Abstract
Cyclin-dependent kinases are critical regulators of eukaryotic DNA replication. We show that the S-phase cyclin Clb5 binds stably and directly to the origin recognition complex (ORC). This interaction is mediated by an “RXL” target sequence, or “Cy” motif, in the Orc6 subunit that is recognized by the “hydrophobic patch” region on Clb5. The Clb5-Orc6 interaction requires replication initiation, and is maintained throughout the remainder of S phase and into M phase. Eliminating the Clb5-Orc6 interaction has no effect on initiation of replication but instead sensitizes cells to lethal overreplication. We propose that Clb5 binding to ORC provides an origin-localized replication control switch that specifically prevents reinitiation at replicated origins.
Keywords: Cell cycle control, DNA replication, cyclin-dependent kinase, origin recognition complex, genomic stability, re-replication
The eukaryotic cell cycle is controlled by oscillations in cyclin-dependent kinase (Cdk) activity (Zachariae and Nasmyth 1999). Cdk activity oscillations are required because critical cell cycle steps are both positively and negatively regulated by Cdk activity. A well-characterized example is DNA replication (Bell and Dutta 2002), in which oscillating Cdk levels control the formation and activation of protein complexes at origins of DNA replication.
In budding yeast, origins are bound throughout the cell cycle by the six-member origin recognition complex (ORC; Diffley et al. 1994). As cells pass from the M to the G1 phase of the cell cycle, Cdc6 and Cdt1 proteins interact with the ORC-bound origin and direct the loading of the six-member Mcm2-7 complex (Cocker et al. 1996; Aparicio et al. 1997; Tanaka et al. 1997; Devault et al. 2002; Tanaka and Diffley 2002). The resulting structure is called the “pre-Replicative Complex” or pre-RC.
As cells enter S phase, elevated levels of Cdk activity stimulate initiation of DNA replication from the pre-RC loaded origins (Schwob et al. 1994). Positive control of replication by Cdk activity in Saccharomyces cerevisiae is primarily mediated by S-phase cyclins Clb5 and Clb6 (Schwob and Nasmyth 1993; Schwob et al. 1994; Donaldson et al. 1998a; Donaldson 2000; Epstein and Cross 2002), although it is not well understood how Cdk activity induces replication. Cdk phosphorylation of the Sld2/Drc1 replication protein has been demonstrated to be essential for replication initiation (Masumoto et al. 2002), but there may be additional activating substrates. During S phase, ORC is normally phosphorylated by Clb5,6, and biochemical studies have identified an interaction between Clb5 and ORC (Weinreich et al. 2001). The function of this interaction has been unclear. In Xenopus, cyclin E is recruited to origins of replication through Cdc6 to activate replication (Furstenthal et al. 2001a,b), although the targets for cyclin E-directed Cdk activity are also unclear.
Continued high Cdk activity also blocks any new pre-RC formation until the end of M phase, thus restricting replication to a single round per cell cycle (Dahmann et al. 1995; Cocker et al. 1996). Interestingly, the S phase-promoting Clb5,6 kinases are competent at this negative control as well (Dahmann et al. 1995). In S. cerevisiae, Cdk activity inhibits replication in at least three different ways. Cdk phosphorylation of Cdc6 results in its SCF-dependent proteolysis (Drury et al. 2000; Nguyen et al. 2001; Archambault et al. 2003), and phosphorylation of the Mcm2-7 complex leads to its exclusion from the nucleus (Labib et al. 1999; Nguyen et al. 2000). Cdk phosphorylation of Orc2 and Orc6 also helps prevent re-replication (Nguyen et al. 2001). In Schizosaccharomyces pombe, the mitotic cyclin Cdc13 interacts directly with the Orc2 subunit (Leatherwood et al. 1996), and with origins of replication in M phase (Wuarin et al. 2002). Ablating this interaction allows pre-RC reloading and re-replication within a single cell cycle (Wuarin et al. 2002). In other eukaryotes, related mechanisms allow Cdk inhibition of origin reloading (Kearsey and Cotterill 2003). Cdc2-cyclin A associates with ORC, and may regulate replication, for example by inducing Orc1 degradation (Romanowski et al. 2000; Mendez et al. 2002).
The molecular basis for target recognition by cyclin-Cdk complexes is not completely understood. Substrate-targeting regions may enhance Cdk phosphorylation of different targets. The “hydrophobic patch” region of cyclins interacts with known targets containing the “RXL” or “Cy” motif (Chen et al. 1996; Russo et al. 1996; Schulman et al. 1998; Adams et al. 1999; Brown et al. 1999; Takeda et al. 2001; Wohlschlegel et al. 2001). The hydrophobic patch is required for efficient function of the yeast S-phase cyclin Clb5 (Cross and Jacobson 2000), but importantly, the hydrophobic patch mutation does not interfere with activation of Cdc28 kinase activity toward nonspecific substrates. This suggests the hypothesis that the hydrophobic patch is required for specific target interaction, but endogenous binding targets for the Clb5 hydrophobic patch are unknown.
Here we report that the Clb5 hydrophobic patch mediates an interaction with an RXL sequence in the Orc6 subunit of ORC in budding yeast. Orc6-Clb5 interaction occurs at origins of replication only after initiation has occurred. This interaction is maintained at origins during the remainder of S phase. Mutation of the Orc6 RXL motif strongly increases vulnerability of cells to induction of lethal re-replication, while having no effect on the ability of cells to initiate replication. We propose that the association of Clb5 with ORC occurs only after the pre-RC is dismantled following replication, and that bound Clb5 specifically protects replicated origins from reinitiation.
Results
Identification of Orc6 as an RXL-containing Clb5-binding partner
The functional importance of the Clb5 hydrophobic patch (Cross and Jacobson 2000) suggests that this region must interact with targets that are important for Clb5-regulated pathways. By a range of methods detailed below, we found that the Orc6 subunit of ORC interacts with Clb5, in a manner dependent on an RXL sequence in Orc6, and on the hydrophobic patch region of Clb5. A weaker dependence of the interaction on Cdk phosphorylation sites in Orc6 (Nguyen et al. 2001) was also detected in these assays.
Two-hybrid analysis
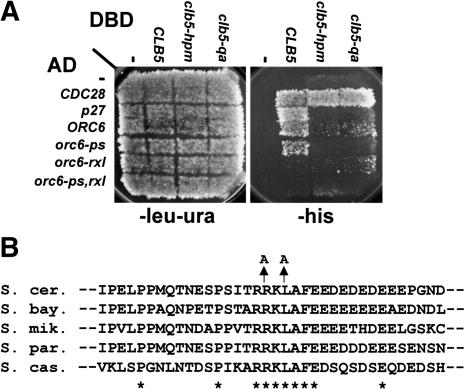
To identify proteins that interacted with Clb5 in a hydrophobic-patch-dependent manner, we carried out a two-hybrid screen using two different bait constructs: a Clb5-Gal4 DNA-binding domain (Clb5-DBD) and a Clb5-DBD with a triply mutated hydrophobic patch (M196A L201A W204A, or “hpm”; Schulman et al. 1998; Cross and Jacobson 2000). From a library of yeast DNA segments fused to the Gal4 activation domain (AD; James et al. 1996), we identified an Orc6-AD clone that activated transcription with Clb5-DBD but not with Clb5-hpm-DBD (Fig. 1A). Orc6-Clb5 interaction was almost as strongly dependent on the Clb5 hydrophobic binding pocket as p27-Clb5 interaction (Fig. 1A; Cross and Jacobson 2000). The Q241A mutation, which inactivates the function of the hydrophobic patch (Cross and Jacobson 2000), also blocks Clb5-Orc6 interaction. Cdc28-AD interaction with wild-type and mutant Clb5-DBD was comparable.
Figure 1.
Two-hybrid analysis of Clb5-Orc6 interaction. (A) Clb5 interaction with Orc6 is dependent on the Clb5 hydrophobic patch and an “RXL” motif in Orc6. Interaction of Clb5, Clb5-hpm, and Clb5-qa (Q241A) with CDC28-AD, p27-AD (Cross and Jacobson 2000), the ORC6-AD clone identified from the GAD-fusion library (James et al. 1996) and its mutagenized derivatives (“ps”: S106A, S116A, S123A, T146A; “rxl”: R178A L180A; “ps,rxl”: combined ps, rxl) is detected by growth on -his (James et al. 1996). Interaction of Clb5-DBD with Cdc28-AD is hydrophobic-patch-independent, whereas interaction with p27-AD is hydrophobic-patch-dependent (Cross and Jacobson 2000). Similar results were obtained using a second reporter (data not shown). (B) The “RXL” motif is conserved in budding yeasts. Alignment of ORC6 homologs from S. cerevisae, Saccharomyces bayanus, Saccharomyces mikatae, Saccharomyces paradoxus, and Saccharomyces castellii (Cliften et al. 2003; Kellis et al. 2003) in the vicinity of R178; complete conservation is indicated by *. The location of the R178A, L180A mutation is shown above.
Two types of mutations on Orc6-AD interfered with the interaction with Clb5-DBD. We identified a candidate “RXL” consensus sequence from Orc6 at positions 178-180 (RKL) that is completely conserved among budding yeast species (Fig. 1B; Cliften et al. 2003; Kellis et al. 2003). Mutation of RKL to AKA (Orc6-rxl-AD) largely eliminated the two-hybrid interaction with Clb5 (Fig. 1A). Because Orc6 is phosphorylated by Clb5/Cdc28 (Weinreich et al. 2001), we tested whether mutations in these sites altered the interaction with Clb5. Simultaneous mutation of four Cdk consensus phosphorylation sites (S106, S116, S123, T146) in Orc6 (Nguyen et al. 2001) reduced but did not eliminate the Clb5-Orc6 two-hybrid interaction (Fig. 1A).
Orc6-AD showed preferential interaction with Clb5-DBD when compared with a mitotic B-type cyclin. Orc6-AD interacted more weakly with Clb2-DBD than with Clb5-DBD, and Clb2-Orc6 interaction did not require the Orc6 RXL or the Clb2 hydrophobic patch (Supplementary Fig. S1). Thus the hydrophobic patch-Orc6-RXL interaction may be specific to Clb5, consistent with the idea that this region of cyclins confers cyclin-specific binding (Cross and Jacobson 2000; Takeda et al. 2001; Wohlschlegel et al. 2001).
Direct interaction of Clb5/Cdc28 with ORC bound to origin DNA
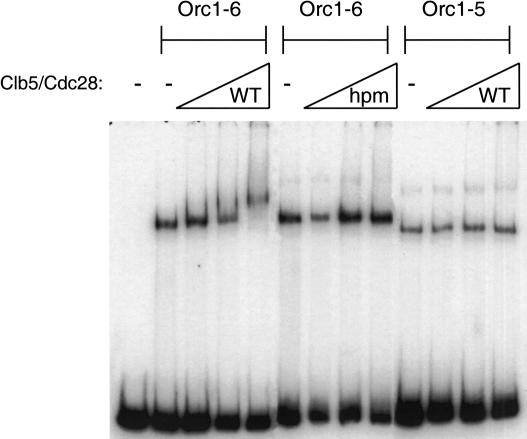
Clb5/Cdc28 complexes purified from baculovirus-infected insect cells interacted with an ORC-origin DNA complex in an electrophoretic mobility shift assay (Fig. 2). Consistent with the two-hybrid analysis, this interaction required the Clb5 hydrophobic patch and the presence of Orc6 in ORC. Although the wild-type and hpm-mutant Clb5/Cdc28 complexes had equal activities using histone H1 or Sic1 as substrates, ORC subunits were phosphorylated much less efficiently by Clb5-hpm/Cdc28 (Supplementary Fig. S2). These observations show that the Clb5-Orc6 interaction is direct and can occur when ORC is bound to origin DNA.
Figure 2.
In vitro interaction between Clb5/Cdc28 and Orc6. Clb5/Cdc28 binds an ORC/origin DNA complex in a hydrophobic-patch- and Orc6-dependent manner. Twenty nanograms of ORC shifted a radiolabeled RI/HIII fragment of ARS1. Addition of wild-type but not hydrophobic patch-mutant Clb5/Cdc28 supershifts the ORC/origin DNA complex, but not an ORC/origin DNA complex lacking Orc6. The amounts of Clb5/Cdc28 added were 80, 330, and 1320 ng, although the Clb5 was substoichiometric. The higher band present in the ORC/origin DNA lanes lacking Clb5/Cdc28 represents a multimer of ORC or Orc1-5. Addition of wild type (WT) but not hydrophobic patch mutant (hpm) Clb5/Cdc28 supershifts the ORC/origin DNA complex (Orc1-6), but not an ORC/origin DNA complex lacking Orc6 (Orc1-5).
Copurification of Orc6 and Clb5 from yeast cells
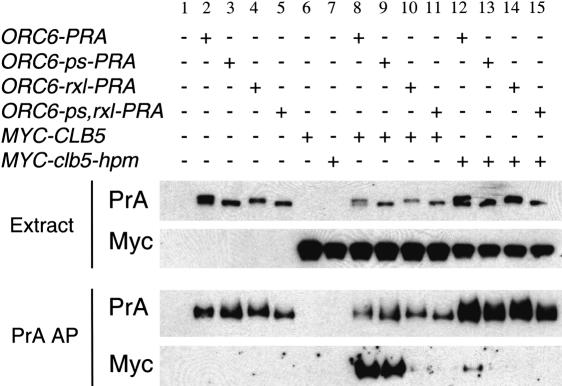
Affinity purification of protein A-tagged Orc6 resulted in copurification of myc-tagged Clb5, when each protein was expressed from its endogenous promoter (Fig. 3). A similar association could be observed when extracts that independently expressed Orc6-PrA and Clb5-Myc were mixed (data not shown), suggesting that the Clb5-Orc6 interaction is quite efficient. Consistent with the results above, this binding was dependent on both the Clb5 hydrophobic patch and the Orc6 RXL sequence (Fig. 3, lanes 10,12-15). In contrast, we observed a weak interaction between Clb2 and Orc6 in this assay, which showed little or no dependence on the Orc6 RXL motif or the phosphorylation sites (Supplementary Fig. S3). These results show that Clb5-Orc6 interaction can occur at endogenous expression levels, with native functional proteins (both epitope-tagged Orc6 and Clb5 complement; data not shown).
Figure 3.
Orc6 and Clb5 are copurified from yeast cells with specific sequence requirements. Cells expressing the indicated fusions (PrA or Myc-tagged, as indicated above) from their endogenous promoters, alone or in combinations, were subjected to Protein A affinity purifications and to Western blotting analysis (as in Materials and Methods). (Extract) Total cell extract; (PrA AP) after IgG-Sepharose purification of the Protein A tag, bound proteins were eluted; (PrA, Myc) detection of the Protein A tag or the Myc epitope by Western blotting. The results show dependence of the Orc6-Clb5 association on the Clb5 hydrophobic patch motif, the Orc6 RXL motif, and weak dependence on Orc6 Cdk consensus sites (S106, S116, S123, T146).
In vivo binding of Clb5 to origins of replication
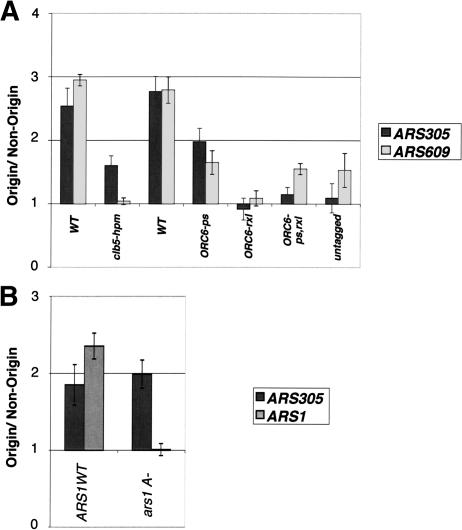
We asked if the Clb5-Orc6 interaction occurred when ORC was bound to origins of replication using chromatin immunoprecipitation (ChIP) from formaldehyde cross-linked cells (Fig. 4). Clb5 was associated with both early- and late-initiating origins of DNA replication in asynchronously growing cells (Fig. 4A). Clb5-origin association depended on a wild-type ORC-binding site because mutation of the ARS1 ORC-binding site (ars1 A-) eliminated association of Clb5 with ARS1 but not other origins (Fig. 4B). Consistent with the results above, Clb5-origin interaction required the Clb5 hydrophobic patch and the Orc6 RXL. These results show that Clb5-Orc6 interaction can occur in vivo, in the context of fully assembled ORC bound to functional origins.
Figure 4.
In vivo interaction between Clb5 and origins of replication. (A) Clb5 associates with origins of replication in vivo, dependent on the Clb5 HP and Orc6 RXL. All strains except the untagged have an N-terminal 9Myc tag on the genomic copy of Clb5. The first tagged wild-type (WT) strain contains the standard wild-type ORC6 locus; the second wild-type strain contains an ORC6-wt locus marked with LEU2 and HIS3 identically to the ORC6 mutant strains. Asynchronous cultures were analyzed by chromatin immunoprecipitation with a monoclonal anti-Myc antibody. Shown are the results of quantitative PCR of a representative experiment. The error bars represent the error of the PCR. The nonorigin DNA sequence is a sequence halfway between ARS305 and ARS306. (B) Clb5 does not associate with a nonfunctional origin that does not bind ORC. MYC-CLB5 cells with or without a linker scanner mutation in the A element of ARS1 (Marahrens and Stillman 1992) were arrested in G1 with 50 ng/mL α-factor and released into S phase at room temperature. The time point shown is 40 min after release, and is representative. Clb5 did not associate with the mutated origin, but did with a wild-type origin (ARS305) in the same strain. DNA was analyzed as in A.
Cell cycle regulation of Clb5 association with origins
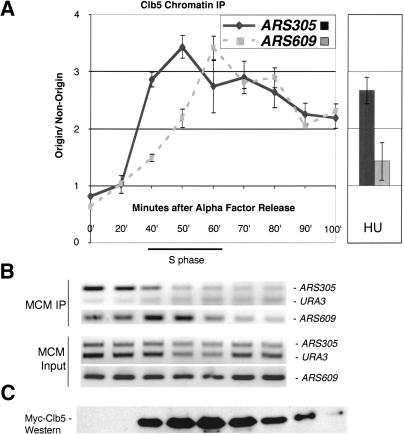
To investigate the timing of Clb5 association with origins of replication, we followed the association during a synchronous passage through S phase. Clb5 first associated with origins at a time that correlated with their time of initiation. The association of Clb5 with an origin that initiated early in the cell cycle (ARS305) was first observed 40 min after release from α-factor, at the same time that MCMs dissociated from that origin (Fig. 5). MCM complex dissociates from the origin upon initiation, and therefore is a temporal marker for initiation (Aparicio et al. 1997; Tanaka et al. 1997). In contrast, Clb5 first associated with an origin that initiated later in S phase (ARS609) ∼60 min after release from α-factor, at the same time that MCMs dissociated from that origin, despite the presence of abundant Clb5 at earlier times. The differences in Clb5 association between different origins did not reflect differences in Orc6 association with origins (data not shown). Interestingly, once associated with a given origin, Clb5 binding persisted until mitosis, even after degradation of a significant pool of Clb5 (Fig. 5).
Figure 5.
Cell cycle regulation of Clb5-origin interaction. Clb5 binds origins of replication at the time that they initiate, and then persists at the origin. (A) Clb5 associates first with the early initiating origin ARS305 and later with the later initiating origin ARS609, and then persists at all of the origins. MYCCLB5 cells were arrested in G1 with α-factor and released into the cell cycle at room temperature. S phase was determined by FACS. Samples were analyzed as in Figure 4, except that the nonorigin sequence used is URA3. In the right half of the panel, the same cells were arrested in 200 mM HU, after early origins had initiated, but before late origins had, and Clb5 associates only with early origins. (B) Timing of association of MCM with origin DNA. Representative gels showing conventional PCR of ARS305 and ARS609, and of URA3. (C) Myc-Clb5 levels were determined by Western blotting and are shown below the PCR.
The above results suggested that Clb5 associates with origins at the time that they initiate replication and then persists at the origin well after initiation is complete. To assess this possibility using a different approach, we arrested cells in hydroxyurea (HU) prior to analysis. This drug causes cells to arrest with high Clb5 levels after early origins have initiated replication but before late origins have done so (Santocanale and Diffley 1998). At the HU arrest point, Clb5 was associated with early origins that had initiated replication, such as ARS305 (Fig. 5A, right), supporting the idea that the Clb5 persists at the origin after initiation. Conversely, Clb5 was not associated with the uninitiated late origin ARS609.
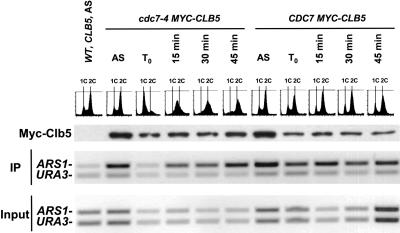
Cdc7 activity is required for Clb5-origin association
The finding that Clb5 did not associate with a late origin in HU-treated cells suggested that initiation might be required for the interaction to occur, but the analysis was restricted to late origins because early origins initiate even in the presence of HU (Santocanale and Diffley 1998). To ask whether Clb5 associated with early origins before initiation, we prevented replication initiation by inactivating the Cdc7 protein kinase using the cdc7-4 temperature-sensitive mutation (Fig. 6). We found that Myc-Clb5 was present in cdc7-4 cells blocked at high temperature, but did not bind significantly to the early origin ARS1. Binding occurred rapidly and concurrently with replication upon release of the block by lowering the temperature. A parallel control with a MYC-CLB5 CDC7-wt strain showed Myc-Clb5 interaction with ARS1 at high and low temperature, indicating that the effect in the cdc7-4 strain was specific to Cdc7 inactivation. Similar results were obtained with a second early origin, ARS305 (data not shown). As seen above (Fig. 5), Clb5-origin interaction persisted even after replication was complete as measured by FACS. Together, our studies of the temporal regulation of Clb5-origin association suggest that replication of each individual origin is required for Clb5 binding, and the association is then retained through the remainder of S phase.
Figure 6.
Cdc7 activation is required for Clb5 origin association. cdc7-4 or CDC7-wt strains (both CLB5-MYC) were grown to log phase in YPD at 23°C, transferred for 3 h to 37°C to inactivate Cdc7 in the cdc7-4 culture, then released in fresh YPD at 23°C. Culture samples were taken at the indicated time points for FACS analysis, anti-Myc Western blotting (corrected volumes for cell equivalence), and anti-Myc chromatin immunoprecipitation.
Mutations that prevent Clb5-origin association do not interfere with replication initiation
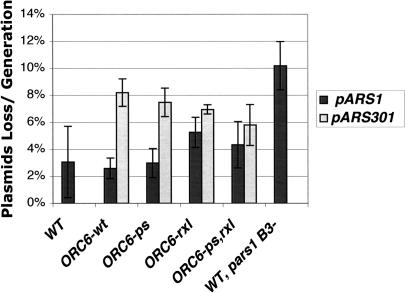
Because Clb5 associated with origins as they commenced initiation, it was possible that this interaction positively regulated initiation. A severe defect in initiation would be expected to interfere with timely passage through S phase; however, ORC6-rxl and ORC6-ps,rxl strains had no defect on the timing or rate of DNA synthesis in synchronized cultures, as judged by FACS analysis (data not shown). Measurement of plasmid stability represents a much more sensitive assay for defects in the initiation of replication. We tested strains containing the same two ORC6 mutations for their ability to maintain an early (ARS1) or a late (ARS301) origin-containing plasmid. In neither case was a significant defect in plasmid stability observed (Fig. 7). Together, these findings argue against a role of the Clb5-Orc6 interaction in the initiation of replication.
Figure 7.
ORC6-rxl cells do not have an initiation defect. Plasmid loss rates/generation for each strain are plotted with error bars representing the standard deviations from at least three plasmid loss experiments. ORC6-wt is the wild-type marked ORC6 strain isogenic to the ORC6 mutant strains, and behaved identically to the parent unmarked strain. (pARS1) pARS1WT; (pARS1B3-) pARS1,757-764 (Marahrens and Stillman 1992).
ORC6 and CLB5 mutations interact with a mutation stabilizing Cdc6
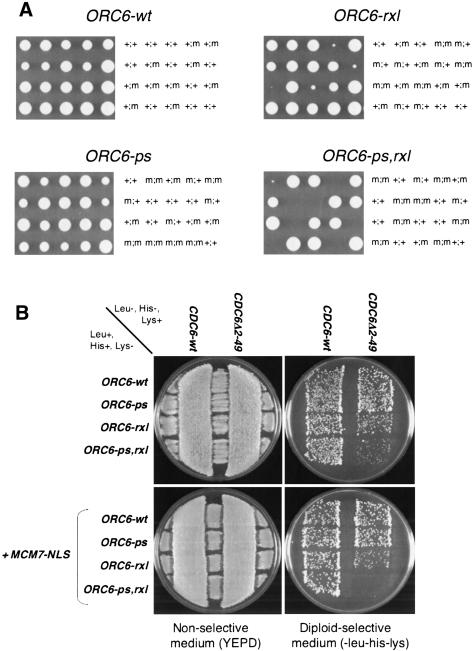
Because the Clb5-Orc6 interaction was unnecessary for initiation of replication, we asked whether the interaction helped protect origins from reinitiating replication within a single cell cycle. Inhibition of re-replication depends on Clb-Cdk phosphorylation of Orc2, Orc6, the Mcm2-7 complex, and Cdc6 (Nguyen et al. 2001). Together these modifications block formation of new pre-RCs. To probe for involvement of the Orc6 RXL-Clb5 HP interaction in control of re-replication, we eliminated elements of this control in the absence of the Orc6 RXL motif. The Cdc6 N terminus is required for Cdk control of Cdc6 abundance (Drury et al. 1997, 2000; Archambault et al. 2003). Combining ORC6-rxl and chromosomal deletion of the Cdc6 N-terminal 49 amino acids resulted in a striking slow-growth phenotype in tetrad analysis (Fig. 8A). This phenotype was enhanced by mutating the Orc6 Cdk phosphorylation sites (ORC6-ps,rxl; Fig. 8A).
Figure 8.
Genetic interactions between dominant mutant alleles of ORC6, CDC6, and MCM7. (A) Interaction between ORC6 alleles and CDC6Δ2-49. ORC6-x::LEU2::HIS3/ORC6 CDC6Δ2-49/CDC6-wt diploid strains were sporulated, and tetrads were dissected on YEPD plates, incubated for 4 d at 30°C, and photographed. Subsequent replica-plating and PCR tests allowed the detection of ORC6 alleles and of CDC6Δ2-49, respectively. Inviable spores were genotyped by assuming 2:2 segregation. The ORC6 allele assayed is indicated above each panel. The presence of the marked ORC6 mutant allele assayed (m) or ORC6-wt (+; left symbol) and the presence of the CDC6Δ2-49 allele (m) or CDC6-wt (+; right symbol) in each colony is indicated. In the control cross (upper left) where the marked ORC6::LEU2::HIS3 allele was ORC6-wt, a + was used for both ORC6 alleles, because the marked and unmarked wild-type alleles behaved identically. (B) Dominance of ORC6-rxl, ORC6-ps,rxl, CDC6Δ2-49, and MCM7-NLS. Strains containing the indicated alleles were mated on YEPD plates (left) and replica-plated on SCD-his-leu-lys for diploid selection (right), and incubated for 2 d at 30°C. Diploids appear at the intersections of the streaks.
The semilethal phenotype was dominant for both CDC6Δ2-49 and ORC6-rxl or ORC6-ps,rxl based on transformation experiments with low-copy plasmids encoding wild-type and mutant CDC6 and ORC6 (data not shown), and on the slow-growth phenotype of CDC6Δ2-49/CDC6 ORC6-ps,rxl/ORC6 diploids (Fig. 8B, top). This dominance is consistent with the synthetic lethality being caused by a gain of function such as would occur if the mutant proteins were escaping negative regulation by the Clb-Cdk system.
The results above with ORC6 mutations suggested that Clb5 interaction with Orc6 might be required for cell viability in the presence of stabilized Cdc6 (Cdc6Δ2-49). Indeed, clb5Δ CDC6Δ2-49 double mutants were inviable in tetrad analysis, and GAL-CLB5 clb5Δ CDC6Δ2-49 strains grew well on galactose but failed to proliferate when plated on glucose to turn off GAL-CLB5 (data not shown). Overall, these genetic findings are consistent with the idea that Clb5-Orc6 interaction is required for viability in the presence of stabilized Cdc6.
Interference with the Orc6-Clb5 interaction stimulates inappropriate re-replication of the genome
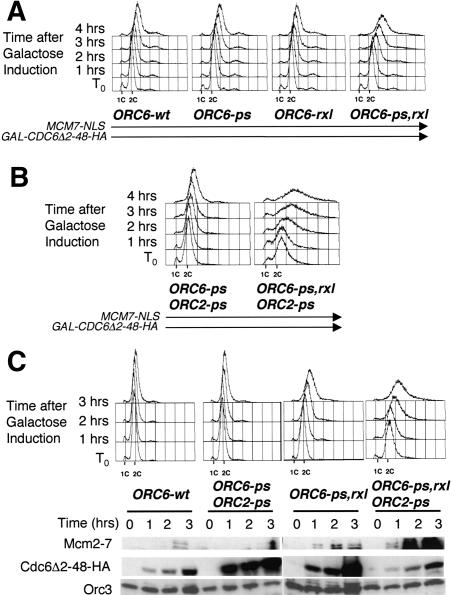
We wanted to test directly whether the lethality associated with combining ORC6 mutations with stabilized Cdc6 was caused by inappropriate re-replication of the genome. We placed CDC6Δ2-48-HA under the control of the GAL1 promoter and assayed for re-replication in strains containing various mutations in ORC6. We also included an MCM7-NLS construct leading to constitutively nuclear Mcm2-7 proteins, because control of MCM complex localization is an independent means of Cdk-dependent re-replication control (Nguyen et al. 2001). To test for inappropriate DNA replication, we arrested these cells in M phase with nocodazole, induced Cdc6Δ2-48-HA expression with galactose, and examined the DNA content by FACS. We found that the ORC6-ps,rxl strain showed a reproducible gain in DNA content beyond 2C, whereas strains with either ORC6-rxl or ORC6-ps maintained ∼ 2C DNA content (Fig. 9A).
Figure 9.
The Orc6 RXL motif helps protect cells from re-replicating. (A) ORC6-ps,rxl MCM7-NLS GAL-CDC6Δ2-48-HA cells can be induced to re-replicate. ORC6 wild-type and mutant strains containing MCM7-NLS and GAL-CDC6Δ2-48-HA were arrested with 15 μg/mL nocodazole in YEP/2% raffinose, and then Cdc6Δ2-48-HA was induced with 2% galactose. (B) ORC2-ps exacerbates the re-replication phenotype (methods same as in A). (C) Pre-RCs form on the chromatin in the ORC6-ps,rxl MCM7-NLS GAL-CDC6Δ2-48-HA re-replicating cells. ORC6-ps,rxl MCM7-NLS cells with or without GAL-CDC6Δ2-48-HA were arrested as above, and then the chromatin pellet was isolated and the DNAse I- and salt-sensitive proteins were extracted and visualized by Western blotting.
Little detectable overreplication was observed in these backgrounds without inclusion of the MCM7-NLS allele (data not shown), despite the significant synthetic lethality observed between the Orc6 mutants and stabilized Cdc6 (Fig. 8A). Possible reasons for the apparent inconsistency between detectable re-replication and lethality are discussed below (see Discussion). This MCM7-NLS allele also led to highly penetrant, dominant induction of lethality in CDC6Δ2-49/CDC6 ORC6-ps,rxl/ORC6 diploids (Fig. 8B, bottom) even without GAL-driven overexpression of Cdc6, confirming by another assay the importance of nuclear localization of the Mcm2-7 complex for strong phenotypes in this system.
Phosphorylation sites on Orc2 were also implicated in control of re-replication (Nguyen et al. 2001). We constructed ORC2-ps ORC6-ps,rxl GAL-CDC6Δ2-48-HA MCM7-NLS strains, with the six predicted Cdk phosphorylation sites in Orc2 mutated, and isogenic controls containing the intact ORC6 RXL sequence, and tested for re-replication in nocodazole-arrested cells (Fig. 9B). ORC2-ps ORC6-ps,rxl GAL-CDC6Δ2-48-HA MCM7-NLS cells showed robust re-replication (in many cells to beyond 4C DNA content) when Cdc6Δ2-48-HA expression was induced (Fig. 9B). The importance of the Clb5-Orc6 interaction for the observed re-replication is emphasized by the comparison with an isogenic strain lacking the ORC6-rxl mutation. Partial re-replication observed in a ORC2-ps ORC6-ps MCM7-NLS GALCDC6Δ2-48-HA strain (Nguyen et al. 2001; Fig. 9B, left, reproducible but relatively weak in our experimental conditions, for unknown reasons) is strongly enhanced by mutation of the ORC6 RXL (Fig. 9B, right). Therefore, the Clb5-Orc6 interaction is sufficient on its own for significant control of re-replication, even with all previously identified controls disrupted.
All of the strains containing inducible, stabilized Cdc6 in combination with ORC mutants demonstrated an immediate loss of viability when switched to galactose-containing media and then plated back onto glucose (Supplementary Fig. S4). The loss of viability correlated approximately with the degree of re-replication observed, suggesting a causal link between the two.
To confirm that the increase in DNA content in these experiments was dependent on new pre-RC formation, we assayed for the presence of pre-RCs in ORC6-ps,rxl MCM7-NLS GAL-CDC6Δ2-48-HA cells arrested in nocodazole (Fig. 9C). We found that Mcm2-7 proteins accumulated in the chromatin-bound fraction of the cells in a manner that closely paralleled the extent of re-replication in the same strains. Thus, the increase in DNA content that we see is likely caused by reinitiation from inappropriately reformed pre-RCs.
Discussion
The Clb5 hydrophobic patch interacts with an RXL sequence in the Orc6 subunit of ORC. This interaction occurs on ORC bound to replication origins, and the interaction occurs only after replication initiation. This interaction has no effect on normal replication initiation, but instead functions as an origin-localized switch to block replication reinitiation specifically at replicated origins.
Substrate targeting of cyclin-dependent kinase activity
Cyclins have a clear biochemical role in activation of cyclin-dependent kinase catalytic subunits, and a less well-defined role in directing kinase activity to particular substrates or regions of the cell (for review, see Miller and Cross 2001). The hydrophobic patch-RXL interaction has been shown to direct cyclin A-Cdk2 activity to substrates such as p107 (see above). Although this interaction was suspected to play a role in function of the B-type cyclin Clb5 based on genetic results (Cross and Jacobson 2000), no RXL-containing targets for any B-type cyclin have been identified. Here we show that the Clb5 hydrophobic patch binds to an RXL sequence in Orc6.
Clb5/Cdc28 bound to replicated origins blocks reinitiation
Maintenance of ploidy is a critical aspect of cell cycle regulation, and perhaps for this reason, multiple mechanisms exist that redundantly ensure this control (Nguyen et al. 2001). We show here that stable binding between Clb5 and Orc6 is an important mechanistic aspect of this control.
A strong overreplication phenotype (Fig. 9) is restricted to the fully deregulated situation with ORC6-ps,rxl, MCM7-NLS, ORC2-ps, and strong overexpression of CDC6. Nevertheless, we observe lethal interactions with subsets of these mutations, such as stabilized Cdc6 combined with the ORC6-rxl mutation (Fig. 8). It is likely that in these strains a low level of reinitiation (e.g., from only a few origins on scattered chromosomes) results in sufficient aneuploidy to cause lethality without increasing DNA content enough for clear detection by FACS. Interestingly, the viability of ORC6-rxl CDC6Δ2-49 strains is completely dependent on the Mec1 DNA-damage-response kinase, strongly suggesting that these strains experience some level of DNA damage possibly resulting from aberrant (extra) replication fork initiation (V. Archambault and F.R. Cross, unpubl.). In the mammalian system, a connection between overreplication and DNA-damage-response kinases has been observed (Vaziri et al. 2003). These findings may help explain the redundancy of control exerted over replication reinitiation; even partial abrogation of this system strongly increases dependence on the Mec1-dependent DNA damage response. We speculate that the DNA damage response may allow cells to repair a limited number of aberrant DNA structures generated by reinitiation.
In fission yeast, the mitotic B-type cyclin Cdc13 binds to ORC. Ablating this interaction results in pre-RC reloading and re-replication (Wuarin et al. 2002). Our findings in budding yeast may reflect an evolutionarily conserved role for origin-localized B-type cyclin-dependent kinase activity in maintaining ploidy, as well as providing a specific molecular mechanism for this association to occur. An interesting difference between our results and those of Wuarin et al. (2002) is that we detect specific binding of the S-phase cyclin Clb5, whereas in the fission yeast studies, the S-phase Cig1 and Cig2 cyclins were reported not to bind to origins, unlike the mitotic cyclin Cdc13. Previous results implicated Clb5 in preventing re-replication despite its S-phase-promoting role (Dahmann et al. 1995), although the mechanism was unclear. We discuss below how our findings accommodate the simultaneous use of Clb5/Cdc28 in the activation and inhibition of DNA replication.
An origin-localized replication control switch
We propose that Clb5 binds to ORC only after replication initiation has occurred (Figs. 5, 6). Consistent with this hypothesis, we find that in HU-arrested cells in which early origins have initiated and late origins have not, we see Clb5 association only at early origins. Similarly, Clb kinases can complete their essential function for replication initiation in the absence of Cdc7 function in yeast (Nougarede et al. 2000), and yet we see no Clb5-origin interaction until after Cdc7 function. This strongly suggests that the Clb5-Orc6 interaction is not involved in the positive replication function of Clb5. Consistent with this, the ORC6-ps,rxl mutation, which eliminates Clb5-Orc6 interaction, does not cause any detectable replication defect using a plasmid maintenance assay (Fig. 7). Because Cdc7 activity is required for Clb5-Orc6 interaction, and Cdc7 acts sequentially at individual origins to drive initiation (Bousset and Diffley 1998; Donaldson et al. 1998b), it is likely that Clb5 binds to individual origins only after initiation has commenced at each origin throughout S phase. Taken together with our observations indicating that the Clb5-Orc6 interaction inhibits pre-RC reformation and re-replication, this mechanism provides an origin-localized replication control switch.
A possible molecular mechanism by which Clb5 binding could be restricted to replicated origins is through steric hindrance of Clb5 association with ORC by pre-RC components. Such a mechanism would allow Clb5/Cdc28 to locally block initiation from origins that have initiated or been passively replicated by a replication fork derived from an adjacent origin (and therefore lack a pre-RC) but not at unreplicated origins in the same cell (which would have a pre-RC). Indeed, we see a tight correlation between the time that Clb5 associates with origins and the time that MCM proteins dissociate from the same sites (Fig. 5).
Our mutational analysis indicates that the Clb5-Orc6 interaction is sufficient on its own for significant control of re-replication, even with all previously identified controls disrupted (Fig. 9). Origin-localized Clb5 could help prevent pre-RC reformation at origins by phosphorylating target proteins (either ORC itself, or other proteins in the vicinity of the initiated origin). Alternatively, Clb5 bound to Orc6 could sterically block binding of one or more pre-RC components. Further work will be required to address these different models, and it is possible that both mechanisms are functioning.
The origin-localized replication control switch model provides an economical mechanism allowing Clb5 to globally promote replication initiation (by a mechanism that does not require origin localization), and at the same time to locally block reinitiation specifically at origins that have already been replicated. Such a mechanism is particularly important when the same kinase is being used in both activation and inhibition as appears to be the case in S. cerevisiae. This contrasts to the postreplicative mitotic cyclin binding to origins described in fission yeast (Wuarin et al. 2002), which can only protect origins from re-replication after the completion of S phase. This difference in CDK regulation of replication may also help to explain the ability of overexpression of replication proteins to drive cells into re-replication in S. pombe but not in S. cerevisiae (Muzi-Falconi et al. 1996; Nishitani et al. 2000). Because Clb5 is present early in S phase and thus can prevent reinitiation at replicated origins even before other origins have fired, it is critical to have a mechanism of Cdk recruitment that allows a distinction between replicated and unreplicated origins.
Materials and methods
Plasmids and strains
Strains (W303 background) and plasmids were prepared using standard laboratory methods (Ausubel 1992). Plasmid pSPB65 contained ORC6 in pRS315, and pSB6-21 contained the ORC6 phosphorylation site mutations S106A, S116A, S123A, and T146A. FC606 and FC608 were identical to SPB65 and SPB6-21 but contained the ORC6-rxl mutation R178A,L180A. The ORC6 coding sequence in SPB65, SPB6-21, FC606, and FC608 was tagged with Protein A as described (Aitchison et al. 1995; Wach et al. 1997) to produce SPB65-A, SPB6-21-A, FC606-A, and FC608-A. The wild-type chromosomal ORC6 locus was tagged similarly. Plasmids pRS406ORC6-wt, pRS406ORC6-rxl, and pRS406ORC6-ps were made by cloning an NotI/XhoI fragment of ORC6 from pSB65, pFC606, and pSPB6.21, respectively, into pRS406WT cut with the same enzymes. pRS406ORC6-ps,rxl was made by amplifying the RXL mutation from pRS406ORC6-rxl with oligonucleotides SB831, TCTATTAC TAGGGCAAAGGCAGCA and SB795, CACACACTAATT GCCATGGGC, and then using this megaprimer to amplify the ORC6 Cdk phosphorylation site mutations from pRS406ORC6-ps with the oligonucleotide SB793, CAAGATGAAGAGGTT GCTAGATGTC. This product was cut with NdeI/NcoI and cloned into pRS406ORC6-wt cut with the same enzymes. ORC6 mutant strains were constructed by cutting plasmids pRS406ORC6-wt, pRS406ORC6-rxl, pRS406ORC6-ps, and pRS406ORC6-ps-,rxl with BsmI and integrating into ySB1028 (orc6::HIS3MX, pSPB66 [pORC6, URA3]), and then streaking onto 5-FOA to lose the pSB66ORC6 plasmid. GAL-CDC6Δ2-48-HA strains were constructed by digesting an RS305-based GAL-CDC6Δ2-48-HA plasmid with EcoRV to target integration to URA3. Copy number was not measured in these integrants, but the results obtained were insensitive to the level of Cdc6Δ2-48-HA produced, as measured by Western analysis in each experiment. Strains containing mutations in ARS1 were made as described previously (Wilmes and Bell 2002). ORC2-ps strains and MCM7-NLS strains were constructed as described with plasmids pJL1095 and pJL1206, respectively, generous gifts from Joachim Li (University of California at San Francisco, San Francisco, CA; Nguyen et al. 2001). See Supplementary Table S1 for strain genotypes.
ORC6-PrA integrated in the chromosome had no significant growth defect, indicating that the Protein A tag did not interfere with function. Similarly, MYC-CLB5 could fully complement the lethal phenotype of clb3,4,5,6 mutants.
Two-hybrid analysis
Full-length CLB5, clb5-hpm, clb5-Q241A (“qa”), and CLB2 were fused to the GAL4 DNA-binding domain in the vector pBDU-C1 (James et al. 1996) as described previously (Cross and Jacobson 2000). GAL4 activation domain (AD) fusion libraries (James et al. 1996) were transformed into PJ69-4A (James et al. 1996) carrying CLB5-DBD, and transformants selected for an His+ Leu+ Ura+ phenotype and subsequently screened for an Ade+ phenotype, essentially as described (James et al. 1996). The His+ and Ade+ phenotypes result from activation of different reporters present in PJ69-4A, eliminating a high background from selecting for the His+ phenotype alone (James et al. 1996). Following CLB5-DBD plasmid loss selected by 5-FOA, the GAD plasmid transformants were mated to PJ69-4α carrying CLB5-DBD or clb5-hpm-DBD plasmids, to identify interactors specifically dependent on the Clb5 hydrophobic patch. Transformants were tested similarly with CLB5-qa-DBD and CLB2-DBD. The ORC6-AD clone (C5I-13) identified in this screen contained the Gal4 AD fused to ORC6 coding sequence starting from I38 and extending to ∼25 codons from the ORC6 C terminus. (This clone was extended to contain the complete ORC6 C terminus with similar results to those presented; data not shown.) The ORC6 phosphorylation site mutations (S106A, S116A, S123A, T146A) present in plasmid SPB6-21 were transferred into C5I-13 by substitution of an NcoI-NdeI fragment. The R178A, L180A mutation was introduced by PCR mutagenesis into either wild-type or phosphorylation-site-mutated ORC6, and the desired coding sequences in the entire mutagenized regions were confirmed.
Clb5/Cdc28 expression and purification
His-HA-Clb5-wt and His-HA-Clb5-hpm were expressed in Fast-Bac baculo transfer vectors (pSB1024 and pSB1023), as was GSTCdc28-HA (pFBCdc28). Baculovirus expression of wild-type and mutant Clb5/Cdc28 was carried out as described previously (Klemm et al. 1997). The proteins were purified from the nuclear extracts by binding and elution from Glutathione Sepharose 4 Fast Flow resin (Amersham Pharmacia), as directed by the manufacturer, except that the binding buffer was 12.5 mM HEPES-KOH (pH 7.5), 0.4 M KCl, 2.5 mM magnesium acetate, 1 mM EDTA, 1 mM EGTA, 0.01% NP-40, 1 mM dithiothreitol, and the elution buffer was the same plus 10 mM reduced glutathione (Sigma-Aldrich).
Kinase assays
The proteins were mixed together on ice in the indicated concentrations in 12.5 mM HEPES-KOH (pH 7.5), 0.4 M KCl, 2.5 mM magnesium acetate, 1 mM EDTA, 1 mM EGTA, 0.01% NP-40, 1 mM dithiothreitol, with 10 μCi γ-32P, 5 μM ATP, and 5 μM magnesium chloride. Reactions were incubated for 20 min at room temperature, and then run on 10% SDS-PAGE, and examined by autoradiography.
Electrophoretic mobility shift assays
ORC/origin EMSAs were performed as described previously (Lee and Bell 1997), except that the level of ATP present in all reactions was 0.5 mM, and the competitor DNA was 50 mg/mL dGdC.
Protein A affinity purifications and Western blotting
Protein A affinity purifications of Orc6-PrA were performed as described previously (Archambault et al. 2003). MYC-CLB5 (wild type and mutant) was expressed from the chromosomal locus. Orc6-PrA, wild type and mutants, were expressed from low-copy plasmids SPB65-A, SPB6-21-A, FC606-A, and FC608-A (see above). These plasmids expressed a somewhat lower level of Orc6-PrA than was observed with chromosomally tagged ORC6; hydrophobic patch-dependent Clb5-Orc6 interaction was also observed in the latter context (data not shown).
Chromatin immunoprecipitation
ChIP was performed as described (Aparicio et al. 1997), with minor modifications. Cells were cross-linked on ice overnight before washing. Cells were broken with a 45-sec pulse with an equal volume of glass beads at setting 6.5 in a FastPrep FP120 machine (Bio 101, Savant). Myc-Clb5 was immunoprecipitated overnight with a 1:250 dilution of 9E11 monoclonal antibody (Genetex). Mcm2-7 were immunoprecipitated overnight with monoclonal antibody AS1.1, which recognizes all six subunits (Schwacha and Bell 2001). Quantitation was performed by running samples in triplicate on a 7000 Sequence Detection System (Applied Biosystems) and comparing to 10-fold dilutions of similarly prepared genomic DNA with TaqMan Universal PCR Mix (Applied Biosystems) and 0.5× Sybr Green (Molecular Probes). The indicated origin/nonorigin DNA was plotted, with the standard deviations of the three reactions. Primer sequences are available upon request.
Re-replication and viability assays
Strains were arrested with 15 μg/mL nocodazole (Sigma-Aldrich) in media containing 2% raffinose for 4 h at room temperature, and then induced after the zero-hour time point with 2% galactose. Samples were taken every hour for FACS, viability, and Western analysis of Cdc6Δ2-48-HA levels (data not shown). For the viability assays, 500 cells were counted at the zero hour, and the same volume of sonicated cells was plated onto glucose media at each time point. The number of colonies was counted after 2 d, and the number of colonies at each time point was divided by the zero-hour time point to determine a percentage viability. DNA content was measured by flow cytometry as described previously (Bell et al. 1993).
Chromatin association assays
Chromatin association was assayed as described previously (Nguyen et al. 2001). Westerns were performed as described above. Mcm2-7 were detected with a 1:2000 dilution of a polyclonal antibody, UMU185. ORC was detected with a 1:1000 dilution of polyclonal antibody. Cdc6Δ2-48-HA was detected with a 1:1000 dilution of Ha.11 (BabCO).
Acknowledgments
We thank S. Chen, P. James, R. Klemm, and J. Li for providing reagents. We thank A. Amon, H. Blitzlau, M. de Vries, A. Dutta, J. Randell, and R. Tanny for comments on the manuscript; J. Li for discussion and communicating unpublished results; M. Rout for useful discussions and for providing laboratory space and support. This work was supported by PHS GM047238 to F.R.C., and PHS GM052339 and Howard Hughes Medical Institute support to S.P.B.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1202304.
Supplemental material is available at http://www.genesdev.org.
References
- Adams P.D., Li, X., Sellers, W.R., Baker, K.B., Leng, X., Harper, J.W., Taya, Y., and Kaelin Jr., W.G. 1999. Retinoblastoma protein contains a C-terminal motif that targets it for phosphorylation by cyclin-cdk complexes. Mol. Cell. Biol. 19: 1068-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison J.D., Rout, M.P., Marelli, M., Blobel, G., and Wozniak, R.W. 1995. Two novel related yeast nucleoporins Nup170p and Nup157p: Complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 131: 1133-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio O.M., Weinstein, D.M., and Bell, S.P. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell 91: 59-69. [DOI] [PubMed] [Google Scholar]
- Archambault V., Li, C.X., Tackett, A.J., Wäsch, R., Chait, B.T., Rout, M.P., and Cross, F.R. 2003. Genetic and biochemical evaluation of the importance of Cdc6 in regulating mitotic exit. Mol. Biol. Cell 14: 4592-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M. 1992. Current protocols in molecular biology. Greene Pub. Associates and Wiley-Interscience, New York.
- Bell S.P. and Dutta, A. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71: 333-374. [DOI] [PubMed] [Google Scholar]
- Bell S.P., Kobayashi, R., and Stillman, B. 1993. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science 262: 1844-1849. [DOI] [PubMed] [Google Scholar]
- Bousset K. and Diffley, J.F. 1998. The Cdc7 protein kinase is required for origin firing during S phase. Genes & Dev. 12: 480-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N.R., Noble, M.E., Endicott, J.A., and Johnson, L.N. 1999. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell Biol. 1: 438-443. [DOI] [PubMed] [Google Scholar]
- Chen J., Saha, P., Kornbluth, S., Dynlacht, B.D., and Dutta, A. 1996. Cyclin-binding motifs are essential for the function of p21CIP1. Mol. Cell. Biol. 16: 4673-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften P., Sudarsanam, P., Desikan, A., Fulton, L., Fulton, B., Majors, J., Waterston, R., Cohen, B.A., and Johnston, M. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301: 71-76. [DOI] [PubMed] [Google Scholar]
- Cocker J.H., Piatti, S., Santocanale, C., Nasmyth, K., and Diffley, J.F. 1996. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature 379: 180-182. [DOI] [PubMed] [Google Scholar]
- Cross F.R. and Jacobson, M.D. 2000. Conservation and function of a potential substrate-binding domain in the yeast Clb5 B-type cyclin. Mol. Cell. Biol. 20: 4782-4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmann C., Diffley, J.F., and Nasmyth, K.A. 1995. S-Phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a prereplicative state. Curr. Biol. 5: 1257-1269. [DOI] [PubMed] [Google Scholar]
- Devault A., Vallen, E.A., Yuan, T., Green, S., Bensimon, A., and Schwob, E. 2002. Identification of Tah11/Sid2 as the ortholog of the replication licensing factor Cdt1 in Saccharomyces cerevisiae. Curr. Biol. 12: 689-694. [DOI] [PubMed] [Google Scholar]
- Diffley J.F., Cocker, J.H., Dowell, S.J., and Rowley, A. 1994. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell 78: 303-316. [DOI] [PubMed] [Google Scholar]
- Donaldson A.D. 2000. The yeast mitotic cyclin Clb2 cannot substitute for S phase cyclins in replication origin firing. EMBO Rep. 1: 507-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson A.D., Raghuraman, M.K., Friedman, K.L., Cross, F.R., Brewer, B.J., and Fangman, W.L. 1998a. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol. Cell 2: 173-182. [DOI] [PubMed] [Google Scholar]
- Donaldson A.D., Fangman, W.L., and Brewer, B.J. 1998b. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes & Dev. 12: 491-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury L.S., Perkins, G., and Diffley, J.F. 1997. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 16: 5966-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2000. The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr. Biol. 10: 231-240. [DOI] [PubMed] [Google Scholar]
- Epstein C.B. and Cross, F.R. 1992. CLB5: A novel B cyclin from budding yeast with a role in S phase. Genes & Dev. 6: 1695-1706. [DOI] [PubMed] [Google Scholar]
- Furstenthal L., Kaiser, B.K., Swanson, C., and Jackson, P.K. 2001a. Cyclin E uses Cdc6 as a chromatin-associated receptor required for DNA replication. J. Cell Biol. 152: 1267-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furstenthal L., Swanson, C., Kaiser, B.K., Eldridge, A.G., and Jackson, P.K. 2001b. Triggering ubiquitination of a CDK inhibitor at origins of DNA replication. Nat. Cell Biol. 3: 715-722. [DOI] [PubMed] [Google Scholar]
- James P., Halladay, J., and Craig, E.A. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsey S.E. and Cotterill, S. 2003. Enigmatic variations: Divergent modes of regulating eukaryotic DNA replication. Mol. Cell 12: 1067-1075. [DOI] [PubMed] [Google Scholar]
- Kellis M., Patterson, N., Endrizzi, M., Birren, B., and Lander, E.S. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241-254. [DOI] [PubMed] [Google Scholar]
- Klemm R.D., Austin, R.J., and Bell, S.P. 1997. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell. 88: 493-502. [DOI] [PubMed] [Google Scholar]
- Labib K., Diffley, J.F., and Kearsey, S.E. 1999. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat. Cell Biol. 1: 415-422. [DOI] [PubMed] [Google Scholar]
- Leatherwood J., Lopez-Girona, A., and Russell, P. 1996. Interaction of Cdc2 and Cdc18 with a fission yeast ORC2-like protein. Nature 379: 360-363. [DOI] [PubMed] [Google Scholar]
- Lee D.G. and Bell, S.P. 1997. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol. Cell. Biol. 17: 7159-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y. and Stillman, B. 1992. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255: 817-823. [DOI] [PubMed] [Google Scholar]
- Masumoto H., Muramatsu, S., Kamimura, Y., and Araki, H. 2002. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature 415: 651-655. [DOI] [PubMed] [Google Scholar]
- Mendez J., Zou-Yang, X.H., Kim, S.Y., Hidaka, M., Tansey, W.P., and Stillman, B. 2002. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell 9: 481-491. [DOI] [PubMed] [Google Scholar]
- Miller M.E. and Cross, F.R. 2001. Cyclin specificity: How many wheels do you need on a unicycle? J. Cell Sci. 114: 1811-1820. [DOI] [PubMed] [Google Scholar]
- Muzi-Falconi M., Brown, G.W., and Kelly, T.J. 1996. cdc18+ regulates initiation of DNA replication in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. 93: 1566-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V.Q., Co, C., Irie, K., and Li, J.J. 2000. Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr. Biol. 10: 195-205. [DOI] [PubMed] [Google Scholar]
- Nguyen V.Q., Co, C., and Li, J.J. 2001. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411: 1068-1073. [DOI] [PubMed] [Google Scholar]
- Nishitani H., Lygerou, Z., Nishimoto, T., and Nurse, P. 2000. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature 404: 625-628. [DOI] [PubMed] [Google Scholar]
- Nougarede R., Della Setta, F., Zarzov, P., and Schwob, E. 2000. Hierarchy of S-phase-promoting factors: Yeast Dbf4-Cdc7 kinase requires prior S-phase cyclin-dependent kinase activation. Mol. Cell. Biol. 20: 3795-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski P., Marr, J., Madine, M.A., Rowles, A., Blow, J.J., Gautier, J., and Laskey, R.A. 2000. Interaction of Xenopus Cdc2 x cyclin A1 with the origin recognition complex. J. Biol. Chem. 275: 4239-4243. [DOI] [PubMed] [Google Scholar]
- Russo A.A., Jeffrey, P.D., Patten, A.K., Massague, J., and Pavletich, N.P. 1996. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature 382: 325-331. [DOI] [PubMed] [Google Scholar]
- Santocanale C. and Diffley, J.F. 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395: 615-618. [DOI] [PubMed] [Google Scholar]
- Schulman B.A., Lindstrom, D.L., and Harlow, E. 1998. Substrate recruitment to cyclin-dependent kinase 2 by a multi-purpose docking site on cyclin A. Proc. Natl. Acad. Sci. 95: 10453-10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A. and Bell, S.P. 2001. Interactions between two catalytically distinct MCM subgroups are essential for coordinated ATP hydrolysis and DNA replication. Mol. Cell 8: 1093-1104. [DOI] [PubMed] [Google Scholar]
- Schwob E. and Nasmyth, K. 1993. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes & Dev. 7: 1160-1175. [DOI] [PubMed] [Google Scholar]
- Schwob E., Bohm, T., Mendenhall, M.D., and Nasmyth, K. 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79: 233-244. [DOI] [PubMed] [Google Scholar]
- Takeda D.Y., Wohlschlegel, J.A., and Dutta, A. 2001. A bipartite substrate recognition motif for cyclin-dependent kinases. J. Biol. Chem. 276: 1993-1997. [DOI] [PubMed] [Google Scholar]
- Tanaka S. and Diffley, J.F. 2002. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2-7 during G1 phase. Nat. Cell Biol. 4: 198-207. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Knapp, D., and Nasmyth, K. 1997. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell 90: 649-660. [DOI] [PubMed] [Google Scholar]
- Vaziri C., Saxena, S., Jeon, Y., Lee, C., Murata, K., Machida, Y., Wagle, N., Hwang, D.S., and Dutta, A. 2003. A p53-dependent checkpoint pathway prevents rereplication. Mol. Cell 11: 997-1008. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat, A., Alberti-Segui, C., Rebischung, C., and Philippsen, P. 1997. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 13: 1065-1075. [DOI] [PubMed] [Google Scholar]
- Weinreich M., Liang, C., Chen, H.H., and Stillman, B. 2001. Binding of cyclin-dependent kinases to ORC and Cdc6p regulates the chromosome replication cycle. Proc. Natl. Acad. Sci. 98: 11211-11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes G.M. and Bell, S.P. 2002. The B2 element of the Saccharomyces cerevisiae ARS1 origin of replication requires specific sequences to facilitate pre-RC formation. Proc. Natl. Acad. Sci. 99: 101-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel J.A., Dwyer, B.T., Takeda, D.Y., and Dutta, A. 2001. Mutational analysis of the Cy motif from p21 reveals sequence degeneracy and specificity for different cyclin-dependent kinases. Mol. Cell. Biol. 21: 4868-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin J., Buck, V., Nurse, P., and Millar, J.B. 2002. Stable association of mitotic cyclin B/Cdc2 to replication origins prevents endoreduplication. Cell 111: 419-431. [DOI] [PubMed] [Google Scholar]
- Zachariae W. and Nasmyth, K. 1999. Whose end is destruction: Cell division and the anaphase-promoting complex. Genes & Dev. 13: 2039-2058. [DOI] [PubMed] [Google Scholar]