Abstract
Imperfect maintenance of genome integrity has been postulated to be an important cause of aging. Here we provide support for this hypothesis by demonstrating that the disruption of PASG (lsh), a SNF2-like factor that facilitates DNA methylation, causes global hypomethylation, developmental growth retardation and a premature aging phenotype. PASG mutant mice display signs of growth retardation and premature aging, including low birth weight, failure to thrive, graying and loss of hair, reduced skin fat deposition, osteoporosis, kyphosis, cachexia, and premature death. Fibroblasts derived from PASG mutant embryos show a replicative senescence phenotype. Both PASG mutant mice and fibroblasts demonstrate a markedly increased expression of senescence-associated tumor suppressor genes, such as p16INK4a, that is independent of promoter methylation, but, instead, is associated with down-regulation of bmi-1, a negative regulator of p16INK4a. These studies show that PASG is essential for properly maintaining DNA methylation and gene expression patterns that are required for normal growth and longevity. PASG mutant mice provide a useful model for the study of aging as well as the mechanisms regulating epigenetic patterning during development and postnatal life.
Keywords: PASG, lsh, SMARCA6, methylation, genomic instability, aging, senescence
Understanding the biological mechanisms underlying aging and cancer predisposition remains a fundamentally important goal in biomedicine. The limited life span of most cells is considered to be the result of a process termed cellular or replicative senescence (Campisi 1996; Sherr and DePinho 2000), which is believed to be due to physiologic responses to a variety of cellular stresses, including DNA damage and replication (Hayflick 1965; Hasty et al. 2003), telomere erosion (Sharpless and DePinho 2004), and aberrantly activated oncogenes (Lundberg et al. 2000; Campisi 2001). This physiological response appears to trigger growth inhibition via activation of tumor suppressor genes, such as p16INK4a, p19ARF, p53, and p21 (Alcorta et al. 1996; Kiyono et al. 1998; Stein et al. 1999; Ramirez et al. 2001; Randle et al. 2001; Rheinwald et al. 2002; Schmitt et al. 2002; Tyner et al. 2002). Recent data show that bmi-1 is required for the maintenance of tissue homeostasis in some tissues partly because it represses genes that induce cellular senescence and cell death (Jacobs et al. 1999; Itahana et al. 2003; Lessard and Sauvageau 2003; Park et al. 2003, 2004). Furthermore, although the importance of epigenetic modification of DNA and chromatin in normal development and in cancer formation has been established (Li et al. 1992; Okano et al. 1999; Li 2002; Robertson 2002; Eden et al. 2003; Gaudet et al. 2003; Jaenisch and Bird 2003), the relationship to aging or senescence has not been extensively investigated (Issa 1999; Bandyopadhyay and Medrano 2003; Dunn 2003; Richardson 2003).
Decreased 5-methyl-cytosine content of genomic DNA has been observed in aging cells in culture and in animals (Wilson and Jones 1983; Wilson et al. 1987; Barbot et al. 2002). Treatment of cells in culture with DNA methyltransferase (DNMT) inhibitors, such as 5-aza-2′-deoxycytidine, results in a shortened life span compared to untreated control cells (Holliday 1986; Fairweather et al. 1987). DNMT inhibition by antisense oligonucleotides leads to DNA hypomethylation and a senescence-like phenotype, mediated by increased expression of the p16INK4a and p21 tumor suppressor genes in human fibroblasts and cancer cells (Fournel et al. 1999; Young and Smith 2001). Disruption of DNA methyltransferases in primary mouse embryo fibroblasts (MEFs) and human cancer cells is also associated with significantly reduced cell proliferation and aberrant gene expression (Jackson-Grusby et al. 2001; Rhee et al. 2002). These in vitro data suggest that decreasing the level of 5-methyl-cytosine in DNA may be an important molecular mechanism leading to senescence, a concept proposed by Holliday nearly 20 years ago (Holliday 1985). However, in vivo studies investigating premature aging in the context of genomic hypomethylation remain largely unexplored.
Recent studies have linked the ATP-dependent SWI/SNF chromatin remodeling proteins to the regulation of epigenetic modification of the genome, which, in turn, contributes to embryogenesis, cellular proliferation, and oncogenesis through the modulation of transcription, replication, and genome stability (Jeddeloh et al. 1999; Gibbons et al. 2000; Meehan et al. 2001; Li 2002; Robertson 2002; Jaenisch and Bird 2003). PASG (proliferation associated SNF-2-like gene) or lsh (lymphocyte-specific helicase) encodes a SNF-2-like protein that has been shown to contribute to global genome methylation (Lee et al. 2000; Dennis et al. 2001). The term lsh was originally based on findings that suggested it to be expressed as a lymphocyte-specific helicase (Jarvis et al. 1996). Subsequent studies showed that PASG (lsh) is expressed ubiquitously in rapidly dividing cells or tissues (Raabe et al. 2001) and is linked to cell proliferation (Geiman and Muegge 2000; Raabe et al. 2001; Fan et al. 2003). The genetic locus has been termed SMARCA6 (Lee et al. 2000). Lsh-/- mice, in which exons 6 and 7 containing helicase domains I, Ia, and part of II were deleted, displayed perinatal mortality with only one out of 480 mice surviving to 5 d postnatally (Geiman et al. 2001). Newborns were noted to be 22% lower in body weight than wild-type littermates and were noted to have renal tubular abnormalities (Geiman et al. 2001). However, because the null animals died within hours of being born, additional conclusions were not able to be made. To further investigate the functional consequences of PASG (lsh) on epigenetic patterning as well as on normal growth and development, we generated a mouse carrying a hypomorphic mutation of PASG by deleting exons 10, 11, and 12 containing helicase domains III, IV, and part of II using homologous recombination. The results demonstrate that targeted disruption of PASG not only leads to both genomic hypomethylation and re-expression of repressed genes, but also to premature aging that is associated with decreased proliferation and increased replicative senescence along with altered gene expression patterns, particularly the senescence-related genes such as p16INK4a and bmi-1.
Results
Targeted disruption of PASG and abnormal genomic methylation
Mice with disruption of PASG expression were generated through homologous recombination by deleting exons 10-12, which included the conserved helicase domains II, III, IV (Fig. 1A). Successful targeting of PASG was verified by Southern blot (Fig. 1B), Northern blot hybridization (Fig. 1C), and by Western blot analysis (Fig. 1D). Although a truncated form of mRNA with predicted length (456-bp cDNA was deleted) was observed in the heterozygous (PASG+/-) and homozygous (PASG-/-) mice (Fig. 1C), a short form of PASG protein with deletion of 152 amino acids was not observed by Western blots using a rabbit antibody against the C-terminal sequences of PASG, which was not in the deletion region of the protein (Fig. 1D), suggesting that the truncated protein was possibly not stable or below the level of detection. To further test this possibility by enriching for PASG, we immunoprecipitated PASG with anti-lsh antibody and then did a Western blot of the immunoprecipitated protein with a rabbit antibody against the C-terminal sequences of PASG (Raabe et al. 2001). These results demonstrated that homozygous and heterozygous mice expressed a truncated PASG protein of the predicted size, although its expression level was low (Fig. 1E).
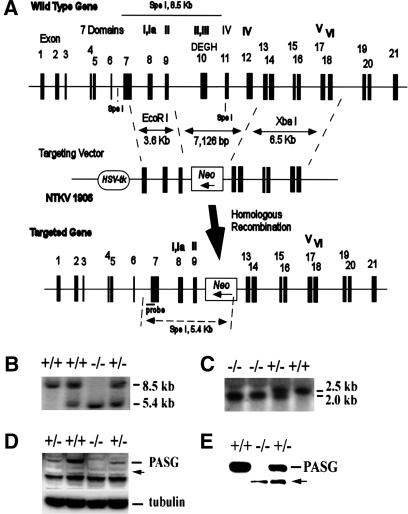
Figure 1.
Targeted disruption of PASG by homologous recombination. (A) Genomic map of the targeting vector and predicted wild-type and disrupted alleles. The black boxes with numbers above them represent the 21 PASG exons. Roman numbers indicate the putative seven-helicase domains with domain I containing the ATP-binding site and domain II containing the DExH box. Probe indicates the 5′ genomic probe external to the targeted region used for Southern blot analysis. (B) Southern blot analysis shows the expected targeted disruption of the PASG gene. Genomic DNA was digested with SpeI and hybridized with 5′ genomic probe. The 8.5-kb fragment indicates the wild-type allele and a 5.4-kb fragment represents the mutant allele. (C) Northern blot analysis of the mRNA purified from E14.5 embryos shows a short form of the PASG transcript (2.0 kb) in heterozygous and homozygous mice. The probe is a 1.0-kb cDNA fragment from the C terminus of PASG ORF. (D) Western analysis of PASG protein shows that the truncated PASG protein is not detected in PASG-/- animals and PASG+/-. The arrow indicates the position of predicted short form of PASG. Nuclear extracts from MEFs were electrophoretically separated, transferred for Western blotting, and exposed to a rabbit antiserum raised against a selected peptide in the C-terminal region of PASG as described in Materials and Methods. The molecular mass of the wild-type and predicted short form of PASG is 95.2 kD and 77.4 kD, respectively. (E) Immunoprecipitation of PASG followed by Western blot analysis showed a low level of truncated PASG protein was expressed in PASG-/- and PASG+/- MEFs. The arrow indicates the predicted short form of PASG. MEF cell lysate was immunoprecipitated with anti-lsh antibody and Western analysis was done with a rabbit antiserum raised against a selected peptide in the C-terminal region of PASG as described in Materials and Methods.
To quantify the methylation level in these PASG-/- mice, we directly measured 5-methyl-cytosine in genomic DNA using MspI digestion, 5′-end labeling, and thin-layer chromatographic separation of methyl-cytosine and cytosine (Fig. 2A). The percentage of methylcytosine in total body DNA of embryonic day 13.5 (E13.5) embryos is 54% and 36% in wild-type and PASG-/- embryos respectively (a 33% decrease). Similar results are observed in the E17.5 liver DNA from wild-type and PASG-/- embryos, showing a 56% and 32% level of methyl-cytosine, respectively (a 43% decrease). The methyl-cytosine content in heterozygous mice is similar to wild-type mice (data not shown).
Figure 2.
PASG-/- embryos and mice show genomic hypomethylation, gene re-expression, growth retardation, and early death. (A) Quantitation of methyl-cytosine in genomic DNA by thin layer chromatography shows decreased levels in PASG-/- embryonic DNA at gestation days 13.5 and 17.5. Equal amounts of genomic DNA from E13.5 body or E17.5 liver were digested with MspI, labeled at 5′ ends, redigested with nuclease P1 to generate 5′-deoxymononucleotides, which were separated by thin layer chromatography and quantitated using a PhosphorImager. The ratio of methyl-cytosine to total cytosine indicates the level of methylation at all CCGG sites. HpaII digests should not generate methyl-cytosine spots and they serve as controls, indicating the specificity of the assay. (B) Methylation-sensitive restriction enzyme digestion with HpaII using a probe for minor satellite centromeric DNA repeats in thymus from mice at day 15 after birth shows hypomethylation in PASG-/- mice. (C) Methylation-sensitive restriction enzyme assay with HpaII using a probe for IAP in kidneys from mice at postnatal day 15. The low molecular weight fragments indicate demethylated restriction sites, whereas high molecular weight fragments indicate methylated restriction sites. (D,E) Northern blot analysis of IAP expression in PASG-/- mice and MEFs shows re-expression of IAP transcripts in mutant mice (D; day 15 after birth) and fibroblasts (E) compared to no expression in control animals or cells. (F) Survival curve of 63 PASG-/- mice from 30 litters. (G) The growth curve of PASG-/- embryos shows growth retardation that becomes statistically significant by about gestation day 15.5. Each data point is derived from at least three litters. (H) The growth curve of the surviving PASG-/- mice demonstrates profound postnatal growth retardation. (I) Glucose levels are 50% lower in PASG-/- mice compared to heterozygotic mutant or wild-type animals during postnatal life. Blood glucose levels were determined by applying whole blood to “one touch” strips (Ultra, Lifespan). Each data point presents at least three mice.
To assess if methylation of specific DNA sequences was also affected in PASG mutant mice, we examined the methylation status of minor satellite centromeric and intracisternal A-particle (IAP) repetitive sequences using methylation-sensitive restriction enzymes (Fig. 2B,C). In both instances, significant hypomethylation is observed in PASG-/- mice (Fig. 2B,C). Northern blot analysis further demonstrates that the level of IAP expression is induced and expressed at high levels in PASG-/- mice and MEFs compared to heterozygotic and wild-type mice, in which the IAP gene is methylated and transcriptionally repressed (Fig. 2D,E). This finding shows that DNA hypomethylation due to disruption of PASG expression can lead to transcriptional derepression.
PASG-/- mice exhibit growth retardation and premature aging
Heterozygous mice are indistinguishable from their wild-type (PASG+/+) littermates and ∼20.5% (42 out of 205) of the offspring born from crosses of PASG+/- mice were PASG-/-, which is slightly lower than the expected 25% and possibly due to the cannibalism of dead offspring by the mothers immediately after birth. Approximately 60% of the PASG-/- mice die shortly after or within a few days after birth, with the remaining 40% surviving up to several weeks (Fig. 2F). Although morphologically normal, newborn PASG-/- mice are significantly smaller than their wild-type and heterozygous littermates. A difference in weight is first noted at E12.5 and increases by E15.5 (p ≤ 0.05; Fig. 2G,H). PASG-/- newborn mice have a 25% lower body weight compared to normal littermates, but by 2 wk of age, their body weight is 70% lower than wild-type animals (Fig. 2H). As limiting glucose availability may restrict the ability of growth factors to maintain cellular viability (Vander Heiden et al. 2001), serum glucose levels were serially measured following birth in PASG-/- mice and control littermates. We found that hypoglycemia is observed in postnatal PASG-/- mice (Fig. 2I) with serum glucose concentrations in newborn and postnatal day 15 animals being 52% (p ≤ 0.002) and 50% (p ≤ 0.0003) less than that of their littermates, despite evidence of adequate nursing and milk present in their stomach. In addition to low birth weight and growth retardation (Figs. 2G,H, 3A), postnatal life is characterized by an early onset of phenotypic changes characteristic of aging, including graying hair and balding, reduced fat deposition, unstable gait, cachexia, and kyphosis (Figs. 3B-D).
Figure 3.
PASG-/- mice age prematurely. (A) Newborn mice. PASG-/- mice (arrows) appear morphologically normal except for their smaller size. (B) PASG-/- mice (shown at day 17) rapidly develop gray hair and balding. (C) PASG-/- mice at day 15 show loss of body mass, muscle atrophy, cachexia, and a profound decrease in adipose tissue. (D) Surviving PASG-/- mice at 15 d of age show kyphosis.
Histopathological examination shows diffuse alveolar atelectasis and lack of aeration of the smaller airways in many of the mutant newborn mice (Fig. 4A). These results suggested decreased expansion of the lungs during the perinatal period, possibly leading to respiratory failure and death of these mice. Because reduced expression of surfactant proteins may lead to alveolar atelectasis, we examined the expression of surfactant proteins A, B, C, and D by Northern blot analysis but found no significant reduction of any of these surfactant-encoding transcripts in the lungs of E17.5 embryos, newborn, or postnatal day 14 PASG-/- mice (data not shown). Other organs, such as thymus, spleen, brain, liver, stomach, intestines, and kidney, appear grossly normal in PASG-/- newborn mice. By postnatal day 14, the skin of PASG-/- mice becomes significantly abnormal, exhibiting hyperkeratosis of the epidermis and the nearly complete absence of subcutaneous adipose tissue (Fig. 4B). By 2 wk of age, the thymus is moderately smaller in size and characterized by severe lymphoid depletion, particularly in the cortex (Fig. 4C). Kidneys of PASG-/- mice are slightly smaller in size and reveal tubular degeneration and dilation, epithelial attenuation, and accumulation of eosinophilic intracytoplasmic inclusions (Fig. 4D; Geiman et al. 2001); such changes have been described as age specific (Hasty et al. 2003).
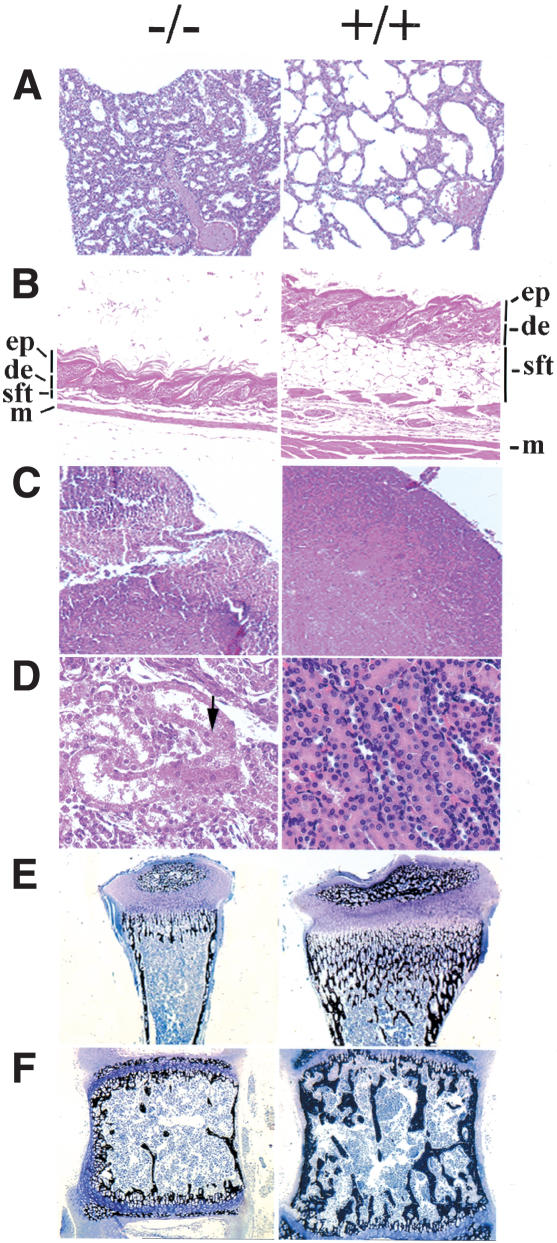
Figure 4.
Histological changes of PASG-/- mice demonstrate multiorgan abnormalities and skeletal defects. (A) Diffuse alveolar atelectasis of the lung from a newborn PASG-/- mouse that contrasts with that of a control littermate lung, which shows normal alveolar expansion (100×). (B) PASG-/- skin exhibits hyperkeratosis and essentially complete absence of subcutaneous fat tissue (100×). (ep) Epidermis; (de) dermis; (sft) subcutaneous fat tissue; (m) muscle. (C) The thymus from a 2.5-week-old PASG-/- mouse is smaller than that of control mice and characterized by severe lymphoid depletion and scattered lymphoid necrosis (100×). (D) Kidneys from PASG-/- mice show tubular degeneration and dilation, epithelial attenuation, and accumulation of age-specific eosinophilic cytoplasmic inclusions (arrow) compared to kidneys from control mice (400×). (E) von Kossa staining of tibiae from 15-day-old mice shows that secondary ossification of epiphyses is delayed in PASG-/- animals. The calcified trabecular bone is fully formed in wild-type mice, but only calcified cartilage is observed in mutant mice. Both cortical and trabecular volume are diminished in PASG-/- mice (25×). (F) von Kossa staining of a thoracic vertebral body from 15-day-old animals demonstrates diminished cortical and trabecular volume in the PASG-/- mice (25×).
A significant delay in bone development along with osteopenia and osteoporosis are observed in the PASG-/- mice. Mutant mice display shorter femurs and tibiae (Fig. 3D) with decreased bone mineral density and smaller epiphyses compared to their wild-type littermates (Figs. 3D, 4E,F). Secondary ossification in the tibial epiphyses is also delayed in PASG-/- mice (Fig. 4E). Calcified cartilage is primarily observed in the PASG-/- mice whereas normal calcified trabecular bone is observed in wild-type mice (Fig. 4E,F). Both cortical and trabecular volumes are diminished in the tibiae and thoracic vertebral bodies of null mice compared to their wild-type littermates (Fig. 4E,F).
The PASG-/- genotype is associated with replicative senescence in vivo and in vitro and increased karyotypic instability in vitro
To determine if the organs from PASG-/- mice displayed molecular markers of senescence, β-galactosidase expression in different organs was measured (Dimri et al. 1995). PASG-/- mice strongly express β-galactosidase in many of their tissues, as demonstrated in kidneys (Fig. 5A) and thymus (Fig. 5B). No significant expression of β-galactosidase is observed in tissues from age-matched, normal littermates. These results show that PASG-/- tissues undergo early onset senescence. Similarly, MEFs derived from PASG-/- embryos show flattened and enlarged morphology, characteristic of a senescence phenotype (Fig. 5C,D), and significantly stronger expression of senescence-associated β-galactosidase compared to the wild-type MEFs (Fig. 5C).
Figure 5.
Increased expression of the senescence biomarker, β-galactosidase, and decreased incorporation of BrdU in PASG-/- mice and/or MEFs. (A,B) β-Galactosidase positive cells are found in PASG-/- tubules of the renal cortex (A) and the thymus (B). Tissues from normal littermates did not show increased expression of this marker of senescence. (C) PASG-/- MEFs show increased β-galactosidase-positive cells compared to wild-type MEFs (passage 5). (D) PASG-/- MEFs showed a flattened and enlarged morphology compared to the normal morphology of the wild-type MEFs (passage 5), consistent with a senescence phenotype. (E) PASG-/- thymus cells from 14-day-old mice show significantly decreased incorporation of BrdU compared with PASG+/+ littermates (6.1 ± 0.8% in PASG-/- mice compared with 15.5 ± 1.4% in PASG+/+ mice, p < 0.001, n = 4).
To determine if the growth retardation and in vivo senescence phenotypes are due to reduced cell proliferation, we examined the BrdU incorporation in mice. Examination of thymus from 14-day-old mice showed significantly lower incorporation of BrdU in PASG-/- mice as compared to their littermates (Fig. 5E).
To better define possible mechanisms leading to the phenotypic changes in mutant animals, we examined the growth characteristics of primary fibroblasts from mutant and wild-type embryos. Early passage MEFs from mutant embryos grow approximately twice as slowly as either PASG+/- or PASG+/+ fibroblasts (Fig. 6A). To quantify the proliferative capacity of PASG deficient MEFs, we calculated population doubling capacity over time. MEFs from PASG-/- embryos show persistent, decreased proliferative capacity after passage 6. However, this decreased proliferative capacity can be rescued by SV40 Large T-antigen transformation (Fig. 6B). Both Large T-antigen transformed wild-type and PASG-/--derived MEFs are able to form tumors in nude mice (four tumors out of eight sites injected with 106 PASG-/- cells and three out of eight using wild-type cells), with the latter MEFs maintaining the same degree of DNA hypomethylation (data not shown). These results demonstrate that PASG-/- MEFs have a significantly decreased proliferation potential, which can be restored possibly by inactivation of p53/Rb pathways. Another characteristic of senescent cells is increased resistance to ionizing radiation. To assess the role of PASG in DNA-damage repair, we exposed MEFs to increasing amounts of ionizing radiation. PASG-/- cells show increased resistance to radiation compared to wild-type MEFs, consistent with a senescent phenotype (Fig. 6C).
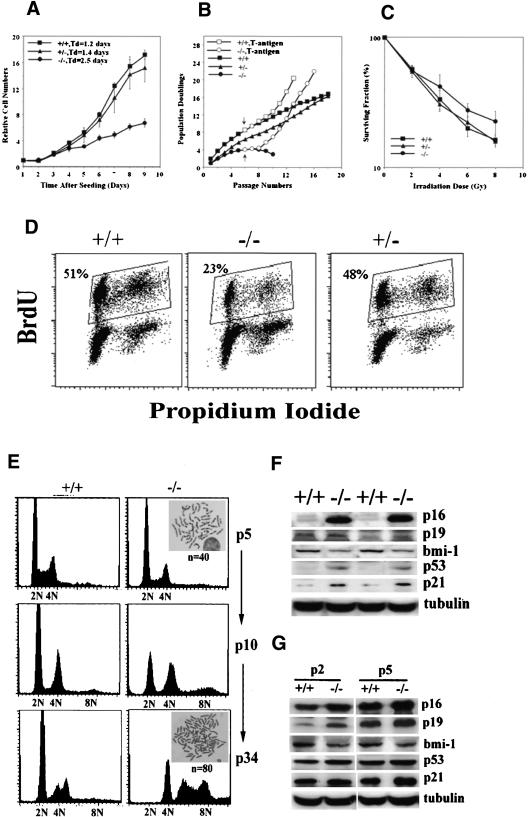
Figure 6.
PASG-/- phenotype is associated with decreased growth potential, replicative senescence, and altered expression of p16INK4a, p19ARF, bmi-1, p53, and p21. (A) Growth kinetics of MEFs (passage 1) demonstrate that the time to increase to fivefold the initial number of plated cells is 3.6, 3.8, and 6.0 d for wild-type, heterozygous, and homozygous mutant MEFs, respectively. The average of two cell lines of each genotype done in triplicate is shown. (B) Population doublings are lower in PASG-/- MEFs, which, however, can be rescued by SV40 Large T-antigen. The arrow indicates the time of transfection with SV40 large T-antigen containing retroviral supernatants. The average of two cell lines of each genotype done in triplicate is shown. (C) PASG-/- MEFs (passage 3) are more resistant to γ-irradiation than control MEFs. The number of surviving cells relative to unirradiated cultures of each genotype is plotted as a function of the dose of radiation. The average of two cell lines of each genotype done in triplicate is shown. (D) PASG-/--derived MEFs show a significant decrease of cycling cells. MEFs (passage 5) were continuously labeled with BrdU for 48 h and then analyzed by flow cytometry to determine the percentage of actively cycling cells. The average of two cell lines of each genotype is shown. (E) Aneuploidy is increased in PASG-/- MEFs over time. PASG-/- MEFs display diploid (n = 40) from early passages, but profoundly abnormal DNA content and increased chromosome numbers from late passages compared to comparatively stable wild-type MEFs. (F) Increased level of p16INK4a, p53, and p21 and decreased level of bmi-1 are observed in the kidney of PASG-/- mice. (G) Increased level of p16INK4a and p19ARF and decreased level of bmi-1 are observed in PASG-/- MEFs at passage 2 (P2) and passage 5 (P5).
To analyze the effect of PASG deficiency on cell cycle, we measured the percentage of replicating cells in an asynchronously growing MEF cell population following BrdU exposure for 48 h. Compared to control MEFs, 50% fewer PASG-/- MEFs are actively progressing through the cell cycle (Fig. 6D). The integrity of cell cycle checkpoints was assessed in PASG deficient MEFs (passage 3) after pulse labeling with BrdU for 2 h. The relative distribution of cells in G1, S, and G2/M is similar for PASG-/- MEFs and controls immediately after the short BrdU pulse labeling; both the PASG-/- and PASG+/+ MEFs re-enter S phase by 8 h (Table 1). Following treatment with 6 Gy of γ-irradiation, both PASG-/- and control cells exhibit a radiation-induced delay in S phase and a G2/M arrest followed by recovery from the initial block and re-entry into G1 by 8 h postirradiation (Table 1). These results demonstrate that G1/S and G2/M DNA damage checkpoints are essentially intact in PASG-/--deficient cells, and that PASG-deficient cells complete the cell cycle in a similar fashion to control cells. The results further establish that the reduced proliferation capacity is a result of the lower replicative capacity of PASG-/- MEFs, not due to defects in cell cycle progression. To determine if apoptosis contributes to the reduced proliferative capacity, MEFs were double stained with FITC-annexin V and propidium iodide. No significant increase in apoptotic population is observed in PASG-/- MEFs compared to wild-type cells (data not shown). Therefore, apoptosis is not a major determinant factor in the reduced proliferative capacity of MEFs and growth retardation of mutant mice.
Table 1.
Cell cycle progression in PASG−/− and control MEFs
| Time in hours after BrdU labeling
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unirradiated
|
Irradiated
|
|||||||||||||||
| Genotype | 0 | 2 | 4 | 6 | 8 | 12 | 24 | 0 | 2 | 4 | 6 | 8 | 12 | 24 | 48 | |
| GI (%) | +/+ | 18 | 6 | 21 | 50 | 65 | 77 | 64 | 14 | 2 | 1 | 7 | 24 | 62 | 48 | 62 |
| +/− | 17 | 6 | 19 | 49 | 64 | 68 | 57 | 11 | 0 | 2 | 7 | 22 | 55 | 65 | 63 | |
| −/− | 15 | 4 | 26 | 53 | 69 | 72 | 58 | 17 | 4 | 2 | 6 | 20 | 65 | 60 | 61 | |
| S(%) | +/+ | 56 | 52 | 14 | 10 | 12 | 14 | 17 | 60 | 58 | 9 | 7 | 5 | 7 | 13 | 13 |
| +/− | 62 | 52 | 20 | 11 | 12 | 20 | 19 | 64 | 63 | 10 | 2 | 3 | 9 | 17 | 11 | |
| −/− | 57 | 47 | 18 | 8 | 10 | 17 | 20 | 53 | 63 | 12 | 8 | 3 | 5 | 13 | 12 | |
| G2(%) | +/+ | 26 | 41 | 66 | 40 | 23 | 9 | 20 | 27 | 43 | 91 | 88 | 71 | 30 | 39 | 25 |
| +/− | 21 | 42 | 62 | 41 | 24 | 12 | 24 | 25 | 37 | 90 | 91 | 75 | 37 | 18 | 26 | |
| −/− | 28 | 49 | 58 | 40 | 22 | 11 | 25 | 31 | 33 | 87 | 88 | 77 | 31 | 26 | 27 | |
MEFs were analyzed after labeling for 2 h with BrdU followed by 6 Gy exposure to γ-irradiation. The top row represents hours following the BrdU pulse. The percentage of G1, S, and G2/M-phase cells among the cycling population (BrdU-positive cells) was quantified using flow cytometry analysis. Data are from one of two experiments that gave similar results.
The association of the replicative senescence phenotype with genomic instability was investigated by determining the DNA content of MEFs at different passages by labeling cells with propidium iodide followed by flow cytometric analysis. DNA profiles show that at passage 5, both wild-type and mutant MEFs maintain essentially normal chromosomal DNA content. However, at passage 10, a significant proportion of PASG-/- MEFs begin to show an abnormally high DNA content and chromosomal number (Fig. 6E). At passage 34, PASG-/- MEFs escape from senescence-mediated proliferative arrest (data not shown) and display greater than 4N DNA content (Fig. 6E). This genomic instability was further investigated by determining karyotypes from passages 5 and 34. Forty karyotypes of each genotype from two independent cell lines were examined. At passage 5, more than 95% of both wild-type and mutant karyotypes are diploid. At passage 34, a major fraction (70%) of wild-type karyotypes is diploid and the remaining 30% are aneuploid with chromosome numbers ranging from 60 to 80. However, the percentage of cells and the degree of aneuploidy is significantly increased in karyotypes from PASG-/- MEFs in which more than 90% had abnormal chromosome numbers ranging from 60 to 120 (Fig. 6E). Of note, chromosomes in PASG-/- MEFs appear morphologically normal without obvious Robert-sonian translocations. The absence of PASG function thus may lead to increased genomic instability, possibly by impairing segregation of chromosomes. Although shortened telomere length is also associated with senescence (Sharpless and DePinho 2004), when we measured telomere length using fluorescence in situ hybridization, no significant difference was observed in telomere content from PASG-/--derived bone marrow cells (day 15 after birth) or MEFs (passage 5) compared to controls (data not shown). Examination of in vivo DNA content from tissues sources such as thymus and bone marrow does not reveal aneuploidy. Several possibilities may explain this result. For example, it is possible that a small percentage of cells may develop aneuploidy but be eliminated by in vivo mechanisms, or that the life span of PASG-/- animals is not long enough to allow escape from senescence and expansion of abnormal clones or that additional gene mutations are required, which are more likely to occur under culture conditions.
Increased accumulation of senescence-associated tumor suppressor genes and decreased bmi-1 expression characterize PASG-/- animals and MEFs
Senescence is the final phenotypic state displayed by cells in response to various physiologic types of stress resulting in decreased cell proliferation and often mediated by increased expression and activation of tumor suppressor genes such as p16INK4a, p19ARF, p53, and p21 (Lundberg et al. 2000; Campisi 2001). Prominent among these is p16INK4a (Lundberg et al. 2000). Consistent with this observation, we found the expression of p16INK4a is significantly increased in PASG-/- mice compared to wild-type mice in which its expression is barely detectable (Fig. 6F). The level of p53 and p21 expression was also up-regulated in PASG-/- mice compared to wild-type mice (Fig. 6F), whereas p19ARF was slightly increased in PASG-/- mice despite its low expression level in the wild-type mice (Fig. 6F). To determine whether up-regulation of p16INK4a expression was induced because of promoter CpG demethylation, we sequenced the 5′ promoter region of p16INK4a from methylation-specific PCR products of bisulfite modified DNA as described previously. Twenty-five CpG sites were analyzed and shown to be unmethylated in both PASG-/- and wild-type animals (data not shown). The up-regulation of p16INK4a could not thus be attributable to hypomethylation, and alternative mechanisms of p16INK4a expression were therefore examined.
The expression of Ets and Id1, regulators of p16INK4a transcription (Ohtani et al. 2001), was examined by Western blot analysis, which demonstrated no significant expression differences of these gene products between PASG-/- and wild-type mice (data not shown). However, bmi-1, another important negative regulator of p16INK4a, which is also associated with changes in cell proliferation and senescence (Jacobs et al. 1999; Park et al. 2004), is reduced by ∼50% in tissues from PASG-/- mice compared to control animals (Fig. 6F).
Consistent with these in vivo results, the expression of p16INK4a is also significantly increased in PASG-/- MEFs (Fig. 6G), along with a concomitant decrease in bmi-1. The expression of p19ARF was higher in PASG-/- MEFs compared to wild-type MEFs, whereas the change of p53 and p21 expression level was less pronounced (Fig. 6G), Thus, the disruption of PASG expression and the subsequent replicative senescent phenotype is closely linked to changes of tumor suppressor gene expression. The induction of p16INK4a expression in PASG-/- mice may, in part, be mediated by the reduced expression of the negative inhibitor, bmi-1.
Discussion
Senescence is a state in which a cell no longer has the ability to proliferate and is accompanied by changes in abnormal gene expression and morphology. To maintain tissue homeostasis during the life time of an animal, stem cells have developed strict regulatory mechanisms to self-renew, differentiate, and prevent premature senescence (Park et al. 2004). In this study, PASG-/- mice exhibit a phenotype characteristic of early onset of aging. Decreased incorporation of BrdU, accumulation of senescence-associated tumor suppressor genes, and increased senescence-associated β-galactosidase as well as the age-related phenotypes in vivo are characteristic of premature aging. However, we cannot rule out that other factors, such as proliferation defects, growth retardation, abnormal lung development, and hypoglycemia, might also contribute to the overall phenotype of PASG-/- animals.
In this study, we demonstrate that targeted disruption of PASG, a SNF2-like factor, leads to genomic hypomethylation and altered gene expression that are linked to developmental growth retardation and premature aging in mice as well as to replicative senescence in PASG-/- MEFs and tissues. These results are the first to link the phenotype of growth retardation and premature aging in mice with epigenetic deregulation due to disruption of a SNF2-like factor, PASG. Although lsh-/- mice were shown to have low birth weight in the previous model, the essentially uniform mortality within hours of being born prevented any detailed postnatal analysis (Geiman et al. 2001). It is unclear why a significant percentage of our PASG-/- mice survive for several weeks of life, but one possibility is that a low level of truncated PASG protein may partially rescue the embryonic null phenotype seen in the previous report (Geiman et al. 2001). The detection of such a truncated mRNA and protein product expressed at a low level supports this conclusion. Furthermore, this targeted deletion of PASG removes the conserved SNF2 helicase domains II, III, and IV, demonstrating that this region of PASG is essential for its normal function. Nevertheless, the extended survival of our PASG-/- animals provides an important opportunity to study in detail the in vivo functions of PASG postnatally. Our PASG-/- model furthermore allows us to demonstrate that the observed growth retardation and premature aging are characterized by a profound up-regulation of tumor suppressor genes such as p16INK4a, p19ARF, p53, and p21 and down-regulation of bmi-1. Among them, p16INK4a up-regulation is most prominent; this increased expression is not due to alterations in promoter methylation, but, instead, is associated with the decreased expression of bmi-1, a negative regulatory of the p16INK4a promoter. Our findings that the premature aging in PASG-/- mice is associated with replicative senescence significantly extend previous work (Wilson and Jones 1983; Holliday 1985; Wilson et al. 1987) and, importantly, links senescence to altered epigenetic patterning and aberrant gene expression.
The maintenance of genomic integrity has emerged as a major factor in cell viability and longevity, with abnormalities leading to telomere shortening or DNA damage and repair being closely linked to senescence, growth retardation, and aging (Hasty et al. 2003; Sharpless and DePinho 2004). Genome instability in patients with Werner syndrome, due to mutations in the WRN DNA helicase, is associated with early onset of aging and cancer predisposition (Hasty et al. 2003). Furthermore, errors in replicating DNA methylation patterns may destabilize genome integrity (Eden et al. 2003; Gaudet et al. 2003) and result in abnormal gene expression, reduced cellular proliferation (Jackson-Grusby et al. 2001), and abnormal embryonic development (Li et al. 1992; Okano et al. 1999). For example, global DNA hypomethylation in DNMT1-deficient ES cells and in mice with a hypomorphic DNMT1 (Dnmt1chip/-) genotype has been shown to contribute to chromosomal instability and an increased incidence of tumor formation (Chen et al. 1998; Eden et al. 2003; Gaudet et al. 2003). Immunodeficiency, chromosome instability, and facial abnormalities characterize the ICF syndrome, a consequence of mutations in the DNMT3b gene (Xu et al. 1999). Similar findings have been observed in animals and MEFs with disruption of Suv39h histone methyltransferases (Peters et al. 2001) and mSds3, an essential component of the functional mSin3/HDAC corepressor complex (David et al. 2003). These important findings demonstrate that normal epigenetic regulation patterns are critical for maintaining genome integrity. Together with the centromeric hypomethylation that we observed in PASG-/- mice and MEFs, these observations strongly suggest that hypomethylation and genome instability may represent fundamental mechanisms capable of leading to premature senescence.
Age-related global hypomethylation has been observed in vitro (Wilson and Jones 1983) and in vivo (Singhal et al. 1987; Wilson et al. 1987; Drinkwater et al. 1989; Barbot et al. 2002). In contrast to the overall reduction in DNA methylation, individual genes may undergo a gradual increase in methylation during the normal process of aging (Issa 1999; Dunn 2003). Genome-wide hypomethylation and site-specific hypermethylation during normal aging can also parallel the findings in cancer cells (Jones and Baylin 2002; Dunn 2003). It has been hypothesized that mechanisms contributing to senescence may protect young organisms from acute death and cancer but predispose them to malignant transformation of somatic cells in later life (Campisi 1996, 2003). This hypothesis has been confirmed in mice lacking Brca1, which is involved in DNA-damage repair and breast cancer (Cao et al. 2003). Aberrant hypomethylation has also been shown to contribute to genome instability and the induction of tumors in the Dnmtchip/- mice (Eden et al. 2003; Gaudet et al. 2003). In this regard, it is important to note that a 75-nt, in-frame deletion of a conserved motif in the C-terminal half of PASG, known to result in dominant negative mutants in similar yeast proteins, has been observed in ∼40%-60% of acute leukemias in patients, suggesting a potential role for PASG in leukemogenesis (Lee et al. 2000).
Although silencing of p16INK4a expression in tumors is often mediated by promoter hypermethylation (Jones and Baylin 2002), the regulation of p16INK4a expression in senescent cells is poorly understood. Our results showed no differences in p16INK4a promoter region methylation between PASG-/- and wild-type animals as well as mutant or wild-type fibroblasts. Instead, the results demonstrated a 50% reduction in bmi-1 expression in PASG-/- tissues. Bmi-1, a member of the Polycomb/trixthorax group (Pc-G/trx-G) proteins, has been shown to be a negative regulator of p16INK4a expression and is a member of a highly conserved system for regulating epigenetic memory and transcriptional repression during development and cell proliferation (Jacobs et al. 1999; Bird 2002; Lessard and Sauvageau 2003; Orlando 2003; Park et al. 2003, 2004). The mechanisms by which PASG regulates bmi-1 expression remain to be determined.
By leading to deregulation of normal gene expression patterns, altered epigenetic patterning is likely to be an important, underlying, molecular mechanism of senescence. Of further interest, altered regulation of transcriptional repressors, such as bmi-1, may provide an alternative mechanism to changes in DNA methylation in regulating the expression of downstream genes playing essential roles in establishing and/or maintaining a replicative senescence phenotype (Park et al. 2004). Other mechanisms may also independently or cooperatively contribute to the observed senescence phenotype in PASG-/- animals, such as altered chromatin modification, structure and function (Peters et al. 2001; David et al. 2003). Although these observations have important implications for understanding the mechanisms of aging and possibly cancer predisposition, they also raise fundamental questions concerning therapeutic strategies directed toward modifying epigenetic patterns in cancer (Karpf and Jones 2002; Schmitt et al. 2002; Kahlem et al. 2004).
Materials and methods
Generation of mice deficient in PASG
The PASG targeting vector was constructed using a 3.65-kb EcoRI genomic fragment from 129/SvJ library that was cloned into the 5′ arm of pKO Scrambler NTKV-1906 Vector (Stratagene); a 6.5-kb XbalI genomic fragment was cloned into the 3′ arm of the vector. With this vector, a 7.126-kb genomic DNA, containing exons 10, 11, and 12 and including helicase domains II, III, and IV, was replaced by the 1.6-kb pGKneo cassette used as a positive selection marker (Fig. 1A).
The targeting vector was electroporated into 129/SvJ ES cells, which were then selected in G418/GCV-containing medium. Targeting frequency was 4/298 (1.3%). Four positive clones were injected separately into C57BL/6J blastocysts to generate chimeric mice. Two clones successfully transmitted the mutant PASG allele to progeny through the germline after the chimeric animals were crossed with normal C57BL/6J mice. Heterozygotic animals were maintained on a mixed 129/SvJ/C57BL/6J background. Similar phenotypes had been observed in 129/SvJ/Swiss black background (data not shown).
Southern and Northern blot analysis
Ten micrograms of DNA were digested with different restriction enzymes and the products electrophoretically resolved in agarose gels, transferred to nylon membranes and hybridized with specific radiolabeled probes. Fifteen micrograms of total RNA, isolated with Trizol (Invitrogen), were electrophoretically separated in formaldehyde agarose gels, transferred to nylon membranes and hybridized with a radiolabeled probe. The IAP probe is homologous to the gag coding region (nt 1570-1899, GenBank accession no. M17551; Walsh et al. 1998). The minor satellite centromere probe has been described (Dennis et al. 2001). RNA loading was normalized to GAPDH expression.
DNA methylation and direct DNA sequencing of bisulfite-modified DNA
Direct measurement of methyl-cytosine at CCGG sites was performed as described previously (Li et al. 1992). The p16INK4a promoter (GenBank accession no. U47108) was amplified using a 757-bp MSP product containing 25 CpG sites, which was ligated into a TA cloning vector and sequenced as previously described (Frommer et al. 1992). Twenty clones were analyzed from each of two mice. Sense primer was from nt 88 to 112: AGTTGTGTATAGAATTTTAGTATTGA; antisense was from nt 845 to 819: CCATACTACTCCAAATAACTCTCCTC.
Mouse embryo fibroblast assays
Sacrificed embryos from gestation day 13.5 were mechanically dissociated and trypsinized followed by filtration through a 100 μM mesh. MEFs (2 × 104 cells per well) were plated in triplicate into 24-well plates and the cells trypsinized, stained with trypan blue, and counted every 24 h. Proliferation kinetics were measured using the 3T9 assay for MEFs, with 9 × 105 cells being plated in 6-cm tissue culture wells and passaged every third day. Population doubling for each passage was computed using the following formula: log2 (cells harvested/cells seeded). For pulse labeling with BrdU (10 μM, Sigma), 5 × 105 MEFs (passage 3) were plated into 6-cm dishes and BrdU added directly to the media for 2 h after the MEFs had attached (Nussenzweig et al. 1996). For continuous labeling, MEFs (passage 5) were plated with 100 μM BrdU and labeled for 48 h (Frank et al. 1998). Cells were fixed with 70% ethanol and stained with FITC-conjugated anti-BrdU antibodies and propidium iodide according to the manufacturer (BD Pharmagen). A Becton and Dickson FACS machine was used to acquire and analyze a minimum of 10,000 events using the Cellquest program. Radiation sensitivity was measured by treating MEFs with 0, 2, 4, 6, or 8 Gy of γ-irradiation from a 137Cr source with a dose rate of 0.7 Gy/min. After 5 d in culture, MEFs were collected, stained with trypan blue, and counted. SV40 Large T-antigen transformation was carried out by infection with viral supernatant from the producer cell line ϕ2-SV40 (Jackson-Grusby et al. 2001). Karyotypes of MEFs were analyzed from colchicine-arrested and Giemsa-stained metaphase chromosome spreads by standard methods.
BrdU labeling of mice
BrdU (100 mg/kg body weight) was injected intraperitoneally into the 14-day-old mice. The mice were killed 2 h after injection; the single cell suspensions from the thymus were stained with anti-BrdU antibody (BD Pharmagen) and analyzed with Becton and Dickson FACS machine.
Histological analysis
Tissues were fixed in 10% neutral buffered formalin, paraffin-embedded, cut into 5-μm sections, and stained with hematoxylin and eosin. Long bones and vertebral bodies were removed and fixed in PLP fixative (2% paraformaldehyde, 0.075 M L-lysine, 0.01 M sodium periodate). The decalcified distal ends of femurs, proximal ends of the tibiae, and vertebral bodies were embedded in LR White acrylic resin (London Resin Company Ltd.) and 1-μm sections cut on an ultramicrotome. These sections were stained for mineral content with the von Kossa staining procedure and counterstained with toluidine blue (Miao et al. 2001). β-Galactosidase activity was measured at pH 6.0 as previously described (Dimri et al. 1995). Quantitative fluorescence in situ hybridization was used to determine telomere lengths (Meeker et al. 2002).
Immunoblot analysis and immunoprecipitation
Whole-cell protein was extracted from organs or cultured cells as described previously (Li et al. 1992). Nuclear protein extracts were prepared using the NE-PER Nuclear and Cytoplasmic Extraction Reagents Kit according to the manufacturer's protocol (Pierce). Western blot analysis was performed according to standard procedures using ECL detection (Amersham). PASG protein was detected using a 1:2000 dilution of a rabbit antiserum raised against a peptide sequence in the C-terminal region of PASG that is not in the deletion region (Raabe et al. 2001). The following primary antibodies were used: p16 (M-156, Santa Cruz), p19ARF (NOVUS), bmi-1 (H-99, Santa Cruz), p21 (Ab-6, Oncogene), p53 (Ab-7, Oncogene), Ets1 (C-20, Santa Cruz), Ets2 (C-20, Santa Cruz), and Id1 (C-20, Santa Cruz). Horseradish peroxidase-conjugated anti-rabbit or anti-mouse antibodies (Promega) were used as secondary antibodies.
MEFs cells were collected and lysed in Triton X-100 lysis buffer (150 mM NaCl, 50 mM Tris-HCl, 1% Triton X-100, proteinase inhibitors). PASG protein was immunoprecipitated with anti-lsh antibody (N-16) and detected with a rabbit antiserum raised against a peptide sequence in the C-terminal region of PASG.
Acknowledgments
We thank Drs. Steven Baylin and Kam-Wing Jair for critical review and helpful discussion, Dr. Jeffrey Whitsett for providing cDNAs encoding lung surfactant proteins, Dr. Angelo DiMarzo for providing critical guidance with the telomere measurements, Dr. R. Jaenisch for SV40 Large T producer cell line, Ms. Jessica Hicks for telomere in situ hybridization, and Drs. Xiao-Nan Li, Wen-Yong Chen, and Ming-Zhou Guo for expert technical assistance. We thank the Children's Cancer Foundation, the Higgin's Scholar Foundation, and the National Institutes of Health for their support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1176104.
References
- Alcorta D.A., Xiong, Y., Phelps, D., Hannon, G., Beach, D., and Barrett, J.C. 1996. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc. Natl. Acad. Sci. 93: 13742-13747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay D. and Medrano, E.E. 2003. The emerging role of epigenetics in cellular and organismal aging. Exp. Gerontol. 38: 1299-1307. [DOI] [PubMed] [Google Scholar]
- Barbot W., Dupressoir, A., Lazar, V., and Heidmann, T. 2002. Epigenetic regulation of an IAP retrotransposon in the aging mouse: Progressive demethylation and de-silencing of the element by its repetitive induction. Nucleic Acids Res. 30: 2365-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. 2002. DNA methylation patterns and epigenetic memory. Genes & Dev. 16: 6-21. [DOI] [PubMed] [Google Scholar]
- Campisi J. 1996. Replicative senescence: An old lives' tale? Cell 84: 497-500. [DOI] [PubMed] [Google Scholar]
- ____. 2001. Cellular senescence as a tumor-suppressor mechanism. Trends Cell. Biol. 11: S27-S31. [DOI] [PubMed] [Google Scholar]
- ____. 2003. Cancer and ageing: Rival demons? Nat. Rev. Cancer 3: 339-349. [DOI] [PubMed] [Google Scholar]
- Cao L., Li, W., Kim, S., Brodie, S.G., and Deng, C.X. 2003. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes & Dev. 17: 201-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.Z., Pettersson, U., Beard, C., Jackson-Grusby, L., and Jaenisch, R. 1998. DNA hypomethylation leads to elevated mutation rates. Nature 395: 89-93. [DOI] [PubMed] [Google Scholar]
- David G., Turner, G.M., Yao, Y., Protopopov, A., and DePinho, R.A. 2003. mSin3-associated protein, mSds3, is essential for pericentric heterochromatin formation and chromosome segregation in mammalian cells. Genes & Dev. 17: 2396-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis K., Fan, T., Geiman, T., Yan, Q., and Muegge, K. 2001. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes & Dev. 15: 2940-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri G.P., Lee, X., Basile, G., Acosta, M., Scott, G., Roskelley, C., Medrano, E.E., Linskens, M., Rubelj, I., Pereira-Smith, O., et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. 92: 9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinkwater R.D., Blake, T.J., Morley, A.A., and Turner, D.R. 1989. Human lymphocytes aged in vivo have reduced levels of methylation in transcriptionally active and inactive DNA. Mutat. Res. 219: 29-37. [DOI] [PubMed] [Google Scholar]
- Dunn B.K. 2003. Hypomethylation: One side of a larger picture. Ann. N.Y. Acad. Sci. 983: 28-42. [DOI] [PubMed] [Google Scholar]
- Eden A., Gaudet, F., Waghmare, A., and Jaenisch, R. 2003. Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300: 455. [DOI] [PubMed] [Google Scholar]
- Fairweather D.S., Fox, M., and Margison, G.P. 1987. The in vitro lifespan of MRC-5 cells is shortened by 5-azacytidine-induced demethylation. Exp. Cell Res. 168: 153-159. [DOI] [PubMed] [Google Scholar]
- Fan T., Yan, Q., Huang, J., Austin, S., Cho, E., Ferris, D., and Muegge, K. 2003. Lsh-deficient murine embryonal fibroblasts show reduced proliferation with signs of abnormal mitosis. Cancer Res. 63: 4677-4683. [PubMed] [Google Scholar]
- Fournel M., Sapieha, P., Beaulieu, N., Besterman, J.M., and MacLeod, A.R. 1999. Down-regulation of human DNA-(cytosine-5) methyltransferase induces cell cycle regulators p16(ink4A) and p21(WAF/Cip1) by distinct mechanisms. J. Biol. Chem. 274: 24250-24256. [DOI] [PubMed] [Google Scholar]
- Frank K.M., Sekiguchi, J.M., Seidl, K.J., Swat, W., Rathbun, G.A., Cheng, H.L., Davidson, L., Kangaloo, L., and Alt, F.W. 1998. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature 396: 173-177. [DOI] [PubMed] [Google Scholar]
- Frommer M., McDonald, L.E., Millar, D.S., Collis, C.M., Watt, F., Grigg, G.W., Molloy, P.L., and Paul, C.L. 1992. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. 89: 1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet F., Hodgson, J.G., Eden, A., Jackson-Grusby, L., Dausman, J., Gray, J.W., Leonhardt, H., and Jaenisch, R. 2003. Induction of tumors in mice by genomic hypomethylation. Science 300: 489-492. [DOI] [PubMed] [Google Scholar]
- Geiman T.M. and Muegge, K. 2000. Lsh, an SNF2/helicase family member, is required for proliferation of mature T lymphocytes. Proc. Natl. Acad. Sci. 97: 4772-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiman T.M., Tessarollo, L., Anver, M.R., Kopp, J.B., Ward, J.M., and Muegge, K. 2001. Lsh, a SNF2 family member, is required for normal murine development. Biochim. Biophys. Acta 1526: 211-220. [DOI] [PubMed] [Google Scholar]
- Gibbons R.J., McDowell, T.L., Raman, S., O'Rourke, D.M., Garrick, D., Ayyub, H., and Higgs, D.R. 2000. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat. Genet. 24: 368-371. [DOI] [PubMed] [Google Scholar]
- Hasty P., Campisi, J., Hoeijmakers, J., van Steeg, H., and Vijg. J., 2003. Aging and genome maintenance: Lessons from the mouse? Science 299: 1355-1359. [DOI] [PubMed] [Google Scholar]
- Hayflick L. 1965. The limited in vitro lifetime of human diploid cell stains. Exp. Cell Res. 37: 614-636. [DOI] [PubMed] [Google Scholar]
- Holliday R. 1985. The significance of DNA methylation in cellular aging. In Molecular Biology of Aging (eds. A.D. Woodhead, A.D. Blackett, and A. Hollaender), pp. 269-283. Plenum Press, New York. [DOI] [PubMed]
- ____. 1986. Strong effects of 5-azacytidine on the in vitro lifespan of human diploid fibroblasts. Exp. Cell Res. 166: 543-552. [DOI] [PubMed] [Google Scholar]
- Issa J.P. 1999. Aging, DNA methylation and cancer. Crit. Rev. Oncol. Hematol. 32: 31-43. [DOI] [PubMed] [Google Scholar]
- Itahana K., Zou, Y., Itahana, Y., Martinez, J.L., Beausejour, C., Jacobs, J.J., Van Lohuizen, M., Band, V., Campisi, J., and Dimri, G.P. 2003. Control of the replicative life span of human fibroblasts by p16 and the polycomb protein Bmi-1. Mol. Cell. Biol. 23: 389-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Grusby L., Beard, C., Possemato, R., Tudor, M., Fambrough, D., Csankovszki, G., Dausman, J., Lee, P., Wilson, C., Lander, E., et al. 2001. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 27: 31-39. [DOI] [PubMed] [Google Scholar]
- Jacobs J.J., Kieboom, K., Marino, S., DePinho, R.A., and van Lohuizen, M. 1999. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397: 164-168. [DOI] [PubMed] [Google Scholar]
- Jaenisch R. and Bird, A. 2003. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 33 Suppl.: 245-254. [DOI] [PubMed] [Google Scholar]
- Jarvis C.D., Geiman, T., Vila-Storm, M.P., Osipovich, O., Akella, U., Candeias, S., Nathan, I., Durum, S.K., and Muegge, K. 1996. A novel putative helicase produced in early murine lymphocytes. Gene 169: 203-207. [DOI] [PubMed] [Google Scholar]
- Jeddeloh J.A., Stokes, T.L., and Richards, E.J. 1999. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22: 94-97. [DOI] [PubMed] [Google Scholar]
- Jones P.A. and Baylin, S.B. 2002. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3: 415-428. [DOI] [PubMed] [Google Scholar]
- Kahlem P., Dorken, B., and Schmitt, C.A. 2004. Cellular senescence in cancer treatment: Friend or foe? J. Clin. Invest. 113: 169-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpf A.R. and Jones, D.A. 2002. Reactivating the expression of methylation silenced genes in human cancer. Oncogene 21: 5496-5503. [DOI] [PubMed] [Google Scholar]
- Kiyono T., Foster, S.A., Koop, J.I., McDougall, J.K., Galloway, D.A., and Klingelhutz, A.J. 1998. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 396: 84-88. [DOI] [PubMed] [Google Scholar]
- Lee D.W., Zhang, K., Ning, Z.Q., Raabe, E.H., Tintner, S., Wieland, R., Wilkins, B.J., Kim, J.M., Blough, R.I., and Arceci, R.J. 2000. Proliferation-associated SNF2-like gene (PASG): A SNF2 family member altered in leukemia. Cancer Res. 60: 3612-3622. [PubMed] [Google Scholar]
- Lessard J. and Sauvageau, G. 2003. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 423: 255-260. [DOI] [PubMed] [Google Scholar]
- Li E. 2002. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3: 662-673. [DOI] [PubMed] [Google Scholar]
- Li E., Bestor, T.H., and Jaenisch, R. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915-926. [DOI] [PubMed] [Google Scholar]
- Lundberg A.S., Hahn, W.C., Gupta, P., and Weinberg, R.A. 2000. Genes involved in senescence and immortalization. Curr. Opin. Cell. Biol. 12: 705-709. [DOI] [PubMed] [Google Scholar]
- Meehan R.R., Pennings, S., and Stancheva, I. 2001. Lashings of DNA methylation, forkfuls of chromatin remodeling. Genes & Dev. 15: 3231-3236. [DOI] [PubMed] [Google Scholar]
- Meeker A.K., Gage, W.R., Hicks, J.L., Simon, I., Coffman, J.R., Platz, E.A., March, G.E., and De Marzo, A.M. 2002. Telomere length assessment in human archival tissues: Combined telomere fluorescence in situ hybridization and immunostaining. Am. J. Pathol. 160: 1259-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao D., Bai, X., Panda, D., McKee, M., Karaplis, A., and Goltzman, D. 2001. Osteomalacia in hyp mice is associated with abnormal phex expression and with altered bone matrix protein expression and deposition. Endocrinology 142: 926-939. [DOI] [PubMed] [Google Scholar]
- Nussenzweig A., Chen, C., da Costa Soares, V., Sanchez, M., Sokol, K., Nussenzweig, M.C., and Li, G.C. 1996. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature 382: 551-555. [DOI] [PubMed] [Google Scholar]
- Ohtani N., Zebedee, Z., Huot, T.J., Stinson, J.A., Sugimoto, M., Ohashi, Y., Sharrocks, A.D., Peters, G., and Hara, E. 2001. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature 409: 1067-1070. [DOI] [PubMed] [Google Scholar]
- Okano M., Bell, D.W., Haber, D.A., and Li, E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247-257. [DOI] [PubMed] [Google Scholar]
- Orlando V. 2003. Polycomb, epigenomes, and control of cell identity. Cell 112: 599-606. [DOI] [PubMed] [Google Scholar]
- Park I.K., Qian, D., Kiel, M., Becker, M.W., Pihalja, M., Weissman, I.L., Morrison, S.J., and Clarke, M.F. 2003. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423: 302-305. [DOI] [PubMed] [Google Scholar]
- Park I.K., Morrison, S.J., and Clarke, M.F. 2004. Bmi1, stem cells, and senescence regulation. J. Clin. Invest. 113: 175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A.H., O'Carroll, D., Scherthan, H., Mechtler, K., Sauer, S., Schofer, C., Weipoltshammer, K., Pagani, M., Lachner, M., Kohlmaier, A., et al. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107: 323-337. [DOI] [PubMed] [Google Scholar]
- Raabe E.H., Abdurrahman, L., Behbehani, G., and Arceci, R.J. 2001. An SNF2 factor involved in mammalian development and cellular proliferation. Dev. Dyn. 221: 92-105. [DOI] [PubMed] [Google Scholar]
- Ramirez R.D., Morales, C.P., Herbert, B.S., Rohde, J.M., Passons, C., Shay, J.W., and Wright, W.E. 2001. Putative telomere-independent mechanisms of replicative aging reflect inadequate growth conditions. Genes & Dev. 15: 398-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randle D.H., Zindy, F., Sherr, C.J., and Roussel, M.F. 2001. Differential effects of p19(Arf) and p16(Ink4a) loss on senescence of murine bone marrow-derived preB cells and macrophages. Proc. Natl. Acad. Sci. 98: 9654-9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee I., Bachman, K.E., Park, B.H., Jair, K.W., Yen, R.W., Schuebel, K.E., Cui, H., Feinberg, A.P., Lengauer, C., Kinzler, K.W., et al. 2002. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416: 552-556. [DOI] [PubMed] [Google Scholar]
- Rheinwald J.G., Hahn, W.C., Ramsey, M.R., Wu, J.Y., Guo, Z., Tsao, H., De Luca, M., Catricala, C., and O'Toole, K.M. 2002. A two-stage, p16(INK4A)- and p53-dependent keratinocyte senescence mechanism that limits replicative potential independent of telomere status. Mol. Cell. Biol. 22: 5157-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson B. 2003. Impact of aging on DNA methylation. Ageing Res. Rev. 2: 245-261. [DOI] [PubMed] [Google Scholar]
- Robertson K.D. 2002. DNA methylation and chromatin—Unraveling the tangled web. Oncogene 21: 5361-5379. [DOI] [PubMed] [Google Scholar]
- Schmitt C.A., Fridman, J.S., Yang, M., Lee, S., Baranov, E., Hoffman, R.M., and Lowe, S.W. 2002. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell 109: 335-346. [DOI] [PubMed] [Google Scholar]
- Sharpless N.E. and DePinho, R.A. 2004. Telomeres, stem cells, senescence, and cancer. J. Clin. Invest. 113: 160-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C.J. and DePinho, R.A. 2000. Cellular senescence: Mitotic clock or culture shock? Cell 102: 407-410. [DOI] [PubMed] [Google Scholar]
- Singhal R.P., Mays-Hoopes, L.L., and Eichhorn, G.L. 1987. DNA methylation in aging of mice. Mech. Ageing Dev. 41: 199-210. [DOI] [PubMed] [Google Scholar]
- Stein G.H., Drullinger, L.F., Soulard, A., and Dulic, V. 1999. Differential roles for cyclin-dependent kinase inhibitors p21 and p16 in the mechanisms of senescence and differentiation in human fibroblasts. Mol. Cell. Biol. 19: 2109-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner S.D., Venkatachalam, S., Choi, J., Jones, S., Ghebranious, N., Igelmann, H., Lu, X., Soron, G., Cooper, B., Brayton, C., et al. 2002. p53 mutant mice that display early ageing-associated phenotypes. Nature 415: 45-53. [DOI] [PubMed] [Google Scholar]
- Vander Heiden M.G., Plas, D.R., Rathmell, J.C., Fox, C.J., Harris, M.H., and Thompson, C.B. 2001. Growth factors can influence cell growth and survival through effects on glucose metabolism. Mol. Cell. Biol. 21: 5899-5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C.P., Chaillet, J.R., and Bestor, T.H. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20: 116-117. [DOI] [PubMed] [Google Scholar]
- Wilson V.L. and Jones, P.A. 1983. DNA methylation decreases in aging but not in immortal cells. Science 220: 1055-1057. [DOI] [PubMed] [Google Scholar]
- Wilson V.L., Smith, R.A., Ma, S., and Cutler, R.G. 1987. Genomic 5-methyldeoxycytidine decreases with age. J. Biol. Chem. 262: 9948-9951. [PubMed] [Google Scholar]
- Xu G.L., Bestor, T.H., Bourc'his, D., Hsieh, C.L., Tommerup, N., Bugge, M., Hulten, M., Qu, X., Russo, J.J., and Viegas-Pequignot, E. 1999. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402: 187-191. [DOI] [PubMed] [Google Scholar]
- Young J.I. and Smith, J.R. 2001. DNA methyltransferase inhibition in normal human fibroblasts induces a p21-dependent cell cycle withdrawal. J. Biol. Chem. 276: 19610-19616. [DOI] [PubMed] [Google Scholar]