during grain development. Perturbing H2A.Z occupancy, through higher temperature or genetically, strongly reduces yield. Thus, we provide a molecular understanding of the pathways through which high temperature impacts on yield. These findings may be useful for breeding crops resilient to thermal stress.
Background
Members of the Pooideae grass family, including wheat, barley, oat and rye, are a major source of human nutrition. The phenology of these crop plants, and the yield and quality of grain produced are significantly influenced by temperature [1,2], making them vulnerable to climate change [3,4].
The effects of temperature at various stages of cereal development have been extensively studied, and optimal temperatures determined for phenological phases from sowing and emergence through to grain development (reviewed in [5]). During vegetative stages, the effects of temperature on growth are evident by the rise in leaf extension rates that occur as temperature increases [6,7]. During generative stages, the influence of temperature on leaf extension rate increases, suggesting that monocot plants have varying degrees of thermal sensitivity depending on their developmental stage [7]. This is evident during late reproductive stages, where the effects of thermal stress are significantly stronger at anthesis and stages thereafter, compared to the double ridge stage, which is the earliest morphological sign of a reproductive plant [8]. Importantly, this includes a major effect of increasing temperature during endosperm development, with growth at moderately high temperatures of 27°C to 32°C reducing the duration of grain filling without a compensatory increase in the rate of grain filling, resulting in significantly reduced yield [9-12]. Increased temperatures also affect the transcriptome of developing grain, resulting in grain at elevated temperatures having a more advanced developmental age [13-15]. Taken together, these results indicate there is a genome-wide mechanism that integrates thermal information into the transcriptome of developing grain.
In Arabidopsis thaliana, H2A.Z-nucleosomes play a key role in mediating the effects of ambient temperature on the transcriptome[16]. H2A.Z-nucleosomes are frequently found at positions surrounding the transcription start site (TSS) [17-22]. Occupancy of H2A.Z-nucleosomes at the TSS restricts access of transcriptional machinery into the gene body, and is reduced as temperature increases [16]. The reduced occupancy occurs irrespective of a given gene's transcriptional response to increased temperature, indicating eviction of H2A.Z is caused by exposure to warmer temperature and not simply a consequence of a higher transcription rate [16]. The developmental phenotypes that occur when Arabidopsis plants are exposed to warmer temperatures, including accelerated flowering, are constitutively present at cooler temperatures in genotypes compromised in their ability to incorporate H2A.Z into chromatin [16,23-26]. H2A.Z-nucleosomes therefore provide a genome-wide mechanism by which the transcriptome can be coordinated with temperature to fine-tune development in response to the environment.
To understand how crop plants respond to warmer temperatures we have used Brachypodium distachyon, a model Pooid grass and close relative of wheat and barley, which is a good exemplar of cereal biology and grain development [27,28]. We assessed the effects of temperature on plant phenology and H2A.Z-nucleosomes of thermally responsive genes, and find that they are more pronounced in developing grain compared to vegetative seedlings. Grain from transgenic plants deficient in H2A.Z deposition resemble those of wild-type plants grown at higher temperature. Our results suggest that H2A.Z-nucleosomes are responsible for the increased thermal sensitivity of reproductive grain-filling tissue compared to vegetative tissue in monocot crop plants.
Results
Warmer ambient temperature is not sufficient to induce flowering in Brachypodium
Flowering is a major phase transition in plants. In A. thaliana, the floral transition is highly responsive to ambient temperature, with growth at 27°C sufficient to overcome late flowering in short photoperiods [29]. Flowering in natural accessions of B. distachyon responds to a shift from long days (LD) to short days (SD) [30]. In our growth conditions, the accession Bd21 did not flower even after 150 days when grown in SD (14 h light/10 h dark) at 22°C, indicating it is a long-day accession. Brachypodium also did not flower in SD at 27°C, indicating that, unlike Arabidopsis, increased ambient temperatures are unable to substitute for long day induction of flowering (Figure 1). The effect of increased temperature on flowering was further investigated in LD (20 h light/4 h dark), as well as after transfer from SD to LD. No acceleration of flowering was observed at 27°C compared to 22°C when plants were grown constantly in LD, with flowering at both temperatures occurring 22 days after germination (Figure 1a). Previous studies have suggested that temperate grasses are more thermally responsive during reproductive stages than vegetative development [6,7]. To test if this is the case in Brachypodium, we assessed the response to increased temperature following floral initiation. Accordingly, plants were grown at 22°C in SD until the emergence of leaf 7, and then transferred to LD for 2 days, which is sufficient for the induction of flowering by transcriptional activation of Flowering Locus T (Figure S1 in Additional file 1). Thereafter, plants were either maintained at 22°C or transferred to 27°C, also in LD. By limiting the plant's exposure to warmer temperatures following its commitment to reproductive development, head emergence was found to occur 4 days earlier at 27°C compared to 22°C (Figure 1a). These results indicate that increased temperatures can accelerate flowering in the presence of favorable photoperiods, but are not alone sufficient to trigger flowering.
Figure 1.
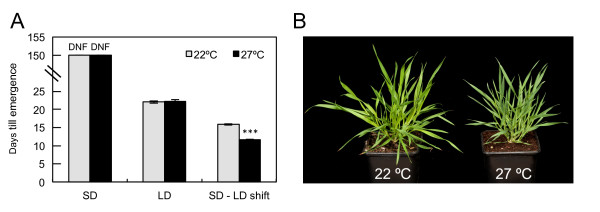
Higher ambient temperature is not sufficient to induce flowering in Brachypodium distachyon . (a) Plants were grown in one of three photoperiod conditions: SD (14 h light/10 h dark), LD (20 h/4 h) or in LD after shifting from SD. In each condition, plants were grown at either 22°C (grey) or 27°C (black). DNF (did not flower) indicates the non-flowering phenotype of plants grown in SD for 150 days. Values are the mean ± standard error of ten plants. (***P < 0.001). (b) SD grown plants after 50 days at constant temperature of 22°C and 27°C.
This lack of responsiveness of vegetative plants to 27°C could be because Brachypodium has evolved to respond to a different range of temperatures than Arabidopsis, or because it is differentially responsive to temperature depending on its developmental phase. To assess the latter possibility, we assayed the effects of high temperature on grain development, because it is a late-reproductive stage that is particularly sensitive to thermal stress [2,11]. Plants were grown at 22/17°C day/night cycles until the onset of endosperm development [28], and then maintained at this temperature or shifted to 27/22°C. These temperature treatments were used because growth between 27 and 30°C is recognized as a moderately high thermal stress that adversely affects yield in wheat and barley, compared to control conditions at 20 to 24°C [11,12,15,31]. At 27°C, grain weight initially increased faster than at 22°C, such that after 4 days at 27°C, grain was 14% heavier than at 22°C (P < 0.005), indicating an accelerated rate of grain development (Figure 2a,b). However, grain-filling ceased 10 days after pollination (DAP) at 27°C while it continued until 16 DAP at 22°C, which caused a significant reduction in final fresh and dry weights for grain grown at 27°C. The reduced duration of grain-filling was confirmed with mature grain weight measurements that show a 16% decrease (P < 0.001) at the higher temperature (Figure 2c). Therefore, grain development is significantly affected by increased ambient temperature, with higher temperatures accelerating the rate of grain development so that there is a reduced developmental window during which grain fill occurs, causing a reduction in yield.
Figure 2.
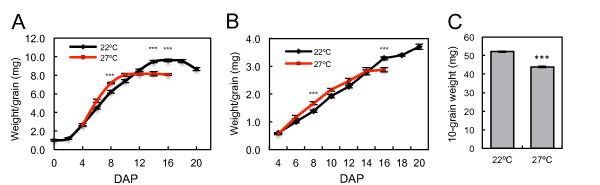
Increased ambient temperature reduces grain yield in Brachypodium. (a,b) Fresh weight (a) and dry weight (b) of developing grain from plants grown at either 22/17°C (black line) or 27/22°C (red line) (day/night temperatures). Data are the replicate of 5 replicate plants, including measurements of at least 20 seed. Values are the mean ± standard error (**P < 0.01). (c) Final yield measurements in units of weight per 10 grain for plants transferred from 22/17°C to 27/22°C throughout grain-filling, then transferred back to 22/17°C at 16 days after pollination, compared to plants grown constantly at 22/17°C (***P < 0.001). Data are from 20 biological repeats. Values are the mean ± standard error.
The Brachypodiumtranscriptome responds to changes in ambient temperature
Having observed that reproductive tissue appears to be more developmentally responsive to increased temperature than vegetative plants, we sought to test if this is reflected in the regulation of transcription and chromatin state by temperature. To identify marker genes up-regulated in response to increased ambient temperature, we assayed the transcriptional responses of vegetative plants shifted from 12°C to 22°C or 27°C for 2 and 24 hours, using whole genome microarrays and quantitative real-time PCR (qRT-PCR), to obtain rapid and longer-term transcriptomic responses to temperature change. In Brachypodium seedlings, 9% of all expressed transcripts were either significantly up- or down-regulated at least two-fold after the shift from 12°C in at least one of the temperature treatments (two-way ANOVA, temperature effect P ≤ 0.05; Figure 3a-c; Figure S2a-b in Additional file 1). Importantly, as in other systems [32], transfer to 27°C is below the threshold for inducing most heat shock genes, while HSP70, HSF23 and HSP90 are strongly induced (Figure 3d). Bradi4g32941 and Bradi1g32990 were also selected as genes for further analysis as they too were up-regulated at 27°C (Figure 3e). This induction occurs both in response to short- and medium-term shifts as well as in plants grown continuously at the warmer temperature, suggesting that these genes are responding to absolute temperature, and are not dependent on a temperature change for their expression to be induced (Figure 3e,h). We also identified genes whose transcript levels were down-regulated, such as Bradi2g14220, Bradi5g00970, Bradi2g48450 and Bradi4g17230 (Figure 3f), as well as genes that were unaffected by increased temperature, including Bradi3g31120 and Bradi1g47790 (Figure 3g). These results indicate that while the higher temperature regime does not trigger flowering in short photoperiods, the transcriptome of young seedlings remains thermally responsive.
Figure 3.
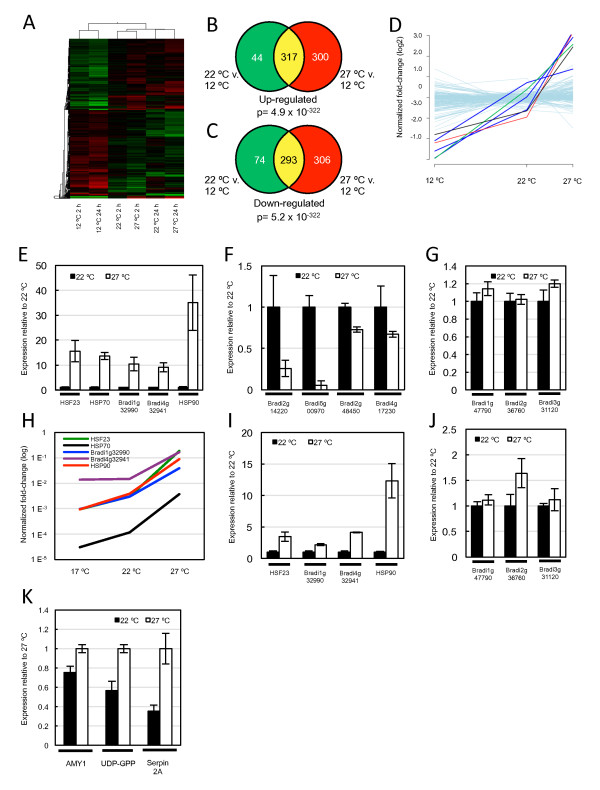
The Brachypodiumtranscriptome responds to changes in ambient temperature. (a) Transcript profiling experiment shows a robust response to changes in ambient temperature in vegetative seedlings. The heat map depicts all differentially expressed genes (DEGs) with at least two-fold change in any of the temperature treatments as determined from two-way ANOVA (P for temperature effect ≤ 0.05). Expression levels of up-regulated genes are in shades of red and of down-regulated genes in shades of green. (b,c) Venn diagrams of total numbers of up-regulated (b) or down-regulated (c) DEGs in vegetative seedlings after 24 h shift to either 22°C (green), 27°C (red), or in both temperatures (yellow). The two-tailed P-values for the significance of the overlap represented under the Venn diagrams have been calculated using Fisher's exact test. (d)HSF23 (green line), HSP70 (black line) and HSP90 (red line) are induced strongly with increasing temperature, in contrast to other heat-shock genes (grey) that do not respond significantly over the temperature range assessed. (e-g) Quantitative real-time PCR (qRT-PCR) analysis of genes that are up-regulated by increasing temperature (e), down-regulated (f) or show constant expression (g) in vegetative seedlings 24 h after temperature-shift. (h)qRT-PCR analysis of up-regulated genes in plants grown constantly at either 17°C, 22°C or 27°C. (i,j)qRT-PCR analysis of genes in developing grain that are up-regulated by increasing temperature (i), or remain constant within the temperature range (j). (k)qRT-PCR analysis of genes that are up-regulated by temperature with known roles in developing grain. Data are from at least three biological replicates.
To determine if this transcriptional response to temperature is tissue specific, we analyzed the expression of these marker genes in developing grain grown at either 22/17°C (day/night) or 27/22°C. We assessed the transcriptional response during endosperm development as this is the stage of grain development in wheat that responds strongly to increased temperature [11,15]. Hence, at 6 to 8 DAP, plants were either shifted to 27/22°C or maintained at 22/17°C, and grains were harvested after 24 h. We found that HSF23, HSP90, Bradi4g32941 and Bradi1g32990 were also up-regulated in developing grain (Figure 3i) and that Bradi3g31120 and Bradi1g47790 transcript levels remained constant between the two temperature treatments (Figure 3j). Expression levels of HSP70 were very low and did not appear to be thermally regulated in developing grain, which is similar to results in Arabidopsis where HSP70 was not temperature-regulated in seeds [33]. We could not detect expression of the genes that weredown-regulated by higher temperature in young seedlings. We also assessed the expression of some genes with important roles during grain development (Figure 3k) shown to be thermally responsive during grain filling in wheat [13], including beta-amylase (AMY1) and UDP-glucose pyrophosphorylase (UDP-GPP) from the starch metabolism pathway, and Serpin 2A, which has a role in plant defense [13]. These genes were up-regulated in grain grown at 27/22°C, relative to 22/17°C, which is consistent with observations in wheat. These results indicate that the developing grain of Brachypodium responds transcriptionally to increased ambient temperature, and that some genes display a shared transcriptional response to thermal stress in young seedlings and developing grain.
H2A.Z-nucleosomes are more thermally responsive in developing grain than vegetative seedlings
The histone variant H2A.Z has an important role in regulating the ambient temperature transcriptome of Arabidopsis, and loss of H2A.Z incorporation into nucleosomes surrounding TSSs promotes thermally sensitive developmental responses [16,26]. We therefore investigated the behavior of H2A.Z-nucleosomes in young seedlings and developing grain to determine if these nucleosomes might explain the altered developmental responsiveness of the two tissue types to increased temperature. Nucleosome positions surrounding the TSS were predicted using software prediction algorithms and confirmed using micrococcal nuclease (MNase) digestion and subsequent histone 3 (H3) chromatin-immunoprecipitation (ChIP) (Figure 4a,b; Figure S3 in Additional file 1). For HSF23 and HSP70, we used quantitative-PCR (qPCR) with primer pairs tiled across the promoter region surrounding the TSS, which revealed the appearance of two characteristic peaks that are suggestive of -1 and +1 nucleosomes (Figure 4a,b). As the software prediction algorithms accurately predicted nucleosome positions for HSF23 and HSP70, we used the software to identify the likely +1 nucleosomes of the other thermally responsive genes, which were confirmed by qPCR on DNA isolated from H3 ChIP experiments [34] (Figure 4c-e).
Figure 4.
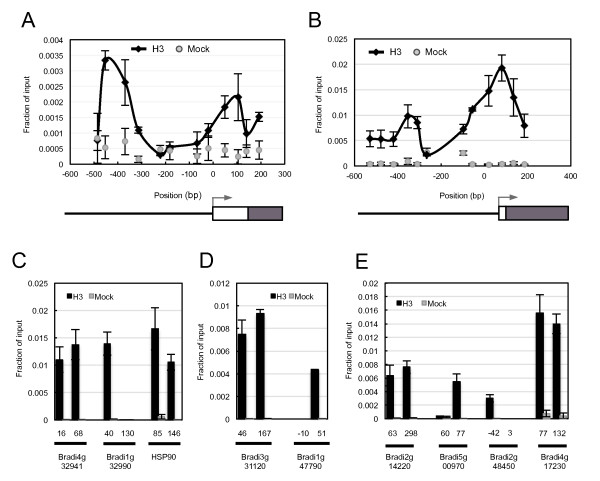
Nucleosome positioning by H3 ChIP analysis. (a,b)ChIP of cross-linked H3 at the promoter sites of HSF23 (a) and HSP70 (b) reveal well-positioned -1 and +1 nucleosomes. The x-axis indicates the central position of each amplicon relative to the TSS. In each schematic, the promoter (solid line), 5'UTR (white box), exons (black box) and TSS (arrow) are shown. (c-e)ChIP of cross-linked H3 for genes that were detected to be up-regulated (c), constant (d) or down-regulated (e) in response to temperature reveal sites that show strong enrichment of H3 at probable +1 nucleosome sites according to in silicosequence analysis (see Materials and methods). The x-axis indicates the central position of each amplicon relative to the TSS. Values from H3 and mock reactions are shown in black and grey, respectively. Values are the mean ± standard error of three biological replicates.
To investigate if H2A.Z is incorporated into these nucleosomes, we surveyed the Brachypodium genome for H2A.Z homologues. Phylogenetic analysis revealed the presence of three H2A.Z genes in Brachypodium: BdHTA1, BdHTA9 and BdHTA11 (Figure 5a; Figure S4 in Additional file 1). Transcript analysis in different tissue types, including young seedlings and developing grain, showed that BdHTA1 and BdHTA9 are strongly expressed, while we could not detect transcripts of BdHTA11 (Figure 5b). We therefore selected BdHTA9, based on its homology to the H2A.Z orthologue investigated in Arabidopsis (AtHTA11) and its expression in seedlings and developing grain, for further analysis. To study the dynamics of BdHTA9 in response to temperature, we created a BdHTA9 3XFLAG form expressed under its native promoter. ChIP analysis using anti-FLAG resin on Brachypodium seedlings grown at 17°C revealed the presence of HTA9 at the presumptive -1 and +1 nucleosomes of both HSF23 and HSP70, and +1 nucleosomes of the other thermally responsive genes (Figure 5c,d). These results are consistent with localization studies of H2A.Z in other organisms, which show incorporation into nucleosomes surrounding the TSS [17-22].
Figure 5.
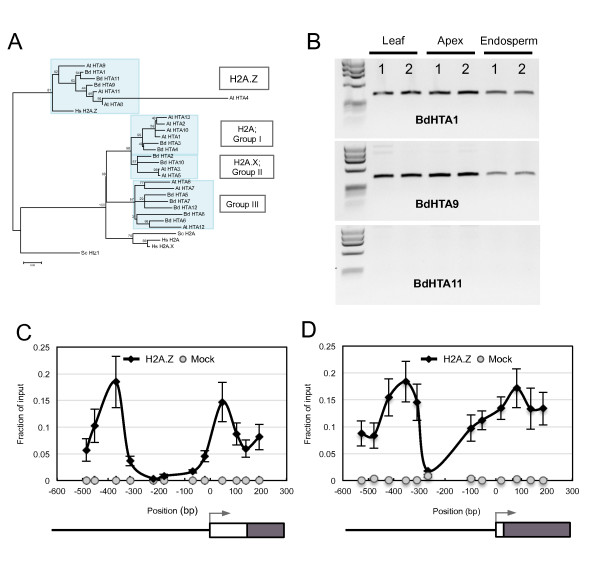
Identification and nucleosome positioning of BdHTA9 in Brachypodium. (a) An unrooted maximum-likelihood phylogenetic tree of HTA proteins in Brachypodium, Arabidopsis, humans and yeast, constructed using MEGA5 with 100 bootstrap replicates, summarizes the evolutionary relationship among the HTA proteins and the separation in four phylogenetic subfamilies. Branches are drawn to scale with scale bar representing the number of substitutions per site. (b) Reverse-transcriptase PCR analysis of BdHTA1, BdHTA9 and BdHTA11 from leaf, apex and endosperm tissue of Bd21. Two biological replicates are shown. (c,d)ChIP analysis of HTA9:3XFLAG (H2A.Z) at 17°C shows H2A.Z is enriched at the -1 and +1 nucleosomes of HSF23 (c) and HSP70 (d). The x-axis indicates the central position of each amplicon relative to the TSS. In each schematic, the promoter (solid line), 5'UTR (white box), exons (black box) and TSS (arrow) are shown. Mock reactions (grey) were performed on identical tissue from wild-type plants. Values are the mean ± standard error of three biological replicates.
To determine if eviction of H2A.Z-nucleosomes occurs in vegetative seedlings, as has been described in Arabidopsis[16], we performed nucleosome analysis and ChIP for BdHTA9 3X FLAG from plants maintained at 17°C or shifted to 27°C. For all genes and nucleosome sites analyzed, we did not detect a decrease in H2A.Z and nucleosome occupancy between 17°C and 27°C (Figure 6a-c; Figure S5 in Additional file 1). This appears to be independent of transcriptional status, since it was observed equally for genes up-regulated, down-regulated or with constant expression after shifting to 27°C (Figure 6a-c).
Figure 6.
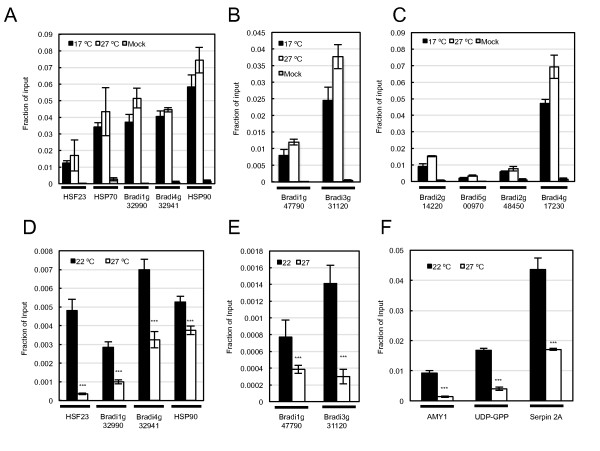
Occupancy of H2A.Z-nucleosomes is reduced at higher ambient temperatures in developing grain but not in vegetative seedlings. (a-c)ChIP analysis of HTA9:3XFLAG (H2A.Z) at 22°C and 27°C in vegetative seedlings at +1 nucleosomes of genes whose expression was up-regulated (a), remained constant (b) or down-regulated (c) upon an increase in temperature. (d-f)ChIP analysis of HTA9:3XFLAG (H2A.Z) at 22/17°C and 22/27°C in developing grain at +1 nucleosomes of genes whose expression was up-regulated (d) in both seedlings and grain, or remained constant (e) upon an increase in temperature. (f)ChIP analysis of HTA9:3XFLAG (H2A.Z) at 22/17°C and 27/22°C for genes with roles in grain development. Mock reactions (grey) were performed on identical tissue from wild-type plants. ***P < 0.001.
This apparent stability of H2A.Z-nucleosomes to higher temperature in vegetative plants could be a general feature of chromatin in Brachypodium, or it could be an indicator of temperature responsiveness being dependent on developmental phase. To test this latter possibility, we examined the thermal responsiveness of H2A.Z-nucleosomes in developing grain. To ensure that any altered molecular response to temperature was not a consequence of a difference in the physical temperature of the two tissue types, thermal imaging was obtained for both organs in each treatment (Figure S6 in Additional file 1). This analysis showed that there was no difference in physical temperature between vegetative seedlings and developing grain (Figure S6 in Additional file 1). We then investigated H2A.Z-nucleosome behavior under the same conditions used for transcript analysis. BdHTA9 3X FLAG plants were grown at 22/17°C until 6 DAP, and then shifted to 27/22°C or maintained at 22/17°C, with grains harvested after 24 h. ChIP and nucleosome analysis showed that there is a striking reduction in HTA9 occupancy at 27/22°C, relative to 22/17°C, for all genes analyzed, including AMY1, UDP-GPP and Serpin 2A (Figure 6d-f). We also observed a comparable reduction in nucleosome levels (Figure S7 in Additional file 1). These results show that in the endosperm of developing grain there is a considerable increase in the mobility of H2A.Z at the +1 nucleosomes of these genes, compared to the same nucleosome sites in vegetative seedlings. In developing grain the transcript levels of BdHTA9 did not change with temperature, indicating that the increased mobility of H2A.Z in grain does not occur as a consequence of reduced BdHTA9 transcription (Figure S8 in Additional file 1). This result is consistent with the absence of large-scale phenological changes in vegetative plants at 27°C, while a considerable acceleration in grain development is seen at this higher temperature.
Grain from ARP6 RNAi lines phenocopy responses to increased temperature
To determine whether loss of H2A.Z-nucleosomes at higher temperature is the cause of altered grain development, or simply a correlated event, we sought to perturb H2A.Z-nucleosome occupancy independently of temperature. H2A.Z is inserted into chromatin through the highly conserved SWR1 complex [24,35,36]. We disrupted the activity of SWR1 using RNA interference (RNAi) silencing of a key conserved component, ACTIN RELATED PROTEIN 6 (ARP6), of which there is only one copy in the Brachypodium genome (Bradi2g10130). Using three independent transgenic lines with reduced expression of ARP6, we observed a reduction in seed weight (60%, P < 0.001) and overall yield per plant (30 to 50%, P < 0.01) that was comparable with the reduced yield observed in wild-type plants grown at 27/22°C (Figure 7b,c). We also observed an increased rate of floret sterility and grain abortion in the ARP6 RNAi lines (Figure 7c; Table S1 in Additional file 1), which is consistent with observations in wheat where high temperature treatment of early developing grain, prior to the onset of endosperm development, promotes grain abortion and reduction in grain number [37-39]. Other than the phenotypes associated with grain development, the ARP6 RNAi lines appeared phenotypically normal and displayed a flowering-time consistent with that of wild-type plants, including an inability to flower in SD (Figure S9 in Additional file 1). These results show that developing grain of ARP6 RNAi plants grown at 22/17°C simulate grain harvested from wild-type plants grown at 27/22°C.
Figure 7.
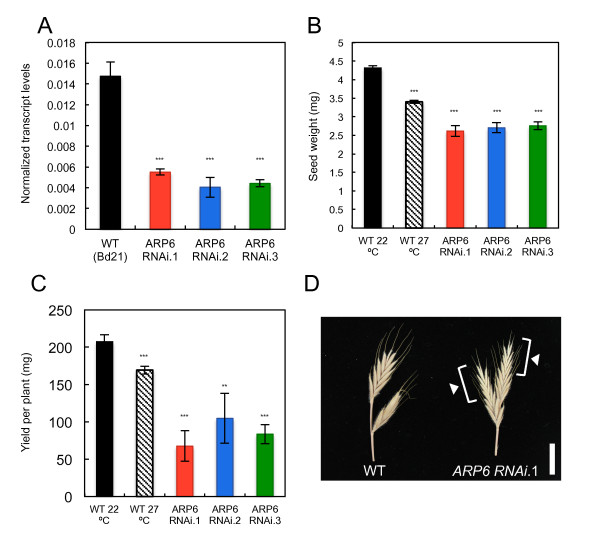
Seeds from plants with reduced expression of ARP6 phenocopy seed from plants grown at higher temperature. (a) Relative expression of ARP6 in wild-type (WT; Bd21) and three independent ARP6 RNAi transgenic lines (T1 generation); ***P < 0.001. (b,c) Seed weight measurements (b) and yield per plant (c) in WT (Bd21) at 22/17°C and 27/22°C, as well as three independent transgenic lines at 22/17°C (T1 generation). Data are the mean ± standard error of at least 15 grains (**P < 0.01; ***P < 0.001). (d) A representative spike from WT Bd21 and ARP6 RNAi.1 displaying the empty florets (white arrowheads) that contained aborted grain. Scale bar, 1 cm.
To determine if the grain development phenotypes observed in ARP6 RNAi plants may be caused by the mis-regulated expression of thermally responsive genes, we compared the transcript levels of thermally induced genes in wild-type and transgenic plants grown at 22/17°C. We used qRT-PCR to assess the relative expression of genes that were up-regulated in developing grain upon transition from 22/17°C to 27/22°C. These results show that reduced activity of ARP6 causes elevated transcript levels for genes that were induced by increased temperature in wild-type plants (Figure 8). Some variation in the behavior of transcript levels between RNAi lines may reflect residual ARP6 activity. These results indicate that when measured transcriptionally, the developing grain of ARP6 RNAi lines grown at 22/17°C resemble those of wild-type plants grown at warmer temperatures.
Figure 8.
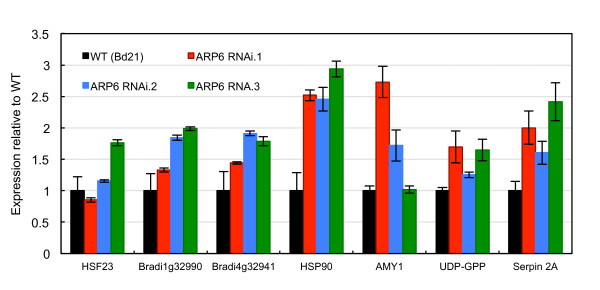
Genes up-regulated by increased ambient temperature are up-regulated in seeds of ARP6 RNAi transgenic lines at 22°C. Expression of genes that are up-regulated in developing grain by transfer from 22/17°C to 27/22°C (Figure 3) are up-regulated in grain of three ARP6 RNAi transgenic lines (T1 generation) grown at 22/17°C. Values are the mean ± standard error of 4 replicates, each containing 6 grain at 8 DAP.
Discussion
Ambient temperature has a large effect on plant development, reflected in changes to the transcriptome[32,40]. In A. thaliana, H2A.Z-containing nucleosomes are necessary to correctly express the temperature transcriptome[16]. We therefore sought to determine whether H2A.Z-mediated perception of ambient temperature is conserved in monocot plants, and whether the response to increased temperature varies between developing grain and vegetative seedlings.
We have observed that chromatin sites containing H2A.Z-nucleosomes are more responsive to increases in ambient temperature in developing grains than vegetative seedlings. These molecular observations support phenological changes reported in this and other studies, which show that increased temperatures cause a greater response in plants progressing through late-reproductive development compared to those of vegetative stages. For example, the base temperature used to calculate the rate of development using thermal time in wheat is not constant throughout plant development, with the base temperatures for terminal spikelet to anthesis, and anthesis to maturity being considerably higher than that of the vegetative stages [41-43]. A differential response of vegetative and reproductive tissue to moderate and high temperature has also been observed in other plants, such as orchids and strawberry [44,45]. Similarly, it has been shown in Arabidopsis that cold temperature treatment causes approximately ten-fold more genes to be induced in seeds than in seedlings [33]. One hypothesis for the distinct responses of vegetative and reproductive tissues is that it reflects an adaptation to low temperatures during early developmental stages and to warmer conditions during reproductive stages [42,43]. Our findings suggest that plants may be able to modulate responsiveness to temperature in a tissue- and lifecycle-dependent manner by controlling the stability of H2A.Z-nucleosomes. To determine how direct the link between H2A.Z-nucleosomes is to the control of grain development in response to temperature, it will be necessary to identify the loci regulating this process and the binding dynamics of H2A.Z-nucleosomes to their promoters.
The effect of temperature on floral initiation has been an active area of research in plant development. Here we find that increased temperature alone cannot promote flowering in non-inductive photoperiods, indicating that Brachypodium is an obligate long-day plant and that increased ambient temperature cannot compensate for photoperiod as a floral inductive signal. These observations are consistent with other studies, where it has been shown in wheat and barley that increased temperature in SD conditions does not promote floral initiation, but in fact delays the onset of reproductive development [46,47]. Similarly, in strawberry, olive, rice flowers, perennial grasses and orchids moderately high temperatures delay or completely inhibit floral induction [44,45,48-50]. Taken together, these observations suggest that some plants, including annual temperate grasses such as Brachypodium, have adopted different strategies to respond to increased ambient temperature compared to Arabidopsis, where flowering is accelerated by high temperature.
The results presented in this study indicate that the transcriptional responses of ripening grain to increased temperature, and the accompanied accelerated rate of development and reduction in grain weight, are at least partially mediated by H2A.Z-nucleosomes. Recent studies in wheat and barley have investigated the transcriptional events initiated by moderate and high increases in temperature during grain development [13-15]. These studies have shown that the transcriptome of a developing grain is highly responsive to increased ambient temperatures [14] and that the developmental age of the grain is more advanced at warmer temperatures, as measured transcriptionally [15]. Therefore, it may be possible to reduce the negative effects of thermal stress on grain weight and quality by nullifying the transcriptional response of developing grain to increasing temperatures. This is a concept that has already been investigated in rice, where attenuating the transcriptional activity of α-amylases, which are normally up-regulated by increased temperatures, has been used to ameliorate the deleterious effects that high temperatures have on grain quality [51]. Given the broad role for H2A.Z-nucleosomes in regulating the ambient temperature transcriptome in Arabidopsis seedlings, it is possible that a large proportion of the transcriptional responses initiated by increased temperature in developing grain are coordinated by H2A.Z-nucleosomes. Therefore, reducing the responsiveness of H2A.Z-containing nucleosomes in developing grain may help improve yield and grain quality for plants grown in high temperatures. This hypothesis is supported by findings in Arabidopsis that plants which lack H2A.Z have fertility defects [26], as well as the results of this study that decreased activity of ARP6 causes reduced seed weight and increased expression of thermally responsive genes in plants grown at lower temperatures.
A key question for future research will be to understand the mechanism underlying the differential response of H2A.Z-nucleosomes to increased temperature in reproductive and vegetative tissue. Histone modification is one possible explanation for the more pronounced effect of thermal stress on H2A.Z behavior in nucleosomes of developing grain compared to vegetative seedlings. Recent results in humans have shown that while H2A.Z is localized at sites surrounding the TSS of both active and poised gene promoters, acetylated H2A.Z is only found at TSSs of active genes [52]. In yeast, acetylation of H2A.Z was found to correlate with genome-wide gene activity [53], and confer with nucleosome destabilization and an open conformation of chromatin [54]. It is possible, therefore, that differences in the degree of H2A.Z acetylation may account for tissue-specific variation in thermal responsiveness. In addition to the H2A.Z-nucleosome dependent regulation of transcription, we also observe warm temperature induction of gene expression in seedlings, where no significant decreases in H2A.Z-nucleosomes are seen. This is consistent with work in Arabidopsis showing that H2A.Z-nucleosomes do not account for all the transcriptional responses to warmer temperature [16]. The identification of these parallel temperature responsive pathways will be of interest.
The results presented in this work, as well as many phenological studies, suggest that Brachypodium and other grasses are more responsive to warmer temperatures as they enter the generative phase of their lifecycle. This may be because the initiation of flowering represents a point of no return, since the reproductive program must be completed. Higher temperatures would then represent an environmental cue associated with the likely onset of temperature stress and drought. Accelerating the developmental transition through this vulnerable stage to the resilient desiccated grain state may therefore be an adaptive response to anticipated abiotic stress. Conversely, activating flowering in response to warmer temperatures independently of photoperiod would risk exposing the inflorescence and the developing grains to frost [55]. This study highlights the value of complementing work in Arabidopsis with monocot systems to assess the direct applicability of Arabidopsis research to other systems, especially for traits that are important for crop yield, such as temperature perception.
Conclusions
In common with other monocot plants, Brachypodium displays a marked difference in temperature responsiveness depending on the developmental stage. We show that the process of grain-fill is particularly sensitive to warmer temperatures. Our data indicate that H2A.Z-nucleosomes are necessary for coordinating the sensitivity of Brachypodium to increased temperature during grain development. Perturbing H2A.Z occupancy, through higher temperature or genetically, strongly reduces yield. Thus, we provide a molecular understanding of the pathways through which high temperature impacts on yield. These findings may be useful for breeding crops resilient to thermal stress.
Materials and methods
Plant material and growth conditions
All experiments were carried out using Brachypodium distachyon . Material included wild-type Bd21 and transformed lines containing BdHTA9 with a carboxy-terminal 3X FLAG tag, including native promoter with all exons and introns, and transformed lines containing a RNAi construct containing sequence complementary to ACTIN RELATED PROTEIN 6 (ARP6) (nucleotides 360 to 508 of open reading frame). Transformation was performed as described in [56]. All plants were grown in growth cabinets (Microclima MC1000E, Snijders Scientific, Tilburg, Netherlands) with 70% relative air humidity and 350 µmol m-2 s-1 PPFD (Photosynthetic Photon Flux Density).
For flowering-time experiments, plants were grown in one of three photoperiod regimes: i) SD (14 h light/10 h dark); ii) LD (20 h/4 h); or iii) transferred from SD to LD at the seven leaf stage. Plants were grown at constant temperatures of either 22°C or 27°C. For analysis of temperature effects on grain development, plants were grown in SD at 22/17°C (day/night temperatures) until the seven-leaf stage to synchronize development, and shifted to LD at the same temperature. After head emergence, date of pollination was monitored for the main stem. At 5 DAP, plants were either maintained at 22/17°C or shifted to 27/22°C and developing grains were collected at two-day intervals. At 16 DAP, plants grown at 27/22°C were shifted back to 22/17°C until maturity to measure final yield. Dry weight of developing grain was measured after desiccating grain at 60°C for 10 days. For each time point and temperature, a total of 20 seed were measured that were collected from 5 plants.
For transcript analysis of vegetative plants, seedlings were grown at 17°C in SD until emergence of the third leaf, and transferred to 12°C for 48 h before shifting to either 22°C or 27°C. Samples were collected at 2 h and 24 h post-shift. Transcript analysis of developing grain was performed only on grain harvested from the main stem. Grain were collected from plants grown at 22/17°C until the end of 6 DAP, which were then transferred to 27/22°C or maintained at 22/17°C. Samples were collected 24 h after shift. Each replicate contained pools of 6 grain, from which the glumes had been removed.
Transcript analysis
Transcript analysis was performed on total RNA extracted using the Plant RNeasy Plant Mini kit (Qiagen, Crawley, West Sussex, UK). Single-stranded cDNA was amplified for microarray analysis using the Ambion WT Expression Kit (Ambion Life Technologies, Paisley, Renfrewshire, UK), labeled and hybridized to the custom Brachypodium microarray (Affymetrix, Santa Clara, California, USA) using the GeneChip WT terminal labeling and hybridization kit (Affymetrix). Microarray data were analyzed using GeneSpringGX v.11 (Agilent, Santa Clara, California, USA). For each time-point after the temperature shift (2 h and 24 h) and temperature treatment (12°C, 22°C and 27°C), two to three biological replicates were analyzed. Samples taken before the shift at 12°C 0 h were considered to be equivalent to the 12°C 24 h treatment for the purpose of the statistical analysis. Raw data were normalized using the RMA algorithm and filtered by expression level using the cut-off value of ≥20 in at least one of the treatments to define the group of genes expressed in vegetative seedlings. The statistical significance of changes in the transcripts with expression levels above the cut-off level was assessed using two-way ANOVA with temperature and time as main factors. The P-values were adjusted for multiple testing by the Benjamini and Hochberg's method at a false discovery rate of ≤0.05. The transcripts significantly affected by temperature as the main factor (corrected P ≤ 0.05) and displaying ≥2-fold change in any of the temperature-time combinations were reported as differentially expressed genes (DEGs). Hierarchical clustering and heat mapping were used to visually display DEGs. The significance of the overlap between the lists of DEGs in different temperature-time treatments represented in Venn diagrams were tested using Fisher's exact test. The DEGs were annotated using the bradinet platform of web tools [57], and are provided as a list in Table S5 in Additional file 2). The data for these experiments have been deposited in ArrayExpress (E-MEXP-3918).
For transcript analysis by qRT-PCR, cDNA was synthesized using the Fermentas First Strand cDNA synthesis kit (Thermo Scientific, St. Leon-Rot, Germany). qRT-PCR was performed using SYBR Green I (Roche, Burgess Hill, West Sussex, UK) and a Roche LightCycler 480. All qRT-PCR data points are the average of three biological replicates, with two technical replicates performed in each reaction. See Table S2 in Additional file 3 for oligonucleotide sequences used for qRT-PCR. Expression of candidate genes was normalized against SamDC and UBC18[58].
Nucleosome positioning and ChIP
Nucleosome positioning and ChIP were performed essentially as described [16], with minor modifications. For positioning of nucleosomes in vegetative plants, chromatin from seedlings grown at 17°C was cross-linked using 1% formaldehyde. Chromatin was fragmented using 0.2 units of micrococcal nuclease (Sigma, Gillingham, Dorset, UK) as described [16]. Nucleosome positions were identified by immunopurification using the H3 antibody (Abcam, ab1791, Cambridge, Cambridgeshire, UK)). Relative enrichment of associated DNA fragments was analyzed by qPCR. Nucleosome occupancy was determined as a fraction of uncut chromatin DNA, with data plotted against the HSF23 and HSP70 promoters. HSP70 was selected for detailed nucleosome positioning because it is the homologous gene used for transcript and H2A.Z-nucleosome analysis in A. thaliana[16], and HSF23 was selected because it is expressed at a level proportionate to ambient temperature within the range 12 to 27°C. For in silico predictions of +1 nucleosome sites, 1 kb and 250 bp of sequence upstream and downstream of the TSS, respectively, were used to query the online nucleosome position prediction software (version 3.0) on the Segal lab website [34]. The two most probable sites were selected for qPCR analysis of H3 ChIP DNA, with amplicons that provided the strongest signal used for further analysis. The center of the amplicon indicates its position relative to the TSS. Oligonucleotide sequences are provided in Table S3 in Additional file 4.
Similar methods as outlined above were used to determine nucleosome and H2A.Z dynamics at different temperatures. Nucleosome and H2A.Z dynamics were assessed using chromatin isolated from seedlings that were grown at 17°C until 10 days after germination, and then either maintained at 17°C or shifted to 27°C for 24 h. Chromatin was digested with MNase, and for nucleosome analysis, mononucleosome-sized fragments were gel purified and used in qPCR. For H2A.Z dynamics, ChIP was performed on MNase digested chromatin using the ANTI-FLAG M2 Affinity Gel (Sigma), and histone-DNA complexes eluted using the 3X Flag peptide (Sigma). For assays of nucleosome and H2A.Z occupancy in developing grain, plants were grown at 22/17°C until 6 DAP. Plants were then either maintained at 22/17ºC or shifted to 27/22°C and developing grains (1 g per ChIP assay) were collected after 24 h (7 DAP), with grain harvested only from the main stem. Native ChIP assays were performed as described above, with the following modifications. Ground tissue was washed twice in extraction buffer I, and three times in extraction buffer II to remove metabolic contaminants. Following MNase digestion, two rounds of centrifugation were performed on uncut chromatin DNA and digested chromatin DNA prior to reverse cross-linking and immunopurification, respectively, to further remove contaminants. Nucleosome and H2A.Z occupancy was determined as a fraction of input DNA that was diluted ten-fold for qRT-PCR. Oligonucleotide sequences are provided in Table S3 in Additional file 4. All ChIP assays were performed at least three times, and two technical replicates were performed for each qRT-PCR.
Sequence alignment and phylogenetic analysis
Brachypodium HTA sequences were identified by BLAST search [59] from public databases using Arabidopsis HTA protein sequences as bait (Table S4 in Additional file 5). Multiple sequence alignment was performed using webPRANK[60], and manually corrected using GENEDOC [61]. Phylogenetic trees were constructed in MEGA5 [62] using the maximum likelihood algorithm with the following parameters: JTT substitution model, Gamma distributed rates and partial deletions. Bootstrap values are based on 100 replicates for testing the significance of the nodes.
Seed weight measurements from ARP6 RNAi lines
For wild-type (Bd21) and each transgenic ARP6 RNAi line, seed weight measurements were determined for 15 seeds harvested from 3 plants (5 seeds from each replicate plant). Seeds were harvested from basal florets of spikelets from the main spike, and the lemma and palea were removed before weighing. For yield per plant measurements, all seeds were harvested from three replicate plants.
Statistical analysis
When not described specifically, differences between treatments were tested by Student's t-test. Results in figures are shown as means ± standard error.
Abbreviations
bp: base pair; ChIP: chromatin-immunoprecipitation; DAP: days after pollination;H2A.Z: histone 2A variant Z; H3: histone 3; LD: long day; MNase: micrococcal nuclease; qPCR: quantitative-PCR; qRT-PCR: quantitative real-time PCR; RNAi: RNA interference; SD: short day; TSS: transcription start site; UTR: untranslated region.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SAB, MK, JF and PAW participated in the design of the study and drafted the manuscript. SAB and JF performed the molecular experiments and plant phenotype analysis. MK performed the statistical analysis of microarray data, sequence alignments and constructed the phylogenetic tree. SAB and PAW conceived the project. All authors read and approved the final manuscript.
Supplementary Material
Supplementary figures.
Table S5. The differentially expressed genes (DEGs), defined as significantly affected by temperature as the main factor (P ≤ 0.05) and displaying ≥2-fold change in at least one of the temperature-time treatments.
Table S2.
Table S3.
Table S4.
Supplementary methods and figure legends.
Contributor Information
Scott A Boden, Email: scott.boden@csiro.au.
Monika Kavanová, Email: kavanova@wzw.tum.de.
E Jean Finnegan, Email: jean.finnegan@csiro.au.
Philip A Wigge, Email: philip.wigge@slcu.cam.ac.uk.
Acknowledgements
The authors would like to thank Rene Dreos for help with the microarray experiments, Ben Trevaskis for the plasmid used for RNA interference, Silvia Alves for technical assistance with Brachypodium transformation and Steve Swain and David Weiss for valuable discussions. This work was funded by grants from the European Union to SAB (PIIF-GA-2009_237398) and MK (PIEF-GA-2008-220431). The laboratory of PAW was supported by core funding from the John Innes Centre, an institute of the Biotechnology and Biological Sciences Research Council. PAW is the recipient of a European Research Council grant to study the mechanisms of temperature perception in plants (ERC 243140) and is funded by the Gatsby Foundation.
References
- Wardlaw IF, Dawson IA. The tolerance of wheat to high temperatures during reproductive growth. II Grain development. Aust J Agric Res. 1989;14:15–24. doi: 10.1071/AR9890015. [DOI] [Google Scholar]
- Wallwork MAB, Jenner CF, Logue SJ, Sedgley M. Effect of high temperature during grain-filling on the structure of developing and malted barley grains. Ann Bot. 1998;14:587–599. doi: 10.1006/anbo.1998.0721. [DOI] [Google Scholar]
- Battisti DS, Naylor RL. Historical warnings of future food insecurity with unprecedented seasonal heat. Science. 2009;14:240–244. doi: 10.1126/science.1164363. [DOI] [PubMed] [Google Scholar]
- Lobell DB, Schlenker W, Costa-Roberts J. Climate trends and global crop production since 1980. Science. 2011;14:616–620. doi: 10.1126/science.1204531. [DOI] [PubMed] [Google Scholar]
- Porter JR, Gawith M. Temperatures and the growth and development of wheat: a review. Eur J Agronomy. 1999;14:23–36. doi: 10.1016/S1161-0301(98)00047-1. [DOI] [Google Scholar]
- Peacock JM. Temperature and leaf growth in Lolium perenne III. Factors affecting seasonal differences. J Appl Ecol. 1975;14:685–697. doi: 10.2307/2402182. [DOI] [Google Scholar]
- Parsons AJ, Robson JM. Seasonal changes in the physiology of S24 perennial rygrass (Lolium perenne L.). 1. Response of leaf extension to temperature during the transition from vegetative to reproductive growth. Ann Bot. 1980;14:435–444. [Google Scholar]
- Wollenweber R, Porter JR, Schellberg J. Lack of interaction between extreme high temperature events at vegetative and reproductive growth stages in wheat. J Agron Crop Sci. 2003;14:142–150. doi: 10.1046/j.1439-037X.2003.00025.x. [DOI] [Google Scholar]
- Tashiro T, Wardlaw IF. A comparison of the effect of high temperature on grain development in wheat and rice. Ann Bot. 1989;14:59–65. [Google Scholar]
- Wardlaw IF, Moncur L, Patrick JW. The response of wheat to high temperature following anthesis. II. Sucrose accumulation and metabolism by isolated kernels. Aust J Plant Physiol. 1995;14:399–407. doi: 10.1071/PP9950399. [DOI] [Google Scholar]
- Gooding MJ, Ellis RH, Shewry PR, Schofield JD. Effects of restricted water availability and increased temperature on the grain filling, drying and quality of winter wheat. J Cereal Sci. 2003;14:295–309. doi: 10.1006/jcrs.2002.0501. [DOI] [Google Scholar]
- Wardlaw IF, Moncur I. The response of wheat to high temperature following anthesis. I. The rate and duration of kernal filling. Aust J Plant Physiol. 1995;14:391–397. doi: 10.1071/PP9950391. [DOI] [Google Scholar]
- Altenbach SB, Kothari KM. Transcript profiles of genes expressed in endosperm tissue are altered by high temperature during wheat grain development. J Cereal Sci. 2004;14:115–126. doi: 10.1016/j.jcs.2004.05.004. [DOI] [Google Scholar]
- Mangelsen E, Killian J, Harter K, Jansson C, Wanke D, Sundberg E. Transcriptome analysis of high-temperature stress in developing barley caryopses: Early stress responses and effects on storage compound biosynthesis. Mol Plant. 2011;14:97–115. doi: 10.1093/mp/ssq058. [DOI] [PubMed] [Google Scholar]
- Wan Y, Poole RL, Huttly AK, Toscano-Underwood C, Feeney K, Welham S, Gooding MJ, Mills C, Edwards KJ, Shewry PR, Mitchell RA. Transcriptome analysis of grain development in hexaploid wheat. BMC Genomics. 2008;14:121. doi: 10.1186/1471-2164-9-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;14:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA. H2A.Z is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;14:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Xiaoyong L, Jiang C, Venters BJ, Zanton SJ, Tomsho LP, Glaser RL, Schuster SC, Gilmour DS, Istvan A, Pugh BF. Nucleosome organization in the Drosophila genome. Nature. 2008;14:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5' Ends of both active and inactive genes in euchromatin. Cell. 2005;14:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle CM, McClinic KN, Ercan S, Zhang X, Green RD, Kelly WG, Lieb JD. The genomic distribution and function of histone variant HTZ-1 during C. elegans embryogenesis. PLoS Genet. 2008;14:e1000187. doi: 10.1371/journal.pgen.1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;14:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;14:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Trillo M. EARLY IN SHORT DAYS 1 (ESD1) encodes ACTIN-RELATED PROTEIN 6 (AtARP6), a putative component of chromatin remodelling complexes that positively regulates FLC accumulation in Arabidopsis. Development. 2006;14:1241–1252. doi: 10.1242/dev.02301. [DOI] [PubMed] [Google Scholar]
- Deal RB, Topp CN, McKinney EC, Meagher RB. Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C Expression by the histone variant H2A.Z. Plant Cell. 2007;14:74–83. doi: 10.1105/tpc.106.048447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March-Díaz R, García-Domínguez M, Lozano-Juste J, León J, Florencio FJ, Reyes JC. Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 2007;14:475–487. doi: 10.1111/j.1365-313X.2007.03361.x. [DOI] [PubMed] [Google Scholar]
- Coleman-Derr D, Zilberman D. Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 2012;14:e1002988. doi: 10.1371/journal.pgen.1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opanowicz M, Vain P, Draper J, Parker D, Doonan JH. Brachypodium distachyon: making hay with a wild grass. Trends Plant Sci. 2008;14:172–177. doi: 10.1016/j.tplants.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Opanowicz M, Hands P, Betts D, Parker ML, Toole GA, Mills ENC, Doonan JH, Drea S. Endosperm development in Brachypodium distachyon. J Exp Bot. 2010;14:735–748. doi: 10.1093/jxb/erq309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Sureshkumar S, Lempe J, Weigel D. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2006;14:e106. doi: 10.1371/journal.pgen.0020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz CJ, Doyle MR, Manzaneda AJ, Rey PJ, Mitchell-Olds T, Amasino RM. Natural variation of flowering time and vernalization responsiveness in Brachypodium distachyon. Bioenerg Res. 2010;14:38–46. doi: 10.1007/s12155-009-9069-3. [DOI] [Google Scholar]
- Wardlaw IF. Interaction between drought and chronic high temperature during kernel filling in wheat in a controlled environment. Ann Bot. 2002;14:469–476. doi: 10.1093/aob/mcf219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Wigge PA. Ambient temperature perception in plants. Curr Opin Plant Biol. 2005;14:483–486. doi: 10.1016/j.pbi.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Kendall SL, Hellwege A, Marriot P, Whalley C, Graham IA, Penfield S. Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell. 2011;14:2568–2580. doi: 10.1105/tpc.111.087643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;14:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Keogh M-C, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, Rischards DP, Wu X, Emili A, Hughes TR, Buratowski S, Greenblatt JF. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003;14:1565–1576. doi: 10.1016/S1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu W-H, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;14:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Gibson LR, Paulsen GM. Yield components of wheat grown under high temperature stress during reproductive growth. Crop Sci. 1999;14:1841–1846. doi: 10.2135/cropsci1999.3961841x. [DOI] [Google Scholar]
- Tashiro T, Wardlaw IF. The response to high temperature shock and humidity changes prior to and during the early stages of grain development in wheat. Aust J Plant Physiol. 1990;14:551–561. doi: 10.1071/PP9900551. [DOI] [Google Scholar]
- Bhullar SS, Jenner CF. Responses to brief periods of elevated temperature in ears and grains of wheat. Aust J Plant Physiol. 1983;14:549–560. doi: 10.1071/PP9830549. [DOI] [Google Scholar]
- McClung CR, Davis SJ. Ambient thermometers in plants: From physiological outputs towards mechanisms of thermal sensing. Curr Biol. 2010;14:R1086–R1092. doi: 10.1016/j.cub.2010.10.035. [DOI] [PubMed] [Google Scholar]
- Del Pozo AH, García-Huidobro J, Novoa R, Villaseca S. Relationship of base temperature to development of spring wheat. Exp Agriculture. 1987;14:21–30. doi: 10.1017/S0014479700001095. [DOI] [Google Scholar]
- Slafer GA, Savin R. Developmental base temperature in different phenological phases of wheat (Triticum aestivum). J Exp Bot. 1991;14:1077–1082. doi: 10.1093/jxb/42.8.1077. [DOI] [Google Scholar]
- Angus JF, MacKenzie DH, Morton R, Schafer CA. Phasic development in field crops II. Thermal and photoperiodic responses of spring wheat. Field Crops Res. 1981;14:269–283. [Google Scholar]
- Heide OM. Photoperiod and temperature interactions in growth and flowering of Strawberry. Physiologia Plantarum. 1977;14:21–26. doi: 10.1111/j.1399-3054.1977.tb01486.x. [DOI] [Google Scholar]
- Newton LA, Runkle ES. High-temperature inhibition of flowering of Phalaenopsis and Doritaenopsis orchids. HortScience. 2009;14:1271–1276. [Google Scholar]
- Rawson HM, Richards RA. Effects of high temperature and photoperiod on floral development in wheat isolines differing in vernalisation and photoperiod genes. Field Crops Res. 1993;14:181–192. doi: 10.1016/0378-4290(93)90030-Q. [DOI] [Google Scholar]
- Hemming M, Walford SA, Fieg S, Dennis ES, Trevaskis B. Identification of high temperature responsive genes in cereals. Plant Physiol. 2012;14:1439–50. doi: 10.1104/pp.111.192013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett WP, Hartmann HT. The influence of temperature on floral initiation in the olive. Physiologia Plantarum. 1967;14:430–436. doi: 10.1111/j.1399-3054.1967.tb07183.x. [DOI] [Google Scholar]
- Ofir M, Kigel J. Opposite effects of daylength and temperature on flowering and summer dormancy of Poa bulbosa. Ann Bot. 2006;14:659–666. doi: 10.1093/aob/mcl021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halevy AH, Shlomo E. Environmental factors affecting flowering of rice flower (Ozothamnus diosmifolius, Vent.). Scientia Horticulturae. 2001;14:303–309. doi: 10.1016/S0304-4238(00)00182-5. [DOI] [Google Scholar]
- Hakata M, Kuroda M, Miyashita T, Yamaguchi T, Kojima M, Sakakibara H, Mitsui T, Yamakawa H. Suppression of α-amylase genes improves quality of rice grain ripened under high temperature. Plant Biotechnol J. 2012;14:1110–1117. doi: 10.1111/j.1467-7652.2012.00741.x. [DOI] [PubMed] [Google Scholar]
- Valdes-Mora F, Song JZ, Statham AL, Strbenac D, Robinson MD, Nair SS, Patterson KI, Tremethick DJ, Stirzaker C, Clark SJ. Acetylation of H2A.Z is a key epigenetics modification associated with gene deregulation and epigenetic remodeling in cancer. Genome Res. 2012;14:307–321. doi: 10.1101/gr.118919.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar CB, Xu F, Zhang K, Grunstein M. Acetylation of H2A.Z Lys 14 is associated with genome-wide gene activity in yeast. Gene Dev. 2006;14:711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi T, Dryhurst D, Rose KL, Shabanowitz J, Hunt DF, Ausio J. Acetylation of vertebrate H2A.Z and its effect on the structure of the nucleosome. Biochemistry. 2009;14:5007–5017. doi: 10.1021/bi900196c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcellos H, Single WV. Frost injury in wheat ears after ear emergence. Aust J Plant Physiol. 1984;14:7–15. doi: 10.1071/PP9840007. [DOI] [Google Scholar]
- Alves SC, Worland B, Thole V, Snape JW, Bevan MW, Vain P. A protocol for Agrobacterium-mediated transformation of Brachypodium distachyon community standard line Bd21. Nat Protoc. 2009;14:638–649. doi: 10.1038/nprot.2009.30. [DOI] [PubMed] [Google Scholar]
- bradinet. http://aranet.mpimp-golm.mpg.de/bradinet
- Hong S-Y, Seo P, Yang M-S, Xiang F, Park C-M. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol. 2008;14:112. doi: 10.1186/1471-2229-8-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic Local Alignment Search Tool. J Mol Biol. 1990;14:405–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Löytynoja A, Goldman N. webPRANK: a phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinformatics. 2010;14:579. doi: 10.1186/1471-2105-11-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas K, Nicholas Jnr H, Deerfield D II. GeneDoc: Analysis and Visualization of Genetic Variation. EMBNEWNEWS. 1997;14:14. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Masatoshi N, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;14:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.
Table S5. The differentially expressed genes (DEGs), defined as significantly affected by temperature as the main factor (P ≤ 0.05) and displaying ≥2-fold change in at least one of the temperature-time treatments.
Table S2.
Table S3.
Table S4.
Supplementary methods and figure legends.


