Abstract
Recent studies of Saccharomyces cerevisiae have significantly advanced our understanding of the molecular mechanisms of meiotic chromosome behavior. Structural components of the synaptonemal complex have been identified and studies of mutants defective in synapsis have provided insight into the role of the synaptonemal complex in homolog pairing, genetic recombination, crossover interference, and meiotic chromosome segregation. There is compelling evidence that most or all meiotic recombination events initiate with double-strand breaks. Several intermediates in the double-strand break repair pathway have been characterized and mutants blocked at different steps in the pathway have been identified. With the application of genetic, molecular, cytological, and biochemical methods in a single organism, we can expect an increasingly comprehensive and unified view of the meiotic process.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alani E., Padmore R., Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990 May 4;61(3):419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Alani E., Reenan R. A., Kolodner R. D. Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics. 1994 May;137(1):19–39. doi: 10.1093/genetics/137.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D. K., Park D., Xu L., Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992 May 1;69(3):439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Bishop D. K. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994 Dec 16;79(6):1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Borts R. H., Lichten M., Haber J. E. Analysis of meiosis-defective mutations in yeast by physical monitoring of recombination. Genetics. 1986 Jul;113(3):551–567. doi: 10.1093/genetics/113.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Alani E., Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990 Jun 15;61(6):1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- Collins I., Newlon C. S. Meiosis-specific formation of joint DNA molecules containing sequences from homologous chromosomes. Cell. 1994 Jan 14;76(1):65–75. doi: 10.1016/0092-8674(94)90173-2. [DOI] [PubMed] [Google Scholar]
- Dawson D. S., Murray A. W., Szostak J. W. An alternative pathway for meiotic chromosome segregation in yeast. Science. 1986 Nov 7;234(4777):713–717. doi: 10.1126/science.3535068. [DOI] [PubMed] [Google Scholar]
- De Massy B., Baudat F., Nicolas A. Initiation of recombination in Saccharomyces cerevisiae haploid meiosis. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):11929–11933. doi: 10.1073/pnas.91.25.11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detloff P., White M. A., Petes T. D. Analysis of a gene conversion gradient at the HIS4 locus in Saccharomyces cerevisiae. Genetics. 1992 Sep;132(1):113–123. doi: 10.1093/genetics/132.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresser M. E., Giroux C. N. Meiotic chromosome behavior in spread preparations of yeast. J Cell Biol. 1988 Mar;106(3):567–573. doi: 10.1083/jcb.106.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel-Mitani M., Olson L. W., Egel R. Meiosis in Aspergillus nidulans: another example for lacking synaptonemal complexes in the absence of crossover interference. Hereditas. 1982;97(2):179–187. doi: 10.1111/j.1601-5223.1982.tb00761.x. [DOI] [PubMed] [Google Scholar]
- Egel R. Synaptonemal complex and crossing-over: structural support or interference? Heredity (Edinb) 1978 Oct;41(2):233–237. doi: 10.1038/hdy.1978.92. [DOI] [PubMed] [Google Scholar]
- Engebrecht J., Hirsch J., Roeder G. S. Meiotic gene conversion and crossing over: their relationship to each other and to chromosome synapsis and segregation. Cell. 1990 Sep 7;62(5):927–937. doi: 10.1016/0092-8674(90)90267-i. [DOI] [PubMed] [Google Scholar]
- Friedman D. B., Hollingsworth N. M., Byers B. Insertional mutations in the yeast HOP1 gene: evidence for multimeric assembly in meiosis. Genetics. 1994 Feb;136(2):449–464. doi: 10.1093/genetics/136.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game J. C. Pulsed-field gel analysis of the pattern of DNA double-strand breaks in the Saccharomyces genome during meiosis. Dev Genet. 1992;13(6):485–497. doi: 10.1002/dvg.1020130610. [DOI] [PubMed] [Google Scholar]
- Gilbertson L. A., Stahl F. W. Initiation of meiotic recombination is independent of interhomologue interactions. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):11934–11937. doi: 10.1073/pnas.91.25.11934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldway M., Sherman A., Zenvirth D., Arbel T., Simchen G. A short chromosomal region with major roles in yeast chromosome III meiotic disjunction, recombination and double strand breaks. Genetics. 1993 Feb;133(2):159–169. doi: 10.1093/genetics/133.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyon C., Lichten M. Timing of molecular events in meiosis in Saccharomyces cerevisiae: stable heteroduplex DNA is formed late in meiotic prophase. Mol Cell Biol. 1993 Jan;13(1):373–382. doi: 10.1128/mcb.13.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V., Kaback D. B. Distributive disjunction of authentic chromosomes in Saccharomyces cerevisiae. Genetics. 1991 Mar;127(3):475–488. doi: 10.1093/genetics/127.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N. M., Byers B. HOP1: a yeast meiotic pairing gene. Genetics. 1989 Mar;121(3):445–462. doi: 10.1093/genetics/121.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N. M., Goetsch L., Byers B. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell. 1990 Apr 6;61(1):73–84. doi: 10.1016/0092-8674(90)90216-2. [DOI] [PubMed] [Google Scholar]
- Hollingsworth N. M., Johnson A. D. A conditional allele of the Saccharomyces cerevisiae HOP1 gene is suppressed by overexpression of two other meiosis-specific genes: RED1 and REC104. Genetics. 1993 Apr;133(4):785–797. doi: 10.1093/genetics/133.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N. M., Ponte L., Halsey C. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 1995 Jul 15;9(14):1728–1739. doi: 10.1101/gad.9.14.1728. [DOI] [PubMed] [Google Scholar]
- Ivanov E. L., Korolev V. G., Fabre F. XRS2, a DNA repair gene of Saccharomyces cerevisiae, is needed for meiotic recombination. Genetics. 1992 Nov;132(3):651–664. doi: 10.1093/genetics/132.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks-Robertson S., Petes T. D. High-frequency meiotic gene conversion between repeated genes on nonhomologous chromosomes in yeast. Proc Natl Acad Sci U S A. 1985 May;82(10):3350–3354. doi: 10.1073/pnas.82.10.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johzuka K., Ogawa H. Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics. 1995 Apr;139(4):1521–1532. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaback D. B., Guacci V., Barber D., Mahon J. W. Chromosome size-dependent control of meiotic recombination. Science. 1992 Apr 10;256(5054):228–232. doi: 10.1126/science.1566070. [DOI] [PubMed] [Google Scholar]
- Kaback D. B., Steensma H. Y., de Jonge P. Enhanced meiotic recombination on the smallest chromosome of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989 May;86(10):3694–3698. doi: 10.1073/pnas.86.10.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J. S., Mortimer R. K. A polymerization model of chiasma interference and corresponding computer simulation. Genetics. 1990 Dec;126(4):1127–1138. doi: 10.1093/genetics/126.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Weiner B. M. Potential advantages of unstable interactions for pairing of chromosomes in meiotic, somatic, and premeiotic cells. Cold Spring Harb Symp Quant Biol. 1993;58:553–565. doi: 10.1101/sqb.1993.058.01.062. [DOI] [PubMed] [Google Scholar]
- Kolodner R., Evans D. H., Morrison P. T. Purification and characterization of an activity from Saccharomyces cerevisiae that catalyzes homologous pairing and strand exchange. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5560–5564. doi: 10.1073/pnas.84.16.5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer W., Kramer B., Williamson M. S., Fogel S. Cloning and nucleotide sequence of DNA mismatch repair gene PMS1 from Saccharomyces cerevisiae: homology of PMS1 to procaryotic MutL and HexB. J Bacteriol. 1989 Oct;171(10):5339–5346. doi: 10.1128/jb.171.10.5339-5346.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M., Borts R. H., Haber J. E. Meiotic gene conversion and crossing over between dispersed homologous sequences occurs frequently in Saccharomyces cerevisiae. Genetics. 1987 Feb;115(2):233–246. doi: 10.1093/genetics/115.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wu T. C., Lichten M. The location and structure of double-strand DNA breaks induced during yeast meiosis: evidence for a covalently linked DNA-protein intermediate. EMBO J. 1995 Sep 15;14(18):4599–4608. doi: 10.1002/j.1460-2075.1995.tb00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J., Klein F., Scherthan H. Homologous pairing is reduced but not abolished in asynaptic mutants of yeast. J Cell Biol. 1994 Jun;125(6):1191–1200. doi: 10.1083/jcb.125.6.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J., Scherthan H., Kaback D. B. Physical association between nonhomologous chromosomes precedes distributive disjunction in yeast. Proc Natl Acad Sci U S A. 1994 Jan 4;91(1):331–334. doi: 10.1073/pnas.91.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire M. P. A possible role for the synaptonemal complex in chiasma maintenance. Exp Cell Res. 1978 Mar 15;112(2):297–308. doi: 10.1016/0014-4827(78)90213-6. [DOI] [PubMed] [Google Scholar]
- Maguire M. P. Crossover site determination and interference. J Theor Biol. 1988 Oct 21;134(4):565–570. doi: 10.1016/s0022-5193(88)80058-4. [DOI] [PubMed] [Google Scholar]
- Malone R. E., Bullard S., Lundquist S., Kim S., Tarkowski T. A meiotic gene conversion gradient opposite to the direction of transcription. Nature. 1992 Sep 10;359(6391):154–155. doi: 10.1038/359154a0. [DOI] [PubMed] [Google Scholar]
- Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- Murray A. W. Creative blocks: cell-cycle checkpoints and feedback controls. Nature. 1992 Oct 15;359(6396):599–604. doi: 10.1038/359599a0. [DOI] [PubMed] [Google Scholar]
- Møens P. B., Pearlman R. E. Chromatin organization at meiosis. Bioessays. 1988 Nov;9(5):151–153. doi: 10.1002/bies.950090503. [DOI] [PubMed] [Google Scholar]
- Nag D. K., Petes T. D. Physical detection of heteroduplexes during meiotic recombination in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993 Apr;13(4):2324–2331. doi: 10.1128/mcb.13.4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
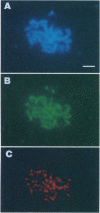
- Nag D. K., Scherthan H., Rockmill B., Bhargava J., Roeder G. S. Heteroduplex DNA formation and homolog pairing in yeast meiotic mutants. Genetics. 1995 Sep;141(1):75–86. doi: 10.1093/genetics/141.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag D. K., White M. A., Petes T. D. Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature. 1989 Jul 27;340(6231):318–320. doi: 10.1038/340318a0. [DOI] [PubMed] [Google Scholar]
- New L., Liu K., Crouse G. F. The yeast gene MSH3 defines a new class of eukaryotic MutS homologues. Mol Gen Genet. 1993 May;239(1-2):97–108. doi: 10.1007/BF00281607. [DOI] [PubMed] [Google Scholar]
- Nicolas A., Treco D., Schultes N. P., Szostak J. W. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature. 1989 Mar 2;338(6210):35–39. doi: 10.1038/338035a0. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Yu X., Shinohara A., Egelman E. H. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993 Mar 26;259(5103):1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- Padmore R., Cao L., Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell. 1991 Sep 20;66(6):1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- Prolla T. A., Christie D. M., Liskay R. M. Dual requirement in yeast DNA mismatch repair for MLH1 and PMS1, two homologs of the bacterial mutL gene. Mol Cell Biol. 1994 Jan;14(1):407–415. doi: 10.1128/mcb.14.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prolla T. A., Pang Q., Alani E., Kolodner R. D., Liskay R. M. MLH1, PMS1, and MSH2 interactions during the initiation of DNA mismatch repair in yeast. Science. 1994 Aug 19;265(5175):1091–1093. doi: 10.1126/science.8066446. [DOI] [PubMed] [Google Scholar]
- Raymond W. E., Kleckner N. RAD50 protein of S.cerevisiae exhibits ATP-dependent DNA binding. Nucleic Acids Res. 1993 Aug 11;21(16):3851–3856. doi: 10.1093/nar/21.16.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayssiguier C., Thaler D. S., Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989 Nov 23;342(6248):396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- Reenan R. A., Kolodner R. D. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics. 1992 Dec;132(4):975–985. doi: 10.1093/genetics/132.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reenan R. A., Kolodner R. D. Isolation and characterization of two Saccharomyces cerevisiae genes encoding homologs of the bacterial HexA and MutS mismatch repair proteins. Genetics. 1992 Dec;132(4):963–973. doi: 10.1093/genetics/132.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco V., de Massy B., Nicolas A. The Saccharomyces cerevisiae ARG4 initiator of meiotic gene conversion and its associated double-strand DNA breaks can be inhibited by transcriptional interference. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):12068–12072. doi: 10.1073/pnas.89.24.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B., Engebrecht J. A., Scherthan H., Loidl J., Roeder G. S. The yeast MER2 gene is required for chromosome synapsis and the initiation of meiotic recombination. Genetics. 1995 Sep;141(1):49–59. doi: 10.1093/genetics/141.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B., Roeder G. S. Meiosis in asynaptic yeast. Genetics. 1990 Nov;126(3):563–574. doi: 10.1093/genetics/126.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose D., Holm C. Meiosis-specific arrest revealed in DNA topoisomerase II mutants. Mol Cell Biol. 1993 Jun;13(6):3445–3455. doi: 10.1128/mcb.13.6.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose D., Thomas W., Holm C. Segregation of recombined chromosomes in meiosis I requires DNA topoisomerase II. Cell. 1990 Mar 23;60(6):1009–1017. doi: 10.1016/0092-8674(90)90349-j. [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald P., Roeder G. S. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell. 1994 Dec 16;79(6):1069–1080. doi: 10.1016/0092-8674(94)90037-x. [DOI] [PubMed] [Google Scholar]
- Ross L. O., Treco D., Nicolas A., Szostak J. W., Dawson D. Meiotic recombination on artificial chromosomes in yeast. Genetics. 1992 Jul;131(3):541–550. doi: 10.1093/genetics/131.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H., Loidl J., Schuster T., Schweizer D. Meiotic chromosome condensation and pairing in Saccharomyces cerevisiae studied by chromosome painting. Chromosoma. 1992 Oct;101(10):590–595. doi: 10.1007/BF00360535. [DOI] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994 Jan 14;76(1):51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- Sears D. D., Hegemann J. H., Hieter P. Meiotic recombination and segregation of human-derived artificial chromosomes in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5296–5300. doi: 10.1073/pnas.89.12.5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selva E. M., New L., Crouse G. F., Lahue R. S. Mismatch correction acts as a barrier to homeologous recombination in Saccharomyces cerevisiae. Genetics. 1995 Mar;139(3):1175–1188. doi: 10.1093/genetics/139.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A., Ogawa H., Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992 May 1;69(3):457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Shuster E. O., Byers B. Pachytene arrest and other meiotic effects of the start mutations in Saccharomyces cerevisiae. Genetics. 1989 Sep;123(1):29–43. doi: 10.1093/genetics/123.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siede W., Friedberg A. S., Dianova I., Friedberg E. C. Characterization of G1 checkpoint control in the yeast Saccharomyces cerevisiae following exposure to DNA-damaging agents. Genetics. 1994 Oct;138(2):271–281. doi: 10.1093/genetics/138.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Treco D., Schultes N. P., Szostak J. W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989 Mar 2;338(6210):87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- Sun H., Treco D., Szostak J. W. Extensive 3'-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991 Mar 22;64(6):1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994 Aug 26;265(5176):1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- Sym M., Engebrecht J. A., Roeder G. S. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993 Feb 12;72(3):365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- Sym M., Roeder G. S. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell. 1994 Oct 21;79(2):283–292. doi: 10.1016/0092-8674(94)90197-x. [DOI] [PubMed] [Google Scholar]
- Sym M., Roeder G. S. Zip1-induced changes in synaptonemal complex structure and polycomplex assembly. J Cell Biol. 1995 Feb;128(4):455–466. doi: 10.1083/jcb.128.4.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Tishkoff D. X., Rockmill B., Roeder G. S., Kolodner R. D. The sep1 mutant of Saccharomyces cerevisiae arrests in pachytene and is deficient in meiotic recombination. Genetics. 1995 Feb;139(2):495–509. doi: 10.1093/genetics/139.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner B. M., Kleckner N. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 1994 Jul 1;77(7):977–991. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988 Jul 15;241(4863):317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- Weinert T. A., Kiser G. L., Hartwell L. H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994 Mar 15;8(6):652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- West S. C. Enzymes and molecular mechanisms of genetic recombination. Annu Rev Biochem. 1992;61:603–640. doi: 10.1146/annurev.bi.61.070192.003131. [DOI] [PubMed] [Google Scholar]
- White M. A., Detloff P., Strand M., Petes T. D. A promoter deletion reduces the rate of mitotic, but not meiotic, recombination at the HIS4 locus in yeast. Curr Genet. 1992 Feb;21(2):109–116. doi: 10.1007/BF00318468. [DOI] [PubMed] [Google Scholar]
- Wu T. C., Lichten M. Factors that affect the location and frequency of meiosis-induced double-strand breaks in Saccharomyces cerevisiae. Genetics. 1995 May;140(1):55–66. doi: 10.1093/genetics/140.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. C., Lichten M. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science. 1994 Jan 28;263(5146):515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- Zenvirth D., Arbel T., Sherman A., Goldway M., Klein S., Simchen G. Multiple sites for double-strand breaks in whole meiotic chromosomes of Saccharomyces cerevisiae. EMBO J. 1992 Sep;11(9):3441–3447. doi: 10.1002/j.1460-2075.1992.tb05423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wettstein D., Rasmussen S. W., Holm P. B. The synaptonemal complex in genetic segregation. Annu Rev Genet. 1984;18:331–413. doi: 10.1146/annurev.ge.18.120184.001555. [DOI] [PubMed] [Google Scholar]