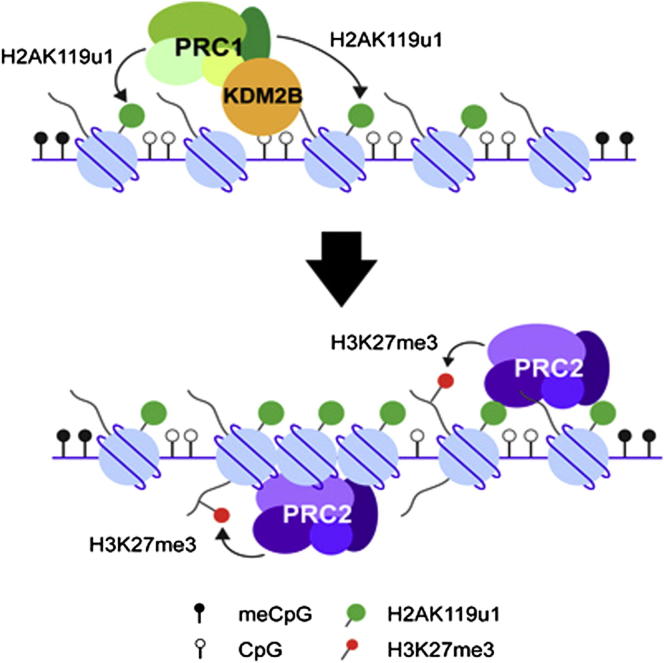
Summary
The mechanisms by which the major Polycomb group (PcG) complexes PRC1 and PRC2 are recruited to target sites in vertebrate cells are not well understood. Building on recent studies that determined a reciprocal relationship between DNA methylation and Polycomb activity, we demonstrate that, in methylation-deficient embryonic stem cells (ESCs), CpG density combined with antagonistic effects of H3K9me3 and H3K36me3 redirects PcG complexes to pericentric heterochromatin and gene-rich domains. Surprisingly, we find that PRC1-linked H2A monoubiquitylation is sufficient to recruit PRC2 to chromatin in vivo, suggesting a mechanism through which recognition of unmethylated CpG determines the localization of both PRC1 and PRC2 at canonical and atypical target sites. We discuss our data in light of emerging evidence suggesting that PcG recruitment is a default state at licensed chromatin sites, mediated by interplay between CpG hypomethylation and counteracting H3 tail modifications.
Graphical Abstract
Highlights
-
•
Absence of DNA methylation recruits Polycomb complexes to pericentric heterochromatin
-
•
H3K9me3 antagonizes activity of PRC2, but not PRC1, at pericentric heterochromatin
-
•
CpG density and antagonism by H3 modifications define genome-wide Polycomb occupancy
-
•
PRC1-mediated H2AK119u1 recruits PRC2 and H3K27me3
Polycomb group proteins are important repressors of developmentally regulated genes, but how these complexes are recruited to their target genes is still largely unknown. In this study, Cooper et al. show that Polycomb group protein recruitment is a combinatorial readout of unmethylated CpG density and antagonism by specific histone tail modifications. Unexpectedly, they also show that monoubiquitylated histone H2A, the modification produced by Polycomb repressor complex 1 (PRC1), is sufficient to recruit PRC2.
Introduction
Polycomb group (PcG) repressor proteins play an important role in developmental gene regulation in multicellular organisms (Ringrose and Paro, 2004). In most cases, they are assigned as components of one of two major multisubunit complexes, Polycomb repressive complex 1 (PRC1) and PRC2, both of which have intrinsic histone-modifying activities (monoubiquitylation of histone H2A lysine 119 [H2AK119u1] and methylation of histone H3 lysine 27 [H3K27me1/2/3], respectively; reviewed in Simon and Kingston, 2013). Vertebrates possess several variant PRC1 complexes, as defined by the presence of the subunit PCGF1-6 (Gao et al., 2012). The histone-modifying activities of PRC1 and PRC2 are of central importance for PcG function (Endoh et al., 2012, Pengelly et al., 2013), although several studies have revealed that PRC1 also mediates repression through alternative mechanisms (Eskeland et al., 2010, Francis et al., 2004, Isono et al., 2013). Canonical PRC1 and PRC2 function in a hierarchical manner, with PRC1 recruitment occurring via binding of the chromodomain of the Cbx Polycomb subunit to PRC2-mediated H3K27me3 (Cao et al., 2002, Wang et al., 2004). More recent studies have demonstrated that variant PRC1 complexes are recruited through PRC2-independent mechanisms (Schoeftner et al., 2006, Tavares et al., 2012).
In vertebrates, cytosine residues at CpG dinucleotides are extensively methylated, with the exception of CpG islands (CGIs), which are found at the promoter region of more than 50% of genes (reviewed in Illingworth and Bird, 2009). DNA methylation is required for normal development, but not for the maintenance of embryonic stem cells (ESCs) in vitro (reviewed in Bestor, 2000). DNA methylation patterns are erased in early embryos and developing germ cells, and are then reestablished by the de novo DNA methyltransferases Dnmt3a and Dnmt3b, and the accessory protein Dnmt3L (reviewed in Reik et al., 2001). Propagation of DNA methylation patterns through DNA replication is dependent on the maintenance DNA methyltransferase, Dnmt1, and the accessory protein Uhrf1 (Bestor, 2000, Sharif et al., 2007).
In Drosophila, PcG recruitment at Hox loci, as well as other targets, is mediated by transcription factor binding at sequences designated as Polycomb response elements (Kassis and Brown, 2013, Mohd-Sarip et al., 2005). In mammalian cells, PcG proteins are recruited to target gene promoters and also to atypical sites, specifically the inactive X chromosome (de Napoles et al., 2004, Plath et al., 2003, Silva et al., 2003), and paternal pericentric heterochromatin (PCH) domains in early mouse embryos (Puschendorf et al., 2008, Santos et al., 2005). There is evidence supporting a role for sequence-specific factors in PcG recruitment in mammals (Woo et al., 2010, Woo et al., 2013), and also suggesting a role for noncoding RNA (reviewed in Brockdorff, 2013). Additionally, several studies have demonstrated a link between unmethylated CpG residues and PcG recruitment/binding. Specifically, PcG occupancy correlates closely with CpG density in CGIs at target gene promoters (Lynch et al., 2012, Mendenhall et al., 2010). Moreover, depletion of DNA methylation has been correlated with changes in patterns of PRC2 localization (Brinkman et al., 2012, Reddington et al., 2013). Linked to these observations, recent studies have demonstrated that recruitment of the variant PRC1 complex PCGF1-PRC1 is dependent on binding of the KDM2B subunit to unmethylated CpG residues via a CXXC-zinc finger domain (Farcas et al., 2012, He et al., 2013, Wu et al., 2013). Interestingly, a chromatin immunoprecipitation sequencing (ChIP-seq) analysis of KDM2B demonstrated occupancy, albeit at a reduced level, at all CGIs, including those associated with active genes (Farcas et al., 2012). Based on these findings, we have proposed that PcG complexes bind CGIs as a default, with patterns of occupancy being defined on the one hand through reinforcement by positive-feedback mechanisms, and on the other hand by antagonizing chromatin modifications (e.g., H3K4me3 mediated by Trithorax group factors) at CGIs of active genes (Klose et al., 2013).
Here, we show that both PRC1 and PRC2 activities are redirected to noncanonical targets, PCH domains, and other CpG-rich sites, notably gene exons, in embryonic stem cells (ESCs) in which DNA methylation is depleted, and that this is inhibited by the presence of preexisting histone tail modifications, notably H3K9me3 and H3K36me3. Thus, PcG occupancy patterns are a combinatorial readout of unmethylated CpG density and antagonism by specific chromatin modifications. We find that tethering PRC1 proteins to PCH in wild-type (WT) cells is sufficient to establish both H2AK119u1 and PRC2-mediated H3K27me3. Moreover, we show that the observed PRC2 recruitment is directly linked to deposition of H2AK119u1. These observations define a mechanism that potentially explains the recruitment of both PRC1 and PRC2 to unmethylated CpG sites in the vertebrate genome.
Results
Loss of DNA Methylation Recruits PcG Complexes to PCH
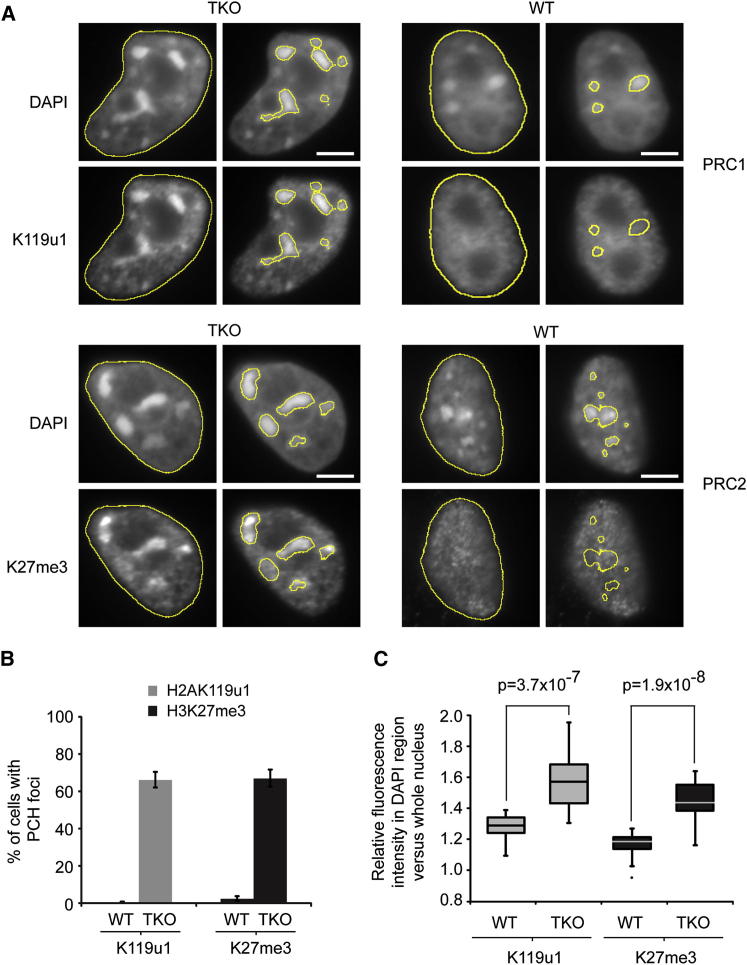
PCH in mouse comprises large blocks of major and minor satellite repeat sequences (Figure S1A) in which the chromatin is modified with H3K9me3, H4K20me3, and associated binding proteins, notably heterochromatin protein 1 (Probst and Almouzni, 2011). Atypical localization of PcG complexes and associated modifications occur on paternal PCH in early embryos (Santos et al., 2005) and have been linked to absence of H3K9 methylation (Puschendorf et al., 2008). Taking into consideration emerging evidence for an antagonistic effect of DNA methylation on PcG activity (Brinkman et al., 2012, Hagarman et al., 2013, Lynch et al., 2012, Reddington et al., 2013), we speculated that programmed DNA demethylation of the paternal genome in fertilized zygotes (Mayer et al., 2000, Oswald et al., 2000) may also play a role. To test this idea, we performed an immunofluorescence (IF) analysis to detect PRC1-mediated H2AK119u1 and PRC2-mediated H3K27me3 in Dnmt1/Dnmt3a/Dnmt3b triple knockout (Dnmt TKO) embryonic stem cells (ESCs), in which DNA methylation is depleted to 0.1%–0.4% of the levels seen in WT cells (Tsumura et al., 2006). We observed both H2AK119u1 and H3K27me3 foci colocalizing with DAPI-dense PCH domains in a large proportion of Dnmt TKO, but not WT, ESCs (Figures 1A, 1B, and S1B), demonstrating that CpG hypomethylation is sufficient to recruit PcG complexes. An unbiased quantitative analysis revealed that Dnmt TKO ESCs have a significant enrichment of H3K27me3 and H2AK119u1 in DAPI-dense domains relative to the whole nucleus (Figure 1C). A western blot analysis indicated that the global levels of PcG proteins and associated histone modifications were similar in WT and Dnmt TKO ESCs (Figure S1C).
Figure 1.
PcG Complexes Localize to PCH in Response to Loss of DNA Methylation
(A) IF of Dnmt TKO and WT cells stained for H2AK119u1 (PRC1) or H3K27me3 (PRC2), and DNA stained with DAPI. Yellow lines show total nuclear area (left) and PCH domains (right) based on DAPI staining defined by the ImageJ threshold algorithm Triangle.
(B) Graph showing the percentage of cells with PCH foci positive for H2AK119u1 or H3K27me3 in WT or Dnmt TKO cells. Bars show average (n > 200 DAPI foci) ± SD (n = 3).
(C) Box and whisker plot showing quantification of fluorescence intensities at PCH domains. The total nuclear area and PCH domains were defined as shown by the yellow lines in (A). Fluorescence intensity (H2AK119u1 or H3K27me3 staining) was calculated within the defined PCH domains in either WT or Dnmt TKO cells and is expressed relative to the fluorescence intensity within the total nuclear area of the cell (n = 20, p values Student’s t test, unpaired).
Scale bars represent 5 μm. See also Figure S1.
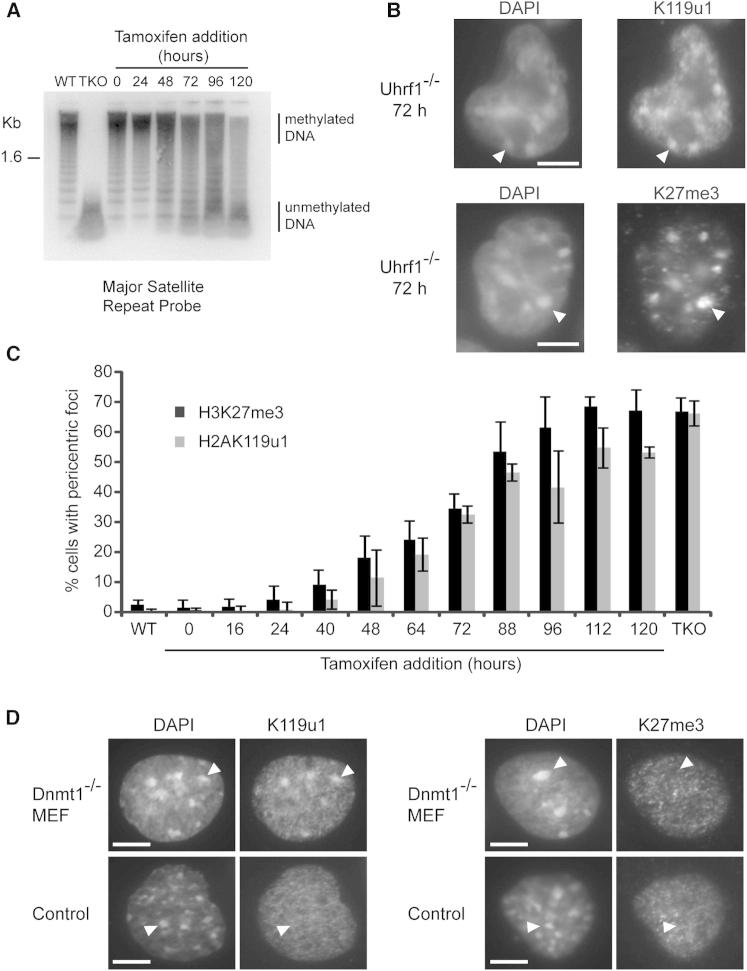
We determined the kinetics of H3K27me3 and H2AK119u1 deposition at PCH domains using ESCs with a tamoxifen-inducible knockout allele for Uhrf1, a factor required for the maintenance of DNA methylation (Sharif et al., 2007). Similarly to Dnmt TKO ESCs, both H3K27me3 and H2AK119u1 localized to PCH in constitutive Uhrf1−/− ESCs, but not in Uhrf1flox/flox controls (Figures S2A and S2B). We therefore treated Uhrf1flox/flox ESCs with tamoxifen and assayed depletion of DNA methylation at major satellite repeats by Southern blot (Figure 2A), and acquisition of H3K27me3 and H2AK119u1 at PCH by IF (Figure 2B) over time. The results demonstrate a close correlation between the rate of depletion of DNA methylation and the deposition of H3K27me3/H2AK119u1 at PCH (Figure 2C). We obtained similar findings using a tamoxifen-inducible knockout ESC model for the maintenance DNA methyltransferase Dnmt1 (Figures S2C and S2D). These findings substantiate that DNA hypomethylation results in H3K27me3/H2AK119u1 deposition at PCH in ESCs, and moreover suggest a direct causative link.
Figure 2.
PcG Complexes Are Dynamically Acquired at PCH upon Loss of DNA Methylation
(A) Southern blot showing loss of DNA methylation over time upon loss of Uhrf1. Genomic DNA from WT, Dnmt TKO, or tamoxifen-induced deletion of Uhrf1 in conditional knockout cells (0–120 hr) was digested with the methylation-sensitive enzyme HpyCH41V and the blot was probed for major satellite repeats.
(B) IF of conditional knockout Uhrf1−/− cells (72 hr, tamoxifen treatment) stained for H2AK119u1 or H3K27me3. Arrowheads indicate an example of staining within PCH.
(C) Graph showing the percentage of cells with H3K27me3 or H2AK119u1 foci during the Uhrf1−/− deletion time course. Bars show average (n > 200 cells) ± SD (n = 3).
(D) IF of Dnmt1−/− (and control) MEF cells stained for H2AK119u1 and H3K27me3. Arrowheads indicate a single PCH domain.
Scale bars represent 5 μm. See also Figure S2.
We also analyzed Dnmt1−/− mouse embryonic fibroblasts (MEFs), which were previously shown to be extensively depleted of DNA methylation (Lande-Diner et al., 2007), to determine whether localization of H2AK119u1 and H3K27me3 to PCH occurs in a differentiated cell type. As illustrated in Figure 2D, H2AK119u1 was indeed found at PCH, but, interestingly, H3K27me3 foci were not detectable (Figure 2D). This observation indicates that PCH domains in differentiated cells have distinct characteristics (also see below).
H3K9me3 at PCH Antagonizes PRC2, but Not PRC1, Activity
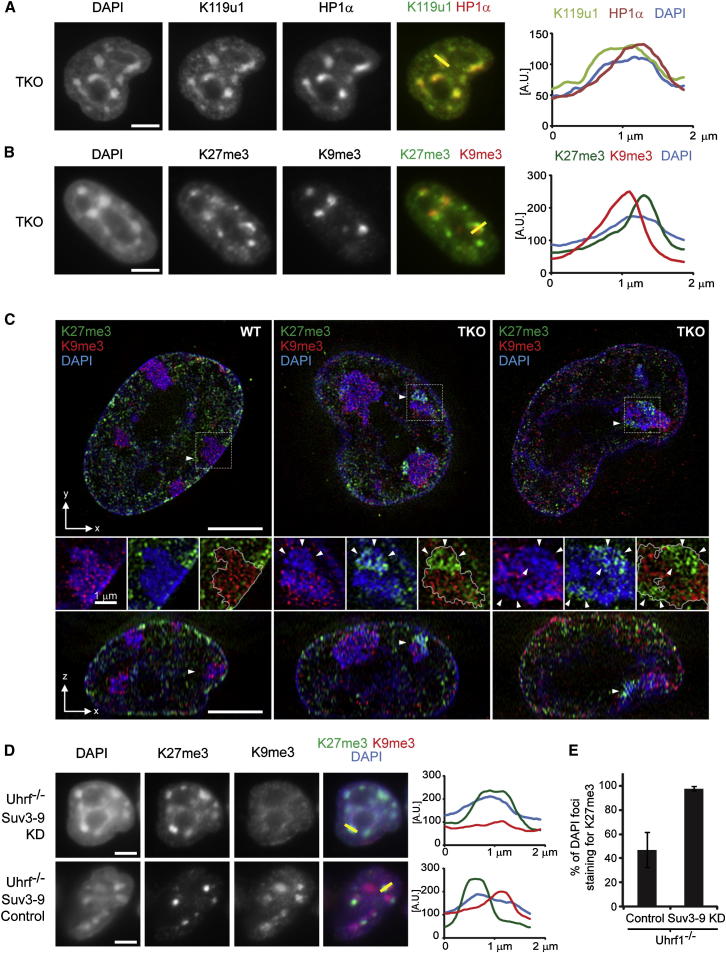
Although both H3K27me3 and H2AK119u1 were seen at PCH in DNA methylation-depleted ESCs, we noted that the frequency of H3K27me3 foci was lower than the H2AK119u1 foci (Figures S3A and S3B), indicating that PCH may be somewhat refractory to PRC2 activity. Previous studies noted the appearance of H3K27me3 at PCH when H3K9me3, a histone tail modification that is normally concentrated at these sites, is depleted (Peters et al., 2003, Puschendorf et al., 2008). We therefore investigated the interplay between H3K9me3 and both H3K27me3 and H2AK119u1 in methylation-deficient Dnmt TKO compared with WT ESCs. As illustrated in Figures 3A and S3C, H2AK119u1 and HP1α (which binds to H3K9me3) show extensive overlap with one another, encompassing the entire PCH domains. In contrast, H3K27me3 and H3K9me3 appear nonoverlapping, although both lie within DAPI-stained PCH domains (Figure 3B). We verified these observations using superresolution 3D structured illumination microscopy (3D-SIM), a technique that offers significantly enhanced spatial resolution compared with conventional fluorescence microscopy (Schermelleh et al., 2010). Representative examples shown in Figure 3C illustrate that in Dnmt TKO ESCs, H3K27me3 and H3K9me3 are enriched in distinct subdomains within PCH clusters.
Figure 3.
H3K9me3 Antagonizes PRC2, but Not PRC1
(A) IF of Dnmt TKO cells costained for H2AK119u1 and HP1α. The graph shows a profile plot of fluorescence intensity (arbitrary units [A.U.]) across a single PCH domain (2 μM) defined by DAPI and marked on the merge image as a yellow bar.
(B) As in (A), but costained for H3K27me3 and H3K9me3.
(C) Superresolution 3D-SIM images of Dnmt TKO and WT cells stained with H3K27me3 (green), H3K9me3 (red), and DAPI (blue). Upper panels show a single xy plane and bottom panels show an orthogonal xz plane. Arrowheads indicate the cutting plane. Middle panels show a single PCH region (delineated with a white line), and merges show overlap of each color. Arrowheads mark H3K27me3 staining within PCH in Dnmt TKO cells.
(D) Stable knockdown of Suv3-9h1/h2 or a scrambled control in Uhrf1−/− cells costained for H3K27me3 and H3K9me3. Profile plots as in (A) across a single PCH domain, marked on the merge image as a yellow bar.
(E) Graph showing the percentage of DAPI foci staining for H3K27me3 in Uhrf1−/− cells with either scrambled control or stable knockdown (KD) of Suv3-9h1/h2. Bars show average (n > 200 DAPI foci) ± SD (n = 3).
Scale bars represent 5 μm (unless otherwise stated). See also Figure S3 and Table S2.
The mutually exclusive deposition of H3K9me3 and H3K27me3 at PCH foci in ESCs suggests that these histone modifications may antagonize one another. Consistent with this, H3K27me3 foci in Suv3-9h1/h2 double knockout (DKO) ESCs, in which H3K9me3 at PCH is entirely depleted (Peters et al., 2001), were more extensive than in DNA methylation-depleted Dnmt TKO ESCs (Figure S3D). Moreover, in contrast to Dnmt TKO ESCs, H3K27me3 in Suv3-9h1/h2 DKO ESCs was detected in the majority of PCH domains, again similar to what was observed for H2AK119u1 (Figure S3E). To further investigate the apparent antagonism of H3K27me3 by H3K9me3, we used RNAi to deplete Suv3-9h1/h2 (and hence H3K9me3) in DNA methylation-depleted (Uhrf1−/−) ESCs (Figure S3F), and then analyzed H3K27me3 at PCH. The extent of H3K27me3 domains was clearly increased in Suv3-9h1/h2 knockdown cells (Figure 3D), as was their frequency (Figure 3E). Moreover, we observed extensive H3K27me3 and H2AK119u1 domains in WT ESCs following Suv3-9h1/h2 depletion (Figure S3G). Taken together, these results indicate that H3K9me3 and H3K27me3 modifications are mutually antagonistic at PCH domains in ESCs.
Unmethylated CpG Titrates PcG Complexes
We went on to investigate PcG localization genome-wide following depletion of DNA methylation by performing a ChIP-seq analysis of H3K27me3, Suz12 (a core subunit of PRC2), and Ring1B (the catalytic subunit of PRC1) in Dnmt TKO and WT ESCs. Determination of the fold change in Dnmt1 TKO compared with WT ESCs at repetitive sequences, which collectively comprise approximately 45% of the genome, demonstrated enhanced levels of H3K27me3, Suz12, and Ring1B at major satellite sequences and, to a lesser extent, at minor satellite sequences (Table S1; Figure 4A), consistent with the IF data. Other common repeat sequence classes showed minimal fold changes, with a moderate reduction of H3K27me3 at L1 repeats and a slight increase of H3K27me3, Suz12, and Ring1B at SINE repeats (Figure 4A).
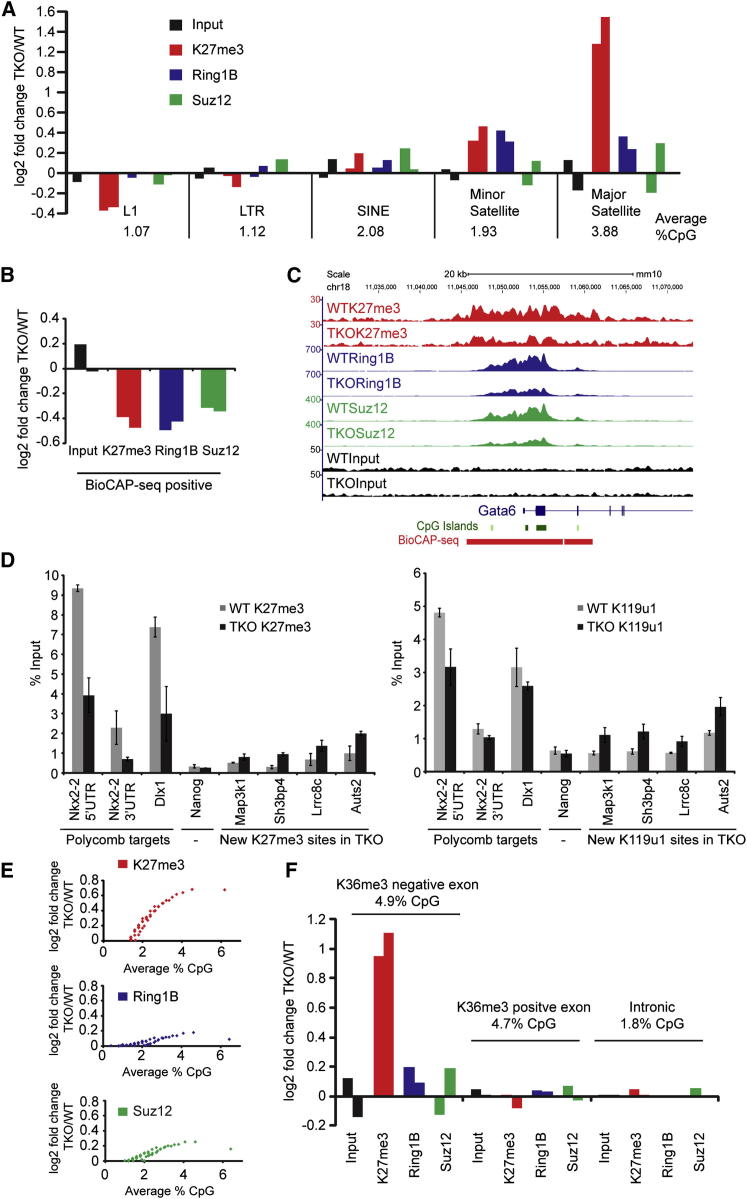
Figure 4.
Loss of DNA Methylation Redistributes PcG Complexes Away from Canonical Sites to New Sites Depending on CpG Content and H3K36me3 Occupancy
(A) Graph showing at the indicated repeat regions the fold change of reads, normalized to total reads, from ChIP-seq analysis of Input, H3K27me3, Ring1B, and Suz12 in Dnmt TKO relative to WT cells. The %CpG of each repeat class is shown. Two bars represent biological repeats.
(B) As in (A), but with fold change at BioCAP-seq positive regions (WT unmethylated CpG regions).
(C) Screen shot of one biological repeat of ChIP-seq of H3K27me3, Ring1B, Suz12, and Input in WT and Dnmt TKO cells.
(D) ChIP-qPCR of H3K27me3 or H2AK119u1 at canonical targets or new sites in WT and Dnmt TKO cells. Bars show average ± SD (n = 3).
(E) Graphs showing a positive fold change in Dnmt TKO versus WT cells using a sliding-window analysis of H3K27me3, Ring1B, and Suz12 ChIP-seq of only BioCAP-seq-negative regions, binned according to CpG content.
(F) As in (A), but with fold change at H3K36me3-positive or -negative exons, or introns.
Recent genome-wide studies have reported aberrant H3K27me3 localization at unique sequences in DNA methylation-deficient ESCs. Specifically, decreased levels occurred at CGIs of canonical PcG target loci (Brinkman et al., 2012, Reddington et al., 2013) and increased levels occurred at sites where DNA methylation levels were relatively high, notably in gene-rich chromosomal domains (Brinkman et al., 2012, Hagarman et al., 2013, Lynch et al., 2012, Reddington et al., 2013). We hypothesized that these opposite effects could be attributable to a titration of PcG complexes to newly acquired unmethylated CpG sites, including major satellite sequences, leading to a reduction in levels at canonical target sites. To investigate this, we first quantified PcG localization at canonical target loci by determining the fold change in Dnmt TKO relative to WT at known CGIs, as defined by biotin CXXC affinity purification sequencing (BioCAP-seq) experiments (Long et al., 2013b). Consistent with previous studies (Brinkman et al., 2010, Reddington et al., 2013), we observed that Dnmt TKO ESCs had reduced levels of H3K27me3, Ring1B, and Suz12 at WT PcG sites (Figure 4B). An example of a single PcG target locus, the Gata6 gene, is shown in Figure 4C, and further validation using ChIP-qPCR for H3K27me3 and H2AK119u1 at selected loci is shown in Figure 4D. We next determined the relationship between PcG occupancy and CpG density at non-CGI sequences by performing a whole-genome sliding-window analysis. We excluded embryonic, somatic, and germ cell CGIs as defined by BioCAP-seq analyis of ESCs, liver, and testis (Long et al., 2013b). Figure 4E shows a positive fold change (new sites) for non-BioCAP-seq regions binned according to CpG content, and reveals a striking correlation between the fold increase for H3K27me3 and the overall CpG content. Such a fold increase, albeit less pronounced, was also seen for Suz12 and Ring1B. We validated increased H3K27me3 at selected new sites by conventional ChIP and also demonstrated a similar increase in H2AK119u1 (Figure 4D). Taken together, these observations support the idea that unmethylated CpG sites are a primary determinant of PcG occupancy in Dnmt TKO ESCs.
A previous analysis of methylation-deficient ESCs documented a gain of H3K27me3 over large gene-rich chromosomal domains (Brinkman et al., 2012). We considered that this could be linked to the higher CpG density that has been reported to occur in gene exons (Lander et al., 2001). Importantly, we determined that the high CpG content of exons is evident even when exon 1, which frequently overlaps with promoter-associated CGIs, is excluded (4.9% CpG compared with 1.8% in introns). We therefore analyzed the fold change in H3K27me3, Suz12, and Ring1B in Dnmt TKO relative to WT ESCs for gene exons (not exon 1) and for introns. As shown in Figure S4A, exons (excluding exon 1), but not introns, show clearly increased H3K27me3 and, to a lesser extent, Ring1B and Suz12 levels, correlating with their relative CpG content.
In vitro analysis has shown that PRC2-mediated H3K27me3 is inhibited when H3K36me2/3 is present on the same H3 tail (Schmitges et al., 2011, Yuan et al., 2011). Given that H3K36me3 is normally associated with the gene bodies of actively transcribed genes, we obtained H3K36me3 profiles for Dnmt TKO ESCs, defined H3K36me3-positive and -negative exons (not exon 1) (Figures S4B–S4D), and then determined PcG acquisition for these two exon categories in Dnmt TKO relative to WT. Consistent with the inhibitory effect of H3K36me3 on PRC2 activity, we observed little or no fold increase of H3K27me3 or the PRC2 subunit Suz12 at H3K36me3-positive exons, despite their high CpG content (4.7%) (Figure 4F). A similar effect was observed for Ring1B. We conclude that PcG redistribution at unique sequences in Dnmt TKO ESCs is a combinatorial readout of the density of unmethylated CpG density coupled to antagonism of PRC2 by H3K36me3 in the gene bodies of active genes.
PRC1 and PRC2 Activity Is Not Dependent on DNA Hypomethylation
Previous studies provided evidence for a direct effect of DNA methylation on binding/recruitment of PRC2 (Bartke et al., 2010, Wu et al., 2010). To investigate this, we assayed the activity of PRC2 (comprising the core subunits Ezh2, Eed, Suz12, and RbAp48) in vitro using chromatin templates reconstituted on a 601 nucleosome positioning sequence with a CpG density of 8.7%, which is similar to that of a CGI (Figures 5A and S5A). To verify the assay conditions, we confirmed that recombinant PRC2 was inhibited using an unmethylated DNA template and nucleosomes reconstituted with H3Kc36me3, relative to unmodified H3, as previously shown (Schmitges et al., 2011; Figure 5B, left panels). We then determined activity using either unmethylated or methylated DNA templates. As shown in Figure 5B (right panels), there was no discernible inhibition of PRC2 using the methylated template. Similarly, H2A ubiquitylation activity in vitro, assayed using a recombinant PRC1 complex comprising Ring1B, Mel-18, and Rybp (Tavares et al., 2012), was unaffected by DNA methylation (Figure 5C). These experiments argue against a direct effect of DNA methylation on the activity of the tested PRC1 and PRC2 complexes.
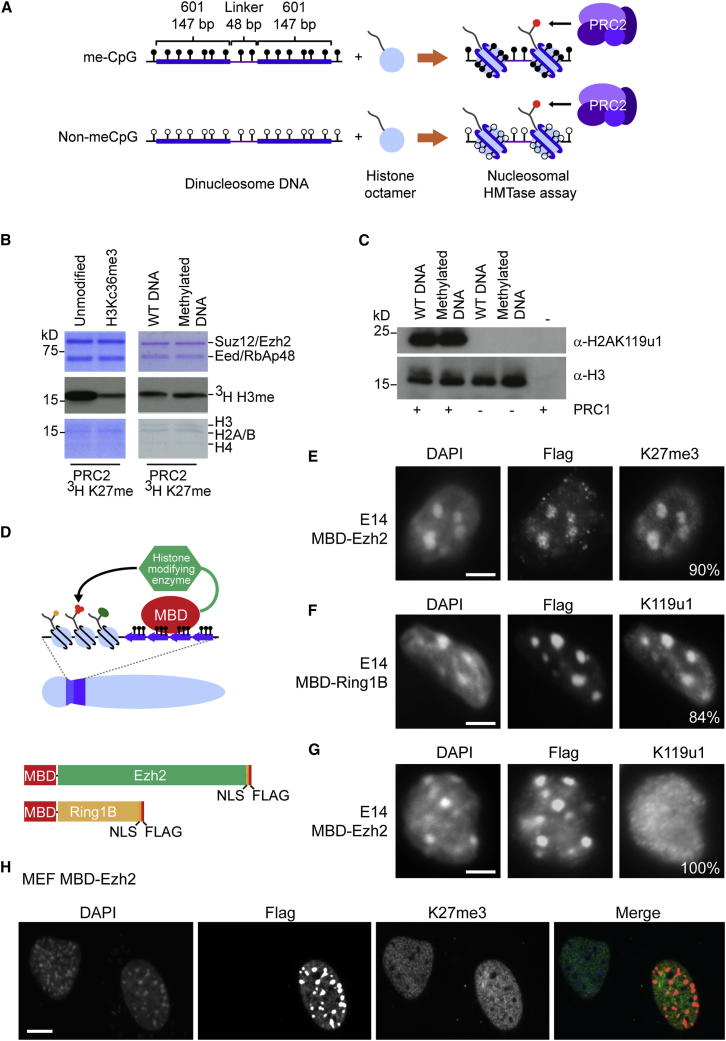
Figure 5.
DNA Methylation Does Not Inhibit PcG Activity Directly
(A) Schematic showing in vitro methylation of 601 positioning DNA and reconstitution of dinucleosomes.
(B) PRC2 histone methyltransferase assay using 3H SAM and either WT/H3Kc36me3 nucleosomes or WT/methylated DNA nucleosomes as substrate. Top and bottom panels: Coomassie-stained gel; middle panel: autoradiography of dried gel.
(C) PRC1 ubiquitylation assay using WT/methylated DNA nucleosomes as substrate, and analyzed by western blot (probed for H2AK119u1 or H3).
(D) Schematic of MBD tethering assay to target proteins of interest to regions of methylated DNA.
(E) MBD-Ezh2-Flag transfected into E14 ESCs stained for Flag and H3K27me3. The percentage of cells that showed the illustrated phenotype is indicated (n > 200).
(F) As in (E), but transfected with MBD-Ring1B-Flag and stained for Flag and H2AK119u1.
(G) As in (E), but transfected with MBD-Ezh2-Flag and stained for Flag and H2AK119u1.
(H) MBD-Ezh2-Flag transfected into MEFs and stained for Flag and H3K27me3.
Scale bars represent 5 μm. See also Figure S5.
We went on to determine whether DNA methylation directly inhibits PcG activity in vivo. To that end, we made use of the MBD1 methyl binding domain (MBD), which binds to densely methylated DNA, including at PCH domains (Hendrich and Bird, 1998, Ng et al., 2000), to tether PcG complexes in WT ESCs with normal levels of DNA methylation (Figure 5D). In initial experiments, we constructed MBD fusion proteins for Ezh2 and Ring1B, the catalytic subunits of PRC2 and PRC1, respectively (Figure 5A). Expression of full-length fusion protein was validated by western blot analysis (Figure S5B). IF with anti-FLAG demonstrated localization of fusion proteins to PCH domains following transient transfection (Figures 5E–5G). The MBD-Ezh2 fusion protein was active, as indicated by the establishment of H3K27me3 at PCH domains (Figure 5E). Similarly, recruitment of an MBD-Ring1B fusion protein to methylated PCH domains in WT ESCs led to H2AK119u1 deposition (Figure 5F). These results argue that CpG methylation does not directly inhibit the activity of either PRC2 or PRC1 complexes.
Establishment of H3K27me3 at PCH by tethering MBD-Ezh2 did not lead to recruitment of H2AK119u1 (Figure 5G). This was unexpected given the prevailing view that H3K27me3 recruits canonical PRC1 complexes via interaction with the chromodomain of the CBX subunit. To test whether the absence of H2AK119u1 is attributable to inhibition of PRC1 activity or recruitment, we transfected either MBD-Ring1B or MBD-Ezh2 into cells stably expressing the canonical PRC1 subunit Cbx7 fused to GFP (GFP-Cbx7). Cbx7-GFP was recruited to PCH when we tethered MBD-Ring1B (because both subunits are in the same complex), but not in response to tethering of MBD-Ezh2 (Figure S5C). We conclude that the chromatin features of PCH in ESCs block binding of Cbx PcG proteins and hence canonical PRC1 to H3K27me3. This contrasts with CGI PcG target sites where H3K27me3 does recruit Cbx-PRC1 (Tavares et al., 2012). Interestingly, MBD-Ezh2 tethering to PCH in MEF cells did not result in H3K27me3 deposition (Figure 5H). This is consistent with the absence of PRC2-mediated H3K27me3 at PCH domains in Dnmt1−/− MEFs (Figure 2D) and indicates that PCH in differentiated MEFs is entirely refractory to PRC2 activity.
KDM2B Recruitment Is Sufficient for H2AK119u1 and H3K27me3 Deposition at PCH
In light of our observations indicating that DNA methylation does not antagonize PcG activity directly, we went on to investigate possible indirect mechanisms. Recent studies have revealed that the zinc finger CXXC (zf-CXXC) domain recruits specific chromatin-modifying factors to CGIs via binding to unmethylated CpG (Long et al., 2013a). One of these factors, KDM2B, is an H3K36me1/2 demethylase that is also a subunit of a variant PRC1 complex, PCGF1-PRC1 (Farcas et al., 2012, He et al., 2013, Wu et al., 2013). We therefore exploited the MBD fusion tethering assay described above to test whether recruitment of KDM2B could account for H2AK119u1 deposition at PCH. We mutated a critical residue in the zf-CXXC domain of KDM2B (Figure 6A) such that the fusion protein could bind to methylated, but not unmethylated, CpG. We found that MBD-KDM2B recruitment was sufficient for H2AK119u1 deposition at PCH (Figures 6B, left, and 6E). Interestingly, we also observed H3K27me3 deposition (Figure 6B, right, and 6E), albeit in a smaller proportion of cells. This was unexpected given that PRC2 is not known to interact with KDM2B /PCGF1-PRC1 (Farcas et al., 2012, He et al., 2013, Wu et al., 2013).
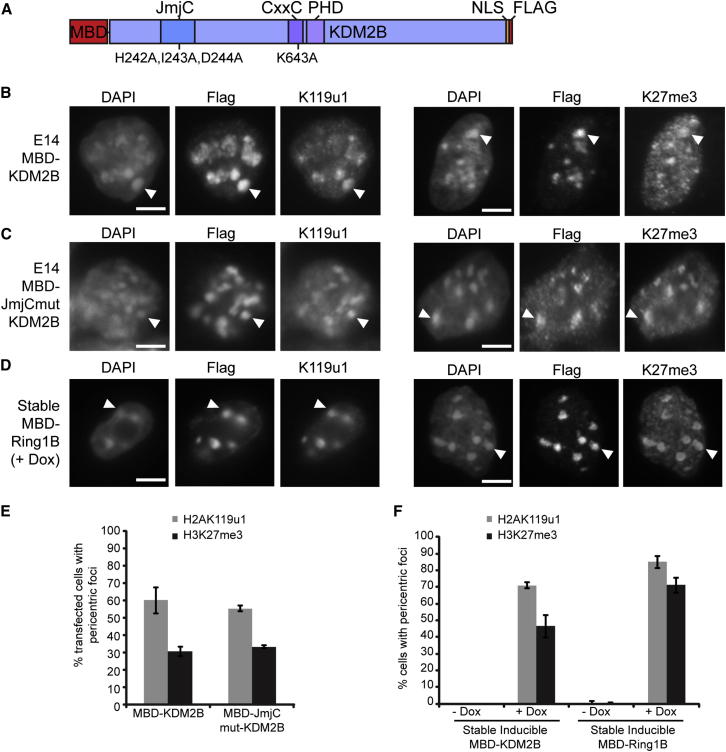
Figure 6.
KDM2B and Ring1B Recruit PRC2 Activity to PCH Domains
(A) Schematic of MBD-KDM2B-Flag indicating the positions of mutations in the JmjC domain (HID-AAA) and the CxxC domain (K-A).
(B) MBD-KDM2B transfected into E14 ESCs stained for Flag and H2AK119u1 (left) and Flag and H3K27me3 (right).
(C) MBD-KDM2B-Flag catalytic mutant (JmjC mutant HID-AAA) transfected into E14 ESCs stained for Flag and H2AK119u1 (left), and Flag and H3K27me3 (right).
(D) Inducible stable line expressing MBD-Ring1B-Flag, stained for Flag and H2AK119u1 3 days after induction with doxycycline (left) and Flag and H3K27me3 (right).
(E) Graph showing the percentage of transfected cells with H2AK119u1 (gray) or H3K27me3 (black) PCH foci in cells transfected with WT MBD-KDM2B and catalytically inactive JmjC domain mutant. Bars show average (n > 200 transfected cells) ± SD (n > 3).
(F) Graph showing the percentage of cells with H2AK119u1 (gray) or H3K27me3 (black) PCH foci in stable MBD-KDM2B and stable MBD-Ring1B either with or without 3 days induction with doxycycline (n > 100 cells) ± SD (n > 3). Arrowheads indicate an example of staining within Flag domains at PCH.
Scale bars represent 5 μm. See also Figures S5 and S6.
KDM2B is a histone demethylase with specificity for H3K36me1/2, a modification that, similarly to H3K4me3 and H3K36me3, inhibits PRC2 in cis on the same histone H3 tail in vitro (Schmitges et al., 2011). To test whether the H3K36me1/2 demethylase activity of KDM2B could account for H3K27me3 deposition, we tethered a catalytically inactive MBD-KDM2B fusion with a mutation in the JmjC domain (Figure 6A). As shown in Figures 6C and 6E, deposition of both H2AK119u1 and H3K27me3 was unaffected, indicating that the demethylase activity of KDM2B is not required for H3K27me3 deposition. Moreover, when we used the MBD domain to tether KDM2A, a closely related H3K36me1/2 demethylase that does not interact with PRC1 components (Blackledge et al., 2010, Farcas et al., 2012), we failed to observe either H2AK119u1 or H3K27me3 (Figures S6A–S6C).
To verify that tethering KDM2B is sufficient for deposition of both H2AK119u1 and H3K27me3, we established a stable ESC line with a doxycycline-inducible MBD-KDM2B transgene (Figure S6D). Consistent with results obtained using transient transfection, we detected deposition of both H2AK119u1 and H3K27me3 at PCH domains (Figures 6F and S6E). To test whether association of KDM2B with other PRC1 subunits is required for H3K27me3 deposition, we established a stable doxycycline-inducible ESC line expressing MBD-Ring1B (Figure S6F). As illustrated in Figures 6D and 6F, MBD-Ring1B localized to PCH domains following tamoxifen induction. This resulted in deposition of H2AK119u1, as expected, but also, importantly, to deposition of H3K27me3. Thus, recruitment of either KDM2B or Ring1B to PCH domains in WT ESCs is sufficient to establish both H2AK119u1 and H3K27me3 histone modifications.
H2AK119u1 Recruits PRC2
Recruitment of PRC2 activity by KDM2B/Ring1B could be due to a protein-protein interaction or, alternatively, to a PRC1-linked modification of underlying chromatin. We were particularly interested in determining whether H2AK119u1 might have a direct role in recruiting PRC2. To test this, we fused the RING finger domains of Ring1B and PCGF4 to generate a minimal catalytic complex that lacks the C-terminal domains required for interaction with other PRC1 subunits (Bezsonova et al., 2009). Previous studies have shown that in vitro, the two RING finger domains dimerize to form an E3 ligase with high specificity for H2AK118/9 on nucleosomal substrates (Bentley et al., 2011, Buchwald et al., 2006, Li et al., 2006). Based on the crystal structure of this complex, which shows that the C terminus of the Ring1B ring finger and the N terminus of PCGF4 ring finger are in close proximity to each other (Bentley et al., 2011), we inserted a short linker to allow expression of the E3 ligase as a single polypeptide (Figure 7A), thereby minimizing the possibility of interactions with endogenous Ring1(A/B) or Pcgf1-6 proteins in cells. Ring1B/Pcgf4 catalytic domain (RPCD) protein was tagged with glutathione S-transferase (GST), expressed, and purified from E. coli. We then verified RPCD activity and specificity by assaying H2A ubiquitylation on reconstituted nucleosomal templates in vitro (Figure S7A). We also designed a catalytically inactive fusion protein by mutating a critical residue in the E2 binding surface of Ring1B (I53A) and in the zinc finger of Pcgf4 (C51G) (Figures 7A and S7A).
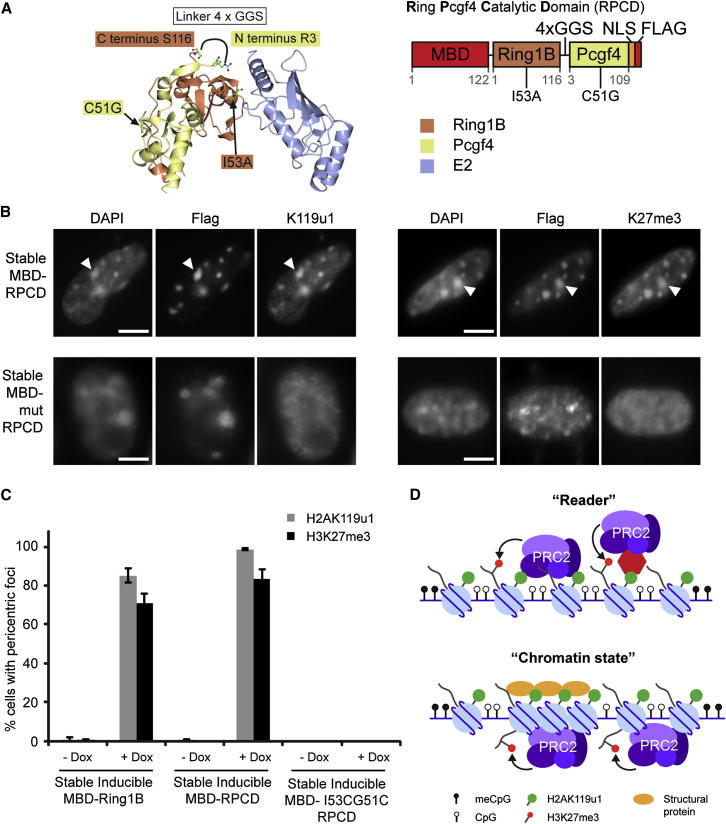
Figure 7.
Ubiquitylated H2A Is Sufficient to Recruit PRC2 to PCH Domains
(A) Structure of the minimal E3 and E2 complex (Bentley et al., 2011), showing the catalytic domain of Pcgf4 (yellow) and Ring1B (orange) interacting with E2 enzyme (blue). RPCD was made by fusing the C terminus (S116) of Ring1B to the N terminus (R3) of Pcgf4 using a 4x GGS flexible linker. Catalytic mutant (mut) RPCD comprised C51G (in Pcgf4) and I53A (in Ring1B).
(B) Inducible stable line expressing MBD-RPCD-Flag or MBD-mut-RPCD-Flag, stained for Flag and H2AK119u1 3 days after induction with doxycycline (left) and Flag and H3K27me3 (right). Arrowheads indicate an example of staining within Flag domains at PCH.
(C) Graph showing the percentage of cells with H2AK119u1 (gray) or H3K27me3 (black) PCH foci in stable MBD-Ring1B-Flag, MBD-RPCD-Flag, and MBD-mut-RPCD-Flag cell lines, either with or without 3 days induction with doxycycline (n > 100 cells) ± SD (n > 3).
(D) Schematic to illustrate potential models of for PRC2 recruitment by H2AK119u1, either by direct/indirect protein interactions “reader” (top) or by “chromatin state” (bottom).
Scale bars represent 5 μm. See also Figure S7.
To test RPCD in vivo, we expressed an MBD-RPCD fusion protein with a FLAG C-terminal tag in ESCs. We verified that the fusion protein did not interact with endogenous PRC1 or PRC2 proteins by conducting a coimmunoprecipitation (coIP) analysis of Rybp, which is associated in complexes with all of Pcgf1-6 (Gao et al., 2012, Tavares et al., 2012), endogenous Ring1B, and the core PRC2 subunit Suz12. As shown in Figure S7B, Rybp coimmunoprecipitated with endogenous and MBD-tagged Ring1B, but not with RPCD. Similarly, neither endogenous Ring1B nor Suz12 coimmunoprecipitated with RPCD.
We went on to establish stable doxycycline-inducible ESCs expressing either active or mutant MBD-RPCD. The expression levels following addition of doxycycline were estimated to be similar to those in the stable MBD-Ring1B cell line described above, as determined by anti-FLAG western blot analysis (Figure S7C). When MBD-RPCD was recruited to PCH domains, we observed efficient deposition of H2AK119u1 and, importantly, H3K27me3 (Figures 7B and 7C). Neither modification could be detected following recruitment of mutant RPCD (Figures 7B and 7C). Recruitment of MBD-RPCD had no discernible effect on the levels of either H3K9me3 or DNA methylation at PCH (Figures S7D and S7E). These results thus demonstrate that H2AK119u1 deposition is sufficient to recruit PRC2. We note that this mechanism could potentially explain the localization of both PRC1 and PRC2 activities at PCH and other CpG-dense sites in DNA methylation-deficient ESCs.
Discussion
A Role for DNA Hypomethylation in Atypical Localization of PcG Proteins to PCH Domains
Localization of PcG complexes to paternal PCH in early mouse embryos has been attributed to the absence of H3K9me3 (Puschendorf et al., 2008). We hypothesized that DNA hypomethylation may also play a role, and consistent with this, we observed a rapid recruitment of PRC1 and PRC2 activities to PCH in ESCs following depletion of DNA methylation, despite the continued presence of H3K9me3. Our results do, however, support a role for H3K9me3 in antagonizing PRC2 activity at PCH. Specifically, we observed mutually exclusive staining for H3K9me3 and H3K27me3, contrasting with the broader staining pattern for H2AK119u1. Moreover, depletion of H3K9me3 by knockdown of Suv39h1/h2 resulted in increased H3K27me3 at PCH. H3K9me3 does not inhibit PRC2 activity in vitro (Schmitges et al., 2011), arguing against direct inhibition of PRC2 activity by H3K9me3 in cis on the same histone tail. With this in mind, we favor the idea that antagonism of PRC2 by H3K9me3 at PCH is an indirect effect, mediated by H3K9me3-binding proteins such as HP1.
Unlike the case with H3K27me3, H2AK119u1 deposition at PCH in Dnmt TKO ESCs is not antagonized by H3K9me3. This raises the question of how H2AK119u1 deposition at PCH occurs in Suv39H1/2 DKO ESCs. Our results suggest that H2AK119u1 deposition is unlikely to be attributable to H3K27me3-mediated recruitment of canonical PRC1 complexes, given that this does not occur when H3K27me3 is directed to PCH in WT ESCs (Figure 5G). A possible explanation comes from the observation that H3K9me3 is required for efficient DNA methylation at PCH (Lehnertz et al., 2003). Thus, DNA hypomethylation in Suv39H1/2 DKO ESCs could recruit PRC1 activity, similar to what is observed in Dnmt TKO ESCs (see below). It should be noted that the absence of canonical PRC1 recruitment at H3K27-methylated PCH is unexpected. We interpret this to indicate that Polycomb Cbx proteins are unable to bind to H3K27me3 due to a specific characteristic or modification at PCH.
We also show that PcG activity/recruitment at PCH is dependent on developmental context. Specifically, in Dnmt1−/− MEF cells, we observed accumulation of H2AK119u1, but never H3K27me3. This was the case even following depletion of H3K9me3 (data not shown). Related to this, in contrast to ESCs, tethering Ezh2 to PCH in WT MEFs did not lead to H3K27me3 deposition. These observations demonstrate that PCH in somatic cells is highly refractory to PRC2 activity. Inhibition of PRC2 may be linked to the fact that PCH chromatin is considerably less dynamic in somatic cells relative to ESCs (Meshorer et al., 2006). Thus, we propose interplay of mutually exclusive chromatin repression machineries, with the equilibrium state being defined in part by chromatin dynamics.
Unmethylated CpG Dictates PcG Occupancy
Our study extends recent reports of a changed distribution of H3K27me3 in DNA methylation-deficient ESCs (Brinkman et al., 2012) and somatic cell lines (Reddington et al., 2013). Specifically, we mapped H3K27me3, Suz12, and Ring1B redistribution in Dnmt TKO ESCs, and also performed an extended analysis to include common repeat elements. These analyses demonstrate that both PRC1 and PRC2 are redistributed away from canonical PcG targets. Moreover, by subtracting ESC PcG target sequences from our analysis, we were able to show that there is a linear relationship between the gain of PcG complexes and the density of unmethylated CpG, notably at exon sequences that are known to have a relatively high CpG density. By taking into consideration the levels of H3K36me3, which was previously shown to inhibit PRC2 activity, we further refined the principles for PcG redistribution following depletion of CpG methylation. Together, our observations indicate that PcG redistribution at unique sequences in Dnmt TKO ESCs is a combinatorial readout of the density of unmethylated CpG and the presence/absence of H3K36me3. We note that modified patterns of DNA methylation could also explain the observed changes in patterns of PcG occupancy in ESCs grown in 2i conditions (Ficz et al., 2013, Habibi et al., 2013, Marks et al., 2012), in Dnmt3a-deficient neural stem cells (Wu et al., 2010), and in cancer cell lines (Varley et al., 2013). In the context of the default model for PcG recruitment (Klose et al., 2013), we suggest that other histone tail modifications, notably H3K4me3, which antagonizes PRC2 activity, must also be taken into account in order to fully explain PcG occupancy patterns in different cell types.
Although we detected gain of both H3K27me3 and H2AK119u1 at new sites in Dnmt TKO ESCs, this was less obvious when we analyzed Suz12 and Ring1B. A possible explanation for this is that at new sites, PcG complexes interact with chromatin with relatively fast dynamics and as such evade capture by formaldehyde crosslinking (Schmiedeberg et al., 2009). Fast dynamics at the relatively short CpG-rich exonic sites could be linked to an absence of positive-feedback mechanisms that stabilize PcG complexes at CGI targets. The enhancement of PcG silencing by polymerization of the SAM domain of the Polyhomeotic subunit of canonical PRC1 complexes (Isono et al., 2013) may represent one such mechanism.
Our results support a view that PcG localization in vertebrate cells is dictated in large part by the distribution of unmethylated CpG sites. This mechanism may also operate in other organisms, such as in A. thaliana, where hypomethylation of DNA at transposons in a met1 mutant or endogenously in the endosperm results in PRC2 recruitment (Deleris et al., 2012, Weinhofer et al., 2010). However, this cannot account for PcG distribution patterns in Drosophila and C. elegans, where there is no DNA methylation. Nevertheless, there is an interesting parallel in C. elegans, where mutation of the H3K36me2/3 methyltransferase MES4 results in PRC2 relocalization from the X chromosome to autosomes in germ cells (Gaydos et al., 2012). Thus, inhibition of PRC2 by specific histone modifications could be used as an alternative mechanism to limit sites of activity and hence define patterns of occupancy of PcG complexes.
Monoubiquitylation of H2A Recruits PRC2
Our findings illustrate that unmethylated CpG is an important determinant of PcG occupancy in mammalian cells. However, although we cannot entirely rule out some contribution, we do not find evidence that this is due to direct inhibition of PRC1/2 enzymatic activity by methylated CpG. Similarly, inhibition of PRC2 by H3K36me2, as predicted on the basis of in vitro studies (Schmitges et al., 2011), does not appear to be a primary determinant of PcG localization in mammalian cells in vivo. Again, we cannot entirely rule out the possibility that H3K36me2 may have some role in limiting PRC2 activity at non-PcG target domains. Our findings do, however, support an alternative model: the sequential recruitment of variant PRC1 to unmethylated CpG, notably via binding of the CXXC domain of the KDM2B protein, and indirect recruitment of PRC2 in response to PRC1-mediated H2AK119u1. Recruitment of PRC2 in response to H2AK119u1 deposition is substantiated by the observation that acquisition of H3K27me3 at PCH shows a linear relationship with H2AK119u1, as seen in a comparison of KDM2B, Ring1B, and RPCD tethering (Figures 6 and 7). Furthermore, only catalytically active RPCD brings in H3K27me3, and in coIP experiments we did not find a direct interaction between RPCD and either PRC1 or, importantly, PRC2 subunits. This is also supported by a mass spectrometry analysis of TetR-RPCD (Blackledge et al., 2014). We found no evidence that RPCD-mediated H2AK119u1 results in DNA hypomethylation, which could potentially account for recruitment of endogenous PRC1/2 to PCH. Consistent with this, recruitment of RPCD to a TetR array that does not have any CpG sites also leads to establishment of H3K27me3 (N.P.B., A.M. Farcas, T. Kondo, H.W. King, J.F. McGouran, L.L.P. Hanssen, S. Ito, S.C., K. Kondo, Y. Koseki, T. Ishikura, H.K. Long, T.W. Sheahan, N.B., B.M. Kessler, H.K., and R.K., unpublished data).
The finding that H2AK119 ubiquitylation is sufficient to recruit PRC2 to chromatin was largely unanticipated, although, interestingly, a prior study noted a loss of H3K27me3 in Ring1A/B DKO ESCs (Endoh et al., 2008). This has been substantiated in a recent study (N.P.B., A.M. Farcas, T. Kondo, H.W. King, J.F. McGouran, L.L.P. Hanssen, S. Ito, S.C., K. Kondo, Y. Koseki, T. Ishikura, H.K. Long, T.W. Sheahan, N.B., B.M. Kessler, H.K., and R.K., unpublished data). Taken together, these observations demonstrate hierarchical recruitment of PRC2 by PRC1 activity. The data are consistent with a primary role for variant PRC1 and H2AK119u1 in determining PcG target sites, although we do not rule out the possibility that PRC2 may also be recruited independently of H2AK119u1.
A key question for future studies is: what is the mechanistic basis for H2AK119u1-mediated recruitment of PRC2? One possibility is that PRC2 acts as a reader of H2AK119u1, either directly or via a cofactor that can bind the modified chromatin (see Figure 7D, top panel). Consistent with this idea, a previous study identified the ZRF1 protein as an H2AK119u1-binding protein (Richly et al., 2010). Alternatively, H2AK119 ubiquitylation may affect chromatin structure in such a way as to facilitate the binding and/or activity of PRC2 complexes (Figure 7D, bottom panel). In this context, it should be noted that H2AK119u1 does not stimulate PRC2 activity on mononucleosome substrates in vitro (Whitcomb et al., 2012). Conversely, a recent study revealed that chromatin compaction stimulates PRC2 (Yuan et al., 2012), consistent with the idea that local chromatin configuration is important.
In summary, our findings provide insights into the link between DNA methylation and PcG recruitment, and are consistent with the recently suggested model in which Polycomb recruitment in vertebrates is a default state at licensed chromatin sites, defined directly or indirectly by CpG methylation status (Klose et al., 2013).
Experimental Procedures
ESC Culture
E14TG2A, Dnmt TKO (Dnmt1−/−, Dnmt3a−/−, Dnmt3b−/−) and matched WT (J1) (Tsumura et al., 2006), conditional Uhrf1 and Dnmt1 knockout cells (Sharif et al., 2007), constitutive Suv3-9h1/h2 DKO and matched WT ESCs (Peters et al., 2003), and Dnmt1−/− MEFs and matched control MEFs (Lande-Diner et al., 2007) were cultivated using established methods. See also Supplemental Experimental Procedures.
Constructs
Amino acids 1–112 of human MBD1, which include the MBD domain and endogenous nuclear localization sequence (NLS) signal, followed by a glycine- and serine-rich flexible linker, were cloned to the N terminus of the protein of interest in pBluescript. An SV40 NLS, followed by a FLAG-tag, was cloned on the C terminus of the protein of interest. These MBD-fusion proteins were then cloned into the mammalian expression plasmid pCAG, in which the MBD-fusion protein was under the control of the constitutive β-actin promoter. Full-length mouse Ezh2, mouse Ring1B, and human KDM2A (K601A) and KDM2B (K643A) were targeted to methylated DNA using the MBD domain. For KDM2A/B, the CxxC DNA binding domains were mutated (K601A and K643A, respectively). The MBD-KDM2B, MBD-Ring1B, MBD-RPCD, and MBD-mutRPCD constructs were also cloned into the pTight vector to allow doxycyline-inducible expression when stably integrated into rtTA2A10 ESCs.
Amino acids 1–116 of Ring1B and 3–109 of Pcgf4 (termed Ring1b Pcgf4 catalytic domain RPCD fusion) were joined using a 4x GGS flexible linker and cloned with an N-terminal GST tag in pGex-6p2 for bacterial protein expression and purification, or with an N-terminal MBD domain in the pCAG vector, for expression in ESCs. Catalytic mutants were made in the JmjC domain of KDM2B (H242A, I243A, D244A) and in RPCD at Ring1B I53A to mutate the E2 interaction domain, and at Pcgf4 C51G to mutate the zinc finger domain, using a site-directed mutagenesis kit (Stratagene). Synthesized core PRC2 subunits (full-length EZH2, EED, SUZ12, and RbAp48) were codon optimized for expression in insect cells (GeneArt) and cloned into pBAC4x-1 (Novagen) using the In-Fusion cloning system (Clontech).
Immunofluorescence
ESCs or MEFs were split onto slides 16 hr before staining at low density (without feeders). Slides were then washed in PBS, fixed with 2% formaldehyde in PBS for 15 min, and permeabilized with 0.4% Triton X-100 in PBS for 5 min. After washing with PBS, the slides were blocked for 30 min in 0.2% fish gelatin (Sigma) in PBS and incubated for 2 hr with primary antibody (diluted in 0.2% fish gelatin and 5% normal goat serum). Slides were washed three times in 0.2% fish gelatin and incubated for 2 hr with Alexa Fluor conjugated secondary antibody (Life Technologies). After two washes in fish gelatin and two washes in PBS, the slides were stained with DAPI (1 μg/ml) and mounted using mounting media (Dako).
The following primary antibodies were used for IF: H2AK119u1 (1:500, rabbit monoclonal, 8240; Cell Signaling Technology), H3K27me3 (1:500, rabbit polyclonal, pAB-069-050; Diagenode), H3K27me3 (1:1,000, mouse monoclonal, 61017; Active Motif), Flag (1:500, mouse M2 monoclonal; Sigma), H3K9me3 (1:500, rabbit polyclonal 39161; Active Motif), H3K4me3 (1:500, rabbit polyclonal, ab8580; Abcam), HP1α (1:500, mouse monoclonal MAB3584; Millipore), and GFP (1:100, mouse monoclonal, sc-9996; Santa Cruz).
Chromatin Immunoprecipitation
ChIP/ChIP-seq was carried out as described previously (Blackledge et al., 2010). Full details are provided in the Supplemental Experimental Procedures. The antibodies for ChIP-seq were H3K27me3 (pAB-069-100; Diagenode), H3K36me3 (AB9050; Abcam), anti-Suz12 (3737S; Cell Signaling Technology), and anti-Ring1B (Atsuta et al., 2001).
Acknowledgments
We thank Andrew Bassett and members of the N.B. and R.K. labs for critical readings of the manuscript. We thank Masaki Okano for Dnmt TKO ESCs, Thomas Jenuwein for Suv3-9 DKO ESCs, and Howard Cedar for p-/p-m- MEFS. This work was funded by grants from the Wellcome Trust (WT081385, WT091911, and WT098024) and the Medical Research Council, UK (G1000902).
Published: May 22, 2014
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.04.012.
Accession Numbers
The European Nucleotide Archive accession number for the ChIP-seq data reported in this paper is ERP005575.
Supplemental Information
References
- Atsuta T., Fujimura S., Moriya H., Vidal M., Akasaka T., Koseki H. Production of monoclonal antibodies against mammalian Ring1B proteins. Hybridoma. 2001;20:43–46. doi: 10.1089/027245701300060427. [DOI] [PubMed] [Google Scholar]
- Bartke T., Vermeulen M., Xhemalce B., Robson S.C., Mann M., Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley M.L., Corn J.E., Dong K.C., Phung Q., Cheung T.K., Cochran A.G. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. EMBO J. 2011;30:3285–3297. doi: 10.1038/emboj.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T.H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- Bezsonova I., Walker J.R., Bacik J.P., Duan S., Dhe-Paganon S., Arrowsmith C.H. Ring1B contains a ubiquitin-like docking module for interaction with Cbx proteins. Biochemistry. 2009;48:10542–10548. doi: 10.1021/bi901131u. [DOI] [PubMed] [Google Scholar]
- Blackledge N.P., Zhou J.C., Tolstorukov M.Y., Farcas A.M., Park P.J., Klose R.J. CpG islands recruit a histone H3 lysine 36 demethylase. Mol. Cell. 2010;38:179–190. doi: 10.1016/j.molcel.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge N.P., Farcas A.M., Kondo T., King H.W., McGouran J.F., Hanssen L.L.P., Ito S., Cooper S., Kondo K., Koseki Y. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman A.B., Simmer F., Ma K., Kaan A., Zhu J., Stunnenberg H.G. Whole-genome DNA methylation profiling using MethylCap-seq. Methods. 2010;52:232–236. doi: 10.1016/j.ymeth.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Brinkman A.B., Gu H., Bartels S.J., Zhang Y., Matarese F., Simmer F., Marks H., Bock C., Gnirke A., Meissner A., Stunnenberg H.G. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res. 2012;22:1128–1138. doi: 10.1101/gr.133728.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N. Noncoding RNA and Polycomb recruitment. RNA. 2013;19:429–442. doi: 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwald G., van der Stoop P., Weichenrieder O., Perrakis A., van Lohuizen M., Sixma T.K. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- de Napoles M., Mermoud J.E., Wakao R., Tang Y.A., Endoh M., Appanah R., Nesterova T.B., Silva J., Otte A.P., Vidal M. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Deleris A., Stroud H., Bernatavichute Y., Johnson E., Klein G., Schubert D., Jacobsen S.E. Loss of the DNA methyltransferase MET1 Induces H3K9 hypermethylation at PcG target genes and redistribution of H3K27 trimethylation to transposons in Arabidopsis thaliana. PLoS Genet. 2012;8:e1003062. doi: 10.1371/journal.pgen.1003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh M., Endo T.A., Endoh T., Fujimura Y., Ohara O., Toyoda T., Otte A.P., Okano M., Brockdorff N., Vidal M., Koseki H. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development. 2008;135:1513–1524. doi: 10.1242/dev.014340. [DOI] [PubMed] [Google Scholar]
- Endoh M., Endo T.A., Endoh T., Isono K., Sharif J., Ohara O., Toyoda T., Ito T., Eskeland R., Bickmore W.A. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet. 2012;8:e1002774. doi: 10.1371/journal.pgen.1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskeland R., Leeb M., Grimes G.R., Kress C., Boyle S., Sproul D., Gilbert N., Fan Y., Skoultchi A.I., Wutz A., Bickmore W.A. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol. Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farcas A.M., Blackledge N.P., Sudbery I., Long H.K., McGouran J.F., Rose N.R., Lee S., Sims D., Cerase A., Sheahan T.W. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife. 2012;1:e00205. doi: 10.7554/eLife.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G., Hore T.A., Santos F., Lee H.J., Dean W., Arand J., Krueger F., Oxley D., Paul Y.L., Walter J. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell. 2013;13:351–359. doi: 10.1016/j.stem.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis N.J., Kingston R.E., Woodcock C.L. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- Gao Z., Zhang J., Bonasio R., Strino F., Sawai A., Parisi F., Kluger Y., Reinberg D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaydos L.J., Rechtsteiner A., Egelhofer T.A., Carroll C.R., Strome S. Antagonism between MES-4 and Polycomb repressive complex 2 promotes appropriate gene expression in C. elegans germ cells. Cell Rep. 2012;2:1169–1177. doi: 10.1016/j.celrep.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi E., Brinkman A.B., Arand J., Kroeze L.I., Kerstens H.H., Matarese F., Lepikhov K., Gut M., Brun-Heath I., Hubner N.C. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell. 2013;13:360–369. doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Hagarman J.A., Motley M.P., Kristjansdottir K., Soloway P.D. Coordinate regulation of DNA methylation and H3K27me3 in mouse embryonic stem cells. PLoS ONE. 2013;8:e53880. doi: 10.1371/journal.pone.0053880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Shen L., Wan M., Taranova O., Wu H., Zhang Y. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat. Cell Biol. 2013;15:373–384. doi: 10.1038/ncb2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B., Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth R.S., Bird A.P. CpG islands—‘a rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Isono K., Endo T.A., Ku M., Yamada D., Suzuki R., Sharif J., Ishikura T., Toyoda T., Bernstein B.E., Koseki H. SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev. Cell. 2013;26:565–577. doi: 10.1016/j.devcel.2013.08.016. [DOI] [PubMed] [Google Scholar]
- Kassis J.A., Brown J.L. Polycomb group response elements in Drosophila and vertebrates. Adv. Genet. 2013;81:83–118. doi: 10.1016/B978-0-12-407677-8.00003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R.J., Cooper S., Farcas A.M., Blackledge N.P., Brockdorff N. Chromatin sampling—an emerging perspective on targeting polycomb repressor proteins. PLoS Genet. 2013;9:e1003717. doi: 10.1371/journal.pgen.1003717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande-Diner L., Zhang J., Ben-Porath I., Amariglio N., Keshet I., Hecht M., Azuara V., Fisher A.G., Rechavi G., Cedar H. Role of DNA methylation in stable gene repression. J. Biol. Chem. 2007;282:12194–12200. doi: 10.1074/jbc.M607838200. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lehnertz B., Ueda Y., Derijck A.A., Braunschweig U., Perez-Burgos L., Kubicek S., Chen T., Li E., Jenuwein T., Peters A.H. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- Li Z., Cao R., Wang M., Myers M.P., Zhang Y., Xu R.M. Structure of a Bmi-1-Ring1B polycomb group ubiquitin ligase complex. J. Biol. Chem. 2006;281:20643–20649. doi: 10.1074/jbc.M602461200. [DOI] [PubMed] [Google Scholar]
- Long H.K., Blackledge N.P., Klose R.J. ZF-CxxC domain-containing proteins, CpG islands and the chromatin connection. Biochem. Soc. Trans. 2013;41:727–740. doi: 10.1042/BST20130028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H.K., Sims D., Heger A., Blackledge N.P., Kutter C., Wright M.L., Grützner F., Odom D.T., Patient R., Ponting C.P., Klose R.J. Epigenetic conservation at gene regulatory elements revealed by non-methylated DNA profiling in seven vertebrates. Elife. 2013;2:e00348. doi: 10.7554/eLife.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M.D., Smith A.J., De Gobbi M., Flenley M., Hughes J.R., Vernimmen D., Ayyub H., Sharpe J.A., Sloane-Stanley J.A., Sutherland L. An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J. 2012;31:317–329. doi: 10.1038/emboj.2011.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A.F., Smith A., Stunnenberg H.G. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W., Niveleau A., Walter J., Fundele R., Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- Mendenhall E.M., Koche R.P., Truong T., Zhou V.W., Issac B., Chi A.S., Ku M., Bernstein B.E. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010;6:e1001244. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E., Yellajoshula D., George E., Scambler P.J., Brown D.T., Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A., Cléard F., Mishra R.K., Karch F., Verrijzer C.P. Synergistic recognition of an epigenetic DNA element by Pleiohomeotic and a Polycomb core complex. Genes Dev. 2005;19:1755–1760. doi: 10.1101/gad.347005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H.H., Jeppesen P., Bird A. Active repression of methylated genes by the chromosomal protein MBD1. Mol. Cell. Biol. 2000;20:1394–1406. doi: 10.1128/mcb.20.4.1394-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald J., Engemann S., Lane N., Mayer W., Olek A., Fundele R., Dean W., Reik W., Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- Pengelly A.R., Copur O., Jäckle H., Herzig A., Müller J. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science. 2013;339:698–699. doi: 10.1126/science.1231382. [DOI] [PubMed] [Google Scholar]
- Peters A.H., O’Carroll D., Scherthan H., Mechtler K., Sauer S., Schöfer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Peters A.H., Kubicek S., Mechtler K., O’Sullivan R.J., Derijck A.A., Perez-Burgos L., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- Plath K., Fang J., Mlynarczyk-Evans S.K., Cao R., Worringer K.A., Wang H., de la Cruz C.C., Otte A.P., Panning B., Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Probst A.V., Almouzni G. Heterochromatin establishment in the context of genome-wide epigenetic reprogramming. Trends Genet. 2011;27:177–185. doi: 10.1016/j.tig.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Puschendorf M., Terranova R., Boutsma E., Mao X., Isono K., Brykczynska U., Kolb C., Otte A.P., Koseki H., Orkin S.H. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat. Genet. 2008;40:411–420. doi: 10.1038/ng.99. [DOI] [PubMed] [Google Scholar]
- Reddington J.P., Perricone S.M., Nestor C.E., Reichmann J., Youngson N.A., Suzuki M., Reinhardt D., Dunican D.S., Prendergast J.G., Mjoseng H. Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes. Genome Biol. 2013;14:R25. doi: 10.1186/gb-2013-14-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W., Dean W., Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Richly H., Rocha-Viegas L., Ribeiro J.D., Demajo S., Gundem G., Lopez-Bigas N., Nakagawa T., Rospert S., Ito T., Di Croce L. Transcriptional activation of polycomb-repressed genes by ZRF1. Nature. 2010;468:1124–1128. doi: 10.1038/nature09574. [DOI] [PubMed] [Google Scholar]
- Ringrose L., Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Santos F., Peters A.H., Otte A.P., Reik W., Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev. Biol. 2005;280:225–236. doi: 10.1016/j.ydbio.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Schermelleh L., Heintzmann R., Leonhardt H. A guide to super-resolution fluorescence microscopy. J. Cell Biol. 2010;190:165–175. doi: 10.1083/jcb.201002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedeberg L., Skene P., Deaton A., Bird A. A temporal threshold for formaldehyde crosslinking and fixation. PLoS ONE. 2009;4:e4636. doi: 10.1371/journal.pone.0004636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitges F.W., Prusty A.B., Faty M., Stützer A., Lingaraju G.M., Aiwazian J., Sack R., Hess D., Li L., Zhou S. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- Schoeftner S., Sengupta A.K., Kubicek S., Mechtler K., Spahn L., Koseki H., Jenuwein T., Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T.A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- Silva J., Mak W., Zvetkova I., Appanah R., Nesterova T.B., Webster Z., Peters A.H., Jenuwein T., Otte A.P., Brockdorff N. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev. Cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Simon J.A., Kingston R.E. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares L., Dimitrova E., Oxley D., Webster J., Poot R., Demmers J., Bezstarosti K., Taylor S., Ura H., Koide H. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148:664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumura A., Hayakawa T., Kumaki Y., Takebayashi S., Sakaue M., Matsuoka C., Shimotohno K., Ishikawa F., Li E., Ueda H.R. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- Varley K.E., Gertz J., Bowling K.M., Parker S.L., Reddy T.E., Pauli-Behn F., Cross M.K., Williams B.A., Stamatoyannopoulos J.A., Crawford G.E. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Brown J.L., Cao R., Zhang Y., Kassis J.A., Jones R.S. Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Weinhofer I., Hehenberger E., Roszak P., Hennig L., Köhler C. H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet. 2010;6:6. doi: 10.1371/journal.pgen.1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcomb S.J., Fierz B., McGinty R.K., Holt M., Ito T., Muir T.W., Allis C.D. Histone monoubiquitylation position determines specificity and direction of enzymatic cross-talk with histone methyltransferases Dot1L and PRC2. J. Biol. Chem. 2012;287:23718–23725. doi: 10.1074/jbc.M112.361824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo C.J., Kharchenko P.V., Daheron L., Park P.J., Kingston R.E. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo C.J., Kharchenko P.V., Daheron L., Park P.J., Kingston R.E. Variable requirements for DNA-binding proteins at polycomb-dependent repressive regions in human HOX clusters. Mol. Cell. Biol. 2013;33:3274–3285. doi: 10.1128/MCB.00275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Coskun V., Tao J., Xie W., Ge W., Yoshikawa K., Li E., Zhang Y., Sun Y.E. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Johansen J.V., Helin K. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol. Cell. 2013;49:1134–1146. doi: 10.1016/j.molcel.2013.01.016. [DOI] [PubMed] [Google Scholar]
- Yuan W., Xu M., Huang C., Liu N., Chen S., Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W., Wu T., Fu H., Dai C., Wu H., Liu N., Li X., Xu M., Zhang Z., Niu T. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science. 2012;337:971–975. doi: 10.1126/science.1225237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.