Abstract
Mitotic exit in Saccharomyces cerevisiae and septation in Schizosaccharomyces pombe are regulated by a conserved signaling network called the mitotic exit and septum initiation networks (SIN), respectively. The network is active on one of the two anaphase B spindle-pole bodies (SPBs). Whereas the inherent asymmetry of growth by budding accounts for elements of the asymmetry in S. cerevisiae, it has been unclear how, or why, the pathway is asymmetric in S. pombe. We show that elements of SPB duplication in S. pombe are conservative, and that the SIN is active on the new SPB. SIN association with the new SPB persists after transient depolymerization of microtubules. The localization of the NIMA-related kinase, Fin1, reveals further complexity in SPB inheritance. Fin1 associates with the SPB bearing the older components in all cells and with the “new” SPB in half of the population. Fin1 only binds the new SPB when this new SPB has arisen from the duplication of an SPB that is two or more cycles old. Thus, each of the four SPBs generated over two consecutive cell cycles are different, because they have distinct fates in the next cell cycle. Fin1 binds the SPB once the SIN is active and the association requires the SIN inhibitors Byr4 and Cdc16. Fin1 physically associates with Byr4. Compromising Fin1 function leads to SIN activation on both anaphase B SPBs and promotes septation, indicating that Fin1 restrains SIN activity on the old SPB.
Keywords: S. pombe, NIMA kinase, Fin1, mitosis, SIN, centrosome, SPB
Commitment to mitosis is promoted by the activation of the Cdk1/cyclinB protein kinase complex called MPF (Nurse 1990). MPF activation promotes a cascade of phosphorylation events that results in the formation of the mitotic spindle. Whereas some of these are due to direct phosphorylation of key substrates by MPF itself, for example, Blangy et al. (1995), the majority arise from the activity of kinases that are activated by MPF. These kinases include polo, aurora, and NIMA-related kinases (Nigg 2001).
Lower eukaryotes characterized to date contain a single NIMA-related kinase, whereas a family of NIMA-related kinases or Neks exist in higher eukaryotes (O'Connell et al. 2003). Aspergillis nidulans NIMA mutants arrest cell cycle progression in G2 with active MPF (Ye et al. 1995). In contrast, deletion of the analogous kinase from either budding or fission yeast is not lethal (Jones and Rosamond 1990; Krien et al. 1998). However, it does delay commitment to mitosis in fission yeast (Krien et al. 1998). Despite the ability of cells to live without these genes, the phenotype of recessive conditional mutants in fission yeast suggests that fission yeast NIMA, Fin1, plays an important role in modulating spindle formation and spindle-pole maturation (Grallert and Hagan 2002). The spindle formation phenotype of conditional fin1 mutants is reminiscent of the consequences of manipulation of one of the human and Xenopus NIMA homologs, Nek2. Nek2 localizes to the centrosome and is required for the maturation of centrosomes in Xenopus egg extracts (Fry 2002). Overproduction of Nek2 or injection of Nek2 antibodies affects the cohesion between centrioles. This is reminiscent of the requirement for the function of the Chlamydomonas NIMA-related kinase, FA2, in breaking the connection between the basal bodies and the flagella (Mahjoub et al. 2002). Mitotic functions, some at centrosomes, are now being ascribed to more Nek molecules (Belham et al. 2003; Swenson et al. 2003), emphasizing the important role played by NIMA kinases in cell cycle control and the function of microtubule organizing centers.
The association of NIMA kinases with the spindle poles in diverse systems resembles the behavior of the more intensively studied mitotic kinases, polo and aurora, which are also found on spindle poles (Nigg 2001). Studies from fission yeast and humans suggest that the activation of polo kinase on the spindle pole is a critical step in regulating the autocatalytic feedback loop that promotes full-scale activation of MPF to drive commitment to mitosis (Jackman et al. 2003; MacIver et al. 2003). This amplification loop highlights the interplay between different mitotic kinases, as both the recruitment of polo kinase to the spindle pole and the activity of this feedback loop in fission yeast are modulated by the NIMA kinase Fin1 (Grallert and Hagan 2002).
Events at the spindle pole also play critical roles in controlling mitotic exit (for review of SIN and MEN, see Bardin and Amon 2001; Simanis 2003). Mitotic exit is controlled by the activity of a conserved regulatory network called the mitotic exit network (MEN) in budding yeast and the septum initiation network (SIN) in fission yeast. The critical events in modulating MEN/SIN activity occur on the spindle-pole body (SPB). In budding yeast, they culminate in the release of the protein phosphatase Cdc14 from the nucleolus, which then dephosphorylates multiple targets, including those that were phosphorylated by Cdk1/cyclin B complexes at earlier stages of mitosis to promote mitotic exit. Although the precise details may differ, an analagous phosphatase exists in fission yeast, and its release from the nucleolus is also modulated by SIN activity (Bardin and Amon 2001; Cueille et al. 2001; Trautmann et al. 2001; Simanis 2003).
The SIN is anchored to the SPB via two scaffold proteins, Cdc11 and Sid4. SIN activity is determined by the status of a small G protein of the ras superfamily, Spg1. SPB-associated Spg1 is in a GDP-bound, inactive state throughout interphase. Upon mitotic commitment, there is a switch in the nucleotide status of the Spg1 associated with both SPBs, and it is in a GTP-bound state on both poles. This persists until early anaphase B, when the Spg1 on one SPB switches to a GDP-bound state. As this coincides with the formation of a GAP complex for Spg1 on this SPB, it is assumed that the GTP is hydrolyzed to create GDP Spg1. This effectively turns off the SIN on one of the two anaphase B SPBs. SIN activity persists on the other SPB until cytokinesis is complete. Active Spg1 binds the protein kinase Cdc7, and this activates the downstream components of the network. Analysis of both the MEN and the SIN highlight the importance of feedback loops in this conserved network. Components that are placed later in the network appear to feed back to either activate or inhibit components that map by genetics and cytology to earlier stages of the network (Pereira et al. 2002; Krapp et al. 2003; for review, see Simanis 2003).
The activation of the MEN/SIN on one of the two anaphase B SPBs highlights a distinction between two, apparently morphologically identical, SPBs. In budding yeast, the identity of the SPB that will host the active MEN is determined by the location of the SPB relative to the mother/bud axis within this highly asymmetric cell (Pereira et al. 2001). The SPB that faces the daughter cell binds the MEN inhibitors Bub2 and Bfa1 (Pereira et al. 2001), whereas the downstream MEN effector Cdc15 is only seen on the SPB in the mother cell prior to mitotic exit (Menssen et al. 2001). The activation of the MEN on the SPB in the daughter cell appears to be modulated by the interaction of astral microtubules with the bud neck and cortex (for review, see Simanis 2003).
In contrast to budding yeast, fission yeast cells divide symmetrically, and yet the SIN is still located asymmetrically on one of the two SPBs. One obvious feature that could generate asymmetric markers within a fission yeast cell is the fact that one end was produced by cytokinesis at the end of the previous cell cycle, whereas the other persists from the previous cycle. As both ends of the cell have a different history, they could send different signals to the SPBs to direct asymmetric activation of the SIN. However, the SIN-active SPB faces these two ends with equal frequency (M. Sohrmann, A. Grallert, I. Hagan, and V. Simanis, unpubl.). In this work, we show that the fission yeast SPB takes more than one cell cycle to mature, so that each of the four SPBs of two daughter cells has a distinct fate in the next cell cycle. This aspect of SPB behavior was revealed by the age dependence of the ability of the SPB to recruit the kinase Fin1. We also show that it is the new SPB that hosts the active SIN in anaphase B and that SIN inhibition on the old SPB is mediated, in part, by the recruitment of Fin1 to that SPB by the SIN GAP component Byr4.
Results
Fin1 associates with the mitotic apparatus in a stage-dependent manner
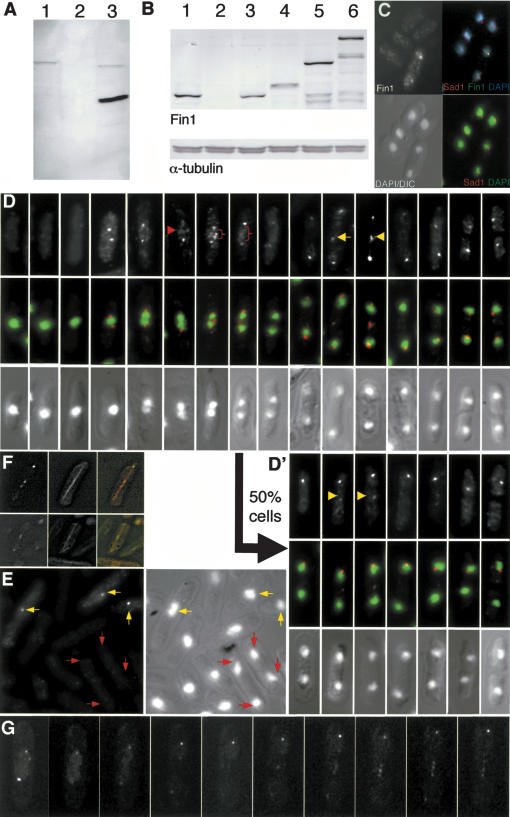
Two polyclonal antibodies were raised against the noncatalytic, C-terminal 441 amino acids of the Fin1. As affinity purification of antibodies from both sera gave the same patterns on Western blots and immunolocalization (data not shown), results with one, which we refer to as αFIN1, are described here. αFIN1 antibodies recognized a single 76-kD band in wild-type extracts (Fig. 1A, lane 1). This band was absent from a fin1.Δ strain from which the fin1+ gene had been deleted (Fig. 1A, lane 2) and an extra band at 46 kD was seen in wild-type cells expressing the 441 C-terminal amino acids (Fig. 1A, lane 3). Costaining cells with αFIN1 and antibodies to the SPB marker Sad1 (Hagan and Yanagida 1995) showed that Fin1 associated with the SPBs of dividing, but not interphase cells (Fig. 1C), stained the region between the separating chromosome masses in early anaphase B (red arrows in Fig. 1D), and gave discrete, but variable, patterns between late anaphase B SPBs (yellow arrows in Fig. 1D′). To check that all of these staining patterns arose from recognition of Fin1 by αFIN1 antibodies, we mixed fin1.Δ nda3+ cells with fin1+ nda3.KM311 cells and stained the mixed culture with αFIN1 antibodies. The β tubulin mutation in the nda3.KM311 cells means that they accumulate in prophase with condensed chromosomes at 20°C (Uemura et al. 1987). Fin1 binds to the SPB in this arrest (data not shown). A signal was only seen on the SPBs following αFIN1 staining in the nda3.KM311-elongated, mononucleated, cells that had condensed chromatin (Fig. 1E, yellow arrows). It was not seen in the anaphase/G1 SPBs of fin1.Δ cells (Fig. 1E, red arrows). This established that the patterns observed after αFIN1 staining could be attributed to the recognition of Fin1. Furthermore, because the background fluorescence was higher throughout the arrested nda3.KM311 cells, this experiment suggests that αFIN1 antibodies can detect a cytoplasmic pool of Fin1. Fin1 associated with the SPB as soon as two distinct SPBs could be detected. In one-half of the population, SPB association persisted until septation was complete (Fig. 1D). In the other, it was lost from one of the two SPBs midway through anaphase B (Fig. 1D′). Whether or not Fin1 associated with one of both of the anaphase B/G1 SPBs, the intensity of the staining increased from the middle of anaphase B to peak just before septation.
Figure 1.
Fin1 distribution in mitosis. (A,B) Western blot analysis of whole-cell extracts probed with Fin1-specific, affinity-purified, polyclonal, Sheep antibodies (αFIN1), or monoclonal antibodies to α-tubulin as indicated. Extracts were prepared from the following strains: (A) fin1+ (lane 1); fin1.Δ (lane 2); fin1+ pRep41fin1 282-722 amino acids (induced; lane 3). (B) fin1+ (lane 1); fin1.Δ (lane 2); fin1.Δ leu1::fin1+ (lane 3); fin1.Δ leu1::fin1.PkC (lane 4); fin1.GFP (lane 5); fin1.4GFP (lane 6). (C) Wild-type cells were stained with αFIN1 and anti-Sad1 antibodies and DAPI as indicated by labeling on the figure. (D,D′) Fin1 staining colocalized with the SPB staining by Sad1. αFIN1 staining varied with cell cycle progression. (Top) αFIN1 staining. (Bottom) DAPI/DIC images. (Middle) Merged αFIN1 and DAPI signals. Staining of the region between the anaphase A chromatin is indicated by the red arrow and brackets, whereas staining between separating anaphase B SPBs is indicated by yellow arrows. No staining was detected in interphase cells. αFIN1 stained the mitotic SPBs in a stage-dependent manner. The arrow from D to D′ indicates that half of the cells proceed from mid-anaphase B with both SPBs staining (D), whereas in the other half of the population, only one SPB stained (D′). In all cells, the intensity of the SPB signal/s increased markedly from the middle of anaphase B to peak just before its disappearance upon cell fission. Even when αFIN1 stained both SPBs, the signal on one was noticeably stronger. The 50:50 ratio of one SPB:two SPB staining of Fin1 was observed in h-, h+ and h90 mating-type backgrounds. (E) fin1+ nda3.KM311 cells were incubated at 20°C for 8 h before being mixed with fin1.Δnda3+ cells and processed for αFIN1 staining. Yellow arrows indicate the prophase arrested nda3.KM311 cells that show strong SPB staining, whereas the red arrows indicate the anaphase B/G1 fin1.Δ cells that did not show the αFIN1 signal that would be expected for cells at this stage of the cell cycle (D,D′). Note the cytoplasmic staining in the nda3.KM311 cells, which is absent from the fin1.Δ cells. In other words, fin1.Δ cells did not show staining with αFIN1 antibodies. (F,G) Fin1.4GFP associated with the central spindle. (F) A Z series of images of Fin1.4GFP and α-tubulin CFP signals from the same living cells was captured by wide-field microscopy and processed by nearest-neighbor deconvolution. The image shows a maximum projection of the processed images. (G) A series of spinning-disk confocal images of a dividing fin1.4GFP cell showing punctate GFP signals along the spindle between the two SPBs.
We also generated strains in which Fin1 could be localized by alternative approaches. Three Pk epitopes were fused to the 3′ end a fin1+ ORF integrated at the leu1+ locus, whereas one and four copies of GFP were fused to the 3′ end of the endogenous fin1+ gene in separate strains (Fig. 1B, lanes 5,6). With two exceptions, localizing the modified proteins with αFIN1 or Fin1.CPk with anti-Pk antibodies gave the same pattern as Figure 1D. These exceptions were the presence of several dots within the nucleus and staining of G2 SPBs (data not shown). These data are consistent with a previous study that reported that both Fin1.HA and ectopically expressed Fin1.GFP associate with G2 SPBs (Krien et al. 2002). It is unclear why the fusion of a tag to Fin1 should influence its localization in this way; however, the critical fact is that the staining between the anaphase B SPBs and the frequency with which Fin1 associated with one or two anaphase B/G1 SPBs was the same in these tagged strains as seen in wild-type cells with αFIN1.
We therefore analyzed fin1.4GFP cells by time-lapse fluorescence microscopy (Fig. 1G). Expression of an α-tubulin-CFP fusion protein (Glynn et al. 2001) did not affect Fin1.4GFP distribution (Fig. 1F,G). Deconvolution of stacks of Z series taken through such cells established that the Fin1.4GFP staining between the anaphase B SPBs arose from the association of Fin1 with the central spindle (Fig. 1F). Like the αFIN1 staining of fixed cells, Fin1.4GFP fluorescence on these anaphase B spindles of living cells was rarely uniform and generally punctate (Fig. 1D,D′,G).
Fin1 leaves SPBs upon completion of septation
Rapidly growing Schizosaccharomyces pombe cells initiate and generally finish S phase before completion of cytokinesis (Nurse et al. 1976). Loss of Fin1 staining from the SPB could therefore be linked to either the completion of S phase or the completion of cytokinesis. We therefore stained cells in which progression through START into S phase of the next cell cycle was blocked by mutation of the transcription factor Cdc10 (Nurse et al. 1976). Fin1 was absent from these arrested cdc10.v50 cells (Marks et al. 1992; data not shown). It was also absent from newly divided cig1.Δ cig2.Δ puc1.Δ cells (Martin-Castellanos et al. 2000), in which G1 phase extends through cytokinesis into the next cell cycle (data not shown). Thus, Fin1 leaves the SPB as a consequence of the completion of septation rather than the transit of START or the initiation of S phase.
When Fin1 associates with one SPB, it always binds to the old SPB
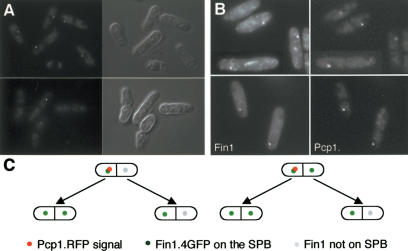
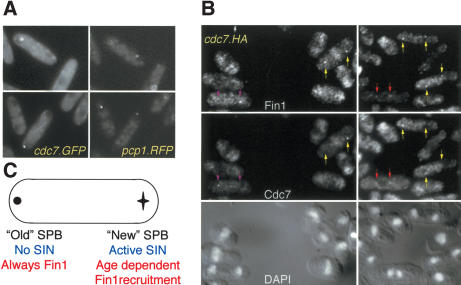
The red fluorescent protein DsRed takes several hours to fold into an actively fluorescing molecule. This property has been exploited to great effect to differentiate between the old and new SPBs in budding yeast (Pereira et al. 2001; Tanaka et al. 2002). In these studies, refeeding starved, Spc42.RFP cells generated a dividing population in which the “old” SPB that persisted throughout the G1 arrest was fluorescent, whereas the “new” SPB that formed in the new cell cycle was not (Pereira et al. 2001). To recapitulate this approach in fission yeast, we fused DsRed to the C terminus of the S. pombe SPB component Pcp1 (Flory et al. 2002) to generate Pcp1.RFP. Very few cells in a pcp1.RFP log-phase culture had fluorescent SPBs, whereas a starvation/refeed regime generated anaphase B/septating cells, in which one SPB was fluorescent and the other was not (Fig. 2A). This asymmetry indicated that partition of actively fluorescing, aged Pcp1.RFP, was conservative. Imaging fin1.4GFP pcp1.RFP cells established that when Fin1 was confined to one of the two anaphase B SPBs, the green and red fluorescent signals were always on the same SPB (Fig. 2B,C, left). Thus, Fin1 always bound the SPB that inherited the old Pcp1.RFP, and so, presumably, was the older of the two SPBs. In half of the cells, Fin1 was also on the RFP-negative SPB, whereas in the other half it was not.
Figure 2.
SPB history determined Fin1 affinity for the SPB. (A) pcp1.RFP cells were starved of nitrogen by switching from supplemented EMM2 (containing ammonium chloride) to EMM-N (no ammonium chloride) for 16 h before resuspension in EMM2. At 8 h, images of the red fluorescence (left) and DIC (right) were taken. The single acentric spots in the largest cells highlight only one of the two anaphase/G1 SPBs. (B) fin1.4GFP pcp1.RFP cells were processed as in A, and the fluorescence signals captured as indicated. When Fin1 was on one of the two anaphase B SPBs, it was always on the same SPB as the Pcp1.RFP signal. (C) fin1.4GFP pcp1.RFP cells were grown on gas-permeable membranes under an EMM2 agarose pad, and cells in which the old SPB could be identified by the RFP signal were followed until both daughter cells had divided to determine the relationship between SPB inheritance and Fin1 recruitment to the SPB. Images were captured in 1-μm steps every 5 min, and each slice monitored to identify the SPB. The cartoon summarizes observations of 15 such pedigrees.
Fin1 affinity for the new SPB is also determined by SPB age and history
We next asked whether the affinity of Fin1 for the new SPB in cells arising from a parent in which both SPBs had stained in the previous division was determined by the history of that SPB. We did this by starving fin1.4GFP pcp1.RFP cells to differentially label the old SPB with fluorescent Pcp1.RFP. Refed cells were then placed underneath an agar pad on an oxygen-permeable membrane, and the SPB inheritance pattern was monitored with a widefield fluorescence microscope. As cells immediately form microcolonies under these conditions (Hagan et al. 1990), we assume that this provides a way of following the fate of cells that inherit particular SPBs without adversely affecting cell growth or division. We first addressed the fate of cells that were produced by division of cells in which only one of the two SPBs was fluorescent. Both anaphase B SPBs bound Fin1.4GFP in the cell that had inherited the old SPB in the previous division, whereas only one of the anaphase B/G1 SPBs bound Fin1.4GFP if the cell had inherited the nonfluorescent new SPB from the previous cell division (Fig. 2C, left). We then followed the fate of cells produced by division of a parent in which both of the anaphase B SPBs had bound Fin1.4GFP. We saw exactly the same relationship between inheritance of the old and new SPB and recruitment of Fin1.4GFP in the next cell cycle (Fig. 2C, right). In other words, old poles always gave rise to cells in which both anaphase B/G1 SPBs recruited Fin1, whereas new poles always gave rise to cells in which Fin1 was recruited to only one SPB, irrespective of whether they had bound Fin1.4GFP in the previous division or not.
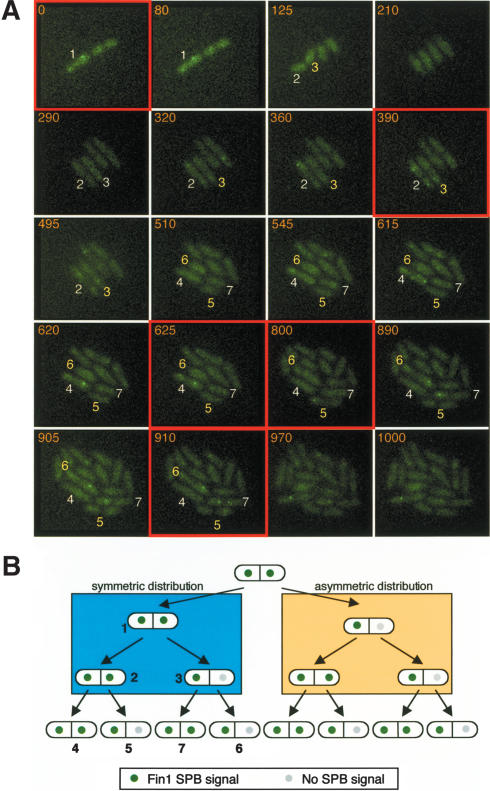
This inheritance pattern was unlikely to arise because of the inclusion of Pcp1.RFP in the SPBs, as it was recapitulated in pedigree analysis of fin1.4GFP cells (Fig. 3). The green dots in the pedigrees in Figure 3B represent SPBs that bound Fin1, gray dots those which did not, and the numbers in Figure 3B refer to the respective cells in Figure 3A. Two distinct inheritance patterns were seen, depending upon whether Fin1 had associated with only one (Fig. 3B, orange block) or both (Fig. 3B, blue block) SPBs in the previous division. When Fin1 had been restricted to one SPB in the previous division (Fig. 3B, orange block), the cell that inherited the Fin1 positive, old, SPB always had Fin1 associated with both anaphase B/G1 SPBs in the subsequent division. In contrast, in cells that had inherited the Fin1 negative, new SPB, Fin1 only associated with one SPB. In the second pattern, progeny arising from a cell in which Fin1 had been associated with both SPBs in the previous cell cycle gave rise to a population that showed a 1:1 ratio of cells with one or both SPBs staining during their divisions (Fig. 3B, blue block).
Figure 3.
Pedigree analysis establishes that Fin1 recruitment to the SPB is determined by SPB history. (A) fin1.4GFP cells were imaged as described in the legend for Figure 2C. Because the cells were sometimes pushed out of the focal plane, individual slices were examined to track the fluorescent SPBs. Stacks were compressed with the maximum projection algorithm to generate the images shown in the figure. The cell number shown in the figure corresponds to the cell number in the cell-fate pedigree in B. In all cases, the cell to the left of the one above arises from the left-hand half of its parent and the cell on the right from the right-hand half. Thus, for cell 3, cell 6 inherited an SPB with which Fin1 had not associated in the previous division, whereas cell 7 inherited an SPB that had been Fin1 positive in the previous cell cycle. The critical panels that highlight the lineage from 1 to 7 are indicated by red boxes. (B) A summary of fin1.4GFP imaging data from eight four-generation pedigrees and five three-generation pedigrees.
The division-dependent maturation of the SPB could occur through progressive accumulation of a marker or molecule until it reached a threshold level, after which it would dictate either that the SPB should bind Fin1, or that both it and its progeny should bind Fin1. Alternatively, the pattern could arise because of a change that is induced by transit of a specific cell cycle stage/s. Forcing cells to speed up transit through a cell cycle would differentiate between these two possibilities, because it would shorten the time between cell cycle events, while maintaining the order of these cell cycle transitions. We therefore scored the frequency with which Fin1 stained one or two anaphase B/G1 SPBs in wee1.50 cells, in which the size control over commitment to mitosis is removed by heat-induced inactivation of the MPF-inhibiting kinase Wee1. Removal of this size control accelerates cells into mitosis (Nurse 1975). The ratio of cells with Fin1 on one or both anaphase B/G1 SPBs was monitored every 30 min for 3 h after shift to 36°C, and was unaffected by the acceleration into mitosis (data not shown). This established that it is transit through the cell cycle rather than the precise age of the SPB that determined Fin1 affinity for the SPB.
We draw the following conclusions from this data. A new SPB that is produced by a SPB that was itself a new SPB in the previous division does not bind Fin1 in the cell cycle in which it is generated. It gains the competence to bind Fin1 during, or after, passage through mitosis into the next cell cycle. However, it cannot confer the ability to bind Fin1 upon its daughter SPB in this, the second cell cycle of its existence, but acquires this ability to dictate that its daughter SPB must bind Fin1 after passage into its third cell cycle.
Mutation of SIN components blocks Fin1 association with the SPB
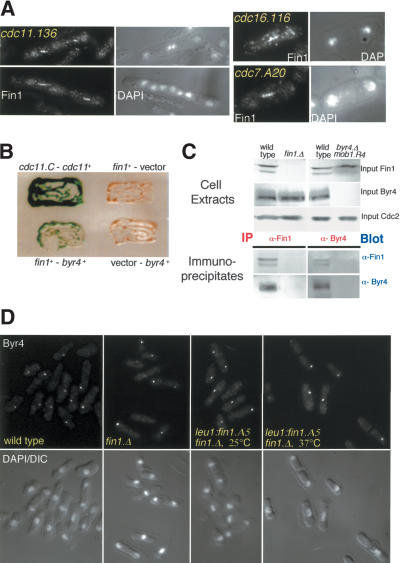
The apparent complexity of the rules governing Fin1 association with the SPB suggested that understanding the molecular basis of this phenomenon would give greater insight into Fin1 function. We therefore asked which cell cycle regulators and SPB components were required for Fin1 association with the SPB. We found that Fin1 association with the SPB required prior activation of MPF, as it did not occur in cdc2.33 or cdc25.22 mutants at the restrictive temperature (Table 1). Fin1 association was not affected by mutation of the SPB components Sad1, Cut12, or Cut11, however, it was blocked by mutation of either cdc11 or sid4 (Table 1; Fig. 4A).
Table 1.
Fin1 staining in sin mutants: where conditional mutants are used, analysis was done at the restrictive temperature
| Strain | Fin1 on SPB | Strain | Fin1 on SPB? |
|---|---|---|---|
| cdc25.22 | − | cdc16.116 | − |
| cdc2.33 | − | cdc16.Δ mob1.R4 | − |
| cdc10.V50 | − | cdc16.Δ sid2.1 | − |
| cut12.1 | + | byr4.Δ germinating spores | − |
| nuc2.663 | + | byr4.Δ mob1.R4 | − |
| cut7.24 | + | spg1.B8 | − |
| sad1.1 | + | cdc7.A20, cdc7.24 | − |
| cut11.1 | + | cdc7.Δ germinating spores | − |
| nda3.KM311 | + | sid1.239 | − |
| cdc11.123, cdc11.136 | cdc14.118 | − | |
| − | sid2.250 | − | |
| sid4.K12 | − | mob1.R4 | − |
Figure 4.
Fin1 binds Byr4. (A) Fin1 failed to associate with SPBs when the SIN was either inactive or hyperactive. cdc16.116 and cdc7.A20 were shifted to 36°C for 60 min, and cdc11.136 for 180 min before being stained with αFIN1 antibodies. The lack of SPB staining was not due to errors in processing, as the central spindle staining was clearly visible. (B) S. cerevisiae transformants containing two-hybrid plasmids transferred to Nitrocellulose and incubated on X-gal-containing filters. Bait and prey plasmids are indicated (the interaction between the cdc11 C terminus and full-length cdc11+ is a positive control [Krapp et al. 2001]). Tests using fin1+ as bait, and spg1+, cdc16+, sid1+, sid2+, mob1+, cdc11+, cdc14+, dma1+, zfs1+, and cdc7+ as prey revealed no significant interaction. A weak interaction above background was noted with sid4+ as prey (data not shown). (C) Immunoprecipitation reactions were performed with αFIN1 or α-Byr4 antibodies and blots of the immunocomplexes were probed with each antibody. The inclusion of the mob1.R4 mutation attenuated the otherwise lethal hyperactivation of the SIN hyperactivity arising from deletion of byr4+, and so, enabled us to demonstrate the specificity of the Byr4 immunoprecipitation reaction with a strain that lacked Byr4, and yet divided normally. (D) Byr4 association with the SPB was unaffected by deletion or mutation of fin1+. Wild-type and fin1.Δ cells were grown at 25°C, whereas fin1.Δ leu1::fin1.A5 cells were grown at 25°C before shift to 37°C for 3 h to compromise Fin1 function.
Fin1 binds to the SPB via association with the Spg1 GAP component Byr4
The dependence of Fin1 upon Sid4 and Cdc11 for recruitment to the SPB suggested that it either bound directly to these scaffold molecules or to the SIN components that they recruit to the SPB (Chang and Gould 2000; Krapp et al. 2001). We therefore screened Fin1 against all known SIN components in the budding yeast two-hybrid assay. The only association we detected was between Fin1 and the component of the Spg1 GAP complex Byr4 (Fig. 4B). Fin1 was found in isolated Byr4 immunocomplexes and Byr4 was found in isolated Fin1 immunocomplexes (Fig. 4C), suggesting that the two-hybrid interaction reflected a genuine association between these two molecules. Fin1 failed to bind to the SPB when byr4+ was deleted (Table 1). The inability of Byr4 to bind to the SPB when its partner molecule, Cdc16, is defective (Li et al. 2000), was mimicked by Fin1, as it did not bind the SPBs of cdc16.116 cells (Fig. 4A). To rule out the possibility that this failure to associate with cdc16 or byr4-defective SPBs was the consequence of a feedback loop arising from a hyperactive SIN, we utilized strains in which either cdc16 or byr4 had been deleted, and cells were kept alive by attenuation of the SIN through mutation of the downstream effector complex Sid2/Mob1 (Cueille et al. 2001; Fournier et al. 2001). Fin1 still failed to localize in these strains (Table 1), suggesting that the inability to bind in cdc16 mutants was not due to the hyperactivation of Sid2 or effectors that function after it. The association of Byr4 with the SPB is independent of the activity of the SIN effectors, Cdc7, Sid2/Mob1, and Sid1/Cdc14 (Li et al. 2000). However, the same was not true of Fin1, as it failed to associate with SPBs when these proteins were rendered inactive in conditional mutants (Table 1; Fig. 4A), suggesting that prior activation of SIN signaling is required for Fin1 to associate with SPBs. Whereas Fin1 required Byr4 to associate with the SPB, the converse was not true, as mutation, deletion, or overexpression of fin1+ did not affect the affinity of Byr4 for the SPB (Fig. 4D; data not shown). We draw two conclusions from these data; first, association of Fin1 with the SPB requires activation of the SIN; second, recruitment of Fin1 to the SPB requires the SIN inhibitor Byr4, with which it interacts.
The SIN is active on the new SPB
The association between Fin1 and a SIN inhibitor suggested that Fin1 may contribute to regulation of SIN activity. We therefore analyzed pcp1.RFP cdc7.GFP cells to ask whether the old or the new SPBs host the active SIN in anaphase B. This revealed that the GFP signal of the active SIN was always on the new SPB (the one that lacked any RFP signal, Fig. 5A). This conclusion was confirmed by indirect immunofluorescence microscopy of a cdc7.HA strain to localize Cdc7 and Fin1 in the same cell (Fig. 5B).
Figure 5.
The SIN was active on the new SPB. (A) pcp1.RFP cdc7.GFP cells were processed as described in the legend for Figure 2A. (B) Cdc7.HA cells were stained with αFIN1 (top) and 12CA5 (bottom) antibodies to reveal the distribution of Fin1 and Cdc7, respectively. Colored arrows indicate the SPBs of the following cell types: (Red) Early anaphase B in which both Fin1 and Cdc7 are on both SPBs; (yellow) late anaphase B cells in which Fin1 is on both SPBs and Cdc7 is only on one; (purple) late anaphase B cells in which Fin1 is on only one SPB and this is opposite pole to the SPB that harbors Cdc7. (C) A cartoon summarizing the findings of Figure 2 and this figure.
In an unperturbed budding-yeast cell cycle, the old SPB is inherited by the bud/daughter, whereas the new one stays in the mother. The SIN equivalent, the MEN, is inhibited on the old SPB that faces the bud and active on the new SPB in the mother cell (Pereira et al. 2001; Menssen et al. 2001). Transient depolymerization of microtubules abolishes this bias, and the new SPB enters the bud as frequently as the old. Despite this randomization of SPB inheritance, the MEN inhibitors Bfa1 and Bub2 are still only found on the SPB that faces the bud tip (Pereira et al. 2001). We therefore asked whether a transient depolymerization of microtubules would mimic the effect seen in budding yeast and lead to SIN activation on the old and new SPBs in equal measure. To do this, we differentiated between the SPB that hosted the active SIN from the one upon which the SIN was inactive by following the association of Cdc7.GFP with the SPB. The Cdc7.GFP fusion protein that marks the SPB that hosts the active SIN. 25 μg ml-1 CBZ was added to refed cdc7.GFP pcp1.RFP cells as they progressed through mitosis. At 15 min, the CBZ containing medium was replaced by drug-free medium, and the location of the SIN relative to the old SPB was determined a further 10 and 15 min later. At these times, the mitotic cells that had reformed a spindle after the removal of the drug were in anaphase B. The SIN remained on the new SPB. Thus, in contrast to the situation with the budding yeast MEN in S. pombe, it is the age of the SPB rather than its association with the cortex via microtubules that determines which SPB hosts the active SIN (Fig. 5C).
The asymmetry of SIN inactivation on one SPB is dependent on Fin1 function
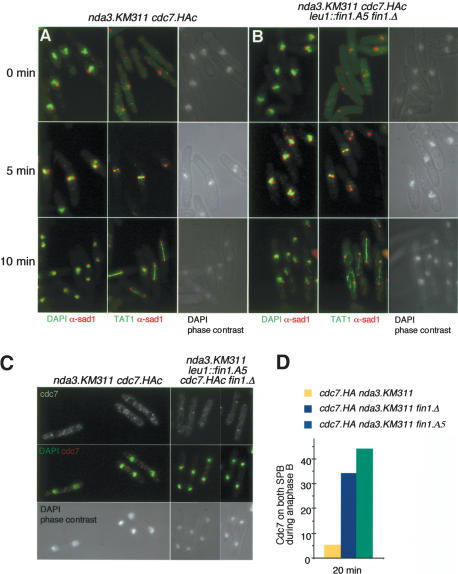
The association between Fin1 and a SIN inhibitor and its recruitment to the SPB, upon which the SIN is inhibited (Sohrmann et al. 1998; Cerutti and Simanis 1999), suggested that Fin1 may regulate SIN activity. However, we were unable to address this possibility by simply inactivating Fin1 in a temperature-sensitive mutant and asking whether SIN activation remained asymmetric, because these mutants are unable to form a mitotic spindle after shift to the restrictive temperature (Grallert and Hagan 2002). The mutant cells would not, therefore, be able to get into the anaphase state where we could ask the question. To circumvent this problem, we asked whether fin1 mutants could undergo a normal anaphase if they had been allowed to enter mitosis at the permissive temperature? We exploited the exquisite reversibility of nda3.KM311β-tubulin mutant. nda3.KM311 cells accumulate in prophase without microtubules at 20°C. Upon rewarming to 36°C, a spindle reforms, and within 10 min, most cells are in anaphase (Fig. 6A) (Uemura et al. 1987). If the fin1.A5 execution point were after that for nda3.KM311, then fin1.A5 nda3.KM311 mutants should also execute a normal anaphase after accumulating in prophase at 20°C. They did (Fig. 6B). We could therefore ask whether Fin1 function affected the symmetry of SIN activation in anaphase of a fin1.A5 nda3.KM311cdc7.HA strain that had been subjected to the arrest/release regime. Whereas Cdc7.HA was asymmetric on the nda3.KM311 cdc7.HA anaphase B spindles, inactivation of Fin1 in the fin1.A5 nda3.KM311 cdc7.HA strain resulted in recruitment of Cdc7.HA to both anaphase B SPBs in 44% of cells (Fig. 6C,D) compared with 5.5% in the fin1+ control. This showed that Fin1 function was required to ensure the asymmetry of SIN activation on the SPBs.
Figure 6.
Compromising Fin1 function derepresses Spg1 on old SPBs. (A,B) Fin1 is not required for spindle formation following release from a prophase arrest imposed by microtubule depolymerization through transient arrest with the cold-sensitive nda3.KM311 mutation (i.e., the fin1- execution point is before that of nda3-). (A) nda3.KM311 cdc7.HA cells were incubated at 20°C for 8 h to inactivate the β tubulin product of the nda3 gene before being rewarmed to 37°C. Samples were processed to visualize the microtubules; SPBs and chromatin are as indicated. (B) Cells were devoid of microtubules at the restrictive temperature (0°C), but spindles formed within 5 min of return to the permissive temperature, and cells were in anaphase B 10 min after the shift to 37°C. Identical behavior was seen in nda3.KM311 cdc7.HA fin1.Δleu1::fin1.A5 cells. (C) The analysis was repeated and cells stained to show the distribution of Cdc7 with 12CA5 antibodies at 20 min after return to the permissive temperature. (D) The frequency with which Cdc7 was seen on both anaphase B/G1 SPBs was scored at the indicated time in the strains shown in A and in a nda3.KM311 cdc7.HA fin1.Δ strain that had been treated in the same way as the nda3.KM311 cdc7.HA fin1.Δleu1::fin1.A5 strain. Analogous results were obtained with strains in which cdc7+ was tagged with GFP rather than the 12CA5 epitope (data not shown).
Loss of Fin1 function promotes septation
Whereas the previous experiment established that Fin1 function contributes to the attenuation of Spg1 function on the old SPB, it did not establish whether this attenuation also influenced the strength of the positive signal that passed through the SIN network. We used two approaches to address this possibility.
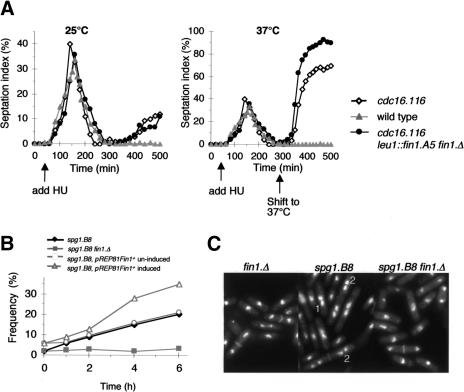
Inactivation of Cdc16 in S-phase-arrested cells induces septation in a proportion of the cells in the culture (Cerutti and Simanis 1999). We examined the effects of this protocol on a fin1.A5 cdc16.116 mutant to assess whether the simultaneous loss of Fin1 and Cdc16 function were additive. If they were, it would indicate that Fin1 normally attenuated the flux through the SIN. Hydroxyurea was added to cultures of small G2 cells 1 h after elutriation at 25°C. After a normal mitosis, the block to replication led to an S-phase checkpoint-dependent arrest in the next cell cycle. Increasing the temperature to 37°C-induced septation in 68% of cdc16.116 cells, but 91% in cdc16.116 fin1.A5 cells (Fig. 7A).
Figure 7.
Fin1 attenuates the signaling flux through the SIN. (A) Small G2 cells were isolated from wild-type, cdc16.116, and cdc16.116 leu1::fin1.A5 fin1.Δ cultures at the permissive temperature, and HU was added to these cultures 1 h after isolation from the centrifuge. A further 4 h later, each culture had completed a synchronous round of cell division, and cells arrested cell cycle progression in S phase of the next cell cycle. The culture was split in two, and the temperature of one-half was shifted to 37°C to induce septation at this 300-min time point. Samples were fixed throughout the experiment and processed for calcofluor staining to score the septation index. A greater degree of septation was induced in the culture, in which Fin1 function was compromised by the shift to 37°C. Arrows indicate the times at which HU was added or the temperature shifted to 37°C. (B,C) spg1.B8, fin1.Δspg1.B8 and spg1.B8 pREP81fin1+ cells were grown to early log phase in minimal medium at 25°C before the temperature was shifted to 30°C. The spg1.B8 pREP81fin1+ strain had been grown in minimal medium + thiamine until 16 h before the temperature shift, when it was removed so that expression of fin1+ would be induced around the time of the shift (Grallert et al. 2002). (B) Samples were removed at the indicated time points, stained with DAPI and calcofluor, and the frequency with which cells that exhibited the paired back-to-back nuclei (type 1 in C) or tetranucleated cell (type 2 in C) was plotted against time (n = 200 for each time point). (C) DAPI/Calcofluor-stained cells 6 h after the shift to 30°C. (1) Cells in which the back-to-back configuration of the two nuclei shows that these cells failed to do cytokinesis in the previous cell cycle and are now in G2 of the next cell cycle; (2) tetranucleated cells that have also clearly failed to do cytokinesis after mitosis in previous cell cycles.
We based our second approach on the reasoning that if Fin1 acts as a negative regulator of SIN signaling, then loss of Fin1 function might be expected to rescue SIN mutants at semipermissive temperature, when signaling is partially compromised. Conversely, increased expression of fin1+ would be predicted to further attenuate SIN signaling, thereby enhancing weak septation defects. We therefore compared the efficiency with which spg1.B8 cells septated at 30°C with that of spg1.B8 fin1.Δ and spg1.B8 cells in which the expression of Fin1 was induced from the plasmid pREP81fin1+ 14 h before the shift to the restrictive temperature. The number of cells in which two nuclei adopted the “back-to-back” (Fig. 7C, 1) and tetra-nucleate cell (Fig. 7C, 2) phenotypes that typify cells in which septation has failed in the previous cell cycle was plotted against time to generate Figure 7B. Loss of Fin1 function reduced the severity of the septation defect, whereas the deficiency in septation was enhanced by increasing Fin1 levels.
We conclude that Fin1 plays a role in attenuating signaling by the SIN, and that the alteration in Cdc7 asymmetry upon loss of Fin1 function reflects a role for Fin1 in repressing SIN activity on the old SPB.
Discussion
Conservative segregation of components during duplication of the fission yeast SPB
We have labeled the fission yeast SPB with a protein that takes several generations to fold into a conformation that fluoresces. A starvation/refeed protocol resulted in a population of cells in which only one of the two anaphase B SPBs was fluorescent. This suggests that elements of the duplication of the SPB are conservative. Conservative segregation of SPB components is consistent with the defects seen in temperature-sensitive alp6, alp4, cut12, fin1, and cut11 mutants, where one SPB works, while the other does not (Bridge et al. 1998; West et al. 1998; Vardy and Toda 2000; Grallert and Hagan 2002). It would seem logical that the functional SPB is one that assembled at the permissive temperature, whereas the nonfunctional one is likely to be composed of material that formed at the restrictive temperature when the defective proteins do not fold or function.
Analysis of a population of synchronized cells by electron microscopy (Ding et al. 1997) suggested that the SPB duplicates by fission of a large precursor into two smaller structures that then grow to the size of the parental SPB before mitosis. At first sight, our data might contradict this view; however, it could be possible that the entire population of folded, fluorescent Pcp1, and the anchor for Fin1/Byr4 are partitioned to one SPB during this fission event. Different regions of a single SPB may have different structures. The SPB component Cut12 can be detected along the entire surface of the SPB that abuts the outer face of the nuclear envelope, but an N-terminal epitope was only detected to one side of this face (Bridge et al. 1998). Alternatively, the fission yeast SPB may actually duplicate by a conservative mechanism in which a new SPB forms de novo each cycle, and that intermediates in SPB duplication have simply not been detected by the electron microscopic studies conducted to date. Further developments in the field will doubtless resolve this. The mechanics of SPB duplication are, however, not pertinent to the findings we report here, as our main findings relate to the demonstration that the Pcp1.RFP and Fin1.GFP inheritance patterns establish distinct fates for specific SPBs.
The SIN is active on the new SPB in anaphase B/G1
Simultaneous imaging of the old SPB and a marker for the active SIN showed that the active SIN was on the new SPB. The controls that modulate the analogous pathway in budding yeast, the MEN, are more complex. In an unperturbed budding yeast mitosis, the old SPB migrates into the bud, whereas the new SPB stays in the mother. The MEN inhibitors, Bub2 and Bfa1, always associate with the bud-bound old SPB, whereas, until mitotic exit, the Cdc15 kinase that marks the active MEN is only seen on the SPB of the mother cell (Menssen et al. 2001; Pereira et al. 2001). Transient microtubule depolymerization resets the inherent orientation in this system, and either SPB subsequently enters the bud with equal frequency. In these cells, the MEN inhibitors reside on the SPB that faces the bud tip, irrespective of its age (Pereira et al. 2001). This shows that it is a combination of the specific architecture of the cell and the association between the astral microtubules and the bud cortex/neck/tip that dictates which SPB will host the active MEN, rather than the age of the SPB. Perhaps the distinction between the anchorage of the MEN and SIN arises from the use of an additional SPB anchor in fission yeast. The MEN is anchored to the budding yeast SPB via Nud1, whereas the fission yeast Nud1 homolog, Cdc11, cooperates with a second scaffold protein Sid4 to anchor SIN components to the S. pombe SPB (Gruneberg et al. 2000; Krapp et al. 2001; Tomlin et al. 2002).
The S. pombe SPB takes at least one and a half cell cycles to mature
We find the NIMA kinase Fin1 on the central spindle, the SPB, and the region between the separating anaphase chromatin masses. The affinity of these sites for Fin1 is stage dependent. The region between the chromatin masses only stains in early anaphase cells, whereas the central spindle stains toward the end of anaphase B. The SPB association shows that each of the four daughter cells in a population are distinct because they have a different fate in the subsequent cell cycle.
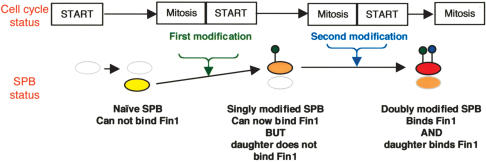
Fin1 always bound the old SPB and associated with the new SPB in half of the cells in the population. The affinity of the new SPB for Fin1 was dependent upon the history of the parent SPB that gave rise to this new SPB. If the parent SPB that produced this new SPB had itself been a new SPB in the previous cell cycle (i.e., the parent had only ever experienced one mitosis/START), then the new SPB did not bind Fin1. If, on the other hand, the parent SPB had produced at least one daughter already and gone through two or more mitosis/START transitions, then the new SPB bound Fin1.
Various models can be proposed to account for this behavior. Perhaps the simplest is best illustrated by considering the fate of a totally naïve SPB (i.e., a new SPB that arose from the duplication of a parent that had, itself, been produced as a new SPB in the previous cell cycle; Fig. 8). This naïve SPB is unable to bind Fin1 in the cell cycle in which it is created. Upon passage through mitosis or START, it is modified so that it will now bind Fin1 in the next anaphase B. In addition to promoting the recruitment of Fin1 to the anaphase B SPB this change, or a modification that is conducted in parallel, primes the SPB to undergo a second change during passage through the next round of mitosis/START. This second change means that this mature SPB can dictate that the SPB it will now produce will be able to bind Fin1 in the subsequent anaphase (Fig. 8). Were such a simple model to underlie the changes that we find, then phosphorylation of an SPB component by Cdk1/cyclin B or mitotic kinases, such as polo or aurora, may account for one or both of the SPB modifications. Whether this is the case or not, the modification that determines that a partner SPB will bind Fin1, it is not maintained through sporulation as only one of the two SPBs stained in germinating spores (data not shown).
Figure 8.
A model in which SPB modifications that are coupled to transit through the cell cycle account for the Fin1 inheritance patterns.
Fin1 as a new component of the SIN
The association between Fin1 and the SIN inhibitor Byr4 is important for Fin1 association with the SPB, as it is unable to bind when Byr4 is missing from the SPB as a result of deletion of the gene encoding byr4 itself or its partner in the GAP complex cdc16 (Cerutti and Simanis 1999). However, as Fin1 associates with the new SPB that hosts the SIN in half of the cells in the population, and Byr4 does not bind this SPB (Cerutti and Simanis 1999; Li et al. 2000), Fin1 must associate with another SPB component after, or as a result of, its association with Byr4. In this context, it is important to note that both Byr4 and Fin1 associate with both SPBs early in mitosis, long before the asymmetry of SIN regulation. This may explain why the Fin1 signal was always brightest on the old SPB, as Fin1 may bind to both the unidentified SPB anchor and Byr4 on this SPB. The association between Fin1 and a SIN inhibitor appears to be biologically relevant, as compromising Fin1 function both enhances SIN signaling on the old SPB and promotes septation. Three points suggest that Fin1 is not a core SIN component; it was not identified in screens that exhaustively identified key SIN components (Fournier et al. 2001); cells can survive without it, and alteration of Fin1 function has only modest effects on SIN output. Fin1, therefore, appears to join molecules such as Dma1, Zfs1, and Amn1 as a regulator at the periphery of SIN/MEN control (Murone and Simanis 1996; Beltraminelli et al. 1999; Wang et al. 2003). Perhaps these molecules become critically important in growth states that have not been encountered in the controlled laboratory environment, such as stress and nutrient limitation. Removal of such elements might have only a moderate consequence upon flux through the pathway in standard growth conditions. Alternatively, the SPB may need to mature through distinct steps before it is fully competent to regulate the pathway. In this case, inhibition by molecules such as Fin1 or Amn1 could regulate the pathway on a subset of SPBs and ensure the maximum fidelity of mitotic controls. This view would be supported by the observations that the SIN is only activated on both SPBs in around half of the population following Fin1 inactivation, and that inactivating Cdc16 in S phase induces around half of the cells in a population to septate (Cerutti and Simanis 1999). Finally, the dependence of Fin1 recruitment upon the activity of SIN effectors could suggest that it is part of a feedback control that limits or dampens SIN activity on one or both SPBs once the network is active. As such, it would resemble the mitotic phosphorylation of the scaffold molecule Cdc11, which depends upon the activity of the kinase that it recruits to the SPB, Cdc7 (Krapp et al. 2003).
Parallels between SPB maturation and centriole maturation
The incremental maturation of the fission yeast SPB bears a striking resemblance to the changes in centriolar morphology that accompany maturation from newborn daughters into fully mature mother centrioles in pig epithelial cells (Vorobjev and Chentsov 1982). Mature mother centrioles have appendages that are not seen on their daughters. In the first stage of centrosome duplication, the single centriole pair splits to produce two independent centrioles that seed short procentrioles. These centrioles elongate during G2 to generate two prophase centrosomes that are similar in structure, except that only one of the pairs has a centriole that is equipped with appendages. This is the original mother centriole. The new mother that was produced by the G1 duplication fission event still lacks appendages. They do not appear on this “ex-daughter, now mother” until the transition from metaphase to anaphase B. At this time, this exdaughter, now mother centriole acquires the appendages. The anaphase cell has two pairs of centrioles, and in each of these pairs, the oldest centriole has appendages. Vorobjev and Chentsov concluded that centriole maturation takes one and half-cell cycles in mammalian cells. The analysis of molecular markers of centriole maturation such as CDIB4 and ninein also reveal incremental changes in centriole structure (Lange and Gull 1995; Mogensen et al. 2000). The fact that both centriole and SPB maturation may take similar times, raises the exciting possibility that the underlying mechanisms are related, and that studies of SPB maturation in yeasts may shed light upon the mechanism of centriole maturation in higher systems.
What purpose could centriole/SPB maturation serve?
There is increasing evidence from yeast and higher systems that the decision to commit to mitosis is taken at the centrosome/SPB (Jackman et al. 2003; MacIver et al. 2003). Similarly, the role for yeast SPBs in regulating mitotic exit/septation is beyond question, and parallels are emerging in higher systems (Simanis 2003). The mother centriole plays a critical role in regulating the final stages of cytokinesis. It migrates to touch the midbody, and this contact then promotes cell fission (Piel et al. 2001). It was also recently established that Centriolin, a component of centriole appendages, has a domain that is homologous to a domain found in both the yeast MEN/SIN scaffold proteins, Nud1/Cdc11 (Gromley et al. 2003). This raises the possibility that a control network analogous to the MEN/SIN associates with the appendages.
If a MEN/SIN-related network were to associate with the appendages, it may explain why appendages only form on daughter centrioles upon commitment to anaphase. The MEN modulates mitotic exit in budding yeast in response to interaction between astral microtubules and the cortex. Prior to anaphase, the spindle-assembly checkpoint can regulate mitotic progression, but once anaphase has started, the astral microtubule checkpoint, the MEN, becomes critical. As a MEN-equivalent network would only be important in anaphase B, there would only be a need for appendages at this time and not before. There may be a positive benefit to having only one site at which to coordinate cell cycle controls prior to anaphase, because two or more control points could generate conflicting signals and result in inappropriate cell proliferation. This would be particularly relevant for tumor cells, as many have multiple centrosomes (Nigg 2002). Restoration of normal centriole maturation or the promotion of centrosome maturation when multiple centrosomes are present could be a powerful way to alter the proliferative capacity of such tumor cells, and so may offer novel avenues for therapeutic intervention.
Materials and methods
Strains, molecular biology, and antibody production
Strains are listed in Table 2. Standard fission yeast and molecular biology methods were used throughout (Moreno et al. 1991). PCR tagging used pSM822 and pSM1023 (Pereira et al. 2001; Maekawa et al. 2003) to generate pcp1.RFP and fin1.4GFP. An NdeI site was generated immediately before the STOP codon of fin1+ ORF, a fragment encoding 3-Pk epitopes (Craven et al. 1998) inserted into this site and the 3.3-Kb fragment from -286 to +627 was inserted into pINTA (Petersen and Hagan 2003) to direct disruption of the leu1+ locus with this modified fin1+ gene alongside ura4+. To generate pRep41fin1 282-722 amino acids, an NdeI site was inserted in pfin1.1 (Grallert and Hagan 2002) directly after nucleotide 1157 of the coding sequence (equivalent to residue 281/282) to generate pfin1.1Nde1int. The 1953-bp NdeI/XbaI fragment of pfin1.1Nde1int was inserted into pREP41. To generate fusion protein for immunization, an NdeI site was introduced at the stop codon of pfin1.1Nde1int to generate pfin1.1Ndefrag. The 1323-bp Nde1 fragment from pfin1.1Ndefrag was inserted into pAR3033 (Studier et al. 1990) to generate pfin1FP1, and this was used to generate fusion protein by standard procedures (Studier et al. 1990).
Table 2.
Yeast strains used in this study
| IH163 | 972 h− | Lab Stock |
| IH592 | wee1.50 leu1.32 h− | Nurse 1975 |
| IH3160 | leu1.32 ura4.D18 pRep41 | This study |
| IH3161 | leu1.32 ura4.D18 pRep41fin1282-722aa | This study |
| IH1824 | fin1::ura4+ura4.D18 his2 h+ | Krien et al. 1998 |
| IH2772 | leu1::fin1+:ura4+fin1::ura4+ura4.D18 | This study |
| IH2282 | leu1::fin1PkC:ura4+fin1::ura4+ura4.D18 | This study |
| IH2674 | fin1.GFP:kanR h− | This study |
| IH2673 | fin1.4GFP:kanR ura4.D18 h− | This study |
| IH376 | cdc10.v50 | Marks et al. 1992 |
| IH2527 | cig1::ura4+cig2::ura4+puc1::ura4+ura4.D18 h+ | Martin-Castellanos et al. 2000 |
| IH2675 | pcp1.RFP:kanR h− | This study |
| IH2719 | pcp1.RFP:kanR fin1.4GFP:kanR | This study |
| IH759 | cdc7.HA:ura4+ura4.D18 h− | Schmidt et al. 1997 |
| IH1106 | cdc7.GFP:ura4+ura4.D18 h− | Sohrmann et al. 1998 |
| IH2721 | cdc7.GFP:ura4+pcp1.RFP:kanR ura4.D18 h− | This study |
| IH635 | cdc25.22 ura4.D18 h− | Nurse et al. 1976 |
| IH164 | cdc2.33 h− | Nurse et al. 1976 |
| IH280 | cut12.1 leu1.32 h− | Bridge et al. 1998 |
| IH110 | nuc2.663 leu1.32 h− | Hirano et al. 1988 |
| IH136 | cut7.24 leu1.32 h− | Hagan and Yanagida 1992 |
| IH156 | sad1.1 leu1.32 h− | Hagan and Yanagida 1995 |
| IH385 | cut11.1 leu1.32 h− | West et al. 1998 |
| IH304 | nda3.KM311 leu1.32 h− | Toda et al. 1983 |
| IH2233 | cdc11.123 leu1.32 h− | Nurse et al. 1976 |
| IH2232 | cdc11.136 ura4.D18 h− | Nurse et al. 1976 |
| IH2650 | sid4.K12 leu1.32 ura4.D18 h− | This study |
| 1296 | cdc16.116 leu1.32 h− | Minet et al. 1981 |
| IH3017 | cdc16::ura4+sid2.1 ade6.M210 leu1.32 ura4.D18 h− | Fournier et al. 2001 |
| IH2940 | cdc16::ura4+mob1.R4 ura4.D18 h+ | Fournier et al. 2001 |
| IH3132 | byr4::ura4+/byr4+ade6.M216/ade6.M210 leu1.32/leu1.32 ura4.D18/ura4.D18 h−h+ | Song et al. 1996 |
| IH2992 | byr4::ura4+mob1.R4 leu1.32 ura4.D18 h− | This study |
| IH1557 | spg1.B8 ura4.D18 leu1.32 his2 h+ | Schmidt et al. 1997 |
| IH738 | cdc7.A20 ura4.D18 h+ | Nurse et al. 1976 |
| IH114 | cdc7.24 h− | Nurse et al. 1976 |
| IH2499 | cdc7::ura4+/cdc7+ade6.M216/ade6.M210 leu1.32/leu1.32 ura4.D18/ura4.D18 h−/h+ | Fankhauser and Simanis 1994 |
| IH1469 | sid1.239 leu1.32 ura4.D18 h+ | Balasubramanian et al. 1998 |
| IH111 | cdc14.118 h− | Nurse et al. 1976 |
| IH1470 | sid2.250 leu1.32 ura4.D18 ade6,M216 h+ | Balasubramanian et al. 1998 |
| IH2939 | mob.1.R4 ura4.D18 h− | Salimova et al. 2000 |
| IH1336 | spg1.HA:ura4+leu1.32 ura4.D18 | Sohrmann et al. 1998 |
| IH3162 | spg1.HA:ura4+leu1.32 ura4.D18 pREP41fin1+ | This study |
| IH2918 | cdc7.HA:ura4+nda3.KM311 ura4.D18 h− | This study |
| IH2920 | cdc7.HA:ura4+nda3.KM311 fin1::ura4+ura4.D18 leu1.32 his2 h+ | This study |
| IH2942 | cdc7.HA:ura4+nda3.KM311 fin1::ura4+leu1::fin1.A5:ura4+ura4.D18 | This study |
| IH2780 | cdc16.116 fin1::ura4+ura4.D18 leu1.32 | This study |
| IH2762 | cdc16.116 fin1.ts1 leu1.32 | This study |
| IH2774 | fin1::ura4+spg1.B8 ura4.D18 leu1.32 h− | This study |
| IH3510 | spg1.B8 leu1.32 ura4.d18 pREP81 h− | This study |
| IH3511 | spg1.B8 leu1.32 ura4.d18 pREP81fin1+h− | This study |
Cell biology
Standard Western blotting approaches following TCA precipitation and immunoprecipitation of cell extracts was performed as described previously (MacIver et al. 2003). Standard immunofluorescence procedures were used for tubulin staining with the anti-α tubulin antibody TAT1 (Woods et al. 1989), whereas Fin1 and Byr4 staining was performed following fixation with 1% formaldehyde for 5 min according to standard procedures (Hagan and Asycough 2000). For pedigree analysis, cells were mounted under agarose pads according to Hagan et al. (1990), with the exclusion that PVP was from the medium and use of EMM2 rather than YES. Images were captured on a Zeiss Axiovert 200M fitted with a PIFOC piezo objective driver and a Coolsnap HQ camera (Roper Scientific). Images were captured in a set of 6-×-1-μm slices every 5 min, and processed with Metamorph software (Universal Imaging). Each slice was scored for the presence of SPBs. Images in Figure 1F were processed by the Metamorph nearest-neighbor algorithm. A Perkin Elmer spinning-disk confocal system was used to generate images in Figure 1F and G. Stacks were compressed with the maximum projection algorithm to generate the images for presentation using proprietary software.
Acknowledgments
We thank the CRUK, HFSP, Swiss National Science Foundation, Swiss Cancer League, and ISREC for funding; Elena Cano and Maggie Chambers (Diagnostics Scotland) for technical assistance; Isabel Alvarez for plasmids; Keith Gull for TAT1; and Elmar Schiebel for plasmids and stimulating discussions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.296204.
References
- Balasubramanian M.K., McCollum, D., Chang, L., Wong, K.C.Y., Naqvi, N.I., He, X.W., Sazer, S., and Gould, K.L. 1998. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics 149: 1265-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin A.J. and Amon, A. 2001. Men and sin: What's the difference. Nat. Rev. Mol. Cell Biol. 2: 815-826. [DOI] [PubMed] [Google Scholar]
- Belham C., Roig, J., Caldwell, J.A., Aoyama, Y., Kemp, B.E., Comb, M., and Avruch, J. 2003. A mitotic cascade of NIMA family kinases. Nercc1/Nek9 activates the Nek6 and Nek7 kinases. J. Biol. Chem. 278: 34897-34909. [DOI] [PubMed] [Google Scholar]
- Beltraminelli N., Murone, M., and Simanis, V. 1999. The S. pombe zfs1 gene is required to prevent septation if mitotic progression is inhibited. J. Cell Sci. 112: 3103-3114. [DOI] [PubMed] [Google Scholar]
- Blangy A., Lane, H.A., Dherin, P., Harper, M., Kress, M., and Nigg, E.A. 1995. Phosphorylation by p34cdc2 regulates spindle association of Human Eg5, a kinesin-related motor essential for bipolar spindle formation in-vivo. Cell 83: 1159-1169. [DOI] [PubMed] [Google Scholar]
- Bridge A.J., Morphew, M., Bartlett, R., and Hagan, I.M. 1998. The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control. Genes & Dev. 12: 927-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti L. and Simanis, V. 1999. Asymmetry of the spindle pole bodies and spg1p GAP segregation during mitosis in fission yeast. J. Cell Sci. 112: 2313-2321. [DOI] [PubMed] [Google Scholar]
- Chang L. and Gould, K.L. 2000. Sid4p is required to localize components of the septation initiation pathway to the spindle pole body in fission yeast. Proc. Natl. Acad. Sci. 97: 5249-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven R.A., Griffiths, D.J.F., Sheldrick, K.S., Randall, R.E., Hagan, I.M., and Carr, A.M. 1998. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene 221: 59-68. [DOI] [PubMed] [Google Scholar]
- Cueille N., Salimova, E., Esteban, V., Blanco, M., Moreno, S., Bueno, A., and Simanis, V. 2001. Flp1, a fission yeast orthologue of the S. cerevisiae CDC14 gene, is not required for cyclin degradation or rum1p stabilisation at the end of mitosis. J. Cell Sci. 114: 2649-2664. [DOI] [PubMed] [Google Scholar]
- Ding R., West, R.R., Morphew, M., and McIntosh, J.R. 1997. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell 113: 1461-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C. and Simanis, V. 1994. The Cdc7 protein-kinase is a dosage-dependent regulator of septum formation in fission yeast. EMBO J. 13: 3011-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory M.R., Morphew, M.M., Joseph, J.D., Means, A.R., and Davis, T.R. 2002. Pcp1, an Spc110p related calmodulin target at the centrosome of the fission yeast Schizosaccharomyces pombe. Cell Growth Dif. 13: 47-58. [PubMed] [Google Scholar]
- Fournier N., Cerutti, L., Beltraminelli, N., Salimova, E., and Simanis, V. 2001. Bypass of the requirement for cdc16p GAP function in Schizosaccharomyces pombe by mutation of the septation initiation network genes. Arch. Microbiol. 175: 62-69. [DOI] [PubMed] [Google Scholar]
- Fry A.M. 2002. The Nek2 protein kinase: A novel regulator of centrosome structure. Oncogene 21: 6184-6194. [DOI] [PubMed] [Google Scholar]
- Glynn J.M., Lustig, R.J., Berlin, A., and Chang, F. 2001. Role of bud6p and tea1p in the interaction between actin and microtubules for the establishment of cell polarity in fission yeast. Curr. Biol. 11: 836-845. [DOI] [PubMed] [Google Scholar]
- Grallert A. and Hagan, I.M. 2002. Schizosaccharomyces pombe NIMA-related kinase Fin1, regulates spindle formation and an affinity of Polo for the SPB. EMBO J. 21: 3096-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley A., Jurczyk, A., Sillibourne, J., Halilovic, E., Mogensen, M., Groisman, I., Blomberg, M., and Doxsey, S. 2003. A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J. Cell Biol. 161: 535-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg U., Campbell, K., Simpson, C., Grindlay, J., and Schiebel, E. 2000. Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J. 19: 6475-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I.M. and Asycough, K.R. 2000. Flourescence microscopy in yeast. In Protein localization by fluorescence microscopy (ed. V.J. Allan), pp. 179-206. Oxford University Press, Oxford, UK.
- Hagan I. and Yanagida, M. 1992. Kinesin-related Cut7 protein associates with mitotic and meiotic spindles in fission yeast. Nature 356: 74-76. [DOI] [PubMed] [Google Scholar]
- ____. 1995. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 129: 1033-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I.M., Riddle, P.N., and Hyams, J.S. 1990. Intramitotic controls in the fission yeast Schizosaccharomyces pombe the effect of cell-size on spindle length and the timing of mitotic events. J. Cell Biol. 110: 1617-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Hiraoka, Y., and Yanagida, M. 1988. A temperature-sensitive mutation of the Schizosaccharomyces-pombe gene nuc2+ that encodes a nuclear scaffold-like protein blocks spindle elongation in mitotic anaphase. J. Cell Biol. 106: 1171-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman M., Lindon, C., Nigg, E.A., and Pines, J. 2003. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat. Cell Biol. 5: 143-148. [DOI] [PubMed] [Google Scholar]
- Jones D.G. and Rosamond, J. 1990. Isolation of a novel protein kinase-encoding gene from yeast by oligodeoxyribonucleotide probing. Gene 90: 87-92. [DOI] [PubMed] [Google Scholar]
- Krapp A., Schmidt, S., Cano, E., and Simanis, V. 2001. S. pombe cdc11p, together with sid4p, provides an anchor for septation initiation network proteins on the spindle pole body. Curr. Biol. 11: 1559-1568. [DOI] [PubMed] [Google Scholar]
- Krapp A., Cano, E., and Simanis, V. 2003. Mitotic hyperphosphorylation of the fission yeast SIN scaffold protein cdc11p is regulated by the protein kinase cdc7p. Curr. Biol. 13: 168-172. [DOI] [PubMed] [Google Scholar]
- Krien M., Bugg, S., Palatsides, M., Asouline, G., Morimyo, M., and Connell, M. 1998. A NIMA homologue promotes chromatin condensation in fission yeast. J. Cell. Sci. 111: 967-976. [DOI] [PubMed] [Google Scholar]
- Krien M.J., West, R.R., John, U.P., Koniaras, K., McIntosh, J.R., and O'Connell, M. 2002. The fission yeast NIMA kinase Fin1p is required for spindle function and nuclear envelope integrity. EMBO J. 21: 1713-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange B.M.H. and Gull, K. 1995. A molecular marker for centriole maturation in the mammalian-cell cycle. J. Cell Biol. 130: 919-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Furge, K.A., Cheng, Q.C., and Albright, C.F. 2000. Byr4 localizes to spindle-pole bodies in a cell cycle-regulated manner to control Cdc7 localization and septation in fission yeast. J. Biol. Chem. 275: 14381-14387. [DOI] [PubMed] [Google Scholar]
- MacIver F.H., Tanaka, K., Robertson, A.M., and Hagan, I.M. 2003. Physical and functional interactions between polo kinase and the spindle pole component Cut12 regulate mitotic commitment in S. pombe. Genes & Dev. 17: 1507-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa H., Usui, T., Knop, M., and Schiebel, E. 2003. Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J. 22: 438-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub M.R., Montpetit, B., Zhao, L., Finst, R.J., Goh, B., Kim, A.C., and Quarmby, L.M. 2002. The FA2 gene of Chlamydomonas encodes a NIMA family kinase with roles in cell cycle progression and microtubule severing during deflagellation. J. Cell Sci. 115: 1759-1768. [DOI] [PubMed] [Google Scholar]
- Marks J., Fankhauser, C., Reymond, A., and Simanis, V. 1992. Cytoskeletal and DNA-structure abnormalities result from bypass of requirement for the cdc10 start gene in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 101: 517-528. [DOI] [PubMed] [Google Scholar]
- Martin-Castellanos C., Blanco, M.A., de Prada, J.M., and Moreno, S. 2000. The puc1 cyclin regulates the G1 phase of the fission yeast cell cycle in response to cell size. Mol. Biol. Cell 11: 543-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen R., Neutzner, A., and Seufert, W. 2001. Asymmetric spindle pole localization of yeast Cdc15 kinase links mitotic exit and cytokinesis. Curr. Biol. 11: 345-359. [DOI] [PubMed] [Google Scholar]
- Minet M., Nurse, P., Thuriaux, P., and Mitchison, M. 1981. Uncontrolled septation in a cell division cycle mutant of the fission yeast Schizosaccharomyces pombe. J. Bact. 137: 440-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen M.M., Malik, A., Piel, M., Bouckson-Castaing, V., and Bornens, M. 2000. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: The role of ninein. J. Cell Sci. 113: 3013-3023. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar, A., and Nurse, P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Meth. Enzymol. 194: 795-823. [DOI] [PubMed] [Google Scholar]
- Murone M. and Simanis, V. 1996. The fission yeast dma1 gene is a component of the spindle assembly checkpoint, required to prevent septum formation and premature exit from mitosis if spindle function is compromised. EMBO J. 15: 6605-6616. [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A. 2001. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2: 21-32. [DOI] [PubMed] [Google Scholar]
- ____. 2002. Centrosome aberrations: Cause or consequence of cancer progression? Nat. Rev. Cancer 2: 815-825. [DOI] [PubMed] [Google Scholar]
- Nurse P. 1975. Genetic control of cell size at cell division in yeast. Nature 256: 547-551. [DOI] [PubMed] [Google Scholar]
- ____. 1990. Universal control mechanism regulating onset of M-phase. Nature 344: 503-508. [DOI] [PubMed] [Google Scholar]
- Nurse P., Thuriaux, P., and Nasmyth, K. 1976. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 146: 167-178. [DOI] [PubMed] [Google Scholar]
- O'Connell M.J., Krien, M.J., and Hunter, T. 2003. Never say never. The NIMA-related protein kinases in mitotic control. Trends Cell Biol. 13: 221-228. [DOI] [PubMed] [Google Scholar]
- Pereira G., Tanaka, T.U., Nasmyth, K., and Schiebel, E. 2001. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 20: 6359-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Manson, C., Grindlay, J., and Schiebel, E. 2002. Regulation of the Bfa1p-Bub2p complex at spindle pole bodies by the cell cycle phosphatase Cdc14p. J. Cell Biol. 157: 367-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J. and Hagan, I.M. 2003. S. pombe aurora kinase/survivin is required for chromosome condensation and the spindle checkpoint attachment response. Curr. Biol. 13: 590-597. [DOI] [PubMed] [Google Scholar]
- Piel M., Nordberg, J., Euteneuer, U., and Bornens, M. 2001. Centrosome dependent exit of cytokinesis in animal cells. Science 291: 1550-1553. [DOI] [PubMed] [Google Scholar]
- Salimova E., Sohrmann, M., Fournier, N., and Simanis, V. 2000. The S. pombe orthologue of the S. cerevisiae mob1 gene is essential and functions in signalling the onset of septum formation. J. Cell Sci. 113: 1695-1704. [DOI] [PubMed] [Google Scholar]
- Schmidt S., Sohrmann, M., Hofmann, K., Woolard, A., and Simanis, V. 1997. The Spg1 GTPase is an essential dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes & Dev. 11: 1519-1534. [DOI] [PubMed] [Google Scholar]
- Simanis V. 2003. Events at the end of mitosis in the budding and fission yeasts. J. Cell Sci. 116: 4263-4275. [DOI] [PubMed] [Google Scholar]
- Sohrmann M., Schmidt, S., Hagan, I., and Simanis, V. 1998. Asymmetric segregation on spindle poles of the Schizosaccharomyces pombe septum-inducing protein kinase Cdc7p. Genes & Dev. 12: 84-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Mach, K.E., Chen, C.V., Reynolds, T., and Albright, C.F. 1996. A novel suppressor of ras1 in fission yeast, byr4, is a dosage-dependent inhibitor of cytokinesis. J. Cell Biol. 133: 1307-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F.W., Rosenberg, A.H., Dunn, J.J., and Dubendorff, J.W. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Meth. Enzymol. 185: 60-89. [DOI] [PubMed] [Google Scholar]
- Swenson K.I., Winkler, K.E., and Means, A.R. 2003. A new identity for MLK3 as an NIMA-related, cell cycle-regulated kinase that is localized near centrosomes and influences microtubule organization. Mol. Biol. Cell 14: 156-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T.U., Rachidi, N., Janke, C., Pereira, G., Galova, M., Schiebel, E., Stark, M.J., and Nasmyth, K. 2002. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 108: 317-329. [DOI] [PubMed] [Google Scholar]
- Toda T., Umesono, K., Hirata, A., and Yanagida, M. 1983. Cold-sensitive nuclear division arrest mutants of the fission yeast Schizosaccharomyces pombe. J. Mol. Biol. 168: 251-270. [DOI] [PubMed] [Google Scholar]
- Tomlin G.C., Morrell, J.L., and Gould, K.L. 2002. The spindle pole body protein Cdc11p links Sid4p to the fission yeast septation initiation network. Mol. Biol. Cell 13: 1203-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann S., Wolfe, B.A., Jorgensen, P., Tyers, M., Gould, K.L., and McCollum, D. 2001. Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr. Biol. 11: 931-940. [DOI] [PubMed] [Google Scholar]
- Uemura T., Ohkura, H., Adachi, Y., Morino, K., Shiozaki, K., and Yanagida, M. 1987. DNA topoisomerase-II Is required for condensation and separation of mitotic chromosomes in S. pombe. Cell 50: 917-925. [DOI] [PubMed] [Google Scholar]
- Vardy L. and Toda, T. 2000. The fission yeast γ-tubulin complex is required in G1 phase and is a component of the spindle assembly checkpoint. EMBO J. 19: 6098-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev I.A. and Chentsov, Y.S. 1982. Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 98: 938-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Shirogane, T., Liu, D., Harper, J.W., and Elledge, S.J. 2003. Exit from exit: Resetting the cell cycle through Amn1 inhibition of G protein signaling. Cell 112: 697-709. [DOI] [PubMed] [Google Scholar]
- West R.R., Vaisberg, E.V., Ding, R., Nurse, P., and McIntosh, J.R. 1998. cut11+: A gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelope and bipolar spindle formation in Schizosaccharomyces pombe. Mol. Biol. Cell 9: 2839-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A., Sherwin, T., Sasse, R., Macrae, T.H., Baines, A.J., and Gull, K. 1989. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal-antibodies. J. Cell Sci. 93: 491-500. [DOI] [PubMed] [Google Scholar]
- Ye X.S., Xu, G., Pu, R.T., Fincher, R.R., McGuire, S.L., Osmani, A.H., and Osmani, S.A. 1995. The NIMA protein kinase is hyperphosphorylated and activated downstream of p34cdc2/cyclin B: Coordination of two mitosis promoting kinases. EMBO J. 14: 986-994. [DOI] [PMC free article] [PubMed] [Google Scholar]