Abstract
The regulation of telomerase action, and its coordination with conventional DNA replication and chromosome end “capping,” are still poorly understood. Here we describe a genetic screen in yeast for mutants with relaxed telomere length regulation, and the identification of Pol12, the B subunit of the DNA polymerase α (Pol1)-primase complex, as a new factor involved in this process. Unlike many POL1 and POL12 mutations, which also cause telomere elongation, the pol12-216 mutation described here does not lead to either reduced Pol1 function, increased telomeric single-stranded DNA, or a reduction in telomeric gene silencing. Instead, and again unlike mutations affecting POL1, pol12-216 is lethal in combination with a mutation in the telomere end-binding and capping protein Stn1. Significantly, Pol12 and Stn1 interact in both two-hybrid and biochemical assays, and their synthetic-lethal interaction appears to be caused, at least in part, by a loss of telomere capping. These data reveal a novel function for Pol12 and a new connection between DNA polymerase α and Stn1. We propose that Pol12, together with Stn1, plays a key role in linking telomerase action with the completion of lagging strand synthesis, and in a regulatory step required for telomere capping.
Keywords: Telomere capping, telomere length regulation, DNA polymerase α, DNA replication, Pol12, Stn1
The telomeres of most eukaryotes are comprised of simple tandem DNA repeats, with a TG-rich strand running toward the 3′-chromosome end, and many associated proteins. Telomeres protect chromosomes from end-to-end fusions and exonucleolytic degradation (the “capping” function), and contribute in less well understood ways to overall chromosome stability (for review, see Cervantes and Lundblad 2002). In addition to capping chromosome ends and thus distinguishing them from accidental chromosome breaks, telomeres provide a mechanism to replicate chromosomes without a net loss of terminal sequences. In most eukaryotes this problem is solved by the telomerase enzyme, a specialized reverse transcriptase containing its own template RNA, that extends the TG-rich strand of chromosome ends (for review, see McEachern et al. 2000). Precisely how telomerase action is regulated to bring about a stable average DNA repeat length at chromosome ends is still unclear, but a growing body of evidence suggests that regulation of telomerase action is coordinated with conventional DNA replication through a coupling to the machinery for initiation of lagging-strand synthesis, the polymerase α-primase complex (for review, see Chakhparonian and Wellinger 2003).
In budding yeast, telomeric DNA has an irregular repeat structure (often abbreviated as TG1-3) that is maintained within a fixed range, varying from ∼250-350 bp, as a result of an equilibrium between shortening and elongation (for review, see Dubrana et al. 2001; Shore 2001). The regulation of this equilibrium appears to be extremely important to the cell, because either replicative loss and/or degradation of terminal sequences (Lundblad and Szostak 1989; Singer and Gottschling 1994; Lendvay et al. 1996) or unregulated extension of altered TG repeats has dramatic consequences for cell growth (McEachern and Blackburn 1995; Smith and Blackburn 1999). Maintenance of telomere length about a fixed average value is achieved through a mechanism that appears to measure not the TG-repeat tract length per se, but, rather, the number of Rap1 proteins bound to it (Marcand et al. 1997; Ray and Runge 1999a). The C terminus of Rap1 negatively regulates telomere elongation in cis (Kyrion et al. 1992; Marcand et al. 1996), a function that requires two additional proteins, Rif1 and Rif2, both of which interact physically with this domain of Rap1 (Hardy et al. 1992; Moretti et al. 1994; Wotton and Shore 1997). The inhibition of telomerase addition by Rap1-Rif1/2 complexes increases linearly as a function of telomere tract length (and presumably the number of Rap1-Rif1/2 complexes bound) through a mechanism that is at present unknown. Potential targets for Rap1-Rif1/2 action at the telomere are Tel1 (the yeast ATM homolog) and the Mre11-Rad50-Xrs2 (MRX) complex, which appear to act downstream from Rap1 and Rif proteins in a pathway required for telomerase activation at chromosome ends (for review, see Dubrana et al. 2001; Shore 2001).
A key regulator of telomere capping and replication in yeast is the Cdc13 protein, which binds to the TG-rich single-strand extensions at telomeres (for review, see Evans and Lundblad 2000). Genetic evidence indicates that Cdc13 recruits the telomerase enzyme to telomeres through a direct interaction with the associated Est1 protein (Evans and Lundblad 1999, 2002; Pennock et al. 2001). However, a recent study suggests that the Cdc13-Est1 interaction may, instead, serve to activate telomerase enzyme already bound at or near its site of action (Taggart et al. 2002). In any event, mutants lacking this Cdc13 function (e.g., cdc13-2) senesce because of gradual telomere erosion (Lendvay et al. 1996; Nugent et al. 1996), whereas complete loss of Cdc13 activity leads to extensive degradation of telomeric 5′-ends and an immediate G2/M cell cycle arrest. This essential capping function of Cdc13, which is lost in the conditional-lethal cdc13-1 allele when grown at the nonpermissive temperature (Garvik et al. 1995), involves the delivery of a separate complex to the telomere, containing Stn1 and Ten1 (Grandin et al. 1997, 2001; Pennock et al. 2001). A temperature-sensitive allele of STN1, stn1-13, confers a phenotype very similar to that displayed by cdc13-1 cells, causing G2/M cell cycle arrest and accumulation of telomeric single-stranded DNA at the nonpermissive temperature. In addition, both mutations lead to telomere length increase at the semipermissive temperature (Grandin et al. 1997).
Interestingly, Cdc13 also interacts, either directly or indirectly, with Pol1 (also known as Cdc17), the catalytic subunit of DNA polymerase α (Qi and Zakian 2000). Point mutations in either protein that affect this interaction lead to a slight increase in average telomere length, but cause no other obvious phenotypes. However, mutations that affect the catalytic function of Pol1 and are lethal at elevated temperatures cause considerable and progressive telomere elongation, large increases in TG-specific single-stranded DNA at telomeres, and a loss of telomeric gene silencing (Carson and Hartwell 1985; Adams and Holm 1996; Adams Martin et al. 2000). These and other data have led to the idea that Pol1, or the complete DNA polymerase α-primase complex, plays an important role in the down-regulation of telomerase action at telomeres, perhaps in a concerted regulatory step that ensures the replication of the CA-rich strand at chromosome ends.
Here we describe a novel genetic screen for factors that limit telomere elongation. This screen has revealed new alleles of the POL1, RIF1, and STN1 genes, as well as uncovered a gene not previously known to be involved in telomere length regulation, POL12. The POL12 gene encodes a highly conserved, putative regulatory subunit of the DNA polymerase α-primase complex (hereafter referred to as Polα-primase; Foiani et al. 1994). The pol12-216 allele that we describe here is a separation-of-function mutation that, although clearly defective in telomere length regulation, shares none of the other telomere defects of hypomorphic POL1 or POL12 mutants and displays no apparent defect in DNA replication elsewhere in the genome. Despite this mild telomere defect, the pol12-216 mutation is lethal in combination with stn1-13, unlike all POL1 mutants tested and all but the most severe POL12 ts mutant tested. Consistent with this specific genetic interaction, we present evidence that Pol12 physically interacts with Stn1. These and other data indicate that Pol12, probably in the context of the Polα-primase complex, plays an important role in telomere capping. Finally, genetic epistasis tests suggest that Pol12 might act downstream from several factors known to be involved in a postulated negative-feedback system of telomere length regulation.
Results
A genetic screen for mutants with elongated telomeres
We previously described the generation of novel telomeres in yeast from tandem arrays of Rap1-binding sites as a means of studying the role of Rap1 binding in telomere formation and length regulation (Grossi et al. 2001). These studies demonstrated that misoriented Rap1 site arrays containing up to eight sites, in which the strand running 5′-to-3′ toward the telomere end is C-rich, were counted by the telomere length-sensing machinery and could promote telomere formation, provided that genuine TG1-3 sequence was present distally to the Rap1-binding sites. However, much longer misoriented arrays (containing 16 or 32 Rap1-binding sites) could not be accommodated by the telomere length regulation system, so that a misoriented array of 32 Rap1-binding sites (approximately twice the normal number of Rap1 sites found at a native telomere) contained the complete synthetic array plus an additional terminal sequence that we presumed to be correctly oriented TG1-3 repeat sequence. These data suggested that the terminal sequence is essential for viability, possibly because it is required for telomere capping and/or maintenance.
We hypothesized that telomeres with 32 misoriented Rap1-binding sites are unstable, and tested this idea by first constructing strains disomic for Chromosome VII, where loss of the modified telomere or the whole chromosome on which it resides would have no effect on cell viability. In such strains telomere instability can be seen directly at the level of individual colonies because loss of the ADE2 gene at the marked telomere generates a red (ade2-) sector within the colony. These events occur so frequently that most colonies contain at least one red sector (Fig. 1A, top panel). In contrast, an otherwise identical disome strain, in which the ADE2-marked telomere terminates in correctly oriented Rap1 site arrays and TG repeat sequence, yields almost exclusively white colonies (Fig. 1A, bottom panel). We imagine that the instability of telomeres with excess misoriented Rap1-binding sites is a direct consequence of two specific effects: (1) hyperrepression, in cis, of telomerase action caused by an excess number of bound Rap1 molecules; and (2) subsequent loss of the TG-repeat “buffer” sequence, exposure of the misoriented Rap1-binding sites, and telomere “uncapping.” This model is supported by the observation of a faint ladder of bands of higher mobility than the elongated telomeres on a Southern blot (Fig. 1B). These bands might represent transient intermediates in the telomere loss process.
Figure 1.
Instability of a modified Chromosome VII-L telomere carrying 32-misoriented Rap1-binding sites. (A) Colony color assay using the ADE2 marker to detect VII-L telomere loss from strains disomic for Chromosome VII (left). The wild-type and modified copies of Chromosome VII-L present in test (top left) and control (bottom left) strains are shown. The modified copy of Chromosome VII-L carries either 32 misoriented (black arrows pointing right; test strain, top left) or correctly oriented (black arrows pointing left; control strain, bottom left) Rap1-binding sites, the URA3 and the ADE2 genes integrated at the ADH4 locus. White arrows represent native TG1-3 sequence. The strains are ADE3 and ade2Δ at their chromosomal loci. Examples of isolated colonies generated by either strain on YPD medium are shown on the right. (B) Schematic representation of the modified VII-L telomere (as in A) in strains disomic for Chromosome VII (top). The black arrowhead (pointing right) represents a synthetic Rap1-binding site in the “incorrect” orientation. (H) HindIII. (Bottom panel) The Southern blot analysis of telomeres containing the indicated number of Rap1 sites. Following HindIII digestion, the URA3 probe detects the endogenous ura3-52 locus, a diffuse telomeric band, and a faint ladder of bands in the case of 32 misoriented Rap1-binding sites.
Starting from the assumption that telomere instability in this artificial system results at least in part from repression of telomerase action at the modified telomere, we developed a screen for mutants with relaxed telomere length regulation. The screen is based on the simple idea that such mutants would increase the probability of telomerase action at these modified telomeres and thus increase their stability. We therefore mutagenized the sectoring disome strain shown in Figure 1A and screened for white, nonsectoring colonies in which the rate of telomere loss at the marked disome has presumably been reduced (see Materials and Methods for additional details). Candidates were then screened by Southern blotting for those in which all of the Y′-containing telomeres exhibited a length increase. From an initial screen of ∼104 mutagenized cells, six mutants were identified as having a general telomere elongation phenotype that displayed clear 2:2 segregation in backcrosses to a parental strain of the opposite mating type (data not shown). Two of the mutants (M35 and M228) failed to grow at 37°C and were partially or strongly defective, respectively, in telomeric gene silencing at the permissive temperature of 30°C (Fig. 2A). These two secondary phenotypes, both of which cosegregated with the telomere elongation phenotype (data not shown), are shared with the pol1-1 mutant (Fig. 2A; Carson and Hartwell 1985; Adams Martin et al. 2000). In fact, complementation analysis showed that both M35 and M228 are alleles of POL1 (hereafter referred to as pol1-35 and pol1-228), which encodes the catalytic subunit of DNA polymerase α (Pol1). Three other mutants that showed an elongated telomere phenotype were identified by direct plasmid complementation and segregation analysis as alleles of either RIF1 (M56 and M174) or STN1 (M3; data not shown). Both of these genes have been shown previously to be involved in telomere length regulation, and, in the case of STN1, in an essential telomere protection function (Hardy et al. 1992; Grandin et al. 1997).
Figure 2.
Phenotypes and synthetic interactions of pol12-216 and pol1 mutants. (A) Growth and silencing phenotypes of pol12-216 and pol1 mutants. Tenfold serial dilutions of overnight cultures were spotted on YPD (30°C and 37°C), SC, SC-Ura, and SC + 5-FOA media (30°C). Relevant genotypes are indicated on the left. (B) In-gel hybridization analysis of telomeres from strains grown for several generations at 30°C. (Left) A native agarose gel was hybridized with a CA-rich labeled probe. (Right) The same gel was denatured and rehybridized with a CA-rich probe. (C) Southern blot analysis of telomeres from the indicated single- and double-mutant strains (XhoI digestion and hybridization to a d(TG)/d(CA) probe; see Materials and Methods). All the strains were restreaked at least 10 times on YPD medium at 30°C (left panel) or 23°C (right panel) before telomere length analysis was performed. (D) The same strains (five of which are shown) were spotted on YPD at 23°C and 30°C to test for viability.
The POL12 gene plays a role in telomere length regulation
The telomere elongation phenotype of the remaining mutant (M216) was not complemented by plasmids containing RIF1, RIF2, POL1, CDC13, or STN1. However, we noted that diploids generated by crossing the M216 mutant to an stn1-13 strain displayed poor spore viability following meiosis, and we showed by tetrad analysis (data not shown) that this was likely to be caused by synthetic lethality of the M216 mutation together with stn1-13 (Fig. 3A). We exploited this observation to design a simple genetic screen to clone the wild-type copy of the gene mutated in M216 (see Materials and Methods for details). Two types of plasmids were recovered in this screen, the first of which contained the STN1 gene, as expected. The second class contained POL12, an essential gene encoding the B subunit of the Polα-primase complex (Foiani et al. 1994). No enzymatic activity has been assigned to this polypeptide, which has been implicated in the stabilization and regulation of the catalytic subunit Pol1. The B subunit is common to most eukaryotic DNA polymerases and shows several conserved motifs, particularly in the C-terminal portion of the protein (Collins et al. 1993; Makiniemi et al. 1999).
Figure 3.

The pol12-216 and pol12-109 (unlike pol1 and other pol12 alleles) are lethal in combination with hypomorphic STN1 mutations. Single- or double-mutant strains (right) containing the YCp33-STN1-URA3 plasmid were grown either in the absence (left) or presence (center) of 5-FOA to select for the loss of the plasmid. Strains carrying the stn1-101 or stn1-154 mutations have been deleted at their STN1 chromosomal locus and contain a LEU2 plasmid with the indicated mutant STN1 allele in addition to the YCp33-STN1-URA3 plasmid (see Materials and Methods).
We showed that the M216 strain contains a single point mutation in the POL12 gene that changes a glycine at position 325 to aspartate (see Materials and Methods). We hereafter refer to this mutation as the pol12-216 allele. Although the pol12-216 mutation falls in a short nonconserved region, it might be significant that this stretch of amino acids, located between two conserved sequences, appears to be an ∼6 amino acid insertion found only in the B subunits of Polα-primase complexes, from yeast to humans (Makiniemi et al. 1999).
pol12-216 defines a distinct telomere function required for viability in cells with reduced STN1 activity
Several lines of evidence strongly suggest that the telomere alteration in pol12-216 cells is distinct from that caused by hypomorphic ts-lethal alleles of POL1, and is not caused by a general defect in lagging-strand DNA synthesis. To begin with, pol12-216 displays no obvious cell growth defect at any temperature tested, nor is it defective in telomeric gene silencing, as are pol1-1 and the two new POL1 alleles, pol1-35 and pol1-228 (Fig. 2A). Furthermore, pol12-216 cells have a normal cell cycle profile and display cell cycle-dependent Pol12 and Rad53 phosphorylation patterns that are indistinguishable from wild type (data not shown). Taken together, these data argue strongly against a general replication defect in pol12-216.
A hallmark of the original pol1-1 mutant (also known as cdc17-1 or pol1-17), in addition to progressive telomere elongation at permissive or semipermissive temperatures (Carson and Hartwell 1985), is a striking elevation in single-stranded telomeric DNA corresponding to the TG-rich strand (Adams Martin et al. 2000). This might arise from a defect in lagging-strand synthesis at the telomere, increased degradation of the CA-rich strand, or a combination of both effects. If the pol12-216 mutation were defective in a general Polα-primase function at telomeres, we would also expect it to cause an increase in telomeric single-strandedness. However, ingel hybridization experiments show that the pol12-216 mutant behaves like wild type in this respect, at all temperatures tested (30°C and 37°C; Fig. 2B; data not shown). In contrast, the two new POL1 mutants, pol1-35 and pol1-228, display high levels of TG-rich single-stranded DNA following growth for at least 30-40 generations at 30°C, comparable to that seen in the original pol1-1 mutant (Fig. 2B). Interestingly, pol12-216 and pol1-35 mutations show a strong synthetic interaction, because a double-mutant strain grows very poorly at 30°C, unlike either single mutant (Fig. 2D). Moreover, these two mutations would appear to have a synergistic effect on telomere length regulation, because the double mutant leads to extremely elongated telomeres with a more disperse pattern than either single mutant (Fig. 2C). Both the original pol1-1 allele and pol1-228 show very similar behavior in combination with pol12-216 (data not shown).
The telomere-specific defect of pol12-216 is reminiscent of point mutations in POL1 that alter or abolish its interaction with Cdc13 (Qi and Zakian 2000). We introduced the most severe of these POL1 mutants (pol1-236) into our strain background and constructed a double mutant with pol12-216. We found that pol1-236 alone causes a modest telomere length increase (somewhat smaller than reported previously in a different strain background) and little or no increase in single-stranded DNA at the telomere (Fig. 2B; Qi and Zakian 2000). Interestingly, the pol1-236 pol12-216 double mutant displays a more than additive (synergistic) increase in telomere length and a dramatic increase in single-stranded TG-rich telomeric DNA compared with either single mutant (Fig. 2B). Nevertheless, the pol1-236 pol12-216 double mutant grows normally. These data suggest that the two mutations, present in the same protein complex, considerably alter the function and/or stability of this complex at telomeres, perhaps by affecting different interactions or regulatory events there.
We also wondered whether other mutations in POL12 would behave similarly to pol12-216, and tested a collection of such mutants, all of which are temperature sensitive (ts) for growth. We found that several of these POL12 mutants also display a telomere length increase (Fig. 2C) and that the most severe of these, pol12-109, also shows a large increase in TG-rich single-stranded DNA at telomeres (Fig. 2B).
Synthetic lethality is indicative of a functional interaction between two different genes, thus we were interested in determining the specificity of the synthetic lethal interaction between the pol12-216 and stn1-13 mutations. We first tested the pol1-236 mutation, which, as shown above and previously (Qi and Zakian 2000), has phenotypes similar to those of pol12-216. In contrast to the pol12-216 mutant, though, pol1-236 is not lethal in combination with stn1-13 (Fig. 3A). We have also tested an allele of CDC13 that is defective in the Cdc13-Pol1 interaction, cdc13-50 (Qi and Zakian 2000), and found that it is also viable in combination with stn1-13 (data not shown). In addition, none of three ts lethal mutations in POL1 that we tested (pol1-1 and the new pol1-35 and pol1-228 alleles), nor a mutation in the associated primase gene (pri1-M4; Marini et al. 1997), is synthetically lethal in combination with stn1-13 (data not shown). Taken together, these observations suggest that POL12 has a stronger functional interaction with STN1 than does POL1, despite the fact that Pol1 and Pol12 act in the same protein complex.
To ask whether the stn1-13 allele is specific in its interaction with pol12-216, we tested two other ts lethal alleles of STN1 (stn1-101 and stn1-154) and found that both are also lethal in combination with pol12-216 (Fig. 3B). These observations show that the effect of stn1-13 is not allele-specific and suggest that a general reduction in STN1 activity might be sufficient to cause lethality in combination with pol12-216. Similarly, we tested the specificity of pol12-216 by combining other POL12 mutations with stn1-13. As shown in Figure 3C, we found that two other alleles of POL12 (pol12-103 and pol12-114) are perfectly viable in combination with stn1-13, whereas the pol12-109 allele, which causes a much more severe telomere elongation phenotype than the others, is lethal together with stn1-13. Given that telomere elongation in pol12-114 is very similar to that of pol12-216, we conclude that telomere elongation alone cannot explain the lethality of the pol12-216 stn1-13 combination. Similarly, all of the POL1 mutants tested have longer telomeres than pol12-216 yet are not lethal in combination with stn1-13. At present, it is unclear whether the ts lethal pol12-109 mutation is inviable with stn1-13 because of a general loss of Pol12 activity, or whether its defect is similar to that of pol12-216. In summary, then, our survey of synthetic lethal interactions between various alleles of POL12, STN1, POL1, and CDC13 indicate a striking degree of specificity for the genetic interaction between POL12 and STN1, and support the idea that a specific function of Pol12, altered or abolished by the pol12-216 mutation, rather than the overall level of Polα-primase activity, is critical for viability in cells with reduced STN1 activity.
Pol12 interacts physically with Stn1
The specific genetic interaction between POL12 and STN1 prompted us to test whether the products of these two genes might physically interact. We first tested this idea using the two-hybrid system, and found that full-length Pol12 (or the mutant pol12-216), when fused to the LexA protein, interacts weakly but reproducibly with a fusion of full-length Stn1 to the Gal4 activation domain (data not shown). In contrast, a LexA fusion with the N-terminal portion of Pol12 [LexA-Pol12(1-381)] interacts very strongly with GAD-Stn1, giving a signal ∼160-fold higher than the GAD-alone control and ∼20 higher than the value obtained with GAD-Stn1 in cells expressing LexA alone. Interestingly, the N-terminal LexA-Pol12 hybrid containing the pol12-216 mutation reproducibly interacted less well (approximately twofold) with GAD-Stn1 than the corresponding wild-type hybrid (Fig. 4A). A comparable difference was also observed in plate assays for β-galactosidase activity (data not shown). We do not know the reason for the large difference in activity between LexA-Pol12 and LexA-Pol12(1-381), but doubt that it is explained by expression differences, because the latter hybrid is present in only slightly higher amounts as judged by Western blotting (data not shown). Perhaps deletion of C-terminal Pol12 sequences relieves an inhibitory interaction or simply frees more LexA-Pol12 hybrid from complexes with Pol1 that might function poorly, or not at all, in the activation of the reporter gene. Although the reduced interaction of mutant LexA-pol12-216 with GAD-Stn1 compared with wild type may reflect a genuine interaction defect in pol12-216, we cannot rule out the possibility that this difference is caused by slightly lower expression levels of mutant hybrid compared with its wild-type counterpart.
Figure 4.
Physical interaction between Pol12 and Stn1. (A) Values obtained from liquid β-galactosidase assays to detect two-hybrid interactions between LexA/Pol12 and GAD/Stn1 fusion proteins. (B) In vitro binding of Stn1-HA to GST-Pol12, GST-pol12-216, GST-Pol12 Nt (N terminus), and GST-pol12-216 Nt hybrid proteins. Crude whole-cell extracts were prepared from W303-1B carrying the multicopy plasmid pMC479 (PGAL1-STN1-2HA-6HIS) and grown to exponential phase in selective medium containing either galactose (lanes 1) or glucose (lanes 2), to induce and repress the expression of Stn1, respectively. The extracts were incubated with GST-Pol12 (or any one of the fusion hybrids indicated above) bound to glutathione agarose beads. As an additional control, the W303-1B strain carrying the empty vector pE195, and grown in the presence of galactose, was included (lanes 3).
To corroborate the two-hybrid results described above, we sought to demonstrate an interaction between Pol12 and Stn1 in a biochemical assay. To this end we fused both the wild-type and mutant versions of full-length and N-terminal Pol12 to GST, expressed them in Esch-erichia coli, and purified the resulting hybrid proteins from whole-cell extracts. When bound to glutathione beads, all four GST-Pol12 hybrid proteins specifically retained an HA-epitope-tagged version of Stn1 present in whole-cell yeast extracts made from a strain expressing STN1-HA from the GAL1 promoter (Fig. 4B). In this assay there appears to be no significant difference between the ability of wild-type and mutant Pol12 (either full length or N-terminal fragment) to interact with Stn1-HA. Stn1 seems to be present in very small amounts in whole-cell extracts, and we have been unable to detect binding of the protein to GST-Pol12 when Stn1 is expressed from its native promoter. Taken together, the two-hybrid and biochemical data described here strongly support the idea that Pol12 and Stn1 physically interact in yeast cells. We do not know whether this interaction is direct, or mediated by other proteins.
pol12-216 causes reduced telomere capping in stn1-13 cells
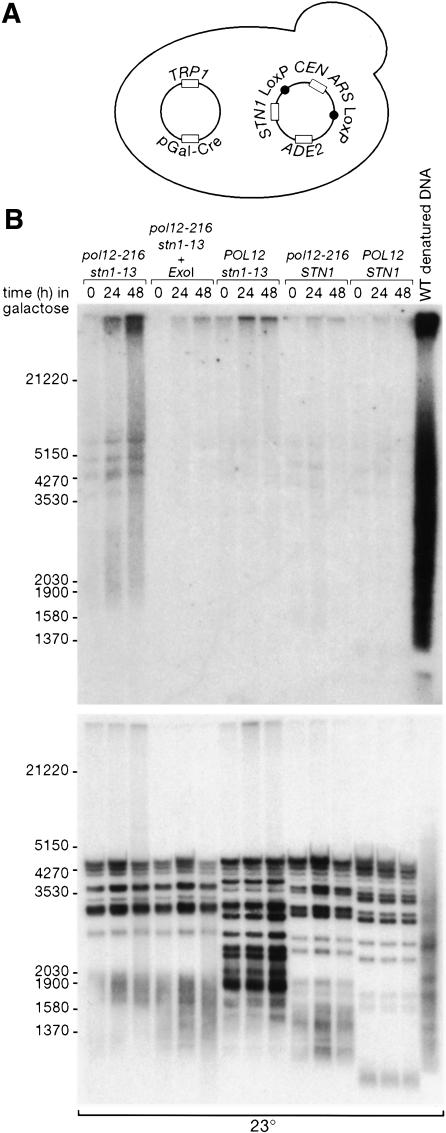
The lethal effect of combining the pol12-216 mutation with stn1-13 and other STN1 mutations, together with evidence for a physical interaction between these two proteins, suggests that Pol12 might normally assist Stn1 in carrying out its essential telomere protection function, and that the pol12-216 protein might be defective in this activity. To test this idea, we developed a strain that allowed us to examine the telomere phenotype of a pol12-216 stn1-13 double mutant. This strain contains both the pol12-216 and stn1-13 mutations at their normal chromosomal loci and a plasmid containing wild-type STN1 to allow growth. The rescuing plasmid was engineered such that its CEN-ARS replicon can be rapidly excised through induced Cre-mediated recombination at flanking LoxP sites (Cohen and Sinclair 2001). In the absence of its replicon, the plasmid is diluted by cell division, thus exposing the lethal pol12-216 stn1-13 double-mutant combination (see Fig. 5A).
Figure 5.
Increase in telomeric single-stranded DNA in pol12-216 stn1-13 double mutants. (A) Schematic representation of the strains used in the experiment. The POL12 and STN1 alleles of each strain are indicated in B. Each strain contains an STN1-ADE2 plasmid in which the CEN/ARS sequence is flanked by two LoxP sites in the same orientation (pGS120). A second plasmid (pSH63) carries the Cre recombinase gene driven by the GAL1 promoter. Growth in galactose at 23°C leads to the “pop out” of the intervening CEN/ARS sequence and consequently to the loss of the pGS120 plasmid. (B) In-gel hybridization analysis of telomeres from POL12 STN1 wild type, pol12-216, and stn1-13 single-mutant and pol12-216 stn1-13 double-mutant strains grown in the presence of galactose for the indicated times. (Top) The native gel was hybridized to a CA-rich labeled probe. (ExoI) ExonucleaseI. (Bottom) The same gel was denatured and hybridized to a CA-rich labeled probe.
Galactose induction of the Cre recombinase at 23°C in the double-mutant strain (but not in otherwise identical pol12-216, stn1-13, or wild-type strains) causes a significant increase in the fraction of large-budded cells in the culture (∼50% of cells at 48 h, compared with 10%-20% for the controls), reminiscent of the G2/M block seen when stn1-13 cells are shifted to the nonpermissive temperature (37°C; Grandin et al. 1997). Significantly, plasmid loss in the double-mutant cells also leads to a marked increase in telomeric single-stranded DNA, an additional phenotype observed in cells lacking STN1 function (Fig. 5B). These data suggest that Pol12 normally cooperates in some way with Stn1 in telomere capping, and that the pol12-216 protein is at least partially defective in this function.
Telomere elongation in pol12-216 mutants requires telomerase, is limited by RAD52, and occurs in the absence of Tel1 or the MRX complex
To determine whether telomere elongation in pol12-216 cells might be caused by a partial deregulation of telomerase-mediated TG-repeat addition, we examined telomere length in pol12-216 mutants lacking in addition a functional telomerase enzyme. As shown in Figure 6A, pol12-216 tlc1Δ double mutants display short telomeres compared with either wild-type cells or pol12-216 single mutants (as does a pol12-216 est1Δ double mutant; data not shown), demonstrating that the telomere elongation seen in pol12-216 cells is telomerase-dependent. As might be expected, pol12-216 tlc1Δ double mutants senesce at a rate similar to that of tlc1Δ single mutants (data not shown), suggesting that pol12-216 is incapable of either rescuing or delaying the death of most telomerase-negative cells following extensive TG-repeat attrition.
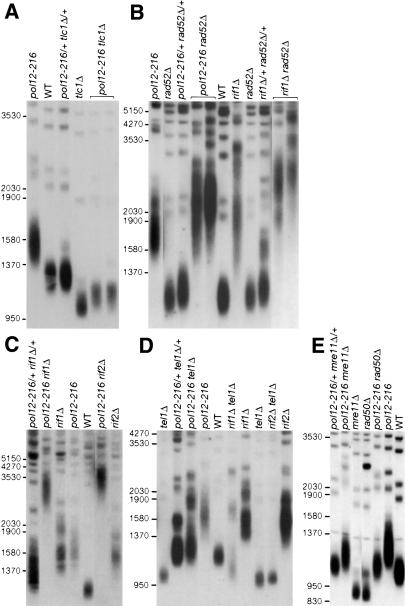
Figure 6.
Genetic interaction of the pol12-216 mutation with TLC1, RAD52, RIF1, RIF2, TEL1, MRE11, and RAD50 deletions. Southern blot analysis of telomeres from the indicated single- and double-mutant strains (as described in Fig. 2C). (A) The tlc1Δ and pol12-216 tlc1Δ double-mutant spores were identified by tetrad analysis, as described in Materials and Methods, and directly inoculated in liquid YPD medium for DNA preparation and Southern blot analysis. (B-E) Diploid parent strains and haploid mutant segregants were restreaked at least 10 times on YPD medium at 30°C prior to DNA preparation (and sporulation, in the case of diploids). Telomeric DNA was hybridized either to a d(TG)/d(CA) probe (A-D) or to a Y′ probe (E), as described in Materials and Methods.
We also examined the effect of RAD52 mutation on the telomere elongation phenotype of pol12-216. RAD52 is required for most types of homologous recombination in yeast, and could in principle play some role in telomere elongation in pol12-216 cells. In fact, both known “survivor” pathways, which are responsible for telomere maintenance in cells lacking active telomerase enzyme, are dependent on RAD52 function. However, we found that deletion of RAD52 actually exacerbates the telomere elongation phenotype of pol12-216 (Fig. 6B), suggesting that Rad52 protein acts to limit telomerase-dependent telomere elongation in the presence of pol12-216. The same is also true for mutation of the RecA homolog Rad51 (data not shown). Significantly, neither RAD52 nor RAD51 mutation has any effect on telomere length in wild-type cells or pol1-1 mutants (Fig. 6B; data not shown), despite the fact that the latter cells display a dramatic increase in telomeric single-stranded DNA that might be expected to promote increased recombination within the TG repeats, as is seen for more centromere-proximal regions (Carson and Hartwell 1985).
Finally, we analyzed the interaction of pol12-216 with mutations in several genes proposed to act in the Rap1 protein “counting” pathway of telomere length regulation (Marcand et al. 1997; Ray and Runge 1999b; see above). Deletion of either RIF1 or RIF2, which encode negative regulators of this pathway that interact directly with Rap1 (Hardy et al. 1992; Wotton and Shore 1997), causes a telomere elongation phenotype that is considerably exacerbated by the presence of the pol12-216 mutation (Fig. 6C). Previous studies have shown that deletion of either TEL1 (the yeast ATM homolog) or the MRX complex genes RAD50 and MRE11 causes severe telomere shortening (Greenwell et al. 1995; Boulton and Jackson 1998), and at least in the case of tel1Δ a loss of Rap1 counting (Ray and Runge 1999a; Brevet et al. 2003). Both Tel1 and MRX are thus thought to be positive regulators of telomerase action, and epistasis analysis would suggest that they act downstream from both RIF1 and RIF2 (Ray and Runge 1999a; Ritchie et al. 1999; Ritchie and Petes 2000; Chan et al. 2001; Diede and Gottschling 2001; Tsukamoto et al. 2001). Significantly, the pol12-216 mutation causes telomere elongation in all three mutant backgrounds (tel1Δ, mre11Δ, and rad50Δ), to slightly longer than wild-type lengths (Fig. 6D,E). As discussed in more detail below, these data suggest that Pol12 either acts downstream of Tel1 and MRX or in a parallel pathway of telomerase regulation.
Discussion
We describe here a novel genetic screen for mutants with relaxed telomere length regulation. The validity of this approach is underscored by the isolation of mutations in several genes known previously to play a role in restricting telomere elongation. Our preliminary results suggest that continued application of this screen might uncover additional, previously unknown factors involved in this process.
The finding reported here that a mutation in POL12 leads to altered telomere length regulation reinforces the notion that the DNA polymerase α-primase complex plays an important role in coordinating telomerase-mediated TG-repeat synthesis with conventional lagging-strand DNA replication at the telomere. It has long been appreciated that full Polα activity is essential for proper telomere length control (Carson and Hartwell 1985). The original POL1 mutant described by Carson and Hartwell (1985) is a hypomorphic ts-lethal allele, presumably affecting the catalytic activity of the enzyme, that displays continual telomere elongation when grown at permissive or semipermissive temperatures, as well as elevated, RAD52-dependent recombination near telomeres. A more detailed examination of the pol1-1 telomere phenotype (Adams and Holm 1996; Adams Martin et al. 2000) has revealed an S-phase-specific increase in single-stranded telomeric TG-repeat DNA, as well as a severe loss of telomeric silencing. Both of these phenotypes precede the telomere length increase seen after temperature up-shift, and are largely independent of telomerase action, suggesting that they are due to a primary defect in telomeric lagging-strand synthesis.
The pol12-216 mutant phenotype described here clearly differs in several important respects from the original pol1-1 mutant and the two new POL1 alleles identified in our screen. To begin with, pol12-216 has no obvious effect on cell growth rate at any temperature tested, and displays normal cell cycle parameters, suggesting that it has little or no effect on the overall activity of Polα-primase. In support of this conclusion, pol12-216 mutants show no increase in telomeric single-stranded DNA, which presumably arises in POL1 mutants because of a reduction in the efficiency of lagging-strand synthesis at the telomere (Adams Martin et al. 2000). Furthermore, pol12-216 does not cause the loss of telomeric heterochromatin seen in POL1 mutants, which could again be a manifestation of reduced lagging-strand synthesis and a consequent reduction in Rap1 binding and Sir protein recruitment (Adams Martin et al. 2000). Finally, pol12-216 leads to a smaller increase in telomere length than the POL1 alleles we have examined, and this increased telomere length set-point in pol12-216 cells is stable, unlike the case for POL1 mutants.
Despite the absence of POL1-like telomere phenotypes in pol12-216, this mutant displays an unusual synthetic lethality in combination with the STN1 mutations. By testing a large number of double-mutant combinations between stn1-13 and various alleles of POL12, POL1, and CDC13, we found that the functional interaction between POL12 and STN1 is highly specific. The most straightforward interpretation of the genetic data is that a Pol12 telomere-specific function, at least partly defective in the separation-of-function pol12-216 mutation, is required for viability in cells with reduced STN1 activity. This genetic analysis also suggests that the critical Pol12 function in question is not related in any simple way to either telomere length regulation or the suppression of single-stranded DNA at the telomere: many mutants with more extreme phenotypes of this sort than pol12-216 are perfectly viable in combination with stn1-13. Both two-hybrid and biochemical experiments support the idea that this specific POL12-STN1 genetic interaction reflects a physical association between the two proteins at telomeres.
To gain insight into why pol12-216 stn1-13 double mutants are inviable, we established experimental conditions to examine the state of such mutants. Despite the technical limitations of the Cre-induced plasmid loss experiment, our data strongly suggest that pol12-216 stn1-13 double mutants rapidly accumulate telomere damage (indicated by increased telomeric single-stranded DNA) and undergo a G2/M cell cycle arrest. The simplest interpretation of this result, which we favor, is that the function missing in the pol12-216 is required for telomere capping in cells containing the stn1-13 mutation, even at the permissive temperature of 23°C. Although the accumulation of single-stranded DNA in the pol12-216 stn1-13 double mutant might be interpreted as a failure of Polα-primase activity to complete C-strand synthesis at the telomere, or an uncoupling of G-strand and C-strand synthesis, we note first that neither single mutant displays a single-stranded TG-specific signal under these conditions (Fig. 5). Furthermore, we would point out that many POL1 mutants display these phenotypes on their own, and yet are viable in combination with stn1-13. We also think that pol12-216 stn1-13 lethality is unlikely to be caused by a telomerase regulation defect. Neither single mutant shows particularly elevated telomere length under the conditions of the experiment (23°C), and the apparently rapid lethality we observe is inconsistent with known effects of unregulated telomerase action in yeast (Kyrion et al. 1992).
How can observations reported here be integrated into the emerging picture (Evans and Lundblad 1999, 2002; Chandra et al. 2001; Pennock et al. 2001; Taggart et al. 2002) of telomerase recruitment/regulation and telomere capping? Although still speculative, our data and other recent studies (Diede and Gottschling 1999; Adams Martin et al. 2000; Qi and Zakian 2000) point to a complex role for Polα-primase function in the coordination of telomerase regulation, lagging-strand synthesis, and telomere end protection. Cdc13 seems to play a central role in this process, interacting with components of both the telomerase (Est1 subunit) and Polα-primase (Pol1 subunit) complexes (Qi and Zakian 2000). The telomeric function of the Cdc13-interacting protein Stn1, and an additional end-protection factor Ten1, are less well defined (Grandin et al. 1997, 2000, 2001) but would appear to be more specifically devoted to end protection, although mutations in either protein can also lead to telomere elongation. Our finding of a genetic and biochemical link between Stn1 and Pol12 suggests that the Polα-primase complex may play a previously unanticipated role in linking the telomerase recruitment and end-protection functions of Cdc13. One possible scenario (Pennock et al. 2001) is that Cdc13 first recruits and/or activates telomerase through an interaction with Est1. This Cdc13-Est1 interaction may then create the conditions for the subsequent recruitment of Stn1. Indeed, Stn1 interacts with cdc13-1 but not with cdc13-2, a mutated protein that leads to senescence because it fails to either recruit (Nugent et al. 1996; Evans and Lundblad 1999; Chandra et al. 2001) or activate (Taggart et al. 2002) telomerase. Cdc13 and Stn1 might then collaborate to recruit the Polα-primase complex by directly binding to its Pol1 and Pol12 subunits, respectively (Qi and Zakian 2000; this work), thus promoting C-strand synthesis and perhaps also contributing to the blockage of further telomerase action. Our results suggest the additional intriguing idea that some as-yet-undefined function of the Polα-primase complex may also contribute to telomere capping, through an activity independent of its RNA/DNA synthetic capability. In strains with reduced Pol1 catalytic activity, we imagine that this capping function is normal (and thus these mutations are not lethal in combination with stn1-13), but that C-strand synthesis proceeds slowly, such that it is not complete until late in G2/M or the following G1 phase (Adams Martin et al. 2000). The coordination of Polα-primase and telomerase function may be a highly conserved feature of telomere replication. For example, studies of the active telomere-formation process in mating cells of the hypotrichous ciliate Euplotes crassus have demonstrated a tight linkage between Polα (and/or δ) action and telomerase addition, as well as a physical association between telomerase and primase (Fan and Price 1997; Ray et al. 2002).
The exacerbation of the pol12-216 telomere elongation phenotype by rad52Δ or rad51Δ suggests, rather surprisingly, that both of these proteins act in pol12-216 cells in such a way as to shorten TG-repeat tract length. This might occur through a mechanism such as telomeric rapid deletion (TRD; Li and Lustig 1996; Bucholc et al. 2001), which has been implicated in the negative regulation of telomere size, particularly in the rapid truncation of overelongated telomeres. The behavior of pol12-216 in combination with rad52Δ stands in contrast to the absence of any effect of rad52Δ on telomere length of either pol1 ts mutants (S. Grossi, A. Puglisi, and D. Shore, unpubl.) or a telomere elongation allele of CDC13, cdc13-5 (Chandra et al. 2001). Interestingly, however, rif1Δ rad52Δ double mutants also display a synergistic telomere elongation phenotype (Fig. 6B; Teng et al. 2000). These results raise the possibility that both rif1Δ and pol12-216 mutations alter telomere structure in some rather unique way such as to favor the action of a RAD52-dependent telomere-shortening pathway.
Results from several studies place RIF1 and RIF2 upstream of TEL1 and the MRX complex in a telomerase regulatory pathway that responds to the number of telomere-bound Rap1 molecules (Ray and Runge 1999a; Ritchie and Petes 2000; Chan et al. 2001; Diede and Gottschling 2001; Tsukamoto et al. 2001). The epistasis data presented here suggest that POL12 either acts downstream from all of these factors, or on a separate regulatory pathway. Without being able to determine the loss-of-function phenotype of POL12 with respect to telomere length regulation, we cannot at present distinguish between these two possibilities. However, the genetic and physical interaction of Pol12 with Stn1 would seem to place Pol12 directly at the telomere end, in a position to respond to regulatory signals that might be generated by Tel1 and/or the MRX complex. A clear challenge for the future will be to understand in more detail how Pol12 contributes to telomere capping and to uncover its precise role in processes that regulate telomere length.
Materials and methods
Yeast strains, media, and plasmids
The yeast strains used in this study are listed in Table 1 and are isogenic to W303 (Thomas and Rothstein 1989), apart from specific point mutations or gene deletion/disruptions, or where otherwise indicated. The Chromosome VII disome strains YSG171 (control strain) and YSG172 (test strain) were constructed by cytoduction (Dutcher 1981), through crosses of YSG174 or YSG175 recipient strains with the donor strain LS15 (Sandell and Zakian 1993), followed by selection on synthetic complete (SC) medium lacking arginine, lysine, and tyrosine and containing 60 mg/L canavanine (SC-Arg-Lys-Tyr + Can). Disome strains were maintained on SC-Lys-Tyr-Ura selective medium. The sir2Δ disome strain YSG219 is isogenic to YSG172 with the exception of the sir2::LEU2 disruption. Mutant strains recovered from the screen were grown in nonselective conditions to allow for the loss of one copy of Chromosome VII and then backcrossed at least three times to W303-1B. Strains carrying either the pol1-1 (cdc17-1; Hartwell 1973) or cdc13-1 mutations were backcrossed once to W303-1A before being used. Strains YAP60 and YAP61 are deleted at their STN1 chromosomal locus and contain either YCp111-stn1-101 (CENLEU2) or YCp111-stn1-154, respectively (Grandin et al. 2001).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| W303-1A | MATaade2-1 trp1-1 can1-100 leu2-3, 112his3-11, 15 ura3-1 rad5-535 | Thomas and Rothstein 1989 |
| W303-1B | MATaade2-1 trp1-1 can1-100 leu2-3, 112 his3-11, 15 ura3-1 rad5-535 | Thomas and Rothstein 1989 |
| LS15/YSG36 | MATakar1-1 aro2 leu2 | Sandell and Zakian 1993 |
| YSG174 | MATaade3::HisG lys5 cyh2 r adh4:: (ADE2, URA3, 32 correctly oriented Raplp sites, 80 bp TG1-3) TelVIIL | This study |
| YSG175 | MATaade3::HisG lys5 cyh2 r adh4:: (ADE2, URA3, 32 misoriented Raplp sites, 80 bp TG1-3) TelVIIL | This study |
| YSG171 | MATaade3::HisG lys5 cyh2 r adh4:: (ADE2, URA3, 32 correctly oriented Raplp sites, 80 bp TG1-3) TelVIIL + ChrVII | This study |
| YSG172 | MATaade3::HisG lys5 cyh2 r adh4:: (ADE2, URA3, 32 Misoriented Raplp sites, 80 bp TG1-3) TelVIIL + ChrVII | This study |
| YSG219 | MATaade3::HisG lys5 cyh2 r sir::HIS3 adh4:: (ADE2, URA3, 32 misoriented Raplp sites, 80 bp TG1-3) TelVIIL + ChrVII | This study |
| YG705 | MATacdc13-1 his7 ural | Hartwell 1973 |
| YPD15 | MATacdc13-50-HIS3 | This study |
| YPD16 | MATacdc13-50-HIS3 | This study |
| YAB342 | mata::loxP est1::KanMX4 ade2::loxP leu2::pGal-HO-LEU2 lys2::loxPmnt::LYS2 RAD5 (pEST1-URA3-CEN) | A. Bianchi (unpubl.) |
| YAB167 | mata::loxP mre11::KanMX4 ade2::loxP leu2::pGal-HO-LEU2 RAD5 | A. Bianchi (unpubl.) |
| YG636 | MATapol1-1 his7 ura1 | Hartwell 1973 |
| YAP11 | MATapol1-236 | This study |
| YSG250 | MAT α pol1-35 TRP1 adh4::ADE2, URA3 TelVIIL | This study |
| YSG453 | MATapol1-228 TRP1 | This study |
| YSG406 | MATapol12-216/13XMYC/SpHis5+ | This study |
| YSG408 | MATapol12-216/13XMYC/SpHis5+ LEU2 | This study |
| YSG361 | MATapol12-103 | This study |
| YSG363 | MATapol12-109 | This study |
| YSG365 | MATapol12-114 | This study |
| CY338 | MATapol12::LEU2 | This study |
| YSG368 | MATapril-M4 | Francesoni et al. 1991; Marini et al. 1997 |
| YSG416 | MATarad50::KanMX4 | This study |
| YSG418 | MATarad51::KanMX4 | This study |
| YSG269 | MATarad52::LEU2 | This study |
| YSG248 | MATarif1-56 ade3::HISG adh4::ADE2, URA3, TelVIIL | This study |
| YSG259 | MATarif1-174 | This study |
| YSG16 | MATarif1::HIS3 | This study |
| YG269 | MATarif2::HIS3 | This study |
| YAP46 | MATastn1::LoxP + YCp-STN1-URA3 | This study |
| YAP60 | MATastn1::LoxP + YCp111-stn1-101-LEU2 | This study |
| YAP61 | MATastn1::LoxP + YCp111-stn1-154-LEU2 | This study |
| YSG252 | MATastn1-3 TRP1 | This study |
| YG147 | MATastn1-13 | This study |
| YG148 | MATastn1-13 | This study |
| YG630 | MATatel1::his3::TRP1 | This study |
| YAB344 | mata::loxP tlc1::KanMX4 ade2::loxP leu2::pGal-HO-LEU2 lys2::loxPmnt2::LYS2 RAD5 (pTLC1-URA3-CEN) | A. Bianchi (unpubl.) |
| YAM21 | MATayku70::KanMX4 adh4::URA3 TelVIIL | This study |
| YSG492 | MATapol12-216/13XMYC/SpHis5+ est1::KanMX4 LEU2 lys2::loxP (pEST1-URA3-CEN) | This study |
| YSG491 | MAT α/mat α::loxP POLI2/pol12-216-13XMYC-SpHis5+ EST1/est1::KanMX4 ade2-1/ade2::loxP LEU2/leu2::pGal-HO-LEU2 LYS2/lys2::loxPMNT2/mnt2::LYS2 RAD5/rad5-535 | This study |
| YSG422 | mat α::loxP pol12-216/13XMYC/SpHis5+ mre11::KanMX4 LEU2 | This study |
| YSG420 | MAT α/mat α::loxP POL12/pol12-216/13XMYC/SpHis5+ MRE11/mre11::KanMX4 LEU2/leu2-3,112 RAD5/rad5-535 | This study |
| YSG475 | MAT α pol12-216/13XMYC/SpHis5+ pol1-1 TRP1 LEU2 | This study |
| YSG474 | MATa/α POL12/pol12-216-13XMYC-SpHis5+ POL1/pol1-1 TRP1/trp1-1 LEU2/LEU2 | This study |
| YSG398 | MAT α pol12-216/13XMYC/SpHis5+ pol1-35 | This study |
| YSG397 | MATa/α POL12/pol12-216-13XMYC-SpHis5+ POL1/pol1-35 TRP1/trp1-1 ADH4/adh4::ADE2, URA3 TelVIIL | This study |
| YSG471 | MATapol12-216/13XMYC/SpHis5+ pol1-228 | This study |
| YSG470 | MATa/α POL12/pol12-216-13XMYC-SpHis5+ POL1/pol1-228 TRP1/trp1-1 LEU2/leu2-3,112 | This study |
| YAP12 | MATapol12-216/13XMYC/SpHis5+ pol1-236 | This study |
| YSG426 | MAT α pol12-216/13XMYC/SpHis5+ rad50::KanMX4 | This study |
| YSG427 | MAT α pol12-216/13XMYC/SpHis5+ rad51::KanMX4 | This study |
| YSG421 | MATa/α POL12/pol12-216-13XMYC-SpHis5+ RAD51/rad51::KanMX4 | This study |
| YSG401 | MAT α pol12-216/13XMYC/SpHis5+ rad52::LEU2 | This study |
| YSG400 | MATa/α POL12/pol12-216-13XMYC-SpHis5+ RAD52/rad52::LEU2 | This study |
| YSG483 | MAT α pol12-216/13XMYC/SpHis5+ rif1::TRP1 | This study |
| YSG482 | MATa/α POL12/pol12-216-13XMYC-SpHis5+ RIF1/rif1::TRP1 | This study |
| YSG486 | MAT α pol12-216/13XMYC/SpHis5+ rif2::HIS3 | This study |
| YSG460 | MAT α pol12-216 stn1-13 + YCp33-STN1-URA3 | This study |
| YSG308 | MATa/α POL12/pol12-216 STN1/stn1-13 | This study |
| YAP44 | MAT α pol12-216/13XMYC/SpHis5+ stn1:: LoxP + YCp-STN1-URA3 | This study |
| YAP62 | MAT α pol12-216/13XMYC/SpHis5+ stn1:: LoxP + YCp33-STN1-URA3 + YCp111-stn1-101-LEU2 | This study |
| YAP63 | MAT α pol12-216/13XMYC/SpHis5+ stn1:: LoxP + YCp33-STN1-URA3 + YCp111-stn1-154-LEU2 | This study |
| YSG481 | MAT α pol12-216/13XMYC/SpHis5+ tel1::his3::TRP1 | This study |
| YSG478 | MATa/α POL12/pol12-216-13XMYC-SpHis5+ TEL1/tel1::his3::TRP1 | This study |
| YSG490 | mat α pol12-216-13XMYC-SpHis5+ tlc1::KanMX4 LEU2 lys2::loxP (pTLC1-URA3-CEN) | This study |
| YSG489 | MAT α/mat α ::loxP POL12/pol12-216-13XMYC-SpHis5+ TLC1/tlc1::KanMX4 ade2-1/ade2::loxP LEU2/leu2::pGal-HO-LEU2 LYS2/lys2::loxP MNT2/mnt2::LYS2 RAD5/rad5-535 | This study |
| YSG415 | MATapol12-216-13XMYC-SpHis5+ yku70::KanMX4 | This study |
| YAP71 | MATapol12-103-KANRstn1-13 | This study |
| YAP72 | MAT α pol12-109-KANRstn1-13 + YCp33-STN1-URA3 | This study |
| YAP73 | MATapol12-114-KANRstn1-13 | This study |
| YPD21 | MATapol1-236 stn1-13 | This study |
| YPD25 | MAT α cdc13-50-HIS3 stn1-13 | This study |
| YSG467 | MAT α pol1-35 rad52::LEU2 TRP1 | This study |
| YSG445 | MATa/α POL1/pol1-35 RAD52/rad52::LEU2 TRP1/trp1-1 ADH4/adh4::ADE2, URA3 TelVIIL | This study |
| YSG466 | MAT α rif1:: HIS3 rad52::LEU2 | This study |
| YSG443 | MATa/α RIF1/rif1::HIS3 RAD52/rad52::LEU2 | This study |
| YG752 | MATarif1::URA3 tel1::HIS3 | This study |
| YSG271 | MAT α rif2::HIS3 tel1::his3::TRP1 | This study |
| YSG309 | MATa/α POL12/pol12-216 CDC13/cdc13-1 | This study |
| YSG456 | MATa/α POL1/pol1-1 STN1/stn1-13 | This study |
| YSG457 | MATa/α PRI1/pri1-M4 STN1/stn1-13 ADH4/adh4::URE3 TelVIIL | This study |
| CTY10-5D | MATaade2-1 trp1-901 leu2-3 his3-200 gal4 gal80 URA3::lexA op-LacZ | Bartel and Fields 1995 |
A wild-type copy of the STN1 gene was first deleted in a diploid strain by transplacement of a PCR-generated cassette (from the pUG73 plasmid; Gueldener et al. 2002) carrying the Kluyveromyces lactis LEU2 gene flanked by two LoxP sites. A heterozygous diploid was cotransformed with plasmids YCp33-STN1 (CEN-URA3; Grandin et al. 2000) and pSH63 (CEN-TRP1), carrying the cre recombinase gene driven by the GAL1 promoter (Cohen and Sinclair 2001) and sporulated. Trp+, Leu+, 5-FOAs spores were grown in 2% galactose to induce the expression of the Cre recombinase gene and the “pop-out” of the KlLEU2 cassette. The stn1Δ + YCp33-STN1-URA3 strain (YAP46) was transformed with either YCp111-stn1-101-LEU2 or YCp-stn1-154-LEU2 and grown on 5-FOA to select for loss of YCp33-STN1-URA3.
The double-mutant strains YAP62 and YAP63 were generated by crossing YAP46 to YSG408. After sporulation of the diploid strain, an His+, 5-FOAs spore colony (YAP44) was transformed with either YCp111-stn1-101 or YCp-stn1-154 plasmids, and Leu+ transformants were grown on 5-FOA medium to test whether they could lose the YCp-STN1-URA3 plasmid and grow. To generate pol12 ts mutant strains, a wild-type copy of the POL12 gene carried by the pMF502 plasmid (CEN-ARS1-TRP1) was amplified by PCR using an error-prone Taq polymerase, and the DNA amplification products were cloned by “gap repair.” Strain CY338, in which deletion of the POL12 gene (pol12::LEU2) is complemented by a POL12-CEN-URA3 plasmid (pFE2), was cotransformed with an SphI-BamHI fragment from pMF502 (missing almost the entire POL12 ORF) and the DNA products of the PCR reaction. Trp+, Ura+, Leu+ transformants were grown on 5-FOA to select for the loss of pFE2 and tested for growth at 25°C and 37°C. POL12 plasmids were rescued from transformants that were able to grow at 25°C but not at 37°C (potential pol12 ts mutants) and used to retransform the CY338 strain to confirm the ts phenotype of Trp+, 5-FOAR, Leu+ transformants.
Temperature-sensitive POL12 alleles were cloned between the SphI-BamHI sites of the pMA10 plasmid, upstream of the KAN gene, conferring resistance to the drug Kanamycin. XbaI-KpnI fragments (carrying pol12 and KAN genes) from these constructs were used to transform the wild-type strain W303-1A. Three of the KanR, temperature-sensitive transformants (pol12-103, pol12-109, and pol12-114) were further analyzed. Double mutants carrying pol12 ts and stn1-13 mutations were generated by crossing either YSG361, YSG363, or YSG365 to YG147 containing YCp33-STN1 (CEN-URA3). Diploids were sporulated and tetrads were analyzed by replica plating on 5-FOA and KAN plates at 23°C and for growth at 37°C. Strains YAP11 and YAP12 were generated by transformation of W303-1A and YSG406, respectively, with the BstXI linearized plasmid pCM1-pol1-236, as previously described (Qi and Zakian 2000).
To construct the YPD15 and YPD16 strains, the cdc13-50 allele was cloned from the pRS314-cdc13-50 plasmid (Qi and Zakian 2000) into the ApaI/BamHI sites of pRS303 (pDM37) and this TthI-linearized plasmid was used to transform W303-1A and W303-1B. Other double-mutant strains were obtained by crossing of single-mutant strains followed by sporulation and tetrad analysis, unless indicated otherwise. Growth and manipulation of yeast were carried out according to standard procedures (Adams et al. 1997). β-Galactosidase expression in the two-hybrid strain CTY10-5D (Bartel and Fields 1995) was measured both by liquid assay (Moretti et al. 1994) and by nitrocellulose filter assay (Breeden and Nasmyth 1987). Plasmids carrying LexA-Pol12 (full-length) fusion hybrids were obtained by cloning XmaI-XhoI DNA fragments containing either POL12 or pol12-216 genes in the XmaI-SalI-digested pBTM116 vector (Bartel and Fields 1995; pGS86 and pGS96, respectively). Plasmids carrying LexA-Pol12 Nt (amino acids 1-381) fusion hybrids were obtained by cloning XmaI-PstI DNA fragments containing either POL12 Nt or pol12-216 Nt genes in the XmaI-PstI-digested pBTM116 vector (pGS84 and pGS97, respectively). The pGAD-STN1 plasmid has been previously described (Grandin et al. 1997).
Spot assays were performed by spotting 3.5-μL aliquots of 10-fold serial dilutions of saturated overnight cultures onto YPD, SC, SC lacking uracil (SC-Ura), and SC + FOA (1 g of 5-fluoroorotic acid [5-FOA]/L) plates. Plates were incubated for 2-3 d at 23°C, 30°C, or 37°C. In the experiment described in Figure 6A, strains YAB342 (est1::KanMX4 + pEST1-URA3) and YAB344 (tlc1::KanMX4 + pTLC1-URA3) were crossed to YSG408 (pol12-216). The heterozygote diploids (YSG491 and YSG489) were grown on SC + 5-FOA medium to select for the loss of the centromeric plasmids pEST1-URA3 and pTLC1-URA3, respectively, and then streaked several times at 30°C before being sporulated, to stabilize telomere length. After sporulation and tetrad dissection, colonies generated by several tetrads with four viable spores were directly inoculated in liquid YPD medium for DNA preparation and Southern blot analysis.
Mutagenesis
EMS mutagenesis was performed according to standard procedures (Adams et al. 1997). In two different experiments, cells were mutagenized to 40% and 20% viability, respectively. Cells were plated on YPD at a density of ∼200 viable cells per plate and allowed to form relatively large colonies so that red sectors could be easily distinguished.
Cloning of POL12
To clone the wild-type copy of the mutant gene in strain M216, we first generated a haploid double-mutant M216/stn1-13 strain (YSG460) kept alive by a URA3 plasmid (YCplac33) carrying the wild-type STN1 gene (Grandin et al. 2000). After transformation with a centromeric LEU2 yeast genomic library (kindly provided by P. Linder, Centre Medicale Universitaire [CMU], Geneva, Switzerland) and selection for Leu+ transformants, we screened for FOAR colonies that had lost the STN1-URA3 plasmid, presumably because they had received a plasmid carrying a wild-type copy of either STN1 or the gene mutated in M216. We recovered plasmids containing STN1, as expected, and others containing a genomic region with POL12, YBL036C, and part of the STU1 gene, from the left arm of Chromosome II. An SacI-NheI DNA fragment carrying POL12 was cloned in the pRS315 (CENVI LEU2) vector between the SacI-XbaI restriction sites (pGS76) and shown to rescue the M216/stn1-13 double mutant strain lethality. The pol12-216 allele was cloned by gap repair using a BssHII-StuI fragment from the pGS76 plasmid to transform the YSG460 yeast strain (pol12-216 stn1-13 + pSTN1-URA3). Recovered plasmids from Leu+ FOAs transformants (unable to lose the pSTN1-URA3 plasmid) contained the pol12-216 allele.
Telomere Southern blot and in-gel hybridization
Yeast DNA was isolated from overnight cultures and 1 μg was digested with the appropriate enzymes. DNA fragments were separated by electrophoresis in 1.5% agarose gels, transferred to HyBond N+ membranes, and hybridized either to a nick-translated d(TG)/d(CA) probe (Amersham Nick Translation Kit), a Y′ probe (Craven and Petes 1999), or to a 1.1-kb URA3 random-primed probe (Amersham Random Priming Kit) by standard procedures. The membranes were then autoradiographed on X-ray film or with a PhosphorImager (Bio-Rad Molecular Imager FX). In-gel hybridization experiments were performed as described previously, using a single-stranded 22-mer CA oligonucleotide probe (Dionne and Wellinger 1996). DNA was isolated from overnight cultures, digested with XhoI and electrophoresed through 0.75% agarose gel. As a control, the same DNA samples were treated for ∼15 h at 37°C with the ExonucleaseI and then analyzed as before. In the experiments described in Figures 2 and 6 (Fig. 6B-D) yeast strains were restreaked at least 10 times at 23°C or 30°C before DNA preparation.
Detection of telomeric single-stranded DNA in pol12-216 stn1-13 double-mutant cells
Strains YSG147 (stn1-13), YSG406 (pol12-216), YSG460 (stn1-13 pol12-216 + pSTN1-URA3), and W303-1A were cotransformed with plasmids pSH63 (a CEN-TRP1 plasmid carrying the cre recombinase gene driven by the GAL1 promoter (Cohen and Sinclair 2001) and pGS120 (pSTN1-ADE2-LoxP-CEN/ARS-LoxP). The pGS120 plasmid was obtained by cloning a BamHI fragment carrying STN1 plus flanking sequences into the BamHI site of pDSSD1 (pADE2-LoxP-CEN/ARS-LoxP; kindly provided by D. Sinclair, Harvard Medical School, Boston, MA). The YSG460 strain carrying both plasmids was then grown on SC medium lacking adenine and tryptophan and containing 5-FOA (SC-Ade-Trp + 5-FOA) to select for loss of the pSTN1-URA3 plasmid. The cotransformed strains were streaked on selective medium (SC-Ade-Trp) at 23°C several times before the experiment was performed, to allow their telomeres to reach a new equilibrium length. Cells were precultured to saturation in SC-Ade-Trp + 2% raffinose, diluted in the same medium to a concentration of ∼3 × 106 cells/mL, at which point galactose was added to a final concentration of 2% to induce expression of Cre recombinase. Adenine was also added to allow cells to grow following loss of the pGS120 plasmid. Samples were collected after 5, 8, 10, 24, and 48 h and analyzed using the in-gel hybridization technique to test for the presence of telomeric single-stranded DNA. Aliquots of the same samples were counted using a hemocytometer and plated on YPD to measure cell viability. Colonies were then replicaplated on SC-Ade medium to determine the fraction of cells that had lost the pSTN1-ADE2 plasmid.
In vitro binding assays
XmaI-XhoI DNA fragments containing either full-length (amino acids 1-705) POL12 (or pol12-216) or N-terminal (amino acids 1-381) POL12 (or pol12-216) were cloned in pGEX-4T-1 (Amersham). Expression and purification of GST, GST-Pol12(1-705), and GST-Pol12(1-381) fusion proteins in E. coli strain BL21, and binding reactions to glutathione-agarose beads, were performed essentially as described (Cuperus et al. 2000). Yeast strain W303-1B, containing either pMC479 (YEp195-PGAL1-STN1-2HA-6HIS, kindly provided by M. Charbonneau, Ecole Normale Supérieure de Lyon, Lyon, France) or YEp195 (empty vector) plasmids, was precultured to saturation in selective medium + 2% raffinose, diluted in selective medium + 2% galactose or glucose and grown to exponential phase. Crude whole-cell extracts were prepared as described (Cuperus et al. 2000), except that the wash buffer was modified (50 mM HEPES at pH 7.5, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1 mM PMSF). Input and bound Stn1-2HA were detected with rat monoclonal anti-HA antibody (kindly provided by J. Ripperger, University of Geneva, Geneva, Switzerland) using the ECL detection system (Amersham).
Acknowledgments
We thank P. Linder and D. Kressler for the gift of a yeast genomic plasmid library; V. Zakian, M. Charbonneau, and D. Sinclair for gifts of plasmids and/or strains; R. Wellinger and K. Mishra for advice on the in-gel hybridization technique; the Schibler laboratory for antibodies; N. Roggli for expert graphics work; and members of the Shore lab for helpful advice throughout the course of this work and comments on the manuscript. Funds and additional support were provided by grants from the Swiss National Fund and the Swiss Federation Against Cancer; by the National Center for Competence in Research (NCCR) program “Frontiers in Genetics” (sponsored by the Swiss National Fund); and by the Canton of Geneva. P.D. was supported in part by an FEBS Short-Term Fellowship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.300004.
References
- Adams A.K. and Holm, C. 1996. Specific DNA replication mutations affect telomere length in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 4614-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A., Gottschling, D.E., Kaiser, C.A., and Stearns, T. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Adams Martin A., Dionne, I., Wellinger, R.J., and Holm, C. 2000. The function of DNA polymerase α at telomeric G tails is important for telomere homeostasis. Mol. Cell. Biol. 20: 786-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel P.L. and Fields, S. 1995. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 254: 241-263. [DOI] [PubMed] [Google Scholar]
- Boulton S.J. and Jackson, S.P. 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17: 1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeden L. and Nasmyth, K. 1987. Cell cycle control of the yeast HO gene: Cis- and trans-acting regulators. Cell 48: 389-397. [DOI] [PubMed] [Google Scholar]
- Brevet V., Berthiau, A.S., Civitelli, L., Donini, P., Schramke, V., Geli, V., Ascenzioni, F., and Gilson, E. 2003. The number of vertebrate repeats can be regulated at yeast telomeres by Rap1-independent mechanisms. EMBO J. 22: 1697-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholc M., Park, Y., and Lustig, A.J. 2001. Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 6559-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson M.J. and Hartwell, L. 1985. CDC17: An essential gene that prevents telomere elongation in yeast. Cell 42: 249-257. [DOI] [PubMed] [Google Scholar]
- Cervantes R.B. and Lundblad, V. 2002. Mechanisms of chromosome-end protection. Curr. Opin. Cell Biol. 14: 351-356. [DOI] [PubMed] [Google Scholar]
- Chakhparonian M. and Wellinger, R.J. 2003. Telomere maintenance and DNA replication: How closely are these two connected? Trends Genet. 19: 439-446. [DOI] [PubMed] [Google Scholar]
- Chan S.W., Chang, J., Prescott, J., and Blackburn, E.H. 2001. Altering telomere structure allows telomerase to act in yeast lacking ATM kinases. Curr. Biol. 11: 1240-1250. [DOI] [PubMed] [Google Scholar]
- Chandra A., Hughes, T.R., Nugent, C.I., and Lundblad, V. 2001. Cdc13 both positively and negatively regulates telomere replication. Genes & Dev. 15: 404-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen H. and Sinclair, D.A. 2001. Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc. Natl. Acad. Sci. 98: 3174-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K.L., Russo, A.A., Tseng, B.Y., and Kelly, T.J. 1993. The role of the 70 kDa subunit of human DNA polymerase alpha in DNA replication. EMBO J. 12: 4555-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven R.J. and Petes, T.D. 1999. Dependence of the regulation of telomere length on the type of subtelomeric repeat in the yeast Saccharomyces cerevisiae. Genetics 152: 1531-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus G., Shafaatian, R., and Shore, D. 2000. Locus specificity determinants in the multifunctional yeast silencing protein Sir2. EMBO J. 19: 2641-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diede S.J. and Gottschling, D.E. 1999. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases α and δ. Cell 99: 723-733. [DOI] [PubMed] [Google Scholar]
- ____. 2001. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr. Biol. 11: 1336-1340. [DOI] [PubMed] [Google Scholar]
- Dionne I. and Wellinger, R.J. 1996. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc. Natl. Acad. Sci. 93: 13902-13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrana K., Perrod, S., and Gasser, S.M. 2001. Turning telomeres off and on. Curr. Opin. Cell Biol. 13: 281-289. [DOI] [PubMed] [Google Scholar]
- Dutcher S.K. 1981. Internuclear transfer of genetic information in kar1-1/KAR1 heterokaryons in Saccharomyces cerevisiae. Mol. Cell. Biol. 1: 245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S.K. and Lundblad, V. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286: 117-120. [DOI] [PubMed] [Google Scholar]
- ____. 2000. Positive and negative regulation of telomerase access to the telomere. J. Cell Sci. 113: 3357-3364. [DOI] [PubMed] [Google Scholar]
- ____. 2002. The Est1 subunit of Saccharomyces cerevisiae telomerase makes multiple contributions to telomere length maintenance. Genetics 162: 1101-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X. and Price, C.M. 1997. Coordinate regulation of G- and C strand length during new telomere synthesis. Mol. Biol. Cell 8: 2145-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M., Marini, F., Gamba, D., Lucchini, G., and Plevani, P. 1994. The B subunit of the DNA polymerase α-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol. Cell. Biol. 14: 923-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesconi S., Longhese, M.P., Piseri, A., Santocanale, C., Lucchini, G., and Plevani, P. 1991. Mutations in conserved yeast DNA primase domains impair DNA replication in vivo. Proc. Natl. Acad. Sci. 88: 3877-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvik B., Carson, M., and Hartwell, L. 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15: 6128-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N., Reed, S.I., and Charbonneau, M. 1997. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes & Dev. 11: 512-527. [DOI] [PubMed] [Google Scholar]
- Grandin N., Damon, C., and Charbonneau, M. 2000. Cdc13 cooperates with the yeast Ku proteins and stn1 to regulate telomerase recruitment. Mol. Cell. Biol. 20: 8397-8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2001. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 20: 1173-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwell P.W., Kronmal, S.L., Porter, S.E., Gassenhuber, J., Obermaier, B., and Petes, T.D. 1995. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell 82: 823-829. [DOI] [PubMed] [Google Scholar]
- Grossi S., Bianchi, A., Damay, P., and Shore, D. 2001. Telomere formation by rap1p binding site arrays reveals end-specific length regulation requirements and active telomeric recombination. Mol. Cell. Biol. 21: 8117-8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueldener U., Heinisch, J., Koehler, G.J., Voss, D., and Hegemann, J.H. 2002. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acid Res. 30: e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy C.F.J., Sussel, L., and Shore, D. 1992. A RAP1-interacting protein involved in silencing and telomere length regulation. Genes & Dev. 6: 801-814. [DOI] [PubMed] [Google Scholar]
- Hartwell L.H. 1973. Three additional genes required for deoxyribonucleic acid synthesis in Saccharomyces cerevisiae. J. Bacteriol. 115: 966-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrion G., Boakye, K.A., and Lustig, A.J. 1992. C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 5159-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvay T.S., Morris, D.K., Sah, J., Balasubramanian, B., and Lundblad, V. 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144: 1399-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. and Lustig, A.J. 1996. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes & Dev. 10: 1310-1326. [DOI] [PubMed] [Google Scholar]
- Lundblad V. and Szostak, J.W. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57: 633-643. [DOI] [PubMed] [Google Scholar]
- Makiniemi M., Pospiech, H., Kilpelainen, S., Jokela, M., Vihinen, M., and Syvaoja, J.E. 1999. A novel family of DNA-polymerase-associated B subunits. Trends Biochem. Sci. 24: 14-16. [DOI] [PubMed] [Google Scholar]
- Marcand S., Buck, S.W., Moretti, P., Gilson, E., and Shore, D. 1996. Silencing of genes at nontelomeric sites in yeast is controlled by sequestration of silencing factors at telomeres by Rap 1 protein. Genes & Dev. 10: 1297-1309. [DOI] [PubMed] [Google Scholar]
- Marcand S., Gilson, E., and Shore, D. 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275: 986-990. [DOI] [PubMed] [Google Scholar]
- Marini F., Pellicioli, A., Paciotti, V., Lucchini, G., Plevani, P., Stern, D.F., and Foiani, M. 1997. A role for DNA primase in coupling DNA replication to DNA damage response. EMBO J. 16: 639-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern M.J. and Blackburn, E.H. 1995. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature 376: 403-409. [DOI] [PubMed] [Google Scholar]
- McEachern M.J., Krauskopf, A., and Blackburn, E.H. 2000. Telomeres and their control. Annu. Rev. Genet. 34: 331-358. [DOI] [PubMed] [Google Scholar]
- Moretti P., Freeman, K., Coodly, L., and Shore, D. 1994. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes & Dev. 8: 2257-2269. [DOI] [PubMed] [Google Scholar]
- Nugent C.I., Hughes, T.R., Lue, N.F., and Lundblad, V. 1996. Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274: 249-252. [DOI] [PubMed] [Google Scholar]
- Pennock E., Buckley, K., and Lundblad, V. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104: 387-396. [DOI] [PubMed] [Google Scholar]
- Qi H. and Zakian, V.A. 2000. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase α and the telomerase-associated est1 protein. Genes & Dev. 14: 1777-1788. [PMC free article] [PubMed] [Google Scholar]
- Ray A. and Runge, K.W. 1999a. Varying the number of telomere-bound proteins does not alter telomere length in tel1Delta cells. Proc. Natl. Acad. Sci. 96: 15044-15049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1999b. The yeast telomere length counting machinery is sensitive to sequences at the telomere-nontelomere junction. Mol. Cell. Biol. 19: 31-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S., Karamysheva, Z., Wang, L., Shippen, D.E., and Price, C.M. 2002. Interactions between telomerase and primase physically link the telomere and chromosome replication machinery. Mol. Cell. Biol. 22: 5859-5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie K.B. and Petes, T.D. 2000. The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics 155: 475-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie K.B., Mallory, J.C., and Petes, T.D. 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 6065-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell L.L. and Zakian, V.A. 1993. Loss of a yeast telomere: Arrest, recovery, and chromosome loss. Cell 75: 729-739. [DOI] [PubMed] [Google Scholar]
- Shore D. 2001. Telomeric chromatin: Replicating and wrapping up chromosome ends. Curr. Opin. Genet. Dev. 11: 189-198. [DOI] [PubMed] [Google Scholar]
- Singer M.S. and Gottschling, D.E. 1994. TLC1: Template RNA component of Saccharomyces cerevisiae telomerase. Science 266: 404-409. [DOI] [PubMed] [Google Scholar]
- Smith C.D. and Blackburn, E.H. 1999. Uncapping and deregulation of telomeres lead to detrimental cellular consequences in yeast. J. Cell Biol. 145: 203-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart A.K., Teng, S.C., and Zakian, V.A. 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297: 1023-1026. [DOI] [PubMed] [Google Scholar]
- Teng S.C., Chang, J., McCowan, B., and Zakian, V.A. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell 6: 947-952. [DOI] [PubMed] [Google Scholar]
- Thomas B.J. and Rothstein, R. 1989. The genetic control of direct-repeat recombination in Saccharomyces: The effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics 123: 725-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y., Taggart, A.K., and Zakian, V.A. 2001. The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 11: 1328-1335. [DOI] [PubMed] [Google Scholar]
- Wotton D. and Shore, D. 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes & Dev. 11: 748-760. [DOI] [PubMed] [Google Scholar]