Abstract
The eukaryotic nucleus is a congested place, and macromolecular crowding is thought to play an important role in increasing the relative concentrations of nuclear proteins, thereby accelerating the rates of biochemical reactions. Crowding is also thought to provide the environment needed for formation of nuclear bodies/subcompartments, such as the Cajal Body (CB) and the Histone Locus Body (HLB), via self-organization. In this chapter, we contrast the theories of stochastic self-organization and hierarchical self-organization in their application to nuclear body assembly, using CBs and HLBs as paradigms. Genetic ablation studies in Drosophila on components of CBs and HLBs, have revealed an order to the assembly of these structures that is suggestive of a hierarchical model of self-organization. These studies also show that function(s) attributed to the nuclear bodies are largely unaffected in their absence, reinforcing an emerging theme in the field that the purpose of these subdomains may be to enhance the efficiency and specificity of reactions.
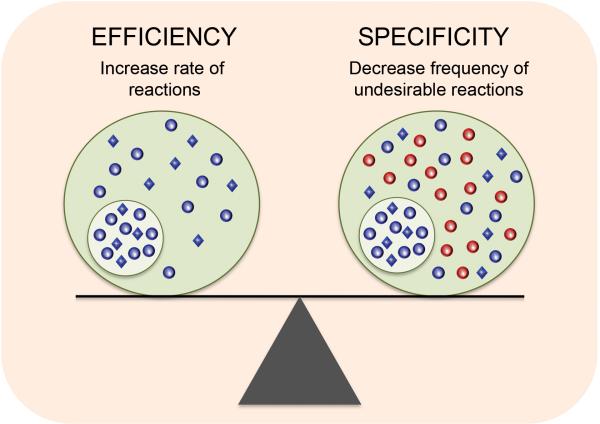
The environment inside a cell is arguably as complex as the one outside the organism itself. Although they perform a plethora of essential activities, the intracellular machineries that execute these functions are not distributed randomly. A salient feature of eukaryotic cells is the division of labor via compartmentalization, which is thought to increase both the rate and the specificity of chemical reactions. Cells thus utilize membrane-bound organelles in order to achieve a balance between productivity and quality control (Fig. 1).
Figure 1.
The yin yang of compartmentalization. In order to achieve greater productivity (efficiency), systems typically must sacrifice on quality control (specificity). Cellular compartmentalization promotes a balance beteween these two opposing forces. Compartments can increase reaction rates by physically bringing together molecules and promoting interactions between them. At the same time, they provide greater specificity by decreasing the frequency of unwanted interactions.
Compartmentalization by the enrichment of factors associated with specific and/or related biological pathways may occur as a “fundamental” property of the cell (as in the case of the mitochondrion, which self-replicates) or as an “emergent” property, wherein compartments form de novo. Irrespective of whether they represent fundamental or emergent properties, each of these compartments may be further subdivided. Though compartments tend to isolate pathways and functions in distinct domains, they must also function in coordination with other compartments to maintain cellular homeostasis (Zhao et al. 2009). As a result of this coordination, even “compartment-specific” factors are often shared among other compartments. One such compartment that contains multiple subcompartments is the cell nucleus (Leonhardt and Cardoso 1995; Lamond and Earnshaw 1998; Stein et al. 2003; Pederson 2010).
The eukaryotic nucleus is an intricate intracellular enclave that orchestrates the essential functions of DNA replication, damage repair, transcription and RNA processing (Cremer et al. 2006; Schneider and Grosschedl 2007). Nuclear subcompartments include chromosome territories, nucleoli, Cajal bodies, histone locus bodies, speckles, paraspeckles, PML Bodies, nuclear stress bodies perinucleolar compartments, etc. (Matera et al. 2009). Although our understanding of the structural constitution and functional attributes of many cytoplasmic compartments is fairly well established, the same cannot be said of a large number of nuclear subdomains that have been identified to date (Spector 2006).
An in-depth analysis of nuclear compartments requires a basic understanding of their intrinsic elements. For any given compartment, one can visualize at least three such elements: (i) Establishing a compartment using basic building-blocks that specify the uniqueness of that compartment, (ii) Targeting of reactants to the compartment. (iii) Clearing the compartment of processed products to make way for a new round of compartment-specific targeting. While these three aspects are relatively well understood for cytoplasmic organelles, the field of nuclear compartments is still evolving in this regard. The Cajal body represents an important paradigm for studies of the structure, function and dynamics of nuclear subdomains.
Cajal bodies and snRNP biogenesis
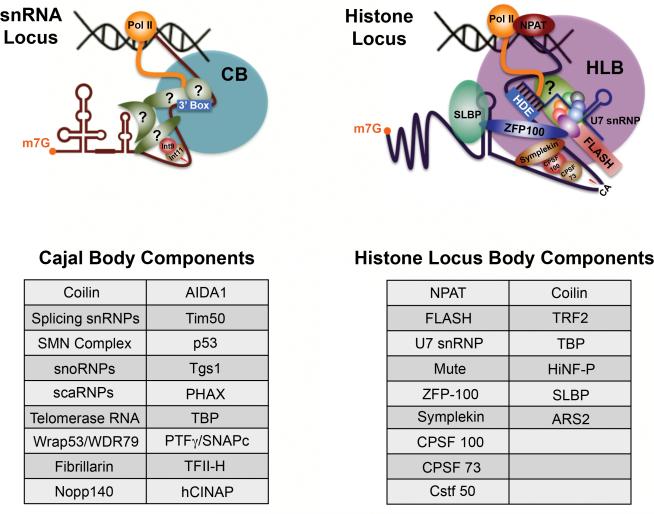
Cajal bodies (CBs) can be viewed as the location where spliceosomal small nuclear ribonucleoprotein (snRNP) biogenesis begins and ends. CBs are conserved nuclear subdomains containing high concentrations of the spliceosomal snRNPs, as well as the CB markers coilin and Survival Motor Neuron (SMN), for review see (Matera and Shpargel 2006; Stanek and Neugebauer 2006; Matera et al. 2009). Transcriptionally active snRNA gene loci are known to associate with CBs in an RNA-dependent manner (Fig. 2; Frey et al. 1999; Frey and Matera 2001; Dundr et al. 2007). Following export to the cytoplasm for SMN-dependent assembly of the core RNPs, the newly-formed snRNPs are imported into the nucleus, targeting to CBs for additional RNA modification and RNP remodeling steps (Carvalho et al. 1999; Sleeman and Lamond 1999; Jady et al. 2003; Nesic et al. 2004; Suzuki et al. 2010). CBs are also thought to be involved in the biogenesis of other classes of RNPs, including small nucleolar (sno)RNPs and telomerase (Filipowicz and Pogacic 2002; Verheggen et al. 2002; Tomlinson et al. 2006). In fact, CB homeostasis appears to depend upon ongoing RNP biogenesis (Shpargel and Matera 2005).
Figure 2.
Comparison of the organization and components of Cajal bodies (CBs) and Histone locus bodies (HLBs). A number of striking structural and functional parallels are apparent. Both CBs and HLBs are known to associate with specific genetic loci (although the association of CBs with snRNA loci is known only in mammals). Factors involved in transcription, processing and export of specialized non-polyadenylated transcripts (snRNAs and histone mRNAs) accumulate in and around these structures in both vertebrates and invertebrates. Nascent pre-snRNA and histone pre-mRNA are shown emerging from RNA polymerase II (Pol II) and processing factors are shown in hypothetical arrangements. A number of putative factors (question marks) are also depicted. A partial list of components of each body is shown below the cartoons, some of which may not be present in every species.
Overexpression of snRNP components, and hence an increase in the kinetics of snRNP biogenesis, leads to the formation of new CBs (Sleeman et al. 2001), whereas inhibition of the snRNP biogenesis pathway negatively affects CB formation (Shpargel and Matera 2005; Girard et al. 2006; Lemm et al. 2006). These observations implicate newly-assembled snRNPs and the SMN complex as primary and essential components required for CB assembly. In contrast, mature snRNPs are not thought to associate with the SMN complex and they ultimately accumulate within the nucleoplasmic speckle compartment. Because speckles fail to recruit other CB components, mature snRNPs are thought to traffic through extant CBs (Stanek et al. 2008), rather than contributing to their assembly. Components of other nuclear pathways including snoRNPs, telomerase RNPs, and small Cajal body (sca)RNPs also appear to traffic through CBs, suggesting that they do not play a cardinal role in CB formation (Matera and Shpargel 2006; Stanek and Neugebauer 2006).
Further evidence for the non-involvement of mature snRNPs in CB formation comes from studies of cell division. Mature snRNPs that are released after nuclear membrane disassembly return back to the newly-formed nuclei (G1 daughters) without accumulating in CBs (Sleeman and Lamond 1999; Sleeman et al. 2003). The CBs that are formed in early G1 daughters are also devoid of SMN (Carvalho et al. 1999), suggesting that the newly-formed snRNPs do not seed formation of CBs, at least in early G1 (Ferreira et al. 1994).
The situation in G1 daughters raises some important nomenclature questions. What constitutes a CB? The nuclear bodies in G1 daughters contain coilin, but lack SMN and splicing snRNPs. Thus they do not constitute classical CBs. However, the ability of coilin to form nuclear bodies in early G1 cells suggests that coilin foci assemble independent of mature snRNPs, SMN and other factors involved in snRNP biogenesis. In contrast, coilin organization into a CB becomes strictly dependent on SMN and ongoing snRNP assembly later in the cell cycle (Sleeman et al. 2001; Shpargel and Matera 2005; Girard et al. 2006; Lemm et al. 2006).
Bridging the dots: An evolutionary perspective
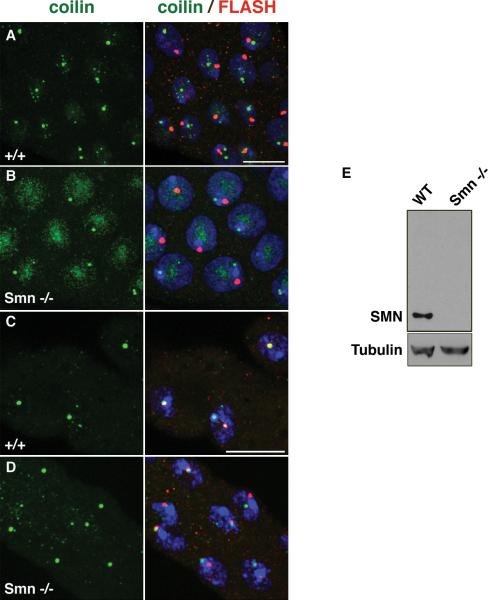
In adult mammalian cells, nuclear SMN and coilin colocalize completely in CBs, however, SMN is not a constitutive CB component in fetal tissues (Young et al. 2000). In mammalian fetal tissues (and certain transformed cell lines) SMN perferentially accumulates in twin structures called Gemini bodies, or gems (Young et al. 2001). The formation of coilin-positive, SMN-negative nuclear bodies in fetal cells (as well as in HeLa early G1 daughters) echoes German naturalist Ernst Häckel's theory of recapitulation. In most tissues of the fly (both larval and adult), SMN is not a constitutive component of CBs. In fact, coilin foci can form in complete loss-of-function Smn mutants (Fig. 3). Thus SMN- and snRNP-independent CB assembly (ontogeny) events in mammalian fetal tissues or early G1 cells recapitulate what happens constitutively in invertebrates (phylogeny). Although SMN itself has a strong influence on CB homeostasis in vertebrate cells, other members of the SMN complex (i.e. Gemins) are not required for SMN recruitment to CBs (Shpargel and Matera 2005; Girard et al. 2006; Lemm et al. 2006). This is presumably because SMN binds directly to coilin (Hebert et al. 2001; Hebert et al. 2002), whereas the Gemins do not. In any case, the evolutionary correlations, similarities and differences included, between humans and flies throws open some unique opportunities to delve into details.
Figure 3.
Drosphila Cajal bodies form in the absence of SMN. (A-D) Salivary glands and Malpighian tubules from wild-type, WT (+/+; A,C) and SmnD/SmnX7 null mutant (−/−; B,D) larvae. Early third instar larvae were stained for CBs with coilin (green) and HLBs with FLASH (red). At this stage, the mutant animals show a complete loss of SMN as detected by Western analysis (E). Note that some nucleolar accumulation of coilin is observed in the salivary glands of mutant animals, similar to that seen in vertebrate cells that have been depleted of SMN (Shpargel and Matera, 2005). Scale bar is 10μm.
The recent identification of structures in Drosophila nuclei that are related to CBs, known as histone locus bodies (HLBs), has helped to better understand the situation in vertebrates. HLBs are so-named because they invariably colocalize with (or are adjacent to) the replication-dependent histone gene cluster (Liu et al. 2006). HLBs are enriched in factors required for histone gene transcription and pre-mRNA 3' end formation, including NPAT, FLASH and the U7 snRNP (Fig. 2). The distinction between CBs and HLBs in flies is more obvious, as the U7 snRNP and other factors involved in histone pre-mRNA processing localize exclusively to HLBs (Liu et al. 2006; Liu et al. 2009). In contrast, though human cancer cells harbor CBs numbering up to 10 per cell, all of which contain the U7 snRNP, only those that are associated with histone gene clusters contain factors required for histone transcription and processing (Frey and Matera 1995; Pillai et al. 2001). In human primary cells, however, U7 snRNP components colocalize exclusively with histone transcription and processing factors (Ghule et al. 2009). The latter situation is reminiscent of that in flies, thus helping to better classify these nuclear bodies into CBs and HLBs.
Cajal bodies and Histone locus bodies
The functional relationship between CBs and HLBs is not well understood, however, there are tantalizing similarities between the non-coding snRNA genes and the replication-dependent histone genes (Fig. 2). Both types of gene are transcribed by RNA pol II (Marzluff 2005; Egloff and Murphy 2008); transcripts are not polyadenylated despite the presence of canonical poly-A signals (Marzluff et al. 2008); both transcripts share similar RNA 3' processing reactions, although they are executed by different machineries (Weiner 2005); their genomic loci associate with distinct nuclear bodies that share dynamic components such as coilin (Frey and Matera 1995; Jacobs et al. 1999). Furthermore, the U7 snRNP, which plays a key role in histone pre-mRNA processing, follows the classical snRNP biogenesis and assembly pathway (Pillai et al. 2003; Azzouz et al. 2005), and may traverse through CBs (Bellini and Gall 1998) prior to localizing within HLBs. Another potential link between CBs and HLBs is the U2 snRNP, which has also been implicated in histone pre-mRNA biogenesis (Friend et al. 2007). The Drosphila system will likely shed more light on the structural and functional relationship between CBs and HLBs, as mutants representing different steps in the snRNP biogenesis and histone premRNA processing pathways are identified.
Cajal body crosstalk and heterogeneity
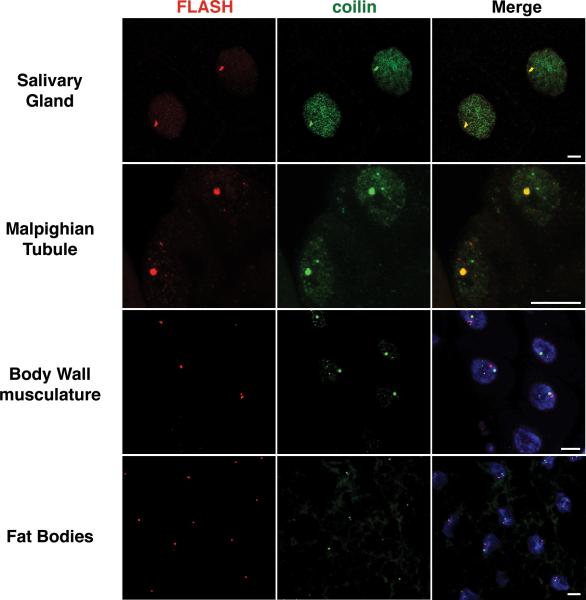
The morphology and composition of nuclear bodies is subject to physiological and developmental changes. This point is nicely illustrated using CBs and HLBs. As described by Liu et al. (Liu et al. 2009), the degree of association between CBs and HLBs changes between tissues and in the course of development (Fig.4). Importantly, this behavior is a characteristic feature of a given tissue or developmental time point. For example, in the Drosphila egg chambers, the CBs and HLBs largely remain as separate bodies during early stages, but coalesce into a single body in later stages, suggesting the existence of regulatory mechanisms.
Figure 4.
The behavior of nuclear bodies in relation to one another, and to other components of the nucleus, is characteristic of the tissue as well as developmental stage. (A and B) CBs (labeled green by coilin) and HLBs (labeled red by FLASH) colocalize in the salivary glands and Malpighian tubules of wandering (late) third instar larvae. In contrast, CBs and HLBs remain separate in salivary glands from early stage larvae (see also Fig. 5A), emphasizing the dependence of nuclear body behavior on developmental time as well as cell type. (C and D) The body wall musculature and fat bodies are shown as examples of tissues where CBs and HLBs remain as separate bodies in wandering third instar larvae. DAPI staining is included in the bottom two panels to mark the nuclei. Scale bar is 10 μm.
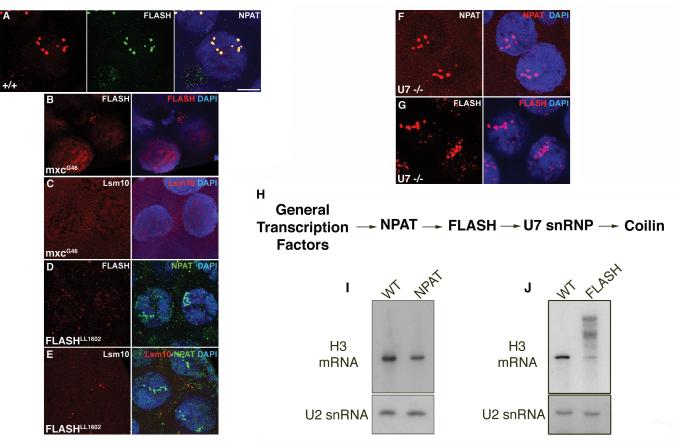
Some of the well-characterized components of vertebrate HLBs include FLASH, NPAT, ARS2, etc. (see Fig. 2). Initial bioinformatic analyses failed to find homologs for FLASH and NPAT in the fruitfly genome. However, Yang et al. (2009) recently identified the Drosphila FLASH ortholog, showing it to be a component of the HLB and essential for proper histone pre-mRNA processing. Subsequently, we and others identified multisexcombs (mxc /CG12124) as the Drosphila NPAT (data not shown and Z. Dominksi, R. Duronio and W. Marzluff pers. comm.). Similar to its mammalian counterpart, Drosphila NPAT colocalizes with FLASH in HLBs in all tissues and developmental stages (Fig. 6A and data not shown). Consistent with previous observations, NPAT- and FLASH-positive HLBs are distinct from CBs, but can colocalize with coilin in a tissue and development specific manner.
Figure 6.
HLB components are assembled in a sequential (hierarchical) manner. (A) WT nurse cells from stage 9 egg chambers stained with FLASH (red) and NPAT/mxc (green) show complete colocalization of the two proteins at the HLB. Lsm10 also localizes to the HLB (Liu et al., 2009 and data not shown) (B and C) In NPAT (mxcG43) mutant nurse cells FLASH (red, B) and Lsm10 (red, C) do not form a nuclear body (HLB). (D) NPAT (green) localizes to HLBs in the absence of FLASH (red) in FLASHLL1602 mutant nurse cells. (E) In contrast, Lsm10 (red), a component of U7 snRNP, fails to localize to HLBs in FLASH mutants. (F and G) In nurse cells from a null mutant of U7 snRNA, both NPAT (red, F) and FLASH (red, G) are present at the HLB. It should be noted that the morphology of the HLBs is affected in all the mutants described. DAPI (blue) shows the nuclei. Scale bar is 10μm. (H) From these observations a hierarchy for the recruitment of proteins to the HLB can be established. (I and J) Northern blots for histone H3 mRNA on ovaries from NPAT (mxcG43) and FLASHLL1602 mutants. NPAT mutants (I) show reduced levels of histone H3 mRNA, but no mis-processing, despite the absence of intact HLBs. In contrast, the loss of a processing and HLB component, such as FLASH, (J) results in mis-processed histone H3 mRNA, suggesting that the HLB per se is not essential for processing.
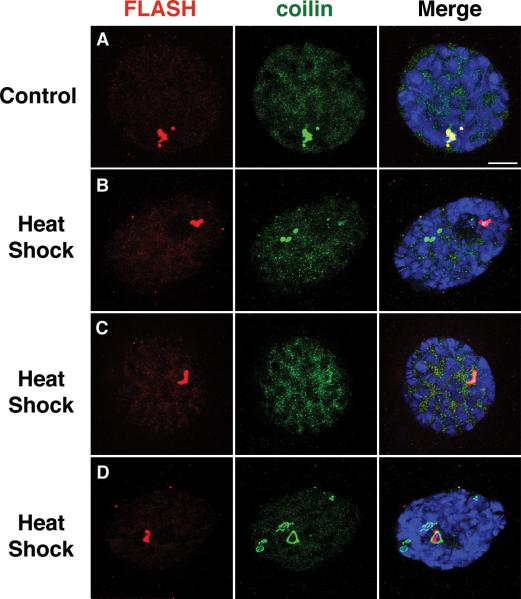
CBs also show sensitivity to physiological alterations and/or environmental assaults. As the temperature decreases, the size of CBs in cultured mammalian cells increases (Carmo-Fonseca et al. 1993). Likewise, with an increase in temperature, the size of the CBs decreases (Carmo-Fonseca et al. 1992). In Drosphila salivary glands from late third instar larvae, the effect of heat shock on coilin is significantly different. Under normal conditions, coilin colocalizes with HLBs in this tissue. Upon heat shock however, coilin becomes delocalized from the HLB and can form ectopic or abnormal structures. This effect of high temperature is specific to CBs, as HLBs remain unaffected (Fig. 5). Collectively, these data suggest that the behavior of nuclear bodies can be defined not just spatiotemporally during development, but also as a matter of physiological state.
Figure 5.
Effects of heat shock on coilin (green) and FLASH (red) in salivary glands from late third instar larvae. In WT salivary glands (without heat shock), coilin and FLASH always colocalize (A). After heat shock at 37°C for 45 min., although localization of FLASH (HLBs) remains unaffected, coilin fails to localize or be recruited to HLBs with (B) or without (C) forming ectopic bodies. Occasionally, residual amounts of coilin can be detected at the HLB. At times, aberrant coilin-positive structures are seen that are not observed under normal conditions (D). Scale bar is 10 μm.
The localization to CBs of proteins expressed in a tissue-specific manner or with tissue-specific functions suggests that the CBs in different tissues vary in their composition. For example, Drosphila ELAV, an RNA binding protein with neuronal specific functions localizes to CBs (Yannoni and White 1997) and scaDm46E3, a small CB associated RNA (scaRNA) of Drosphila germ plasm, localizes to CBs (Tortoriello et al. 2009). Further, the protein product of the gene muscle wasted (mute), involved in Drosphila muscle development, localizes to HLBs, although this localization was also seen in non-muscle cells (Bulchand et al. 2010).
The tissue-specific localization of factors to CBs is not confined to flies. An isoform of Amyloid-β Protein Precursor (AβPP) Intracellular Domain-Associated Protein-1 (AIDA-1d) translocates to CBs upon activation of NMDA receptors at the neuronal synapse (Jordan et al. 2007). Another isoform, AIDA-1c, shows brain-specific expression and localizes to CBs through its interaction with coilin (Xu and Hebert 2005). The localization of membrane-bound Zpr1 and AIDA-1 to CBs is signaling dependent. Zpr1 interacts with the SMN complex upon EGF receptor activation and subsequently localizes to CBs (Galcheva-Gargova et al. 1996; Gangwani et al. 2001). Thus, in addition to tissue-specific differences in the composition, CBs can change their inhabitant proteins in response to cell signaling events.
Self-organization of Nuclear Bodies
“Self-assembly” is a model proposed for the formation of macromolecular machineries (Camazine et al. 2001; Misteli 2001; Misteli 2007; Matera et al. 2009). Self-assembly applies to processes wherein pre-existing components of a disordered system are organized into a structure or pattern as a result of local interactions without external influence. Based on whether the assembly is static or dynamic, it is often classified in two ways. Static self-assembly, or “self-assembly” proper, refers to a state of order that emerges when the system approaches stable equilibrium with minimal free energy. In other words, static self-assembly refers to the aggregation of molecules into stable and well-defined complexes that are essentially glued into place by strong intermolecular interactions. For example, viral capsid proteins can be assembled into stable complexes from their constituents in the absence of auxiliary factors or external influences (Zlotnick 1994). In contrast, dynamic self-assembly, which is also referred to as “self-organization” refers to patterns that emerge by specific local interactions between pre-existing components wherein the system is said to achieve a steady-state, requiring a constant flow of components through the structure. In the context of the cell, self-organization is “the capacity of a macromolecular complex or organelle to determine its own structure, based on the functional interactions of its components” (Misteli 2001). Many nuclear compartments are thought to arise by self-organization. In this chapter, self-assembly refers to static self-assembly and self-organization refers to dynamic self-assembly.
Self-organized patterns are brought about by two different mechanisms. (i) By stochastic means, wherein no specific order to the assembly of its components is necessary. (ii) By an ordered or hierarchical assembly, with factors recruited one after the other, and recruitment of a subsequent component depends on recruitment of the preceding component.
Stochastic self-organization
In the context of nuclear bodies, the stochastic model for self-organization is exemplified by the recent report of Kaiser et al. (2008) who used a genomic tethering assay to analyze individual CB components for their abilities to nucleate formation of a nuclear body. The approach utilizes the lac repressor/operator system to tether a given CB component to a single genomic locus in order to test its ability to recruit the other components and thus self-organize a CB. Though this technical advancement is experimentally elegant, it suffers from a serious drawback. The main problem is that the act of tethering a given CB component to DNA using the lac repressor fusion is tantamount to creating a dominant/gain-of-function mutation (in this case DNA binding). The evidence suggests that the tethering system faithfully recapitulates the normal protein-protein interactions that otherwise take place within a CB (Kaiser et al. 2008), however, it is rather more like performing a biochemical-pulldown experiment in situ than it is a model for nuclear body formation. The stochastic nature of protein-protein interactions within cells is not at issue here. Instead, the question is whether tethering a normally dynamic component to a chromosome creates an artificial situation that does not faithfully reproduce the CB nucleation event.
Stochastic self-organization fails to explain why in some tissues CBs and HLBs are organized specifically as a single ‘organelle’ and while in others as two separate entities (Fig. 4). Implicating the presence or absence of a tissue-specific ‘sticky’ factor to assemble CBs and HLBs as one or two separate nuclear bodies places that sticky protein at the top of a hierarchy. Furthermore, the presence of ‘residual’ CBs in cells derived from coilin knockout mice (Tucker et al. 2001; Jady et al. 2003) suggests that although coilin is required to recruit splicing snRNPs and the SMN complex in order to constitute a proper CB, other CB components (e.g. snoRNPs) self-organize into other nuclear bodies in the absence of coilin. By implication, these residual CBs could act as one of the initial, upstream steps in a hierarchical assembly pathway upon which coilin pulls together a full-fledged CB.
Hierarchical self-organization
In a “hierarchically ordered” assembly pathway, components are recruited in sequential order to an initial stochastic seed, churned out of nuclear chaos, to elaborate a higher order structure. Hierarchical assembly within a stochastic molecular crowd seems self-contradictory at first but is, in fact, complementary. Biological systems have reiteratively used this model in building various macromolecular assemblies such as spliceosomes (including the U4/U6 di-snRNP), ribosomes, the exon-junction complex and cell-matrix adhesion complexes (Nottrott et al. 2002; Zaidel-Bar et al. 2004; Gehring et al. 2009; Staley and Woolford 2009). These macro-molecular assemblies are reportedly built de novo , from scratch, with the initial events being purely stochastic, leading to final higher-order structures built by hierarchically ordered recruitments.
An implication of this model is that disassembly should follow the reverse order of assembly. Contrastingly, in stochastic self-organization, both assembly and disassembly should be random. Though little is known about the disassembly of nuclear bodies, disassembly of mitotic counterparts of nuclear speckles (called mitotic interchromatin granules, or MIGs) is instructive in this regard. Rather than being stochastic, disassembly of MIGs at the final stages of mitosis occurs with each component released and subsequently imported into the daughter nuclei in a reproducible, hierarchical order (Prasanth et al. 2003).
The stochastic self-organization model requires that many, if not all, nuclear body components should be sufficient to nucleate body formation. In contrast, if bodies form hierarchically, only “upstream” components should be able to nucleate formation of a body. The time-honored way to study a biological pathway is not by using gain-of-function mutants, but by biochemical reconstitution or genetic loss-of-function studies. In the paragraphs below, we'll explore what the latter approach has told us.
Genetic dissection of HLB assembly
Null mutations in Drosphila coilin cause CB disassembly but do not affect the organization of HLBs (Liu et al. 2009), consistent with the observation that depletion of coilin by shRNA in human cells does not affect colocalization of HLB markers, FLASH and NPAT (Barcaroli et al. 2006a; Barcaroli et al. 2006b). Thus, coilin has little effect on HLB formation. In fact, HLB-like foci remain detectable even in the background of a histone gene cluster deletion (White et al. 2007). These findings raised the question of whether there is a critical component upon which HLB assembly depends.
The search for an HLB nucleator began by employing mutations in known components of HLBs, including NPAT, FLASH and the U7 snRNP (Fig. 6). In null mutants for U7 snRNA, both FLASH and NPAT could still be detected at the HLB. In FLASH mutants, U7 snRNP was delocalized from the HLB, however, NPAT could still be detected at the body. Finally, a hypomorphic mutation in NPAT (mxcG43) , causes delocalization of both FLASH and the U7 snRNP (as detected by staining for Lsm10). Collectively, the data show that NPAT is a critical factor, without which downstream factors do not accumulate in HLBs (Fig. 6).
Placement of FLASH upstream of U7 snRNP in the heirarchy of HLB formation has interesting implications for the relationship between HLBs and histone processing. For example, based on the biochemical data, it has been proposed that U7 snRNP recruits the N-terminal fragment of human FLASH to histone pre-mRNA during processing (Yang et al. 2009). Assuming FLASH and U7 snRNP recruitment to the histone pre-mRNA is not concomitant, but sequential (U7 snRNP followed by FLASH), this biochemical result places U7 snRNP upstream of FLASH during processing, whereas U7 snRNP is downstream of FLASH in HLB formation. Even if FLASH and U7 snRNP recruitment to the histone pre-mRNA processing machinery is concomitant, then the sequence by which an HLB is built is obviously different from the cascade through which factors are recruited by the histone pre-mRNA processing machinery.
Given the fact that NPAT (mxcG43) mutants are viable (albeit female sterile), the absence of proper HLBs suggests that the structure per se is dispensable for life. Furthermore, hypomorphic mutations in histone stem-loop binding protein (SLBP; Sullivan et al. 2001) and FLASH (our unpublished observations), as well as null mutations in U7 snRNA (Godfrey et al. 2006) are viable. This is thought to be due to the fact that mutations in these factors result in the generation of polyadenylated histone mRNAs, which rescue zygotic viability. Thus, in the absence of a full complement of histone pre-mRNA processing factors, the animals can reach adulthood. These data strongly argue for the dispensability of HLBs, at least in Drosphila .
Histone pre-mRNA processing regulates CB organization
Though the organization of HLBs is independent of coilin or CBs as seen in coilin mutant flies, the two structures associate/colocalize with each other in a developmentally regulated manner. During later stages of Drosphila oogenesis (stages ~9-12), markers for CBs and HLBs completely colocalize, whereas they are typically separate structures during earlier stages (Liu et al. 2009). However, in ovaries from mutants of histone pre-mRNA processing factors, including U7 snRNP, SLBP and FLASH (Fig. 7), coilin is organized in the form of a ‘cloud’ that invariably clusters around the HLBs. In contrast to histone pre-mRNA processing mutants, NPAT hypomorphs (mxcG43) show a pan-nuclear distribution of coilin, with no identifiable CBs (Fig. 7) and no effects on histone H3 pre-mRNA processing, although overall expression is reduced (Fig. 6). Furthermore, no effect on CB organization (in larval stages) was observed in lethal alleles of NPAT (mxcG48). Collectively, the data show that the reorganization of coilin into micro-foci around the HLBs is specific to defects in histone pre-mRNA processing, rather than histone transcription. While no biological reason for the ‘coilin cloud’ phenotype is currently known, these data imply a relationship between the function of histone pre-mRNA processing and CB organization.
Figure 7.
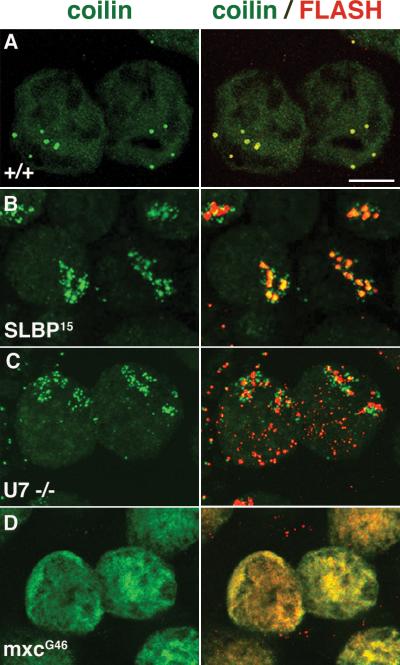
Histone processing defects affect the localization of coilin in Drosphila. (A) Stage 9-10 nurse cells from WT flies stained with coilin (green) to label CBs and FLASH (red) to label the HLBs. The right panels show staining for coilin alone and the left panels show the colocalization of coilin and FLASH. At these stages coilin and FLASH colocalize completely. (B and C) Nurse cells from histone pre-mRNA processing mutants, SLBP and U7 snRNA, respectively, showing the delocalization of coilin into micro-foci around the HLBs (the ‘coilin cloud’ phenotype). Note that, in the U7 mutants, FLASH localizes to HLBs but also shows an abnormal distribution throughout the nucleus. (D) Nurse cells from hypomorphic mutants (mxcG43) in the histone transcription factor, NPAT, show an absence of CBs, with a pan-nuclear distribution of coilin after stages 9-10 of oogenesis. In these mutants, FLASH also fails to accumulate in nuclear foci.
Conclusions
Knockout experiments in mice have shown that coilin and CBs are not absolutely essential for life, but their absence affects overall fitness, with major effects on the fecundity and viability of inbred strains (Tucker et al. 2001; Walker et al. 2009). The Drosphila coilin null mutants display no obvious abnormalities, despite having no CBs (Liu et al. 2009). These observations are surprising in the light of the high conservation of CBs across the evolutionary ladder. However, they suggest that CBs provide added specificity and efficiency to snRNP assembly and modification reactions, rather than being critical for the occurrence of these activities. In this context, the observations made in coilin-depleted zebrafish embryos are more compelling. Depletion of coilin in zebrafish embryos leads to a loss of CBs, and developmental arrest, but can be rescued by the addition of pre-assembled snRNPs (Strzelecka et al. 2010). Perhaps at the early embryonic stages, the demand for snRNPs is sufficiently high that the lack of efficiency in their assembly in the absence of CBs has a negative impact on development. Based on the studies of CBs, we can postulate a similar function for Drosphila HLBs. A mutation in the histone transcription factor NPAT (mxcG43), disrupts its ability to recruit FLASH, a processing factor, to the HLB. Despite the complete delocalization of FLASH (and other downstream HLB components) in this background, histone pre-mRNA 3' end processing is unaffected (Fig. 6). Whether the overall reduction in histone mRNA levels observed in these mutants is due to a bottleneck in pre-mRNA processing or if it is caused by a decrease in transcription initiation has yet to be shown.
Observations from the various model organisms have not only been enlightening in terms of the function of CBs and HLBs, but also in their assembly. It is widely believed that these structures assemble via self-organization, however, whether this occurs in a stochastic or hierarchical manner is a matter of debate. Our studies using Drosphila loss-of-function mutations have revealed the existence of a putative nucleator (NPAT) of HLBs, and a sequence to the recruitment of factors to the HLB. Although the initial seed for assembly of these nuclear bodies may well be stochastic, the subsequent recruitment of factors exemplifies a hierarchical model of self-organization.
Acknowledgements
We thank J. Gall and Z. Dominski for antibodies, the Bloomington and Kyoto stock centers for fly lines and Z. Dominski, R. Duronio and W. Marzluff for communicating results prior to publication. Work in the Matera laboratory was supported by grants from the NIH (R01-NS041617 and R01-GM053034).
References
- Azzouz TN, Pillai RS, Dapp C, Chari A, Meister G, Kambach C, Fischer U, Schumperli D. Toward an assembly line for U7 snRNPs: interactions of U7-specific Lsm proteins with PRMT5 and SMN complexes. J Biol Chem. 2005;280:34435–34440. doi: 10.1074/jbc.M505077200. [DOI] [PubMed] [Google Scholar]
- Barcaroli D, Bongiorno-Borbone L, Terrinoni A, Hofmann TG, Rossi M, Knight RA, Matera AG, Melino G, De Laurenzi V. FLASH is required for histone transcription and S-phase progression. Proc Natl Acad Sci U S A. 2006a;103:14808–14812. doi: 10.1073/pnas.0604227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcaroli D, Dinsdale D, Neale MH, Bongiorno-Borbone L, Ranalli M, Munarriz E, Sayan AE, McWilliam JM, Smith TM, Fava E, Knight RA, Melino G, De Laurenzi V. FLASH is an essential component of Cajal bodies. Proc Natl Acad Sci U S A. 2006b;103:14802–14807. doi: 10.1073/pnas.0604225103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini M, Gall JG. Coilin can form a complex with the U7 small nuclear ribonucleoprotein. Mol Biol Cell. 1998;9:2987–3001. doi: 10.1091/mbc.9.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulchand S, Menon SD, George SE, Chia W. Muscle wasted: a novel component of the Drosophila histone locus body required for muscle integrity. J Cell Sci. 2010;123:2697–2707. doi: 10.1242/jcs.063172. [DOI] [PubMed] [Google Scholar]
- Camazine S, Deneubourg J-L, Franks NR, Theraulaz G, Bonabeau E. Self-organization in biological systems. Princeton University Press; Princeton, NJ: 2001. [Google Scholar]
- Carmo-Fonseca M, Ferreira J, Lamond AI. Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis--evidence that the coiled body is a kinetic nuclear structure. J Cell Biol. 1993;120:841–852. doi: 10.1083/jcb.120.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho T, Almeida F, Calapez A, Lafarga M, Berciano MT, Carmo-Fonseca M. The spinal muscular atrophy disease gene product, SMN: A link between snRNP biogenesis and the Cajal (coiled) body. J Cell Biol. 1999;147:715–728. doi: 10.1083/jcb.147.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer M, Dietzel S, Muller S, Solovei I, Fakan S. Chromosome territories--a functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol. 2007;179:1095–1103. doi: 10.1083/jcb.200710058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Ferreira JA, Carmo-Fonseca M, Lamond AI. Differential interaction of splicing snRNPs with coiled bodies and interchromatin granules during mitosis and assembly of daughter cell nuclei. J Cell Biol. 1994;126:11–23. doi: 10.1083/jcb.126.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr Opin Cell Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- Frey MR, Bailey AD, Weiner AM, Matera AG. Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr Biol. 1999;9:126–135. doi: 10.1016/s0960-9822(99)80066-9. [DOI] [PubMed] [Google Scholar]
- Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci U S A. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey MR, Matera AG. RNA-mediated interaction of Cajal bodies and U2 snRNA genes. J Cell Biol. 2001;154:499–509. doi: 10.1083/jcb.200105084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend K, Lovejoy AF, Steitz JA. U2 snRNP binds intronless histone pre-mRNAs to facilitate U7-snRNP-dependent 3' end formation. Mol Cell. 2007;28:240–252. doi: 10.1016/j.molcel.2007.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galcheva-Gargova Z, Konstantinov KN, Wu IH, Klier FG, Barrett T, Davis RJ. Binding of zinc finger protein ZPR1 to the epidermal growth factor receptor. Science. 1996;272:1797–1802. doi: 10.1126/science.272.5269.1797. [DOI] [PubMed] [Google Scholar]
- Gangwani L, Mikrut M, Theroux S, Sharma M, Davis RJ. Spinal muscular atrophy disrupts the interaction of ZPR1 with the SMN protein. Nat Cell Biol. 2001;3:376–383. doi: 10.1038/35070059. [DOI] [PubMed] [Google Scholar]
- Gehring NH, Lamprinaki S, Hentze MW, Kulozik AE. The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol. 2009;7:e1000120. doi: 10.1371/journal.pbio.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghule PN, Dominski Z, Lian JB, Stein JL, van Wijnen AJ, Stein GS. The subnuclear organization of histone gene regulatory proteins and 3' end processing factors of normal somatic and embryonic stem cells is compromised in selected human cancer cell types. J Cell Physiol. 2009;220:129–135. doi: 10.1002/jcp.21740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard C, Neel H, Bertrand E, Bordonne R. Depletion of SMN by RNA interference in HeLa cells induces defects in Cajal body formation. Nucleic Acids Res. 2006;34:2925–2932. doi: 10.1093/nar/gkl374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey AC, Kupsco JM, Burch BD, Zimmerman RM, Dominski Z, Marzluff WF, Duronio RJ. U7 snRNA mutations in Drosophila block histone pre-mRNA processing and disrupt oogenesis. RNA. 2006;12:396–409. doi: 10.1261/rna.2270406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert MD, Shpargel KB, Ospina JK, Tucker KE, Matera AG. Coilin methylation regulates nuclear body formation. Dev Cell. 2002;3:329–337. doi: 10.1016/s1534-5807(02)00222-8. [DOI] [PubMed] [Google Scholar]
- Hebert MD, Szymczyk PW, Shpargel KB, Matera AG. Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev. 2001;15:2720–2729. doi: 10.1101/gad.908401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs EY, Frey MR, Wu W, Ingledue TC, Gebuhr TC, Gao L, Marzluff WF, Matera AG. Coiled bodies preferentially associate with U4, U11, and U12 small nuclear RNA genes in interphase HeLa cells but not with U6 and U7 genes. Mol Biol Cell. 1999;10:1653–1663. doi: 10.1091/mbc.10.5.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Darzacq X, Tucker KE, Matera AG, Bertrand E, Kiss T. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J. 2003;22:1878–1888. doi: 10.1093/emboj/cdg187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan BA, Fernholz BD, Khatri L, Ziff EB. Activity-dependent AIDA-1 nuclear signaling regulates nucleolar numbers and protein synthesis in neurons. Nat Neurosci. 2007;10:427–435. doi: 10.1038/nn1867. [DOI] [PubMed] [Google Scholar]
- Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonne R, Luhrmann R. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol Cell. 2006;17:3221–3231. doi: 10.1091/mbc.E06-03-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt H, Cardoso MC. Targeting and association of proteins with functional domains in the nucleus: the insoluble solution. Int Rev Cytol. 1995;162B:303–335. doi: 10.1016/s0074-7696(08)62620-0. [DOI] [PubMed] [Google Scholar]
- Liu JL, Murphy C, Buszczak M, Clatterbuck S, Goodman R, Gall JG. The Drosophila melanogaster Cajal body. J Cell Biol. 2006;172:875–884. doi: 10.1083/jcb.200511038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JL, Wu Z, Nizami Z, Deryusheva S, Rajendra TK, Beumer KJ, Gao H, Matera AG, Carroll D, Gall JG. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol Biol Cell. 2009;20:1661–1670. doi: 10.1091/mbc.E08-05-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF. Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr Opin Cell Biol. 2005;17:274–280. doi: 10.1016/j.ceb.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell. 2009;17:639–647. doi: 10.1016/j.devcel.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Shpargel KB. Pumping RNA: nuclear bodybuilding along the RNP pipeline. Curr Opin Cell Biol. 2006;18:317–324. doi: 10.1016/j.ceb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Misteli T. The concept of self-organization in cellular architecture. J Cell Biol. 2001;155:181–185. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Nesic D, Tanackovic G, Kramer A. A role for Cajal bodies in the final steps of U2 snRNP biogenesis. J Cell Sci. 2004;117:4423–4433. doi: 10.1242/jcs.01308. [DOI] [PubMed] [Google Scholar]
- Nottrott S, Urlaub H, Luhrmann R. Hierarchical, clustered protein interactions with U4/U6 snRNA: a biochemical role for U4/U6 proteins. EMBO J. 2002;21:5527–5538. doi: 10.1093/emboj/cdf544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. The Nucleus Introduced. Cold Spring Harb Perspect Biol. 2010 doi: 10.1101/cshperspect.a000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Grimmler M, Meister G, Will CL, Luhrmann R, Fischer U, Schumperli D. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 2003;17:2321–2333. doi: 10.1101/gad.274403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Will CL, Luhrmann R, Schumperli D, Muller B. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 2001;20:5470–5479. doi: 10.1093/emboj/20.19.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Sacco-Bubulya PA, Prasanth SG, Spector DL. Sequential entry of components of the gene expression machinery into daughter nuclei. Mol Biol Cell. 2003;14:1043–1057. doi: 10.1091/mbc.E02-10-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 2007;21:3027–3043. doi: 10.1101/gad.1604607. [DOI] [PubMed] [Google Scholar]
- Shpargel KB, Matera AG. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc Natl Acad Sci U S A. 2005;102:17372–17377. doi: 10.1073/pnas.0508947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman JE, Ajuh P, Lamond AI. snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J Cell Sci. 2001;114:4407–4419. doi: 10.1242/jcs.114.24.4407. [DOI] [PubMed] [Google Scholar]
- Sleeman JE, Lamond AI. Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr Biol. 1999;9:1065–1074. doi: 10.1016/s0960-9822(99)80475-8. [DOI] [PubMed] [Google Scholar]
- Sleeman JE, Trinkle-Mulcahy L, Prescott AR, Ogg SC, Lamond AI. Cajal body proteins SMN and Coilin show differential dynamic behaviour in vivo. J Cell Sci. 2003;116:2039–2050. doi: 10.1242/jcs.00400. [DOI] [PubMed] [Google Scholar]
- Spector DL. SnapShot: Cellular bodies. Cell. 2006;127:1071. doi: 10.1016/j.cell.2006.11.026. [DOI] [PubMed] [Google Scholar]
- Staley JP, Woolford JL., Jr. Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr Opin Cell Biol. 2009;21:109–118. doi: 10.1016/j.ceb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek D, Neugebauer KM. The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma. 2006;115:343–354. doi: 10.1007/s00412-006-0056-6. [DOI] [PubMed] [Google Scholar]
- Stanek D, Pridalova-Hnilicova J, Novotny I, Huranova M, Blazikova M, Wen X, Sapra AK, Neugebauer KM. Spliceosomal small nuclear ribonucleoprotein particles repeatedly cycle through Cajal bodies. Mol Biol Cell. 2008;19:2534–2543. doi: 10.1091/mbc.E07-12-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein GS, Zaidi SK, Braastad CD, Montecino M, van Wijnen AJ, Choi JY, Stein JL, Lian JB, Javed A. Functional architecture of the nucleus: organizing the regulatory machinery for gene expression, replication and repair. Trends Cell Biol. 2003;13:584–592. doi: 10.1016/j.tcb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Strzelecka M, Trowitzsch S, Weber G, Luhrmann R, Oates AC, Neugebauer KM. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat Struct Mol Biol. 2010;17:403–409. doi: 10.1038/nsmb.1783. [DOI] [PubMed] [Google Scholar]
- Sullivan E, Santiago C, Parker ED, Dominski Z, Yang X, Lanzotti DJ, Ingledue TC, Marzluff WF, Duronio RJ. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev. 2001;15:173–187. doi: 10.1101/gad.862801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Izumi H, Ohno M. Cajal body surveillance of U snRNA export complex assembly. J Cell Biol. 2010;190:603–612. doi: 10.1083/jcb.201004109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell. 2006;17:955–965. doi: 10.1091/mbc.E05-09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortoriello G, Accardo MC, Scialo F, Angrisani A, Turano M, Furia M. A novel Drosophila antisense scaRNA with a predicted guide function. Gene. 2009;436:56–65. doi: 10.1016/j.gene.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Tucker KE, Berciano MT, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ, Chan EK, Lafarga M, Conlon RA, Matera AG. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol. 2001;154:293–307. doi: 10.1083/jcb.200104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen C, Lafontaine DL, Samarsky D, Mouaikel J, Blanchard JM, Bordonne R, Bertrand E. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 2002;21:2736–2745. doi: 10.1093/emboj/21.11.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MP, Tian L, Matera AG. Reduced viability, fertility and fecundity in mice lacking the cajal body marker protein, coilin. PLoS One. 2009;4:e6171. doi: 10.1371/journal.pone.0006171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AM. E Pluribus Unum: 3' end formation of polyadenylated mRNAs, histone mRNAs, and U snRNAs. Mol Cell. 2005;20:168–170. doi: 10.1016/j.molcel.2005.10.009. [DOI] [PubMed] [Google Scholar]
- White AE, Leslie ME, Calvi BR, Marzluff WF, Duronio RJ. Developmental and cell cycle regulation of the Drosophila histone locus body. Mol Biol Cell. 2007;18:2491–2502. doi: 10.1091/mbc.E06-11-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Hebert MD. A novel EB-1/AIDA-1 isoform, AIDA-1c, interacts with the Cajal body protein coilin. BMC Cell Biol. 2005;6:23. doi: 10.1186/1471-2121-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Burch BD, Yan Y, Marzluff WF, Dominski Z. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3' end processing of histone pre-mRNAs. Mol Cell. 2009;36:267–278. doi: 10.1016/j.molcel.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannoni YM, White K. Association of the neuron-specific RNA binding domain-containing protein ELAV with the coiled body in Drosophila neurons. Chromosoma. 1997;105:332–341. doi: 10.1007/BF02529748. [DOI] [PubMed] [Google Scholar]
- Young PJ, Le TT, Dunckley M, Nguyen TM, Burghes AH, Morris GE. Nuclear gems and Cajal (coiled) bodies in fetal tissues: nucleolar distribution of the spinal muscular atrophy protein, SMN. Exp Cell Res. 2001;265:252–261. doi: 10.1006/excr.2001.5186. [DOI] [PubMed] [Google Scholar]
- Young PJ, Le TT, thi Man N, Burghes AH, Morris GE. The relationship between SMN, the spinal muscular atrophy protein, and nuclear coiled bodies in differentiated tissues and cultured cells. Exp Cell Res. 2000;256:365–374. doi: 10.1006/excr.2000.4858. [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R, Cohen M, Addadi L, Geiger B. Hierarchical assembly of cell-matrix adhesion complexes. Biochem Soc Trans. 2004;32:416–420. doi: 10.1042/BST0320416. [DOI] [PubMed] [Google Scholar]
- Zhao R, Bodnar MS, Spector DL. Nuclear neighborhoods and gene expression. Curr Opin Genet Dev. 2009;19:172–179. doi: 10.1016/j.gde.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnick A. To build a virus capsid. An equilibrium model of the self-assembly of polyhedral protein complexes. J Mol Biol. 1994;241:59–67. doi: 10.1006/jmbi.1994.1473. [DOI] [PubMed] [Google Scholar]