Summary
Numerous human genome instability syndromes, including cancer, are closely associated with events arising from malfunction of the essential recombinase Rad51. However, little is known about how Rad51 is dynamically regulated in human cells. Here, we show that the breast cancer susceptibility protein BRCA2, a key Rad51 binding partner, coordinates the activity of the central cell-cycle drivers CDKs and Plk1 to promote Rad51-mediated genome stability control. The soluble nuclear fraction of BRCA2 binds Plk1 directly in a cell-cycle- and CDK-dependent manner and acts as a molecular platform to facilitate Plk1-mediated Rad51 phosphorylation. This phosphorylation is important for enhancing the association of Rad51 with stressed replication forks, which in turn protects the genomic integrity of proliferating human cells. This study reveals an elaborate but highly organized molecular interplay between Rad51 regulators and has significant implications for understanding tumorigenesis and therapeutic resistance in patients with BRCA2 deficiency.
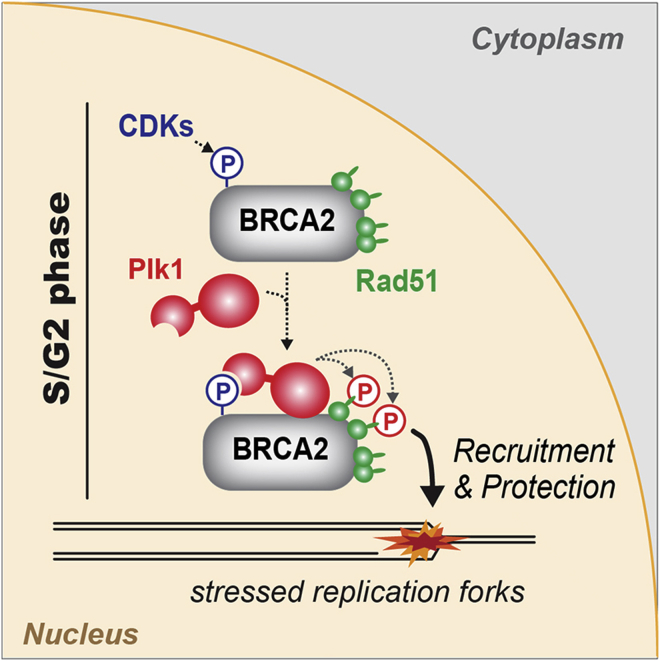
Graphical Abstract
Highlights
-
•
CDK-mediated phosphorylation of BRCA2 triggers Plk1 binding
-
•
Breast cancer-associated BRCA2 mutations abrogate Plk1 binding
-
•
BRCA2, Plk1, and Rad51 form a soluble complex in a cell-cycle-dependent manner
-
•
BRCA2 facilitates Rad51 phosphorylation by Plk1, which promotes genome stability
Malfunction of the essential recombinase Rad51 is closely linked to numerous human genome instability syndromes, including cancer. Here, Yata et al. demonstrate that the breast cancer susceptibility protein BRCA2, a key Rad51 binding partner, binds Plk1 in a CDK-dependent manner and acts as a molecular platform to facilitate Rad51 phosphorylation by Plk1. This phosphorylation is important for the enhancing the association of Rad51 with stressed replication forks and thus provides an elaborate mechanism for the protection of genomic integrity in proliferating cells.
Introduction
Germline mutations in the breast cancer susceptibility gene BRCA2 confer an elevated risk of breast, ovarian, and other cancers (Lancaster et al., 1996, Wooster et al., 1995), as well as developmental defects, childhood brain tumors, and other solid tumors in a subgroup of Fanconi anemia (FA-D1) patients (Howlett et al., 2002). These human disorders linked to BRCA2 deficiency are thought to reflect the functions of BRCA2 in controlling genome integrity, which are primarily mediated through its binding to the Rad51 recombinase, an essential enzyme that plays central roles in faithful repair of DNA double-strand breaks (DSBs) and in protection of stressed replication forks (Hashimoto et al., 2010, Petermann et al., 2010, West, 2003). BRCA2 contains eight evolutionarily conserved Rad51-binding BRC motifs, which facilitate the recruitment of Rad51 to sites of DSBs and the subsequent stimulation of DSB repair by homologous recombination (HR) (Pellegrini and Venkitaraman, 2004). Additionally, an unrelated Rad51 binding site within the BRCA2 C-terminal domain, termed the TR2 motif, plays a critical role in assisting the Rad51-mediated protection of replication forks, especially when DNA synthesis is stalled due to nucleotide depletion, as happens following treatment with hydroxyurea (HU), a potent ribonucleotide reductase inhibitor (Lomonosov et al., 2003, Schlacher et al., 2011).
BRCA2 undergoes extensive phosphorylation by central cell-cycle regulators such as cyclin-dependent kinases (CDKs) (Esashi et al., 2005), suggesting that BRCA2 functions are dynamically regulated during the cell cycle. In particular, CDK-dependent phosphorylation at serine 3291 (S3291) within the TR2 region switches the function of BRCA2 such that it contributes to the removal of residual Rad51 from DNA as cells progress into mitosis (Ayoub et al., 2009, Esashi et al., 2005). Additional CDK-dependent phosphorylation sites have been identified in the BRCA2 N- and C-terminal regions (Esashi et al., 2005), although their molecular and physiological roles remain unknown. BRCA2 is also known to be phosphorylated by polo-like kinase 1 (Plk1) (Lee et al., 2004, Lin et al., 2003), a proto-oncogene product that is well documented as a mitotic regulator. Plk1 typically binds CDK-phosphorylated proteins via its phospho-binding polo-box domain (PBD), and subsequently phosphorylates proteins locally at specific subcellular structures or within the same complex (Barr et al., 2004, Bruinsma et al., 2012, Elia et al., 2003). Significantly, abundant evidence indicates that Plk1 plays multiple cellular functions in time and space through this mechanism, and contributes to the control of not only mitosis but also DNA stress responses and DNA replication (Bruinsma et al., 2012). BRCA2 has been proposed to function closely with Plk1, but it remains unknown whether Plk1 binds directly to BRCA2 and/or provides broader functions in regulating other subunits of the BRCA2 complex to maintain genome stability.
We recently found that Plk1 phosphorylates Rad51 at serine 14 (S14) within a structurally disordered part of the N-terminal domain (Yata et al., 2012), which is connected by a flexible loop to the central ATP-binding core domain. Phosphomimetic mutation of Rad51 at S14 led to no detectable change in its binding to BRCA2 (Yata et al., 2012) or to its ATP-dependent biochemical properties as assessed in vitro by the formation of nucleoprotein filament on single-stranded DNA (ssDNA) and by homologous pairing and strand transfer reactions (F.E., unpublished data). Instead, we found that this phosphorylation stimulates subsequent Rad51 phosphorylation at threonine 13 (T13) by an acidophilic kinase, casein kinase 2 (CK2), which in turn facilitates Rad51 accumulation at DNA damage sites through its phospho-dependent interaction with the Nijmegen breakage syndrome gene product Nbs1, a subunit of the MRN (Mre11-Rad50-Nbs1) damage sensor complex. Strikingly, S14 phosphorylation transiently increases in response to DNA damage, such as ionizing radiation (IR), and promotes efficient HR repair of DSBs, but the mechanism by which damage-induced Rad51 phosphorylation is regulated remains unidentified.
In this study, we investigated the molecular link between BRCA2-mediated and Plk1-mediated Rad51 regulation. Our data provide evidence that CDK-mediated BRCA2 phosphorylation triggers binding of Plk1, which in turn phosphorylates Rad51 within the BRCA2 complex. Furthermore, using genome-wide chromatin immunoprecipitation sequencing (ChIP-seq), isolation of proteins on nascent DNA (iPOND), and a single-molecule DNA fiber technique, we demonstrate that Rad51 phosphorylation by Plk1 is important for replication fork stability. This study uncovers an unexpected molecular mechanism by which BRCA2 coordinates CDK and Plk1 activities to promote Rad51-mediated maintenance of genome stability.
Results
The BRCA2 N Terminus Interacts with Plk1 in a CDK-Dependent Manner
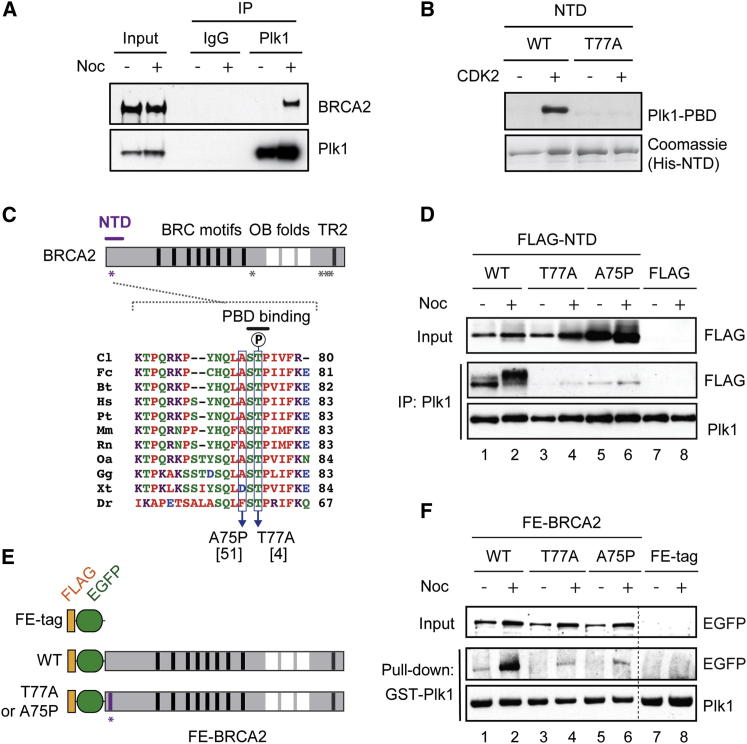
In the course of our investigation into the regulation of BRCA2 during the cell cycle, we found an enhanced association between BRCA2 and Plk1 in human cells following treatment with nocodazole, an anti-tubulin reagent that arrests cells in early mitosis with high CDK activity and is also known to provoke substantial DNA stress after prolonged exposure (Figure 1A) (Dalton et al., 2007, Giunta et al., 2010, Quignon et al., 2007). Furthermore, GFP pull-down analyses using GFP fusions of nine overlapping BRCA2 fragments, which together encompass the whole of BRCA2, revealed that Plk1 association is mediated through the N-terminal domain (NTD; 1–454, B2-1 fragment in Figure S1A) and the C-terminal domain (CTD; 3,189–3,418, B2-9 fragment in Figure S1A), the regions that are efficiently phosphorylated by recombinant CDK2/cyclin A in vitro (Figure S1B; Esashi et al., 2005). In line with these observations, we identified minimal Plk1 PBD-binding motifs S-[pT/pS]-P with predicted CDK target residues within the NTD (T77) and CTD (T3193, S3219, and T3242). In particular, the T77-containing motif is distinctively conserved among vertebrates and is a good match to the optimal PBD-binding motif [P/F]-[φ/P]-φ-[T/Q/H/M]-S-[pT/pS]-[P/X], where φ represents a hydrophobic amino acid (Figure S1C; Elia et al., 2003). As predicted from the amino acid sequence, our far-western blotting analyses confirmed a direct interaction between recombinant Plk1-PBD and a His-NTD fragment phosphorylated in vitro by CDK2/cyclin A, but not a His-NTD variant with an alanine substitution at T77 (T77A; Figure 1B).
Figure 1.
BRCA2 Interacts with Plk1 through Residues Mutated in Breast Cancer
(A) WCEs from untreated or nocodazole (Noc)-treated HeLa cells were used for IP with anti-Plk1 or control rabbit IgG, and coprecipitated BRCA2 was detected with BRCA2 antibody.
(B) Recombinant His-NTD was phosphorylated by recombinant CDK2/cyclin A in vitro and the interactions between Plk1-PBD and BRCA2 fragments were detected by far-western blotting.
(C) BRCA2 domain architecture, highlighting BRC motifs (black bars), oligonucleotide/oligosaccharide binding (OB) folds (white blocks), and the TR2 motif (gray bar). Locations of minimal PBD-binding sequences (asterisks) and the N-terminal BRCA2 fragment, NTD (purple bar) are shown. Bottom panel shows multiple amino acid sequence alignment surrounding the PBD-binding motif within the NTD. Breast cancer-associated mutations at residues A75 and T77, and the putative Plk1-binding region are also indicated. The reported numbers of breast cancer patients (Breast Cancer Information Core) are shown in brackets. Cl, dog; Fc, cat; Bt, cattle; Hs, human; Pt, chimpanzee; Mm, mouse; Rn, rat; Oa, platypus; Gg, chicken; Xt, frog; Dr, fish.
(D) FLAG-tag or FLAG-BRCA2 NTD variants (WT, T77A, and A75P) were transiently expressed in HEK293T cells, and following treatment with nocodazole, interaction with endogenous Plk1 was analyzed by IP with Plk1 antibody.
(E) Schematic diagram of FLAG-EGFP (FE)-tagged full-length BRCA2 variants, representing FLAG (orange) and EGFP (green). An amino acid substitution within the NTD (asterisk) that impairs Plk1 interaction is also indicated.
(F) FE-BRCA2 WT, T77A, A75P variants, or FE-tag were conditionally expressed in HEK293 Flp-In T-REx cells and treated with nocodazole, where indicated. The association of the BRCA2 variants with an exogenously expressed GST fusion of Plk1 was detected following GST pull-down.
See also Figure S1.
Breast Cancer Mutations at the PBD-Binding Site within the BRCA2 NTD Abrogate Plk1 Interaction
Notably, T77A and A75P mutations within the NTD of BRCA2 have been reported in a number of breast cancer patients (Breast Cancer Information Core Database, National Human Genome Research Institute; Figure 1C). Hence, we next examined whether these cancer-associated mutations affect BRCA2-Plk1 interaction in vivo. Remarkably, in contrast to wild-type (WT) FLAG-tagged BRCA2 NTD (FLAG-NTD), which showed a strong in vivo interaction with endogenous Plk1, no such association was detected for FLAG-NTD harboring a T77A or A75P mutation (Figure 1D). We further assessed the impact of a T77A or A75P mutation on the interaction between full-length BRCA2 and Plk1 using cells that coexpress FLAG- and EGFP-tagged BRCA2 (FE-BRCA2) variants and a GST fusion of Plk1 (Figure 1E). Significantly, the association of FE-BRCA2 bearing either a T77A or A75P mutation with GST-Plk1 was largely impaired compared with WT FE-BRCA2 (Figure 1F), demonstrating that these residues within the BRCA2 NTD may serve as the primary phospho-dependent binding site for Plk1.
Encouraged by these findings, we next examined whether BRCA2 is indeed phosphorylated at T77. A phospho-specific antibody was raised against a BRCA2 peptide phosphorylated at T77 (pT77) and the specificity of the pT77 antibody was verified by a peptide dot blot assay (Figure S2A). The pT77 antibody also detected BRCA2 NTD (Figure 2A) or full-length BRCA2 phosphorylated with recombinant CDK2/cyclin A (Figure S2B), confirming that CDK2 directly phosphorylates T77 in vitro. To further assess T77 phosphorylation of BRCA2 in vivo, we affinity purified FLAG-NTD, FE-BRCA2, and their T77A and A75P variants from human cells, and examined their phosphorylation status using the pT77 antibody. The antibody detected WT FLAG-NTD and FE-BRCA2, but not the T77A or A75P variants, showing that the overexpressed NTD fragment and full-length BRCA2 can be phosphorylated at T77 in vivo (Figures S2C and 2B, respectively). These results demonstrate that the cancer-associated T77A and A75P mutations block phosphorylation at residue 77. Importantly, the pT77 antibody also detected endogenous BRCA2 from HeLa cells, but not from EUFA423 (FA-D1) fibroblasts, which express BRCA2 lacking the C-terminal 192 amino acids (Howlett et al., 2002), or from the Capan-1 pancreatic cancer cell line, which has a larger C-terminal deletion of 1,416 amino acids (Goggins et al., 1996; Figures 2C and 2D). These observations suggest that endogenous BRCA2 is phosphorylated at T77 in vivo, but only in the full-length form.
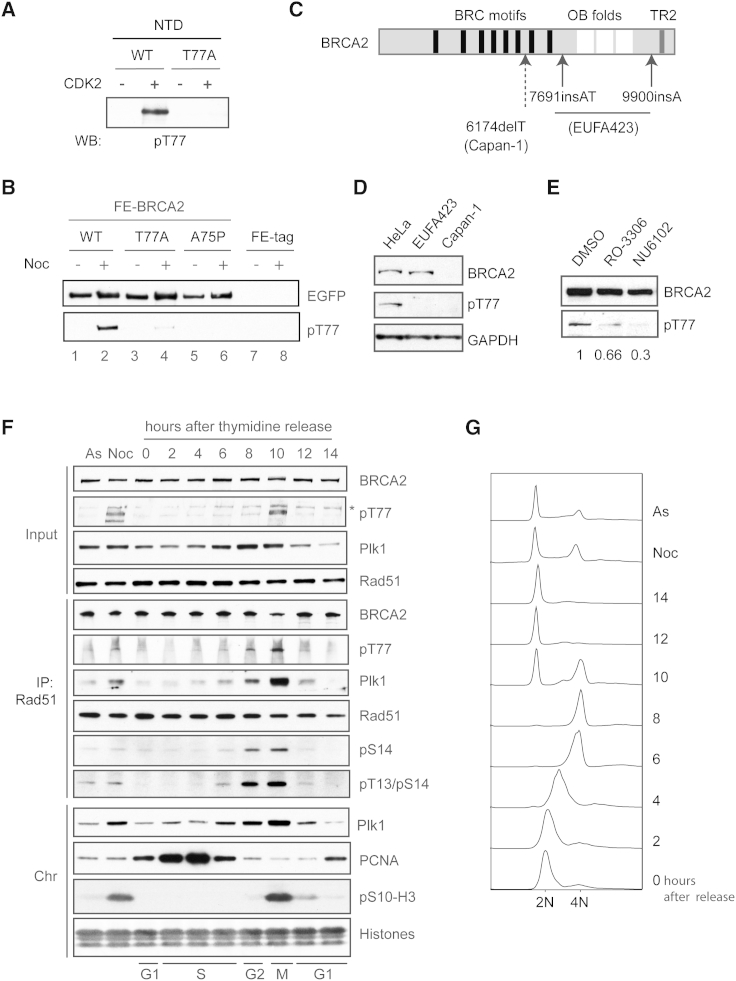
Figure 2.
BRCA2 T77 Is Phosphorylated In Vivo in a CDK-Dependent Manner
(A) Recombinant WT and T77A His-BRCA2 NTD were phosphorylated in vitro with recombinant CDK2/cyclin A, and phosphorylation at T77 was assessed using pT77 phospho-specific antibody.
(B) Western blot analysis of partially purified FE-BRCA2 WT, T77A, and A75P variants using pT77 antibody.
(C) Schematic diagram of full-length BRCA2 showing mutations in Capan-1 and EUFA423 cells indicated by dotted and solid arrows, respectively.
(D) Detection of pT77-BRCA2 in WCEs of HeLa, EUFA423, and Capan-1.
(E) Detection of pT77-BRCA2 after treatment of HeLa cells with CDK inhibitors, 10 μM RO-3306, 60 μM NU6102, or DMSO for 4 hr. Relative signal intensities of pT77 in each sample were quantified against the BRCA2 blot and are indicated under each lane.
(F) Detection of pT77-BRCA2 in synchronized HeLa cells using double thymidine block and release. Rad51 complexes were immunoprecipitated from the soluble fraction and analyzed using the indicated antibodies. To monitor progression through the cell cycle, chromatin-bound PCNA (S phase) (Moldovan et al., 2007) and Histone H3 phosphorylated at Serine 10 (pS10-H3; mitosis) (Hans and Dimitrov, 2001) were detected. The asterisk indicates a nonspecific band.
(G) Cell-cycle distribution analysis of propidium-iodide-stained cells. As, asynchronous cells; Noc, nocodazole-treated cells.
See also Figure S2.
We further assessed whether BRCA2 phosphorylation at T77 is indeed mediated by CDKs in vivo. To this end, HeLa cells were treated with the CDK inhibitors RO-3306, which primarily inhibits CDK1 (Vassilev et al., 2006), or NU6102, which inhibits CDK1 and CDK2 with comparable potency (Davies et al., 2002). pT77 levels were reduced in cells treated with either CDK inhibitor, with a greater reduction following NU6102 treatment, suggesting that BRCA2 T77 is phosphorylated by both CDKs in vivo (Figure 2E). Analysis of synchronized HeLa cells further revealed that T77-phosphorylated BRCA2 (pT77-BRCA2) increased in abundance as cells progressed into mitosis, closely resembling the timing of BRCA2 phosphorylation at S3291 (pS3291-BRCA2) (Figures 2F, 2G, S2D, and S2E) (Esashi et al., 2005). To our surprise, however, pS3291-BRCA2 was barely detectable in the Rad51 immunocomplexes from the soluble fraction (Figure S2D), suggesting that an as-yet-unknown mechanism may interfere with Rad51 binding to the pool of endogenous BRCA2 that is physiologically phosphorylated at S3291, despite the presence of additional Rad51-binding BRC motifs in the middle region. In marked contrast, pT77-BRCA2 substantially retained Rad51 binding, and from late S phase to mitosis formed a soluble complex containing Plk1 and Rad51 phosphorylated at both S14 and T13 (Figure 2F). Furthermore, we observed that a complex containing pT77-BRCA2, Plk1, and S14-phosphorylated Rad51 (pS14-Rad51) became abundant in cells treated with nocodazole (Figure S2F), but was disrupted following brief treatment with RO-3306 (Figure S2G). Together, these data indicate that CDKs phosphorylate BRCA2 at T77 in a cell-cycle-dependent manner, leading to the formation of a protein complex containing Plk1, pS14-Rad51, and pT13/pS14-Rad51.
BRCA2 Facilitates Plk1-Dependent Rad51 Phosphorylation
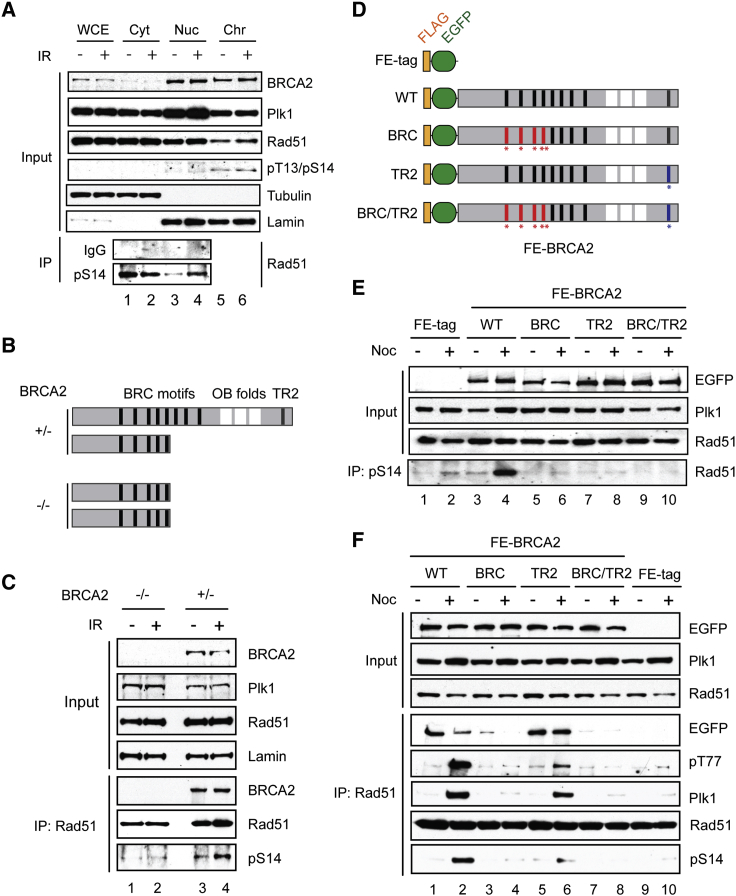
The finding of BRCA2, Plk1, and Rad51 in a common complex (Figures 2F, S2D, S2F, and S2G) led us to explore the possibility that BRCA2 may act as a core regulatory platform that brings Plk1 and Rad51 into close proximity and, in this manner, promotes Plk1-dependent Rad51 phosphorylation at S14 during the cell cycle and/or following genotoxic stress. In line with this idea, we found that BRCA2 interacts exclusively with Plk1 and not with the other Plk family members Plk2 and Plk3, and that the interaction with Plk1 is retained after DNA damage treatment (Figures S2H and S2I). Furthermore, a cellular fractionation analysis of irradiated cells revealed that although pS14-Rad51 was detectable in both the cytoplasmic and nuclear soluble fractions, IR treatment induced a higher level of pS14-Rad51 specifically in the nuclear soluble fraction, which contained the majority of BRCA2 and Plk1 (Figure 3A, lanes 3 and 4). To investigate whether BRCA2 is indeed involved in damage-induced Rad51 phosphorylation in vivo, we evaluated the level of pS14-Rad51 in two isogenic DLD1 cell lines, BRCA2+/− and BRCA2−/−, in which respectively one or both BRCA2 alleles contain a premature stop codon after BRC repeat 5 (Hucl et al., 2008; Figure 3B). Analogous to the data we obtained with HEK293T cells (Figure 3A), we found an IR-induced increase in pS14-Rad51 in the nuclear soluble fraction from DLD1 BRCA2+/− cells (Figure 3C, lanes 3 and 4). In contrast, no substantial increase was detectable in BRCA2−/− cells, even with a higher level of Plk1 compared with DLD1 BRCA2+/− cells (Figure 3C, lanes 1 and 2). Similarly, pS14-Rad51 signal was reduced in BRCA2-defective EUFA423 cells and Capan-1 cells, or when endogenous BRCA2 was downregulated using small hairpin RNA (shRNA) in human cell lines (Figure S5F; K.Y. and F.E., unpublished data).
Figure 3.
BRCA2 Facilitates Plk1-Dependent Rad51 Phosphorylation at S14
(A) HEK293T cells were treated with IR (4 Gy, 20 min recovery), and pS14-Rad51 from the indicated cellular fractions was immunoprecipitated with pS14 antibody and detected with Rad51 antibody. Cyt, cytoplasmic soluble fraction; Nuc, nuclear soluble fraction; Chr, chromatin-bound fraction.
(B) Schematic diagram of the BRCA2 alleles in DLD1 BRCA2−/− and BRCA2+/− cells.
(C) Endogenous Rad51 was immunoprecipitated from DLD1 BRCA2−/− or BRCA2+/− cells before and after irradiation (4 Gy). Thirty minutes after IR treatment, nuclear soluble fraction was prepared and pS14 was detected as in Figure 3A.
(D) Schematic diagram of FE-BRCA2 variants as shown in Figure 1E. Amino acid substitutions within the BRC (red asterisks and bars) and TR2 motifs (blue asterisk and bar) that impair Rad51 interaction are also indicated.
(E) FE-tag, FE-BRCA2 WT, BRC, TR2, or BRC/TR2 variants were conditionally expressed in HEK293 Flp-In T-REx cells and the level of S14 phosphorylation was assessed by IP with pS14-Rad51 antibody.
(F) As in Figure 3E, BRCA2 variants were conditionally expressed in HEK293 Flp-In T-REx cells, and the level of S14 phosphorylation and association of BRCA2 and Plk1 were assessed following Rad51 IP.
See also Figure S3.
As BRCA2 deficiencies resulted in a reduction of pS14-Rad51, we next asked whether elevated expression of BRCA2 would conversely lead to an increase in Rad51 phosphorylation. Strikingly, a higher level of pS14-Rad51 was detected in cells expressing WT FE-BRCA2 compared with cells expressing the FE tag only (Figures 3D and 3E, lanes 1 and 3), such that further IR induction of pS14-Rad51 was obscured under these conditions (data not shown). To further assess the involvement of cell-cycle kinases in regulating BRCA2 functions, we treated these cells with nocodazole, which imposes arrest in early mitosis with high CDK and Plk1 activities, and genotoxic stress (Dalton et al., 2007, Giunta et al., 2010, Quignon et al., 2007). Using this approach, we found an enhanced effect of exogenous FE-BRCA2 expression on the induction of pS14-Rad51 (Figure 3E, lanes 2 and 4).
In order to evaluate more directly whether BRCA2 interaction with Rad51 is important for Plk1-dependent Rad51 phosphorylation, we generated full-length BRCA2 variants with mutations that impair Rad51 binding through either BRC motifs or the C-terminal TR2 (Figures 3D and S3). Such mutations were identified based on the crystal structure of the Rad51-BRC complex and previously reported biochemical studies (Esashi et al., 2005, Lo et al., 2003, Ochiai et al., 2011, Pellegrini et al., 2002, Rajendra and Venkitaraman, 2010) (Figure S3). As expected, mutations within BRC repeats 1–4 led to a substantial reduction of Rad51 binding (Figure S3F, lane 8), whereas the S3291E substitution, which hinders interaction between TR2 and Rad51, largely retained Rad51 binding (Figure S3F, lane 9), likely mediated through the BRC motifs under this condition. Importantly, we found that exogenous expression of FE-BRCA2 variants with the BRC mutations and/or the TR2 mutation failed to increase the level of pS14-Rad51 (Figure 3E, lanes 5–10). We also noticed that although the BRCA2 variant with the TR2 mutation retained Rad51 association, a relative reduction in the amount of pT77-BRCA2 and BRCA2-associated Plk1 was seen following Rad51 IP (Figure 3F, lanes 2 and 6). These observations support the notion that BRCA2 indeed acts as a molecular platform to facilitate Rad51 phosphorylation, and that appropriate interaction among full-length BRCA2, Plk1, and Rad51 is critical for this process.
pS14-Rad51 Accumulates at Sites of Broken DNA
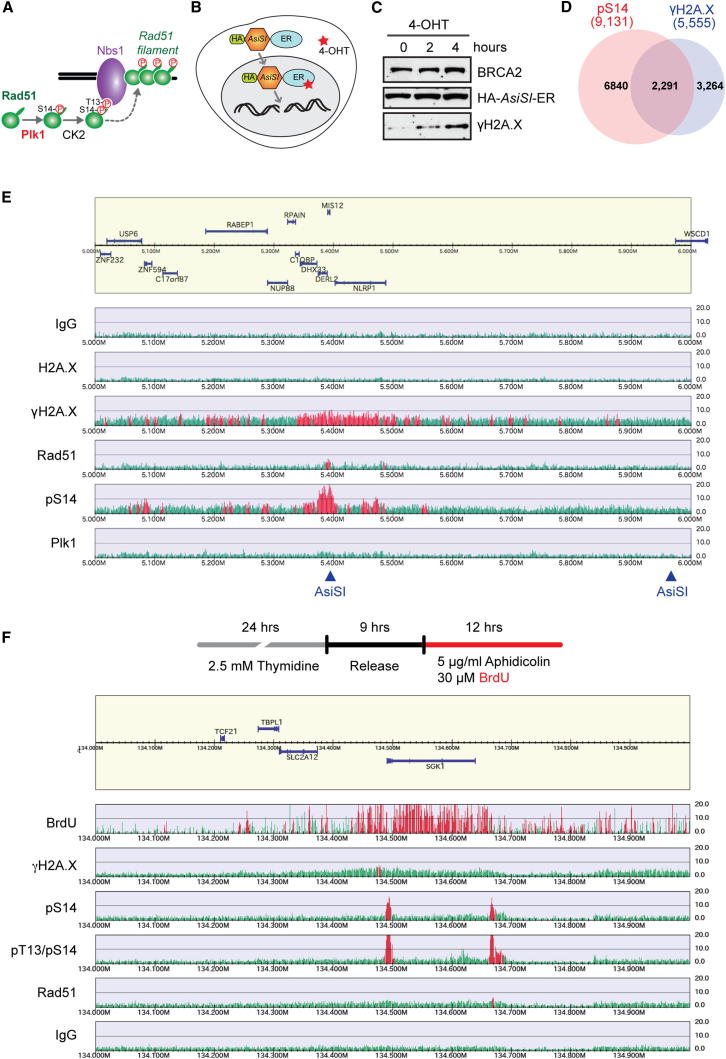
Since Rad51 phosphorylation at S14 requires both the BRC and TR2 motifs of BRCA2, we hypothesized that the cellular functions of BRCA2 in Rad51-mediated genome integrity control, namely, HR repair of DSBs and replication fork protection, are brought about through its activity in facilitating Rad51 phosphorylation by Plk1. In this scenario, the Rad51 S14 site would be expected to be critical both for HR repair and for protection of replication forks. With regard to HR repair, Rad51 S14 has been shown to play a physiologically important role (Yata et al., 2012): S14 phosphorylation triggers subsequent phosphorylation at T13 by CK2, which in turn facilitates Rad51 binding to Nbs1, hence promoting the recruitment of Rad51 to broken DNA and HR repair of DSBs (depicted in Figure 4A). The involvement of pS14-Rad51 in replication fork protection, however, has not previously been addressed.
Figure 4.
Phosphorylated Rad51 Accumulates at Sites of DSB and Replicative DNA
(A) Model showing sequential Rad51 phosphorylation mediated by Plk1 and CK2, and recruitment of Rad51 to stressed DNA via phospho-dependent binding to Nbs1.
(B) Depiction of DSBs induction at AsiSI sites across the genome using U2OS cells stably expressing HA-AsiSI-ER fusion (U2OS AsiSI-ER).
(C) U2OS AsiSI-ER cells were treated with 0.3 mM 4-OHT for the indicated times, and HA-AsiSI-ER and γH2A.X in WCEs were detected using HA and γH2A.X antibodies, respectively.
(D) Venn diagram representing the number of colocalizing peaks for pS14 and γH2A.X quantified from genome-wide ChIP-seq results, using U2OS AsiSI-ER cells treated as in (C).
(E) Representative ChIP-seq profile of DNA-damage-responsive factors at a DSB site within chromosome 17, 5–6 Mb, using U2OS AsiSI-ER cells treated with 4-OHT for 4 hr. Arrowheads indicate locations containing the AsiSI target sequence (GCGATCGC).
(F) Representative ChIP-seq profile following replication stress within chromosome 6, 134–135 Mb, using HeLa cells treated with 5 μg/ml aphidicolin. Nascent DNA is labeled with the incorporation of BrdU.
See also Figure S4.
With this in mind, we assessed the genome-wide distribution of phosphorylated Rad51 on chromatin in relation to DNA breaks by ChIP-seq using Rad51 phospho-specific antibodies. In order to introduce DSBs at defined genomic loci, we exploited U2OS cells stably expressing a fusion of the AsiSI endonuclease with the estrogen receptor ligand-binding domain (AsiSI-ER; Figure 4B; Iacovoni et al., 2010). The AsiSI-ER fusion normally localizes to the cytoplasm, but translocates to the nucleus upon treatment with 4-hydroxy tamoxifen (4-OHT), leading to induction of DSBs at AsiSI sites across the genome (Figures 4C, S4A, and S4B). Significantly, we observed a broad accumulation of pS14-Rad51 at induced DSB sites marked with γH2A.X (Figures 4D, 4E, and S4C), along with more restricted pT13/pS14-Rad51 binding, showing that Rad51 recruited to DSBs retains phosphorylation. Notably, however, only a limited accumulation of Plk1 was detectable at DSBs (Figure S4D), supporting the idea that Rad51 is first phosphorylated at S14 in the soluble nuclear fraction (Figure 3A), followed by cooperative polymerization on chromatin in the pS14 and pT13/pS14 forms.
pS14-Rad51 Accumulates at Stressed Replication Forks
Our ChIP-seq analysis identified pS14-Rad51 and pT13/pS14-Rad51 peaks not only at DSB sites but also at distinct loci where extensive γH2A.X peaks were not detectable (Figure 4D). Given the role of Rad51 in replication fork protection (Hashimoto et al., 2010, Petermann et al., 2010), we reasoned that these signals may reflect association of phosphorylated Rad51 with stalled DNA replication forks. To test this idea, we mapped at high resolution Rad51-binding sites in relation to nascent DNA genome wide. Specifically, HeLa cells were treated with the thymidine analog 5-bromo-2-deoxyuridine (BrdU) and aphidicolin, a potent inhibitor of DNA polymerase α. Loci that had incorporated BrdU were then isolated using a BrdU antibody and identified by next-generation DNA sequencing. In agreement with our hypothesis, pS14-Rad51 and pT13/pS14-Rad51 were detected in the vicinity of BrdU incorporation regardless of the presence of extensive γH2A.X signals (Figure 4F).
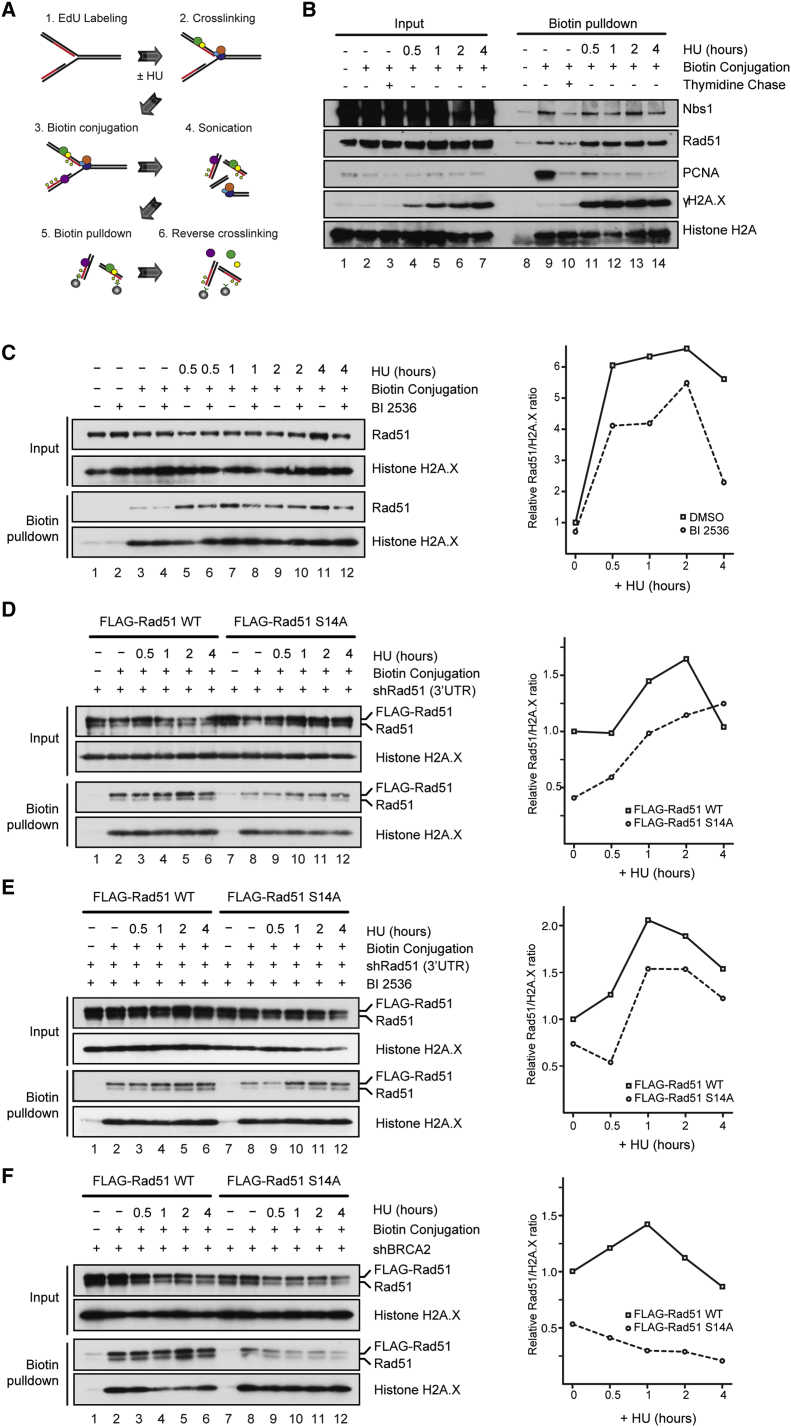
We further analyzed the dynamics of proteins that bound to the nascent DNA following HU treatment using the iPOND method, which allows sensitive detection of events associated with replication forks (Sirbu et al., 2012). It has been demonstrated that HU treatment initially leads to stalled replication forks, which eventually collapse into DSBs following prolonged HU exposure (Petermann et al., 2010, Sirbu et al., 2011). In our iPOND analyses, nascent DNA was first pulse labeled with the thymidine analog 5-ethynyl-2′-deoxyuridine (EdU), and following exposure to HU for increasing lengths of time, proteins associated with labeled DNA were isolated and detected by western blotting (Figure 5A). As shown in Figure 5B, HU treatment led to the rapid removal of proliferating cell nuclear antigen (PCNA) from stalled replication forks, while triggering H2A.X phosphorylation as a consequence of checkpoint activation (Sirbu et al., 2011). In parallel, we found that Rad51 swiftly accumulated at replication forks following HU treatment, whereas Nbs1 association with the forks was mostly unaffected (Figure 5B). Strikingly, when cells were treated with the Plk1 inhibitor BI 2536, reduced Rad51 association with replication forks was readily detectable in the absence of HU-induced stress (Figure 5C, lanes 3 and 4, and S5A), and less efficient Rad51 recruitment and impaired retention of Rad51 at HU-stalled replication forks were found (Figure 5C, lanes 3–12). These observations suggest that Plk1 plays multiple roles to facilitate Rad51 association with replication forks during normal cell proliferation, as well as the recruitment and retention of Rad51 at stressed replication forks.
Figure 5.
S14 Is Important for Rad51 Accumulation at Replication Forks
(A) Depiction of the iPOND experimental procedure.
(B) Proteins associated with replication fork in HEK293T cells with or without 4 mM of HU treatment were isolated by iPOND procedure, and detected with indicated antibodies. In order to discriminate proteins associated with DNA behind replication forks, EdU-labeled cells were subsequently incubated with media containing a low concentration of thymidine (10 μM), and associated proteins were similarly detected (thymidine chase).
(C) HEK293T cells were pretreated with 100 nM of Plk1 inhibitor BI 2536 for 30 min, followed by HU treatment and iPOND analysis as in (B). Right panel shows the quantitation of one representative iPOND experiment, expressed as the relative ratio of the total level of Rad51 against H2A.X.
(D) HEK293 Flp-In T-REx cells conditionally expressing FLAG-tagged Rad51 WT or S14A, along with shRNA targeting the 3′ UTR of endogenous RAD51, were treated with HU, and proteins associated at nascent DNA were analyzed as in (B).
(E) As in (D), except that cells were pretreated with BI 2536 for 30 min as in (C).
(F) As in (D), except that endogenous BRCA2 rather than Rad51 was downregulated by shRNA.
See also Figure S5.
To further assess whether Plk1 facilitates Rad51 association with replication forks by phosphorylating S14, thus promoting its interaction with Nbs1, we performed analogous experiments using cells expressing a FLAG fusion of WT Rad51 or the S14A nonphosphorylatable Rad51 variant (Yata et al., 2012). We moderately downregulated endogenous Rad51 using shRNA targeting the RAD51 3′ UTR. Under these conditions, we found a high level of exogenously expressed WT Rad51 at the replication forks in proliferating cells, and this association peaked at 2 hr after HU treatment (Figures 5D and S5B). In contrast, we observed a significantly lower level of fork-associated S14A Rad51 compared with WT Rad51 (Figure 5D, lanes 2 and 8), and the S14A form accumulated more slowly at HU-stalled replication forks over the 4 hr of the time course (Figure 5D). Importantly, Plk1 inhibitor treatment of these cell lines, which moderately reduced S14 phosphorylation of WT FLAG-Rad51 (Figure S5C), advanced the peak of both WT and S14A Rad51 at stalled replication forks to 1 hr, followed by faster dissociation at later time points (Figures 5E and S5D). These findings suggest that Plk1-mediated Rad51 phosphorylation facilitates its association at nonstressed replication forks and shortly after HU treatment, but that the role of Plk1 in sustaining Rad51 at stalled forks after prolonged HU treatment is independent of S14 phosphorylation.
Our previous study suggested that unregulated Rad51 phosphorylation at S14, as found in cancer cells with a high level of Rad51 or Plk1, bypasses BRCA2-mediated Rad51 recruitment to sites of DNA damage, contributing to the resistance of BRCA2-defective cancer cells to genotoxic stresses (Yata et al., 2012). On the basis of this finding, we next investigated the importance of BRCA2 in Rad51 association with replication forks by downregulating BRCA2 (Figure S5E). With FLAG-Rad51 expressed at a high level, we found that Plk1-mediated Rad51 phosphorylation was indeed detectable, at a lower level, after BRCA2 downregulation (Figure S5F), and that WT Rad51 was transiently recruited to HU-stalled replication forks (Figure 5F, lanes 2–6, and S5G). We also noticed that BRCA2 downregulation led to premature dissociation of WT Rad51 from stalled replication forks 2–4 hr after HU treatment, likely reflecting a previously described BRCA2 function by which it mechanically stabilizes Rad51 nucleoprotein filaments (Schlacher et al., 2011). Remarkably, extensive destabilization of Rad51 S14A at replication forks was found after HU treatment in BRCA2-depleted cells (Figure 5F, lanes 8–12). This observation supports the notion that Plk1-mediated Rad51 phosphorylation and BRCA2 have complementary functions in facilitating Rad51 association with HU-induced stalled replication forks.
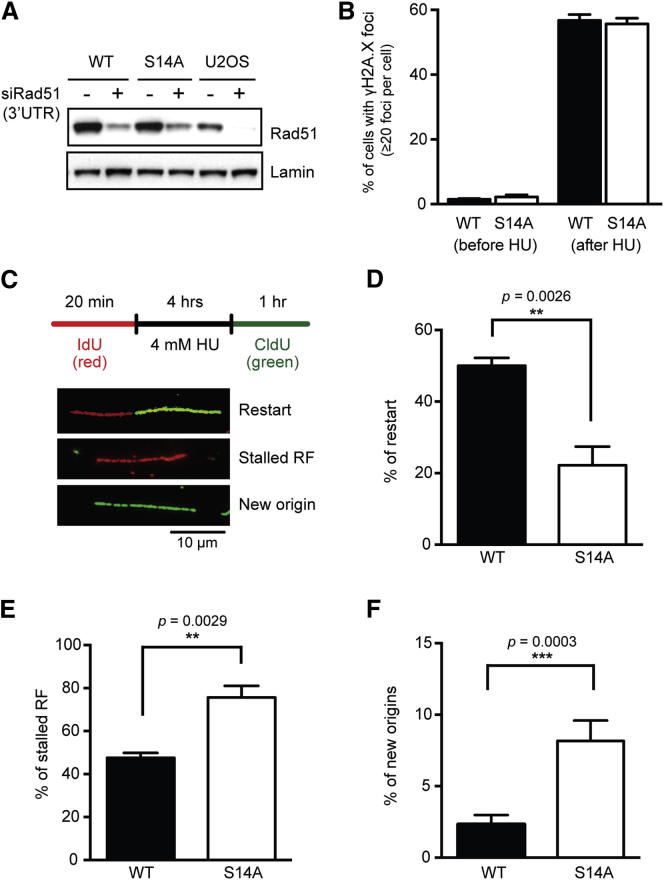
Rad51 S14 Is Important for Efficient Replication Resumption
To further assess the impact of Plk1-dependent Rad51 phosphorylation in response to HU-induced replication stress, we analyzed cellular phenotypes of U2OS cells stably expressing exogenous WT Rad51 or the S14A variant, in which endogenous Rad51 was downregulated with small interfering RNA (siRNA) targeting the RAD51 3′ UTR (Yata et al., 2012) (Figure 6A). Immunofluorescence (IF) microscopy analysis showed, in both cell lines, a similar increase in the number of γH2A.X foci following HU treatment (Figures 6B and S6A). This observation is consistent with the iPOND result showing that equivalent levels of γH2A.X were induced at stalled replication forks of cells expressing either WT or S14A Rad51 (Figures 5D and S5B), and indicates that Rad51 and its S14A variant have little impact on either the replication checkpoint or the DNA damage checkpoint.
Figure 6.
Rad51 S14 Is Important for Replication Restart after Replication Stress
(A) Western blot analysis of U2OS cell lines expressing exogenous Rad51 WT or S14A, after downregulation of endogenous Rad51 using siRNA targeting its 3′ UTR.
(B) Percentage of HU-treated (4 mM for 4 hr) Rad51 WT and S14A U2OS cells containing more than 20 γH2A.X foci. Error bars, SEM (n = 3).
(C) Illustration of the DNA fiber experimental setup, with incorporation of IdU and CldU shown in red and green, respectively. Representative IF images of DNA fibers defined as replication restart (incorporation of both IdU and CldU), stalled replication fork (RF; IdU only), and new origin firing (CldU only) are shown. The bar indicates 10 μm.
(D) Recovery of DNA synthesis was evaluated by the percentage of labeled tracts containing both IdU and CldU against the total number of labeled tracts. Error bars, SEM (n = 3).
(E) Stalled replication forks were evaluated by the percentage of labeled tracts containing only IdU. Error bars, SEM (n = 3).
(F) Firing of new origins after HU treatment was evaluated by the percentage of labeled tracts containing only CldU. Error bars, SEM (n = 3).
In panels (D)–(F), asterisks indicate unpaired two-tailed Student’s t test; ∗∗∗p < 0.001, ∗∗p < 0.01. See also Figure S6.
Appropriate protection of stalled replication forks from nascent DNA degradation facilitates efficient restart of replication when the obstacle is removed (Petermann et al., 2010, Schlacher et al., 2011, Schlacher et al., 2012). We therefore examined the involvement of Rad51 S14 in replication restart using the single-molecule DNA fiber technique (Jackson and Pombo, 1998). Specifically, nascent DNA before and during recovery from an HU-induced replication block was sequentially labeled by incorporation of the thymidine analogs 5-iodo-2′-deoxyuridine (IdU) and 5-chloro-2′-deoxyuridine (CldU), respectively (Figure 6C). This analysis revealed that ∼50% of all DNA fibers contained both IdU and CldU tracts in cells expressing WT Rad51, whereas only ∼20% of fibers contained both tracts in cells expressing S14A Rad51 (Figures 6D and S6B), suggesting that a greater proportion of HU-stalled replication forks failed to restart in cells expressing the S14A Rad51 mutant (Figure 6E). In parallel, we noticed that there was no detectable difference between the cell lines with respect to the speed of DNA synthesis during recovery from HU treatment, as evaluated from the length of tracts or from the CldU/IdU tract ratio (Figures S6C and S6D). These observations indicate that Rad51 S14 phosphorylation promotes efficient resumption of replication after stress, but is not involved in DNA synthesis once replication restarts. Importantly, we also observed significantly higher numbers of DNA fibers containing only CldU tracts, which represent new origins of replication, in cells expressing S14A Rad51 compared with those expressing WT Rad51 (Figures 6F and S6B). Together, these results suggest that although S14A Rad51 is less proficient than WT in promoting the resumption of ongoing DNA replication, new origins are activated to facilitate completion of S phase.
Discussion
A growing body of evidence indicates that either insufficient or excessive Rad51 activity leads to genome instability, which is closely linked to cancer development and therapeutic resistance. Despite its importance, surprisingly little is known about the dynamic regulation of Rad51 during the cell cycle and in response to genotoxic stress in human cells. Here, we provide several lines of evidence that support a model in which BRCA2 coordinates the timely phosphorylation of Rad51 by Plk1, which in turn facilitates Rad51 association both with DSBs and with stalled replication forks and, through this mechanism, contributes to the maintenance of genome integrity.
We have shown that Plk1 binding to BRCA2 is primarily mediated through CDK-dependent phosphorylation at the evolutionarily conserved T77 site. It has been technically challenging to determine which CDK phosphorylates BRCA2 at T77 in vivo, as CDK family members may play redundant functions when a given CDK is dysfunctional (Murray, 2004). Nonetheless, T77 phosphorylation, which increases from late S phase and peaks in mitosis, was efficiently blocked only when CDK1 and CDK2 were both inhibited, strongly suggesting these CDKs function together to phosphorylate BRCA2 at this residue. Notably, T77A and A75P mutations within the PBD-binding motif have been reported in a number of breast cancer patients, indicating a physiologically important role of the BRCA2-Plk1 interaction in cancer prevention. Furthermore, in vivo phosphorylation of endogenous BRCA2 at T77 was detectable only in cells expressing full-length WT BRCA2, and not in BRCA2-defective cells, including EUFA423 cells, which express an equivalent amount of BRCA2 lacking only a short region of the CTD. These observations highlight the importance of full-length BRCA2 in coordinating its appropriate phosphorylation and thus its protein complex formation and associated functions. As has been suggested elsewhere (Thorslund et al., 2010), it is conceivable that the CTD of BRCA2 engages in a physical and/or functional interplay with its NTD. In this context, it is tempting to speculate that CDKs that associate with BRCA2 through cyclin-binding motifs within the CTD (Esashi et al., 2005) might potentially be responsible for BRCA2 phosphorylation within the NTD.
Our study also revealed that the level of Rad51 phosphorylation at S14 in the nuclear soluble fraction was largely dependent on the presence of functional BRCA2, implying that BRCA2 in this fraction has a distinctive role in facilitating Plk1-dependent phosphorylation of Rad51. Our findings raise an interesting question as to how Plk1 bound at the N-terminal region of BRCA2 phosphorylates Rad51, which associates with the middle (BRC motifs) and the C-terminal (TR2) regions of BRCA2. At present, it remains unknown how full-length BRCA2 folds and associates with its binding partners in three dimensions. Nonetheless, we speculate that the BRC motifs might contribute by presenting monomeric Rad51 in a manner that permits its phosphorylation at S14 by Plk1. This idea is supported by previous observations that (1) the BRC motifs directly bind monomeric Rad51 through its central ATPase core domain (Pellegrini et al., 2002); (2) residues between the BRC repeats, but not the motifs themselves, are targeted by Plk1 (Lee et al., 2004); and (3) Plk1-dependent BRCA2 phosphorylation does not interfere with the BRCA2-Rad51 interaction (Lee et al., 2004, Lin et al., 2003). We consider that the C-terminal TR2 motif may contribute to Plk1-mediated Rad51 phosphorylation differently from the BRC motifs. Rather than simply presenting Rad51 for phosphorylation by Plk1 on a BRCA2 platform (as would be the case with the BRC motifs), TR2 may modify the function of the N-terminal region of BRCA2. This view is supported by our observation that T77 phosphorylation levels were reduced in BRCA2 bearing a TR2 mutation or C-terminal truncation. TR2 would therefore indirectly alter the Plk1 interaction with the N-terminal region of BRCA2, leading to a reduced level of Rad51 phosphorylation at S14. In parallel, we also noticed that although the total level of pS14 was significantly reduced in cells expressing the BRCA2 TR2 mutant, pS14-Rad51 was still detectable in the complex containing this BRCA2 variant (K.Y., C.R., and F.E., unpublished data). These observations pose an alternative model in which a TR2-mediated Rad51 interaction contributes to the release of Rad51 from the BRC motif, providing an increased pool of phosphorylated Rad51 that is free from BRCA2; however, further investigation will be required to evaluate this possibility.
In this study, we further demonstrated that the S14 residue in Rad51 is important for protection of nascent DNA at stalled replication forks following HU treatment. We also found that in normally proliferating cells, Rad51 phosphorylation at S14, along with formation of a complex containing Rad51, BRCA2, and Plk1, takes place from late S phase to mitosis. This finding is complementary to previous reports that Rad51 is dispensable for the completion of bulk DNA replication but is essential in G2 for entry into mitosis (Su et al., 2008), and that Rad51 protects ssDNA gaps left behind replication forks from Mre11-mediated degradation (Hashimoto et al., 2010). In earlier work, we showed that phosphorylation of Rad51 at S14 triggers T13 phosphorylation, which then stimulates direct binding of the Nbs1 subunit of the MRN complex (Yata et al., 2012). Of note, Nbs1 was recently shown to protect genomic integrity in unperturbed cells during G2 (Bruhn et al., 2014). We therefore envisage that Plk1-mediated Rad51 phosphorylation provides a universal mechanism to initiate Nbs1-mediated Rad51 recruitment to stressed or fragile DNA to which the MRN complex is bound, including damaged DNA, stalled replication forks, and postreplicative ssDNA. In this way, completion of DNA repair and replication are ensured before a higher level of chromosome condensation takes place in mitosis, thus ensuring the protection of genome integrity.
The role described here for BRCA2 in facilitating Plk1-dependent Rad51 phosphorylation might appear to be at odds with our previous observation that Rad51 bypasses BRCA2 function in HR repair, in a manner that is dependent on Plk1 phosphorylation at S14 (Yata et al., 2012). Indeed, we cannot exclude the possibility that, together with BRCA2, additional factors may help to coordinate the timely phosphorylation of Rad51 following DNA stress. In support of this idea, we found that the pS14 signal was reduced but still detectable in the absence of BRCA2. Also, our iPOND analysis demonstrated that the association of Rad51 with replication forks was fully blocked only when both BRCA2 and Plk1-mediated phosphorylation of Rad51 at S14 were defective. Furthermore, our analyses using ChIP-seq and iPOND detected a partial association of Plk1 with some DSB sites and replication forks. Notably, Plk1 has been shown to interact with several DNA-damage-responsive proteins, such as MDC1, 53BP1, and SLX4 (Lowery et al., 2007, Svendsen et al., 2009, van Vugt et al., 2010), and also with the MCM replication complex at stalled replication forks (Song et al., 2011, Trenz et al., 2008, Tsvetkov and Stern, 2005). Such factors, or elevated Plk1 activity, which is found in many cancers, might be involved in the resistance of BRCA2-defective cells to anticancer drugs that induce DSBs or inhibit replication.
In summary, our work illuminates a dynamic and well-coordinated molecular mechanism whereby the soluble nuclear fraction of BRCA2 links the activities of CDKs and Plk1 to facilitate Rad51-mediated genome stability control. This role of BRCA2 is distinct from the previously demonstrated functions of chromatin-associated BRCA2, which has been proposed to recruit Rad51 directly to sites of stressed DNA. Our study has important implications for understanding the spatiotemporal functions of BRCA2 in proliferating human cells and how patients with BRCA2 deficiencies develop malignancy and therapeutic resistance.
Experimental Procedures
Mammalian Cell Culture
All mammalian cells were cultured at 37°C with 5% CO2. HeLa, HEK293T, U2OS, and EUFA423 cells were cultured in Dulbecco’s modified Eagle’s medium containing streptomycin (0.1 mg/ml), penicillin (100 U/ml), and 10% v/v fetal bovine serum. Capan-1 cells were maintained in the same medium, with the addition of 20% v/v fetal bovine serum. DLD1 BRCA2+/− and DLD1 BRCA2−/− cells (a kind gift from Dr. Scott Kern) were cultured in RPMI with streptomycin (0.1 mg/ml), penicillin (100 U/ml), and 10% v/v fetal bovine serum. HEK293 Flp-In T-REx FE-BRCA2 variant cell lines were maintained in the same medium described above for HEK293T cells, supplemented with 15 μg/ml blasticidin (InvivoGen) and 50 μg/ml hygromycin (InvivoGen). Expression of full-length FE-BRCA2 variants was induced with 2 μg/ml doxycycline for 48 hr prior to analyses. U2OS stable cell lines expressing Rad51 variants were described previously (Yata et al., 2012). DNA transfection was carried out using JetPRIME (Polyplus Transfection) following the manufacturer’s protocols. Where indicated, cells were irradiated at 4 Gy with a 137Cs source (Gravitron RX 30/55; Gravatom) or treated with 200 nM nocodazole (Sigma-Aldrich) for 18 hr. For downregulation of endogenous proteins using siRNA, cells were seeded at a density of 1.5–3 × 105 cells per well in six-well plate format. On the next day when cells were attached on the plates, siRNAs targeting human Rad51 (siRad51) was transfected at a final concentration of 20 nM, using DharmaFECT 1 (Dharmacon). Custom siRad51 was synthesized by Sigma-Aldrich with the sequence provided in Table S1. For negative controls, Mission siRNA Universal Negative Control #1 (SIC001; Sigma-Aldrich) was used at final concentration of 20 nM. Following a further 24 hr incubation, cells were harvested for analyses.
Antibodies
The antibodies used in this study are listed in Supplemental Experimental Procedures. For pT77 antibody, a peptide (NH2)-CNQLAS(pT)PIIFK-(COOH) coupled with KLH was used for rabbit immunization (Biogenes), and phospho-specific antibody was purified as previously described (Yata et al., 2012).
ChIP-Seq Analyses
ChIP samples were prepared from U2OS cells stably expressing AsiSI-ER as described previously (Deardorff et al., 2012, Iacovoni et al., 2010). DNA from whole-cell extracts (WCEs) and ChIP fractions was sequenced on Applied Biosystems SOLiD platforms (SOLiD 3 and 5500), and reads were aligned to the human genome (UCSC hg19) using Bowtie as described previously (Deardorff et al., 2012). The genome was scanned with a 3,000 bp sliding window and peaks were identified by the following criteria: (1) fold enrichment (IP reads / WCE reads) > 3, (2) a one-sided Wilcoxon rank-sum test p value < 1 × 10−4, (3) IP intensity of peak summit / IP read average in genome > 3, and (4) IP intensity of the peak summit bin > 4.0 per million reads mapped for each chromosome. The data set is available from the National Center for Biotechnology Information, Sequence Read Archive (NCBI-SRA; http://www.ncbi.nlm.nih.gov/Traces/sra/).
Cell Treatment for iPOND
HEK293 Flp-In T-REx FLAG-Rad51 variant cell lines were cultured in Dulbecco’s modified Eagle’s medium supplemented with streptomycin (0.1 mg/ml), penicillin (100 U/ml), blasticidin (10 μg/ml; InvivoGen), hygromycin B (100 μg/ml; InvivoGen), and 10% v/v fetal bovine serum. Expression of FLAG-Rad51 WT and FLAG-Rad51 S14A was induced by addition of 2 μg/ml doxycycline (Sigma-Aldrich) overnight to 24 hr prior to iPOND experiments. For endogenous Rad51 or BRCA2 knockdown, pSUPERIOR.puro constructs expressing shRNA targeting human RAD51 mRNA 3′ UTR or human BRCA2 mRNA were respectively delivered to the cells 24 and 48 hr before iPOND, using PEI transfection (Ehrhardt et al., 2006). Where indicated, cells were treated with 100 nM Plk1 inhibitor BI 2536 (Axon Medchem) for 15 min before EdU labeling and throughout EdU labeling and HU chase. For iPOND experiments, we adapted the procedures described in (Kliszczak et al., 2011, Sirbu et al., 2012), with modifications described in Supplemental Experimental Procedures.
IF and DNA Fiber Analysis
IF and DNA fiber analyses were carried out as previously described (Bleuyard et al., 2012, Jackson and Pombo, 1998, Schwab et al., 2010), with modifications described in Supplemental Experimental Procedures.
Author Contributions
K.Y. performed most of the analyses using phospho-specific antibodies, in vitro kinase reaction, far-western blotting, IF, and DNA fiber experiments. J.-Y.B. established and performed the iPOND experiments. F.E. purified and evaluated phospho-specific antibodies, and performed cell-cycle analyses. F.E. and C.R. generated full-length BRCA2 with point mutations, and F.E. established stable HEK293 Flp-In T-REx cell lines expressing BRCA2 variants. C.R. and Y.K. contributed to ChIP-seq sample preparation, and R.N. wrote the program for bioinformatic analyses of the ChIP-seq data. R.A.S. and W.N. contributed to the design and supervision of the DNA fiber experiments. K.S. contributed to the design and supervision of the ChIP-seq study. F.E. conceived and supervised the project. R.N., R.A.S., W.N., and K.S. assisted F.E. in preparation of the manuscript. F.E., K.Y., and J.-Y.B. wrote and edited the manuscript.
Acknowledgments
We thank Dr. Gaëlle Legube (University of Toulouse) for sharing the U2OS AsiSI-ER stable cell line, Dr. Scott Kern (Johns Hopkins Medicine) for DLD1 BRCA2 isogenic cell lines, Prof. Wen-Hwa Lee (University of California, Irvine) for pcDNA3.1-BRCA2, Dr. Simon Boulton (Cancer Research UK, LRI) for pFlp-In T-REx 1xFLAG Gateway vector, Prof. Jane Endicott (Newcastle University) and Dr. Aude Echalier (University of Montpellier) for recombinant CDK2/cyclin A, Prof. Martin Noble (Newcastle University) for CDK inhibitor NU6102, and Dr. Tim Hunt (Cancer Research UK, LRI) for Cyclin A and Cyclin B antibodies. We thank Prof. Chris Norbury (University of Oxford) for a critical reading of the manuscript. This work was funded by a Cancer Research UK Career Development Award and a Wellcome Trust Senior Fellowship to F.E., and F.E. received research grants from the Breast Cancer Campaign and the Edward Penley Abraham Research Fund. R.N. is supported by a Grant-in-Aid for Young Scientists. K.S. is supported by CREST, a Research Program of Innovative Cell Biology by Innovative Technology and Grant-in-Aid for Scientific Research. W.N. is the recipient of an Association for International Cancer Research International Fellowship and a Medical Research Council Senior Non-Clinical Fellowship.
Published: May 15, 2014
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information includes Supplemental Discussion, Supplemental Experimental Procedures, six figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.04.023.
Accession Numbers
The NCBI-SRA accession numbers for the ChIP-seq data reported in this paper are SRP040613 (Figures 4D, 4E, and S4D), SRP040614 (Figure 4F), and SRP040615 (Figure S4C).
Supplemental Information
References
- Ayoub N., Rajendra E., Su X., Jeyasekharan A.D., Mahen R., Venkitaraman A.R. The carboxyl terminus of Brca2 links the disassembly of Rad51 complexes to mitotic entry. Curr. Biol. 2009;19:1075–1085. doi: 10.1016/j.cub.2009.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr F.A., Silljé H.H., Nigg E.A. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- Bleuyard J.Y., Buisson R., Masson J.Y., Esashi F. ChAM, a novel motif that mediates PALB2 intrinsic chromatin binding and facilitates DNA repair. EMBO Rep. 2012;13:135–141. doi: 10.1038/embor.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhn C., Zhou Z.W., Ai H., Wang Z.Q. The essential function of the MRN complex in the resolution of endogenous replication intermediates. Cell Rep. 2014;6:182–195. doi: 10.1016/j.celrep.2013.12.018. [DOI] [PubMed] [Google Scholar]
- Bruinsma W., Raaijmakers J.A., Medema R.H. Switching Polo-like kinase-1 on and off in time and space. Trends Biochem. Sci. 2012;37:534–542. doi: 10.1016/j.tibs.2012.09.005. [DOI] [PubMed] [Google Scholar]
- Dalton W.B., Nandan M.O., Moore R.T., Yang V.W. Human cancer cells commonly acquire DNA damage during mitotic arrest. Cancer Res. 2007;67:11487–11492. doi: 10.1158/0008-5472.CAN-07-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies T.G., Bentley J., Arris C.E., Boyle F.T., Curtin N.J., Endicott J.A., Gibson A.E., Golding B.T., Griffin R.J., Hardcastle I.R. Structure-based design of a potent purine-based cyclin-dependent kinase inhibitor. Nat. Struct. Biol. 2002;9:745–749. doi: 10.1038/nsb842. [DOI] [PubMed] [Google Scholar]
- Deardorff M.A., Bando M., Nakato R., Watrin E., Itoh T., Minamino M., Saitoh K., Komata M., Katou Y., Clark D. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 2012;489:313–317. doi: 10.1038/nature11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt C., Schmolke M., Matzke A., Knoblauch A., Will C., Wixler V., Ludwig S. Polyethylenimine, a cost-effective transfection reagent. Signal Transduct. 2006;6:179–184. [Google Scholar]
- Elia A.E., Rellos P., Haire L.F., Chao J.W., Ivins F.J., Hoepker K., Mohammad D., Cantley L.C., Smerdon S.J., Yaffe M.B. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- Esashi F., Christ N., Gannon J., Liu Y., Hunt T., Jasin M., West S.C. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- Giunta S., Belotserkovskaya R., Jackson S.P. DNA damage signaling in response to double-strand breaks during mitosis. J. Cell Biol. 2010;190:197–207. doi: 10.1083/jcb.200911156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goggins M., Schutte M., Lu J., Moskaluk C.A., Weinstein C.L., Petersen G.M., Yeo C.J., Jackson C.E., Lynch H.T., Hruban R.H., Kern S.E. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360–5364. [PubMed] [Google Scholar]
- Hans F., Dimitrov S. Histone H3 phosphorylation and cell division. Oncogene. 2001;20:3021–3027. doi: 10.1038/sj.onc.1204326. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Ray Chaudhuri A., Lopes M., Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett N.G., Taniguchi T., Olson S., Cox B., Waisfisz Q., De Die-Smulders C., Persky N., Grompe M., Joenje H., Pals G. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Hucl T., Rago C., Gallmeier E., Brody J.R., Gorospe M., Kern S.E. A syngeneic variance library for functional annotation of human variation: application to BRCA2. Cancer Res. 2008;68:5023–5030. doi: 10.1158/0008-5472.CAN-07-6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovoni J.S., Caron P., Lassadi I., Nicolas E., Massip L., Trouche D., Legube G. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D.A., Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliszczak A.E., Rainey M.D., Harhen B., Boisvert F.M., Santocanale C. DNA mediated chromatin pull-down for the study of chromatin replication. Sci Rep. 2011;1:95. doi: 10.1038/srep00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster J.M., Wooster R., Mangion J., Phelan C.M., Cochran C., Gumbs C., Seal S., Barfoot R., Collins N., Bignell G. BRCA2 mutations in primary breast and ovarian cancers. Nat. Genet. 1996;13:238–240. doi: 10.1038/ng0696-238. [DOI] [PubMed] [Google Scholar]
- Lee M., Daniels M.J., Venkitaraman A.R. Phosphorylation of BRCA2 by the Polo-like kinase Plk1 is regulated by DNA damage and mitotic progression. Oncogene. 2004;23:865–872. doi: 10.1038/sj.onc.1207223. [DOI] [PubMed] [Google Scholar]
- Lin H.R., Ting N.S., Qin J., Lee W.H. M phase-specific phosphorylation of BRCA2 by Polo-like kinase 1 correlates with the dissociation of the BRCA2-P/CAF complex. J. Biol. Chem. 2003;278:35979–35987. doi: 10.1074/jbc.M210659200. [DOI] [PubMed] [Google Scholar]
- Lo T., Pellegrini L., Venkitaraman A.R., Blundell T.L. Sequence fingerprints in BRCA2 and RAD51: implications for DNA repair and cancer. DNA Repair (Amst.) 2003;2:1015–1028. doi: 10.1016/s1568-7864(03)00097-1. [DOI] [PubMed] [Google Scholar]
- Lomonosov M., Anand S., Sangrithi M., Davies R., Venkitaraman A.R. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev. 2003;17:3017–3022. doi: 10.1101/gad.279003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery D.M., Clauser K.R., Hjerrild M., Lim D., Alexander J., Kishi K., Ong S.E., Gammeltoft S., Carr S.A., Yaffe M.B. Proteomic screen defines the Polo-box domain interactome and identifies Rock2 as a Plk1 substrate. EMBO J. 2007;26:2262–2273. doi: 10.1038/sj.emboj.7601683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan G.L., Pfander B., Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Murray A.W. Recycling the cell cycle: cyclins revisited. Cell. 2004;116:221–234. doi: 10.1016/s0092-8674(03)01080-8. [DOI] [PubMed] [Google Scholar]
- Ochiai K., Yoshikawa Y., Yoshimatsu K., Oonuma T., Tomioka Y., Takeda E., Arikawa J., Mominoki K., Omi T., Hashizume K., Morimatsu M. Valine 1532 of human BRC repeat 4 plays an important role in the interaction between BRCA2 and RAD51. FEBS Lett. 2011;585:1771–1777. doi: 10.1016/j.febslet.2011.05.027. [DOI] [PubMed] [Google Scholar]
- Pellegrini L., Venkitaraman A. Emerging functions of BRCA2 in DNA recombination. Trends Biochem. Sci. 2004;29:310–316. doi: 10.1016/j.tibs.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Pellegrini L., Yu D.S., Lo T., Anand S., Lee M., Blundell T.L., Venkitaraman A.R. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- Petermann E., Orta M.L., Issaeva N., Schultz N., Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quignon F., Rozier L., Lachages A.M., Bieth A., Simili M., Debatisse M. Sustained mitotic block elicits DNA breaks: one-step alteration of ploidy and chromosome integrity in mammalian cells. Oncogene. 2007;26:165–172. doi: 10.1038/sj.onc.1209787. [DOI] [PubMed] [Google Scholar]
- Rajendra E., Venkitaraman A.R. Two modules in the BRC repeats of BRCA2 mediate structural and functional interactions with the RAD51 recombinase. Nucleic Acids Res. 2010;38:82–96. doi: 10.1093/nar/gkp873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K., Wu H., Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R.A., Blackford A.N., Niedzwiedz W. ATR activation and replication fork restart are defective in FANCM-deficient cells. EMBO J. 2010;29:806–818. doi: 10.1038/emboj.2009.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu B.M., Couch F.B., Feigerle J.T., Bhaskara S., Hiebert S.W., Cortez D. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 2011;25:1320–1327. doi: 10.1101/gad.2053211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu B.M., Couch F.B., Cortez D. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat. Protoc. 2012;7:594–605. doi: 10.1038/nprot.2012.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B., Liu X.S., Davis K., Liu X. Plk1 phosphorylation of Orc2 promotes DNA replication under conditions of stress. Mol. Cell. Biol. 2011;31:4844–4856. doi: 10.1128/MCB.06110-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Bernal J.A., Venkitaraman A.R. Cell-cycle coordination between DNA replication and recombination revealed by a vertebrate N-end rule degron-Rad51. Nat. Struct. Mol. Biol. 2008;15:1049–1058. doi: 10.1038/nsmb.1490. [DOI] [PubMed] [Google Scholar]
- Svendsen J.M., Smogorzewska A., Sowa M.E., O’Connell B.C., Gygi S.P., Elledge S.J., Harper J.W. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorslund T., McIlwraith M.J., Compton S.A., Lekomtsev S., Petronczki M., Griffith J.D., West S.C. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat. Struct. Mol. Biol. 2010;17:1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenz K., Errico A., Costanzo V. Plx1 is required for chromosomal DNA replication under stressful conditions. EMBO J. 2008;27:876–885. doi: 10.1038/emboj.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov L., Stern D.F. Interaction of chromatin-associated Plk1 and Mcm7. J. Biol. Chem. 2005;280:11943–11947. doi: 10.1074/jbc.M413514200. [DOI] [PubMed] [Google Scholar]
- van Vugt M.A.T.M., Gardino A.K., Linding R., Ostheimer G.J., Reinhardt H.C., Ong S.E., Tan C.S., Miao H., Keezer S.M., Li J.J. A mitotic phosphorylation feedback network connects Cdk1, Plk1, 53BP1, and Chk2 to inactivate the G(2)/M DNA damage checkpoint. PLoS Biol. 2010;8:e1000287. doi: 10.1371/journal.pbio.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev L.T., Tovar C., Chen S., Knezevic D., Zhao X., Sun H., Heimbrook D.C., Chen L. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. USA. 2006;103:10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S.C. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- Wooster R., Bignell G., Lancaster J., Swift S., Seal S., Mangion J., Collins N., Gregory S., Gumbs C., Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- Yata K., Lloyd J., Maslen S., Bleuyard J.Y., Skehel M., Smerdon S.J., Esashi F. Plk1 and CK2 act in concert to regulate Rad51 during DNA double strand break repair. Mol. Cell. 2012;45:371–383. doi: 10.1016/j.molcel.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.