Abstract
Previously, we identified multiple in vivo mRNA targets of the maxi-KH/STAR domain protein GLD-1 by their ability to interact with GLD-1 in cytoplasmic extracts and, for all targets tested thus far, GLD-1 functions as a translational repressor. However, here we show that GLD-1 stabilizes the mRNAs of two targets, gna-2 (T23G11.2) and Y75B12B.1. gna-2 mRNA has two upstream open reading frames (uORF), resulting in two premature stop codons. We found that gna-2 mRNA is a naturally occurring mRNA target of nonsense-mediated mRNA decay (NMD) and that the binding of GLD-1 protects gna-2 mRNA from NMD, likely by repressing translation of the uORFs. Therefore, gna-2 mRNA comes under two posttranscriptional controls: (1) translation regulation by a specific translational repressor, GLD-1; and (2) uORF elicited regulation, mainly through NMD. As a result, these two posttranscriptional controls together provide precise temporal and spatial control of gene expression. Consistent with this novel mode of regulation, when GLD-1 mRNA targets acquire premature stop codon mutations, GLD-1 protects them from NMD. Analysis of several mRNA targets containing premature stop codons suggests that in translation repression, GLD-1 either represses ribosome assembly on the target mRNA, or subsequent ribosome elongation to the premature stop codon.
Keywords: Germ line, translation repression, upstream open reading frame, nonsense-mediated mRNA decay, gld-1, gna-2
Regulation of gene expression is central to most cellular processes and occurs at multiple steps, including transcription, splicing, nuclear export, localization, stability, and translation of mature mRNAs. Germ-line and early embryonic development are particularly dependent on posttranscriptional control of maternal mRNAs for temporal/spatial regulation of gene expression because the genome is transcriptionally silent from late meiotic prophase (diakinesis), through the meiotic divisions/fertilization and into early embryogenesis (Wickens et al. 2000; De Moor and Richter 2001). Expression of several maternal mRNAs is regulated by more than one posttranscriptional mechanism. For example, during Drosophila oogenesis and early embryogenesis, the combination of two posttranscriptional controls, localization and translational control, precisely regulates the expression of oscar, bicoid, and nanos mRNAs (Johnstone and Lasko 2001). In most cases, cis-acting elements in the 5′-or 3′-untranslated regions (UTRs) of mRNAs mediate their posttranscriptional regulation. When a specific mRNA harbors more than one cis-acting element, and therefore is under more than one posttranscriptional control, its expression is determined combinatorially; this results in precise temporal and spatial control of gene expression.
One important cis-acting element in the 5′-UTR of several eukaryotic mRNAs is an upstream open reading frame (uORF). uORFs often control the efficiency of translation of the downstream main ORF, primarily by altering the activity of the ribosome (Geballe and Sachs 2000; Morris and Geballe 2000). After translation initiation factors bind at the 5′-cap, recruit the small 40S ribosome subunit, and scan to the AUG of the uORF (uAUG), the scanning initiation complex sometimes fails to recognize the uAUG, and proceeds to the downstream AUG (leaky scanning). However, more often the complex recognizes the uAUG and initiates translation, resulting in the production of nascent peptides that can sometimes interfere with translation termination, as a result, the ribosome stalls at the stop codon (ribosome stalling). After the ribosome completes translation of the uORF, it usually dissociates from the mRNA, resulting in the failure to translate the downstream ORF. Alternatively, the 40S subunit can remain associated with the mRNA and resume scanning (reinitiation). In general, reinitiation is inefficient and only occurs after translation of very short uORFs. In addition, several studies have shown that uORFs also affect mRNA stability through nonsense-mediated mRNA decay (NMD), because the stop codon of the uORF can be regarded as a premature stop codon (Ruiz-Echevarria et al. 1996; Linz et al. 1997). Several examples of mRNAs with one or more uORFs indicate that uORFs can elicit multiple modes of regulation, such as leaky scanning and termination-dependent decay for YAP2 mRNA, which together exert precise control over the amount of protein synthesized (Vilela et al. 1998; Meijer and Thomas 2002; Vilela and McCarthy 2003).
NMD can be viewed as a surveillance system that prevents the production of truncated proteins by degrading mRNAs harboring premature stop codons due to mutations, errors induced during transcription or splicing, or mRNAs transcribed from out-of-frame gene rearrangements in the vertebrate immune system. In addition, NMD also contributes to the fine tuning of normal gene expression by degrading specific mRNAs that have naturally occurring premature stop codons—for example, uORF-containing mRNAs (Jacobson and Peltz 2000; Maquat 2000; Maquat and Carmichael 2001; Wagner and Lykke-Andersen 2002). NMD factors have been identified genetically in yeast and Caenorhabditis elegans. Three yeast (UPF1-3) and seven C. elegans (smg-1-smg-7, suppressor with morphogenic defects on genitalia) genes have been shown to play essential roles in NMD. The UPF1/2/3 proteins (SMG-2/SMG-3/SMG-4 in C. elegans) are conserved from yeast to humans, whereas C. elegans SMG-1, SMG-5, and SMG-7 have orthologs in higher eukaryotes but not in yeast (for review, see Wagner and Lykke-Andersen 2002).
Many studies have provided a working model of NMD. NMD requires translation initiation and procession of the ribosome to the premature stop codon, whereupon factors are recruited to degrade the mRNA. Current data support a model in which mammalian UPF3 is loaded onto the mRNA during splicing by interacting with an exon junction complex (EJC), while UPF2 joins the complex soon after mRNA export. UPF2 and UPF3 are displaced by ribosomes as they traverse the mRNA during the first round of translation. Translation termination at a premature stop codon recruits UPF1, probably by interacting with the eRF1-eRF3 translation termination complex. The incomplete removal of UPF2 and/or UPF3 from downstream mRNA sequences results in the assembly of the NMD complex, consisting minimally of UPF1, UPF2, and UPF3. mRNAs that have premature stop codons, and therefore allow the assembly of this complex, are targeted for rapid degradation (Lykke-Andersen et al. 2000; Mendell et al. 2002; Wagner and Lykke-Andersen 2002).
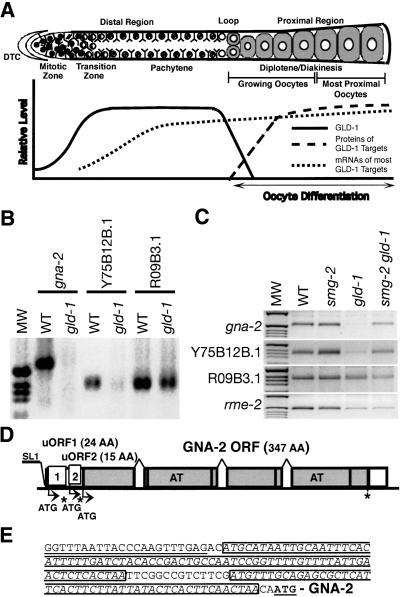
GLD-1 is a member of a family of proteins, including human/mouse Quaking, SAM68, and Drosophila How, that share an ∼200-amino acid region of similarity called the GSG or STAR domain (Jones and Schedl 1995; Vernet and Artzt 1997; Di Fruscio et al. 1998). The conserved region contains a maxi-KH RNA-binding motif that differs from the canonical FMR1/Nova KH motif by the addition of three loops that are conserved only among GSG/STAR proteins and also contains conserved regions N-terminal and C-terminal to the maxi-KH motif (Gibson et al. 1993; Musco et al. 1996; Lewis et al. 2000). The GSG/STAR domain is essential for in vivo function, as missense mutations in nine different conserved residues in this domain alter or eliminate gld-1 function (Francis et al. 1995a; Jones and Schedl 1995). GLD-1 has multiple functions during C. elegans germ cell development including the regulation of meiotic prophase progression of female germ cells, the mitotic versus meiotic switch, and the promotion of the male fate in the hermaphrodite germ line (Francis et al. 1995a,b; Kadyk and Kimble 1998; Hansen et al. 2004a). GLD-1 is exclusively expressed in the germ line and is localized in the cytoplasm of the distal region. The level of GLD-1 is very low in the distal mitotic zone. As germ cells begin meiotic development in the transition zone, the level of GLD-1 increases to its maximum level and stays high through the pachytene stage in the distal region. Levels decrease at the end of pachytene and become undetectable in developing oocytes in the proximal region (Jones et al. 1996; Lee and Schedl 2001). The nonhomogeneous distribution of GLD-1, and its role as a cytoplasmic RNA-binding protein that functions in multiple aspects of germ cell development, indicate that GLD-1 functions to spatially restrict the translation of multiple maternal mRNAs (Fig. 1A).
Figure 1.
(A) Schematic representation of the adult hermaphrodite germ line and qualitative depiction of GLD-1 protein levels (solid line), protein level (long dashed line), and mRNA level (short dashed line) for several GLD-1 targets. (Upper panel) The diagram of a single wild-type adult hermaphrodite gonad arm is drawn linearly instead of its normal reflexed shape for comparison purposes. Our qualitative assessment (Y-axis) of protein levels for GLD-1, protein, and mRNA levels for several GLD-1 targets in the corresponding regions of the germ line (X-axis) are shown in the lower panel. The gonad contains ∼1000 germ nuclei. In the distal region, nuclei are arranged primarily around the periphery of the gonadal tube. Each nucleus is partially enclosed by plasma membranes; although this is a syncytium, each nucleus and its surrounding cytoplasm and membranes is referred as a germ cell (Schedl 1997). (B) The mRNA levels of GLD-1 targets are compared between wild-type adult hermaphrodites and gld-1(q485) adult hermaphrodites using RT-PCR analysis. The first-strand cDNA, which is equivalent to 10 ng of total RNA from animals of the indicated genotypes, was amplified for 30 cycles using gene-specific primers. (C) The mRNA levels of GLD-1 targets are compared between wild-type, smg-2(e2008), gld-1(q485), and smg-2(e2008) gld-1(q485) adult hermaphrodites using RT-PCR analysis. The first-strand cDNA, which is equivalent to 10 ng of total RNA from animals of the indicated genotypes, was amplified for 25 cycles using gene-specific primers. (D) Schematic diagram of gna-2 genomic DNA. The first and second uORFs are indicated by the corresponding boxes 1 and 2 and the GNA-2 ORF by gray boxes. The start codons (ATG) of the uORFs and the GNA-2 ORF are shown by arrows and stop codons by asterisks. Two acetyl transferase domains are indicated by AT within the gray boxes. (E) 5′-UTR sequence of gna-2. gna-2 is trans-spliced by the SL1 leader RNA (underlined). Two uORFs are boxed, and the ATG of the GNA-2 ORF is in bold and underlined.
We previously identified multiple in vivo mRNA targets of GLD-1 that coimmunoprecipitate with GLD-1 from cytoplasmic extracts (Lee and Schedl 2001). These mRNA targets are preferentially expressed in the germ line and, as expected, several of them exhibit essential functions in oocyte differentiation and early embryogenesis. For at least one target, rme-2 mRNA, GLD-1 functions as a translational repressor. RME-2 protein is not expressed in pachytene-stage germ cells in the distal region of wild-type germ line where GLD-1 is abundant in the cytoplasm, but is expressed in developing oocytes in the proximal region where GLD-1 is absent (Fig. 1A). RME-2 is misexpressed in the distal region of gld-1-null germ line. Even though RME-2 protein is not expressed in the distal region of wild-type germ lines, rme-2 mRNA is expressed and accumulates (Lee and Schedl 2001).
In this study, we show that GLD-1 targets can be subjected to an additional control mechanism at the posttranscriptional level. Our results demonstrate the existence of a novel mechanism in which the expression of at least one GLD-1 mRNA target is precisely controlled by the combination of uORF-mediated NMD together with GLD-1-dependent translational repression. Consistent with this mechanism, we find that translational repression by GLD-1 protects mRNA targets that contain nonsense mutations from NMD.
Results
GLD-1 stabilizes gna-2 mRNA, which has two uORFs
In addition to rme-2, we found that GLD-1 represses the translation of two other GLD-1 mRNA targets, oma-1 and oma-2, which display mRNA and protein accumulation patterns similar to rme-2 (Fig. 1A; Supplemental Material). Other groups have shown that tra-2, mes-3, and glp-1 are translationally inhibited by GLD-1 (Jan et al. 1999; Xu et al. 2001; Marin and Evans 2003). Therefore, GLD-1 likely acts as a translational repressor for most of its mRNA targets. Consistent with these findings, the mRNA levels of most GLD-1 targets remain largely unchanged in gld-1-null animals, although they are expressed at slightly lower levels than in wild-type animals as tested by RT-PCR (Fig. 1B,C; data not shown) and by RNA in situ analysis (Lee and Schedl 2001; Supplemental Material). However, two GLD-1 targets, gna-2 (T23G11.2; glucosamine phosphate N-acetyltransferase) and Y75B12B.1 mRNAs, are expressed at very low levels in gld-1-null animals (Figs. 1B,C, 2), suggesting that these messages are degraded in the absence of GLD-1 and that GLD-1 binding likely stabilizes these two mRNA targets.
Figure 2.
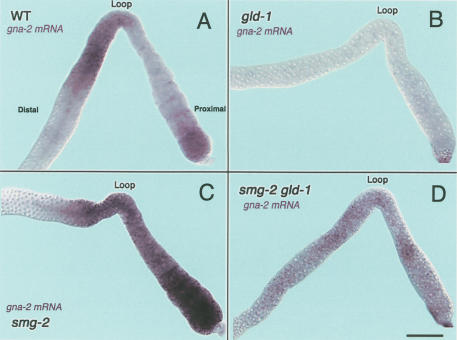
Accumulation of gna-2 mRNA in wild-type, gld-1(q485), smg-2(e2008), and smg-2(e2008) gld-1(q485) adult hermaphrodite germ lines. The distal regions are to the left side of the loop, and the proximal regions are to the right side of the loop. The most distal ends are not shown as they have essentially no signal. (A) gna-2 mRNA (purple staining) in a wild-type adult hermaphrodite gonad is first detected in early meiotic prophase and increases to a high level in late-pachytene-stage germ cells. Around the loop region, gna-2 mRNA is less abundant and is present at very low levels in the proximal region. gna-2 mRNA, however, reaccumulates in the most proximal oocytes. (B) In gld-1-null adult hermaphrodite gonads, there is very little accumulation of gna-2 mRNA throughout the germ line. (C) In smg-2 adult hermaphrodites, gna-2 mRNA is first detected in early meiotic prophase, and increases to a high level in late-pachytene-stage germ cells. The high level of gna-2 mRNA is maintained in the developing oocytes throughout the proximal region. (D) In smg-2 gld-1-null adult hermaphrodites, there is more accumulation of gna-2 mRNA throughout the germ line than in gld-1-null gonads (B). The overall level of gna-2 mRNA appears lower in smg-2 gld-1-null germ lines than in wild-type or smg-2 germ lines, possibly because germ cells in smg-2 gld-1-null germ lines only progress to the early pachytene stage and the accumulation that would normally occur in later pachytene does not occur. Bar, 20 μm.
To investigate the mechanism through which GLD-1 stabilizes the gna-2 mRNA posttranscriptionally, we determined the complete structure of the gna-2 mRNA. The gna-2 mRNA has a 5′ SL1 leader sequence, a 141-nt 5′-UTR, a 1044-nt GNA-2 ORF that encodes 347 amino acids, and an 87-nt 3′-UTR (Fig. 1D; GenBank accession no. AY455927). The GNA-2 protein has two acetyl transferase domains, which likely function in acetylation of carbohydrate moieties of cell surface proteins (Lee and Schedl 2001; W. Johnston and J. Dennis, pers. comm.). From inspection of the gna-2 cDNA, we identified two predicted uORFs of 24 codons (uORF1) and 15 codons (uORF2), respectively, in the 5′-UTR of gna-2 mRNA (Fig. 1D,E). Therefore, the message contains two premature stop codons. uORF1 and uORF2 use different reading frames from each other as well as from the GNA-2 ORF (positions -1 and -2, respectively, relative to the GNA-2 ORF; uORF1, uORF2, and the GNA-2 ORF do not overlap). The presence of premature stop-containing uORFs together with the known role of NMD in targeting such mRNAs suggests a model in which GLD-1 protects gna-2 mRNA from NMD.
gna-2 mRNA is a naturally occurring target of NMD and GLD-1 protects it from NMD
In wild-type adult hermaphrodite germ lines, the bulk of RNA synthesis for oogenesis occurs in pachytene-stage germ cells, with progressively less RNA synthesis as germ cells proceed through the diplotene stage. As chromosomes become highly condensed in late-stage oocytes (diakinesis), almost no de novo RNA synthesis occurs (Gibert et al. 1984; Schisa et al. 2001) and RNA polymerase II becomes inactive (Kelly et al. 2002). Therefore, most maternal transcripts, including GLD-1 targets rme-2, oma-1, and oma-2 mRNAs, are transcribed mainly in pachytene-stage germ cells in the distal region and transported into the common cytoplasmic core. In the proximal region, maternal mRNAs are packaged into growing oocytes. These processes result in extensive RNA accumulation throughout pachytene-stage germ cells and developing oocytes (Fig. 1A; Supplemental Material; Jones et al. 1996; Lee and Schedl 2001).
To investigate how GLD-1 stabilizes gna-2 mRNA, we performed RNA in situ analysis in both wild-type and gld-1-null adult hermaphrodite germ lines. In wild-type germ lines, gna-2 mRNA accumulates in pachytenestage germ cells and the cytoplasmic core in the distal region, where GLD-1 is abundant. GLD-1 levels decrease in the loop region and are undetectable in the proximal region; gna-2 mRNA accumulation mirrors GLD-1 protein accumulation, falling to very low levels in the proximal region (Fig. 2A). Higher levels of gna-2 mRNA, however, are seen in the most proximal oocytes. As a control, no signal is detected with the gna-2 sense probe (Supplemental Material). In gld-1-null germ lines, we detected essentially no accumulation of gna-2 mRNA cytologically (Fig. 2B), consistent with RT-PCR data (Fig. 1B,C). These findings support the model that GLD-1 stabilizes gna-2 mRNA.
The presence of two uORFs in gna-2 mRNA suggests that they may affect mRNA stability and/or be involved in regulating the translation of the GNA-2 ORF. Because gna-2 mRNA exists at low levels in the proximal region of wild-type germ lines, and is essentially absent in gld-1-null germ lines, we speculated that translation termination of the uORFs may destabilize gna-2 mRNA through NMD in the absence of GLD-1. In the distal region of wild-type germ lines, GLD-1 may protect the message from NMD by inhibiting translation of the uORFs. The reaccumulation of gna-2 mRNA in the most proximal oocytes in wild-type germ lines would then suggest that although most gna-2 mRNA is degraded by NMD in the proximal region, a small fraction can escape NMD. To determine if the low levels of gna-2 mRNA in the proximal region is due to NMD, we examined germ lines from animals defective in NMD (smg-2), as well as in smg-2 gld-1 double mutants, for the accumulation of gna-2 mRNA. In smg-2 mutant germ lines, gna-2 mRNA is present at much higher levels throughout the proximal region as compared with wild-type germ lines, showing that the message is stabilized in the absence of NMD (Fig. 2C). We also detected higher accumulation of gna-2 mRNA in smg-2 gld-1 double-mutant germ lines as compared with gld-1 germ lines (Fig. 2D; see also Fig. 1C RT-PCR data). RNA in situ analysis of gna-2 mRNA in other smg mutants, such as smg-3(ma117) and smg-4(ma116), give essentially identical results as those with smg-2(e2008) mutants (Table 1). These data strongly support the idea that gna-2 mRNA is a naturally occurring target of NMD. In the distal region, however, NMD of gna-2 mRNA is blocked by GLD-1. Because NMD is dependent on translation termination at a premature stop codon and GLD-1 acts as a translational repressor, we suggest that GLD-1 protects gna-2 mRNA from NMD by repressing translation of both uORFs of gna-2 mRNA such that the ribosome does not arrive at the stop codon of either uORF. In the absence of GLD-1, such as in growing oocytes in wild-type germ lines and in gld-1-null germ lines, the ribosome likely completes translation of one or both uORFs, and upon arriving at the stop codon, NMD factors are recruited to degrade gna-2 mRNA.
Table 1.
RNA in situ analysis of GLD-1 mRNA targets
| Probe | Genotype | GLD-1 bindingf | mRNA detected | Distal pachytene | Proximal oocytes |
|---|---|---|---|---|---|
| gna-2 | Wild type | 5′-UTR | Wild type | Present | Very low |
| smg-2 (e2008) | Wild type | Present | Present | ||
| smg-3(ma117) | Wild type | Present | Present | ||
| smg-4(ma116) | Wild type | Present | Present | ||
| gld-1(q485) | Wild type | Absent | N/Aa | ||
| smg-2(e2008) gld-1(q485) | Wild type | Present | N/Aa | ||
| rme-2 | Wild type | 5′-end | Wild type | Present | Present |
| rme-2(b1005) | 3′-UTR | Nonsense | Present | Absent | |
| smg-2(2008); rme-2(b1005) | Nonsense | Present | Present | ||
| gld-1(q485) | Wild type | Present | N/Aa | ||
| gld-1(q485); rme-2(b1005) | Nonsense | Absent | N/Aa | ||
| rme-2(RNAi)e | Wild type | Absent | Absent | ||
| smg-2(e2008)e | Wild type | Present | Present | ||
| smg-2(e2008); rme-2(RNAi)e | Wild type | Absent | Absent | ||
| oma-1 | Wild typee | 5′-UTR | Wild type | Present | Present |
| oma-1(te33)d,e | 3′-UTR | Nonsense | Present | Absent | |
| oma-1(te35)d,e | Nonsense | Present | Absent | ||
| oma-1(te33)d, oma-2(te51) | Nonsense | Present | Absent | ||
| gld-1(q485)e | Wild type | Present | N/Aa | ||
| gld-1(q485); oma-1(te33)d, oma-2(te51)e | Nonsense | Absent | N/Aa | ||
| lin-45 | Wild typee | 3′-UTR | Wild type | Present | Present |
| lin-45(dx19)e | Nonsense | Present | N/Ab | ||
| gld-1(q485)e | Wild type | Present | N/Aa | ||
| gld-1(q485); lin-45(dx19)e | Nonsense | Absent | N/Aa | ||
| mes-3 | Wild type | 3′-UTR | Wild type | Present | Present |
| mes-3(bn53)e | Nonsense | Present | Absent | ||
| nos-3 | Wild type | N/Ac | Wild type | Present | Present |
| nos-3(oz235)e | Nonsense | Absent | Absent |
Tumorous germlines of gld-1(q485) and double mutants containing gld-1(q485) do not have developing oocytes.
Germ cells in lin-45(dx19) are all arrested in the pachytene stage; thus, there are no developing oocytes.
NOS-3 protein is expressed throughout the germ line and nos-3 mRMA is also not enriched in a GLD-1 immunoprecipitation (data not shown), indicating that nos-3 mRNA is not a target of GLD-1.
All oma-1 mutants are revertants of oma-1(zu405). Thus, they also contain a zu405 missense mutation in cis.
See Supplemental Material.
GLD-1-binding region.
uORFs of gna-2 mRNA is sufficient to induce NMD
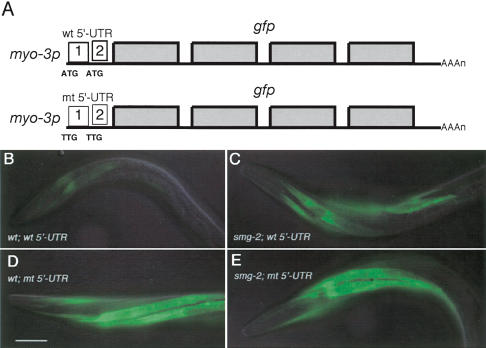
RNA in situ analysis of gna-2 mRNA clearly indicates that gna-2 mRNA becomes a target of NMD that is likely induced by its uORFs. We investigated whether the gna-2 uORFs are sufficient to induce NMD in other cell types by placing the entire 5′-UTR of gna-2, containing the two uORFs, in front of gfp-coding sequences driven by the myo-3 promoter (Fig. 3A; Okkema et al. 1993). When the relative level of GFP expression from this transgene was compared in muscle cells between wild-type and smg-2 mutant animals, it was significantly higher in smg-2 mutants than in wild-type (Fig. 3B,C). This indicates that transgene mRNA containing the gna-2 uORFs is subjected to NMD in wild-type animals. Furthermore, when both uAUGs are mutated to TTGs in this construct (Fig. 3A), the relative level of GFP expression became higher in wild-type animals and was not altered when smg activity was eliminated (Fig. 3D,E). Thus, the gna-2 uORFs are sufficient to induce NMD of transgene mRNA in these cells.
Figure 3.
The uORF of gna-2 mRNA is sufficient to induce NMD of a transgene mRNA in other cell types. (A) Schematic diagram of two transgenes, one with a wild-type gna-2 5′-UTR (wt), the other with a mutated 5′-UTR (mt, both uATGs changed to TTGs), both in front of gfp-coding sequences (gray boxes) and driven by the myo-3 promoter. GFP expression in the anterior body wall muscle of L4 hermaphrodites is shown (head to the left). GFP expression of a transgene with the wild-type gna-2 5′-UTR is significantly lower in the wild-type (B) than the smg-2 mutant (C) background. GFP expression of a transgene with the mutated gna-2 5′-UTR is similar in wild-type (D) and smg-2 mutant (E) backgrounds. Identical expression results are observed with the vulval muscles (data not shown). All images were captured for the same time frame and processed identically. Bar, 20 μm.
GLD-1 binds to the 5′-UTR of gna-2 mRNA independently from the uAUG
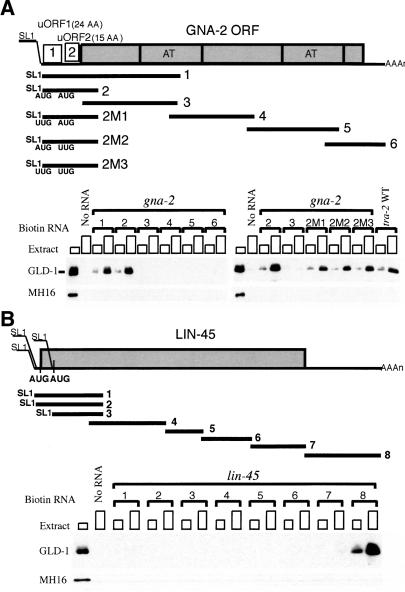
To gain insight into how GLD-1 is regulating the translation and stability of gna-2 mRNA, we identified the GLD-1-binding region on gna-2 mRNA using a biotin-RNA pull-down assay (Nabel-Rosen et al. 1999; Lee and Schedl 2001). Six RNA fragments, corresponding to the entire gna-2 mRNA, were biotin-labeled and allowed to bind GLD-1 in cytoplasmic extracts from wild-type adult hermaphrodites. GLD-1 binds to two fragments, both of which contain the 5′-UTR, but fails to bind the fragments that cover the GNA-2 ORF or 3′-UTR (Fig. 4A). Because the 5′-UTR of gna-2 mRNA contains two uORFs, and GLD-1 protects this mRNA from NMD, it is possible that GLD-1 may recognize the uORF, or the uAUG specifically, either directly or through another protein(s). However, when the first uAUG, the second uAUG, or both uAUGs were substituted to UUG (2M1, 2M2, 2M3, respectively), GLD-1 still bound the fragments (Fig. 4A), suggesting that GLD-1 does not specifically recognize either uAUG. These experiments indicate that GLD-1 binds to the 5′-UTR of gna-2 mRNA and that this binding is independent of the uAUGs.
Figure 4.
GLD-1 binds to the 5′-UTR of gna-2 mRNA and the 3′-UTR of lin-45 mRNA. (A) Schematic diagram of gna-2 RNA probes and GLD-1 binding. Each biotin-labeled RNA (see Materials and Methods) or control sample (No RNA) was allowed to form a complex with increasing amounts of cytosol extract (∼50 μg or ∼150 μg of total protein) from wild-type adult hermaphrodites, and the complex was isolated with streptavidin magnetic beads. The isolated proteins were subjected to Western analysis with anti-GLD-1 Ab. The first lanes show total GLD-1 in cytosol extract (∼30 μg). (B) Schematic diagram of lin-45 RNA probes and GLD-1 binding. lin-45 mRNA is trans-spliced at three different locations (GenBank accession nos. AY455928, AY493413, AY493414). The first and the second SL1 spliced mRNAs (probes 1 and 2, respectively) are predicted to use the same AUG to initiate translation. The third SL1 spliced mRNA (probe 3) likely uses the downstream AUG that is in the same reading frame. MH16 (anti-Paramyosin Ab) was used as a negative control.
GLD-1 and NMD spatially limit the expression of the GNA-2 protein
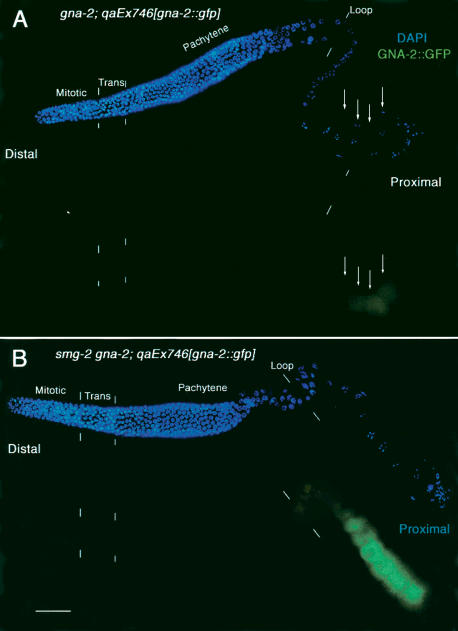
If GLD-1 represses the translation of gna-2 mRNA, then the GNA-2 protein should not be expressed in the distal region of wild-type germ lines, where GLD-1 is abundant. GNA-2 protein should be expressed, however, in the proximal region, where GLD-1 is absent, but only in the most proximal oocytes, where gna-2 mRNA accumulates. To determine the expression pattern of GNA-2, we used a rescuing GNA-2::GFP construct in gna-2 deletion mutant hermaphrodites (kindly provided by W. Johnston and J. Dennis, Mount Sinai Hospital, Toronto, Canada). As expected, the GNA-2::GFP is not expressed in the distal region, where GLD-1 is abundant, and is expressed at very low levels in only the most proximal oocytes (Fig. 5A, arrows). In smg-2 mutant germ lines in which gna-2 mRNA is abundant in oocytes throughout the proximal region (Fig. 2D), GNA-2 protein should be expressed in all of the oocytes and at a higher level than in wild-type germ lines. Consistent with this prediction, we found that GNA-2::GFP is expressed abundantly in the entire proximal region in smg-2 mutants (Fig. 5B). Taken with the gna-2 mRNA accumulation data, these results indicate that the level and location of GNA-2 accumulation are regulated by the combination of translation repression by GLD-1 and mRNA degradation by NMD, which is likely elicited by the uORFs.
Figure 5.
GNA-2::GFP accumulation is regulated by GLD-1 and NMD. Gonad arms, dissected from a gna-2(qa705) adult hermaphrodite that is rescued by gna-2::gfp (A), or from a smg-2(e2008) gna-2(qa705) adult hermaphrodite that is rescued by gna-2::gfp(qaEx746) (B), stained with DAPI to visualize DNA (blue), rabbit anti-GFP (green). Composites show surface views of intact gonads in the distal region and interior focal planes in the proximal region. (A) GNA-2::GFP is not expressed in the distal region, where GLD-1 is abundant in the cytoplasm. In the proximal region, GNA-2::GFP levels are very low and are only present in the most proximal oocytes, as indicated by arrows. The weak staining in the distal region and early proximal region is considered nonspecific background as it is similar to that observed in wild-type germ lines that do not express GFP (data not shown). (B) GNA-2::GFP is not expressed in the distal region, where GLD-1 is abundant in the cytoplasm. However, GNA-2::GFP expression is more abundant in the entire proximal region in the smg-2 mutant. Bar, 20 μm.
GLD-1 protects nonsense-containing mRNA targets from NMD
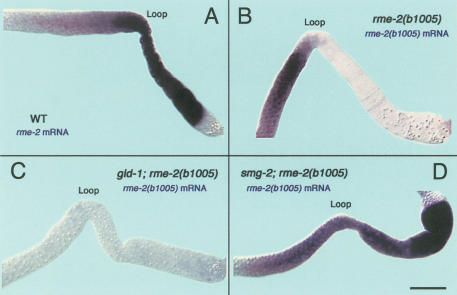
gna-2 mRNA and protein expression patterns in wild-type and several mutant backgrounds indicate that GLD-1 protects gna-2 mRNA from NMD, probably by repressing the translation of the uORFs found in the gna-2 mRNA. If GLD-1-mediated translation repression is able to protect an uORF-containing mRNA from NMD, we would expect that mRNA targets that acquire premature stop codons through mutation should be protected from NMD by GLD-1 present in pachytene-stage germ cells. In the proximal region, where GLD-1 is absent, the mutant mRNAs would be degraded by NMD. To test this idea, we took advantage of alleles of other GLD-1 targets that contain premature stop codons by nonsense mutations including oma-1(te33), oma-1(te35), lin-45(dx19), and mes-3(bn53) (Detwiler et al. 2001; Xu et al. 2001; Hsu et al. 2002), and also rme-2(b1005), which has a nonsense codon due to a small out-of-frame deletion (Grant and Hirsh 1999). GLD-1 binds these mRNA targets and is known to repress the translation of rme-2, oma-1, and mes-3 mRNA (Lee and Schedl 2001; Xu et al. 2001; Supplemental Material). For rme-2, its wild-type mRNA is present in pachytene-stage germ cells in the distal region as well as in developing oocytes in the proximal region. However, we found that rme-2(b1005) mRNA is present only in the distal region, where GLD-1 is abundant, and essentially absent in the proximal region, where GLD-1 is absent (Fig. 6A,B; Table 1). The presence of rme-2(b1005) mutant mRNA in the distal region is dependent on GLD-1 because it is absent in the distal region of gld-1 null; rme-2(b1005) double-mutant germ lines. Furthermore, the absence of rme-2(b1005) mutant mRNA in the proximal region is due to NMD because the message is present in the proximal region of smg-2; rme-2(b1005) double-mutant germ lines (Fig. 6C,D; Table 1). Other nonsense-mutation-containing mRNAs of GLD-1 targets tested here (oma-1, lin-45, and mes-3) are also stabilized in pachytene-stage germ cells in the distal region, where GLD-1 is abundant, and destabilized in developing oocytes, as well as in gld-1-null mutants, even though their wild-type mRNAs are found in both the distal and proximal regions (Table 1; Supplemental Material). These data suggest that NMD is not initiated in the distal region of the germ line because GLD-1 binds the mutant mRNAs and represses their translation.
Figure 6.
Accumulation of rme-2 mRNA in wild-type, rme-2(b1005), gld-1(q485); rme-2(b1005), and smg-2(e2008); rme-2(b1005) adult hermaphrodite germ lines. The distal regions are to the left side of the loop, and the proximal regions are to the right side of the loop. The most distal ends are not shown. (A) In wild-type adult hermaphrodite gonads, wild-type rme-2 mRNA (purple staining) is first detected in early meiotic prophase, in the transition zone, and accumulates to a high level in late pachytene (loop), as well as in the entire proximal region. (B) rme-2(b1005) mRNA has a nonsense codon in exon 6 (out of a total of seven exons). In rme-2(b1005) adult hermaphrodites, rme-2(b1005) mRNA accumulates in a very similar pattern as wild-type rme-2 mRNA in the distal region. However, in the loop and proximal region, rme-2(b1005) mRNA is essentially absent. (C) In gld-1; rme-2 double mutants, rme-2(b1005) mRNA is essentially absent throughout the germ line. (D) In smg-2; rme-2 double mutants, rme-2(b1005) mRNA accumulates to a high level in the distal region as well as in the proximal region. Bar, 20 μm.
However, it is also possible that mRNA targets in the distal region that are bound to GLD-1 are not accessible to any RNA degradation machinery, or that NMD is not active in the distal germ line. To test whether mRNA targets are degradable in the distal region, we performed RNAi by injecting double-stranded rme-2 RNA into wild-type and smg-2(e2008) animals and performed RNA in situ analysis of the resulting progeny. Virtually no rme-2 mRNA is present in the distal or proximal regions (Table 1; Supplemental Material). This indicates that rme-2 mRNA can be degraded in the distal region and that GLD-1 does not sequester its targets from the RNAi degradation machinery. Furthermore, mRNAs that are not translationally repressed by GLD-1 are subject to NMD in the distal germ line. For example, NOS-3 protein is expressed throughout the germ line (Kraemer et al. 1999), and its wild-type mRNA is also expressed throughout the germ line (Table 1; Supplemental Material). However, nonsense-containing nos-3(oz235) mRNA (Hansen et al. 2004b) is essentially absent in both the distal and proximal regions (Table 1; Supplemental Material), indicating NMD is active throughout the germ line. Together, these data strongly support the idea that GLD-1 protects its mRNA targets from NMD in the normal condition (gna-2 mRNA) as well as when mRNA targets acquire nonsense mutations. The ability of GLD-1 to repress translation in the distal region indicates that premature translation termination at nonsense codons does not occur, and as a result, nonsense-containing GLD-1 mRNA targets are not destabilized by NMD.
The GLD-1-binding region can exist in either the 5′-UTR or 3′-UTR to protect its targets from NMD
GLD-1 binds the 5′-UTR of the gna-2 mRNA and protects it from NMD. This raises the possibility that the physical presence of GLD-1 at the 5′-UTR, and not the translation repression activity of GLD-1, is important for its protective function. Therefore, we determined the GLD-1-binding regions of the mRNA targets used in this study. GLD-1 binds to the 5′-end of the coding sequence of rme-2 mRNA (Lee and Schedl 2001) and the 5′-UTR of oma-1 mRNA (Table 1; Supplemental Material). However, GLD-1 binds only to the 3′-UTR of lin-45 mRNA and is still able to protect nonsense-containing lin-45 mRNA from NMD (Fig. 4B; Table 1; Supplemental Material). mes-3 mRNA was previously shown to have GLD-1-binding sites in 3′-UTR (Xu et al. 2001) and nonsense-containing mes-3 mRNA is also protected in the distal region (Table 1; Supplemental Material). These results indicate that the position of the GLD-1-binding region(s) is not an important factor in protecting its targets from NMD and further supports the idea that GLD-1 protects its targets from NMD by binding and repressing translation.
Discussion
GLD-1 has multiple mRNA targets and likely represses translation of most of its targets (Jan et al. 1999; Clifford et al. 2000; Lee and Schedl 2001; Xu et al. 2001; Marin and Evans 2003; Supplemental Material). Therefore, unless GLD-1 targets are regulated by an additional mechanism, target proteins will not be expressed in the distal region of wild-type germ lines, where GLD-1 is present, but will be expressed in the proximal region, where GLD-1 is absent. Here, we show that one GLD-1 target, gna-2 mRNA, which has two uORFs, is subjected to two different controls at the posttranscriptional level. First, GLD-1 binds and represses translation of gna-2 mRNA. The repression by GLD-1 may be independent of the two uORFs in the 5′-UTR of gna-2 mRNA because GLD-1 is able to bind to a mutated 5′-UTR that does not contain uAUGs. Second, NMD destabilizes gna-2 mRNA when it is not repressed translationally by GLD-1, and the destabilization is likely mediated by translation termination of one or both of the uORFs. This is consistent with previous work demonstrating that some mRNAs containing uORFs are targets of NMD (Ruiz-Echevarria et al. 1996; Linz et al. 1997), suggesting that NMD may act as a general regulator of gene expression. Translation repression and destabilization by NMD, however, are not independent of each other, and the interaction between these two controls determines the spatial and temporal expression pattern of the GNA-2 protein. Therefore, gna-2 mRNA provides a novel example in which gene expression is tightly controlled through two mechanisms, translation regulation by a specific translational repressor, GLD-1, and uORF elicited regulation, mainly through NMD. Several mammalian mRNAs encoding proteins with proposed functions in cell growth and differentiation have one or more uORFs (e.g., MDM2, C/EBPα, C/EBPβ, ATF4; Geballe and Sachs 2000; Meijer and Thomas 2002). Thus, the combination of translational repression and NMD described here for gna-2 may be a more general mechanism by which uORF-containing mRNAs are regulated.
uORFs of gna-2 mRNA and translation repression by GLD-1 fine-tune GNA-2 expression
In pachytene cells of wild-type and NMD-deficient (smg-2) mutant germ lines, GLD-1 represses the translation of the GNA-2 ORF. The relative levels of gna-2 mRNA in this region between wild-type and smg-2/-3/-4 mutants are quite similar. This suggests that translation of the uORFs of gna-2 mRNA is likely repressed by GLD-1, and as a result, gna-2 mRNA is stabilized. Once gna-2 mRNA is released from translation repression in the developing oocytes, due to the absence of GLD-1, translation of at least one of the uORFs is likely completed. Translation termination at the uORF stop codon then recruits NMD factors to degrade the gna-2 mRNA in wild-type germ lines because gna-2 mRNA levels are much lower in the proximal region than in the distal region of wild-type germ lines, whereas smg mutant germ lines have much higher levels in the proximal region. However, gna-2 mRNA is not completely absent in the growing oocytes and accumulates at a higher level in the most proximal oocytes (usually two to four oocytes; see Figs. 1A, 2A). We do not know the mechanism by which gna-2 mRNA accumulates in the most proximal oocytes; there may be a specific transport mechanism that preferentially loads certain mRNAs into the most proximal oocytes for translation just prior to maturation/ovulation. Another possibility that NMD itself is less active in the most proximal oocytes, is unlikely because other nonsense mutant mRNAs, including rme-2(b1005), never show increased accumulation in the most proximal oocytes. Together, these data suggest that in the proximal region, where GLD-1 is absent, both uORFs of a small fraction of gna-2 mRNA are not translated, whereas the GNA-2 ORF is translated, possibly because of leaky scanning. Alternatively, after translation termination of the uORF, the ribosome rescans and reinitiates translation of the GNA-2 ORF. Therefore, a small fraction of gna-2 mRNA is not subjected to NMD and is able to accumulate and synthesize GNA-2 protein at the proximal end.
Translation repression and nonsense-mediated mRNA decay
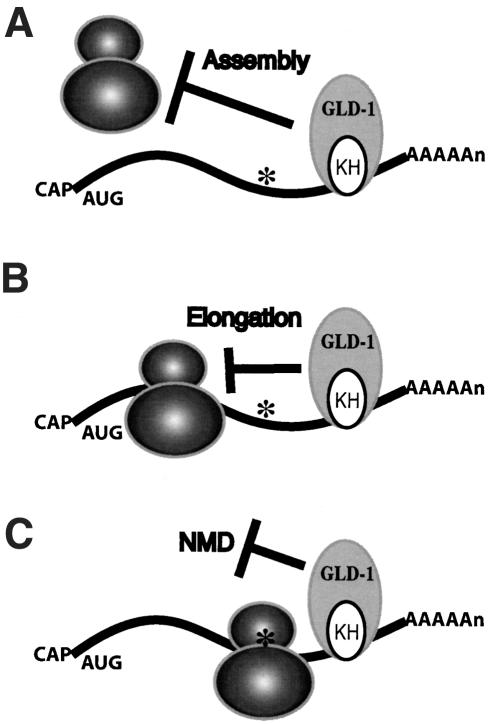
For NMD to occur, the ribosome must proceed to the premature stop codon and recruit factors to degrade the mRNA. This is supported by several findings: (1) inhibition of NMD by drugs and mutations that repress either translation initiation or elongation; (2) inhibition of NMD by the coexpression of a nonsense-suppressing tRNA with the nonsense-containing mRNA; (3) destabilization of polysome-associated nonsense-containing mRNAs immediately after release from stabilization induced by cycloheximide; and (4) interaction of UPF1 with the polypeptide release factors eRF1 and eRF3 (Jacobson and Peltz 2000; Maquat 2000; Maquat and Carmichael 2001; Wagner and Lykke-Andersen 2002). Here we show that translation repression by an endogenous RNA-binding protein blocks NMD of a message that contains uORFs and mutant messages that contain premature stop codons. The translation termination dependency of NMD suggests that when an mRNA is under the control of a translational repressor, and at the same time has a premature stop codon that elicits NMD, the fate of the mRNA will depend on whether or not the ribosome can arrive at the premature stop codon. Becaues NMD requires translation termination at a premature stop codon, this strongly suggests that either GLD-1 represses the assembly of the ribosome on its mRNA targets, or if the ribosome does assemble, then GLD-1 represses the ribosome from elongation to the premature stop codon (Fig. 7). Because the uORFs of gna-2 mRNA are very short, if GLD-1 represses elongation, GLD-1 likely represses very early during elongation. Previous studies suggest that GLD-1 inhibits translation initiation of tra-2 mRNA (Goodwin et al. 1993; Jan et al. 1999; Kuersten and Goodwin 2003); however, it remains possible that GLD-1 inhibits translation elongation of other targets. In addition, our data do not exclude the formal possibility that GLD-1 may block NMD directly after the ribosome arrives at the premature stop codon (Fig. 7).
Figure 7.
Models of how GLD-1 protects its mRNA targets from NMD. (A) GLD-1 protects its mRNA targets that contain premature stop codons (asterisks) by repressing the assembly of the ribosome on its targets. GLD-1 may inhibit any step during translation initiation. (B) Alternatively, if the ribosome is assembled, then GLD-1 may repress ribosome elongation, preventing it from reaching the premature stop codon. (C) It remains formally possible that GLD-1 may repress termination or repress NMD directly after the ribosome arrives at the premature stop codon.
The gna-2 mRNA accumulation pattern presented here, as well as that of other GLD-1 mRNA targets containing nonsense codons, suggest that the expression patterns of wild-type and nonsense-containing mRNAs can be a very reliable test as to whether or not a specific mRNA is a true target of GLD-1. Nonsense-containing mutant mRNAs of GLD-1 targets should be stable in the distal region, where GLD-1 is present, and unstable in the proximal region, where GLD-1 is absent. These mutant targets also should be unstable in a gld-1-null background. However, the wild-type mRNA of these targets should be stable in both the distal region and the proximal region of wild-type and gld-1-null germ lines.
Nonsense-mediated mRNA decay in C. elegans
All eukaryotes examined to date have a system to selectively degrade mRNAs that contain premature stop codons. Genetic analysis of this process in C. elegans identified seven genes (smg-1 to smg-7) that are required for NMD (Hodgkin et al. 1989; Pulak and Anderson 1993; Cali and Anderson 1998; Page et al. 1999; Aronoff et al. 2001). NMD is not essential for viability in either yeast or C. elegans. Although smg mutants are viable and relatively healthy, they do exhibit morphogenic abnormalities. Adult hermaphrodites have protruding vulvae, and adult males have abnormal tail bursae and mate poorly. In addition, their brood sizes are about two-thirds of wild-type under laboratory conditions (Hodgkin et al. 1989; Pulak and Anderson 1993; Cali and Anderson 1998). These defects may simply be caused by the expression of abnormal polypeptides. However, the specificity of these phenotypes suggests that there could be naturally occurring targets of NMD and misregulation of these targets in smg mutant animals may result in these phenotypes. In this study, we demonstrate that gna-2 mRNA is a naturally occurring target of NMD. Previous studies identified aberrantly spliced ribosomal protein or SR protein mRNAs that now contain stop codons as naturally occurring targets of NMD in C. elegans (Morrison et al. 1997; Mitrovich and Anderson 2000). We further demonstrate that gna-2 mRNA becomes degraded by NMD in the proximal region of germ lines, where it is actively translated. Data presented here also support the model that the interaction between translation repression by GLD-1 and uORF-dependent regulation limits the expression of the GNA-2 protein to the most proximal oocytes. It is not known at this time, however, why gna-2 mRNA is under this dual control. Maternal GNA-2 is required as early as the first cytokinesis in the embryo (Lee and Schedl 2001; W. Johnston and J. Dennis, pers. comm.). It is possible that earlier and higher levels of GNA-2 accumulation are detrimental to the germ line. Therefore, this dual control may allow maternal mRNAs to be translated just prior to fertilization, yet prevent the same mRNAs from being detrimentally translated during earlier oogenesis. In the absence of NMD, as shown in smg mutant germ lines, GNA-2 protein is expressed at a higher level and throughout the proximal region. The misexpression of the GNA-2 protein in smg mutant germ lines may be one of the reasons for reduced fertility in NMD-deficient animals.
Materials and methods
Nematode strains and culture
Standard procedures for nematode culture and genetic manipulation were followed with growth at 20°C (Sulston and Hodgkin 1988). C. elegans genes, alleles, and phenotypes relevant to this study are described elsewhere (Francis et al. 1995a; Grant and Hirsh 1999; Hodgkin and Martinelli 1999; Detwiler et al. 2001; Xu et al. 2001; Hsu et al. 2002; Hansen et al. 2004b).
RT-PCR
One microgram of total RNA from wild-type, gld-1(q485), smg-2(e2008), and gld-1; smg-2 adult hermaphrodites were converted into first-strand cDNAs using the 3′-RACE primer and Superscript II (Lifetech), extracted with phenol/chloroform, precipitated with ethanol, and resuspended in 200 μL of TE (pH 8.0). Two microliters of the first-strand cDNA was used as template for PCR with target gene specific primers and the 3′-anchor primer to compare the level of mRNA expression as described (Lee and Schedl 2001). One to three microliters of the first-strand cDNA was used as a template for PCR with SL1 or SL2 primers and gene-specific primers to identify the 5′-end of the gld-1 mRNA targets, including gna-2, lin-45, and oma-1. The 5′-ends were determined by sequencing the RT-PCR product.
Gonad dissection, RNA in situ hybridization, and antibody staining
Dissected gonads from adult hermaphrodites were prepared as described (Francis et al. 1995a; Lee and Schedl 2001). For RNA in situ hybridization, dissected gonads were fixed in 0.25% glutaraldehyde, 3% formaldehyde, 100 mM K2HPO4 (pH 7.2), and processed as described (Jones et al. 1996; Lee and Schedl 2001). Sense- and anti-sense DNA probes of gna-2, rme-2, oma-1, lin-45, mes-3, and nos-3 were synthesized with digoxigenin-11-dUTP by repeated primer extension. Control sense probes gave little or no signal (Supplemental Material; data not shown). For antibody staining, dissected gonads were fixed with 3% formaldehyde, 100 mM K2HPO4 (pH 7.2) for 1 h, and postfixed with 100% methanol (-20°C) for more than 5 min. Antibody incubations and washes were performed as described (Jones et al. 1996; Lee and Schedl 2001). Affinity-purified rabbit polyclonal anti-GFP Ab (kindly provided by P. Silver, Dana-Farber Cancer Institute, Boston, MA) was used at 1:1000 dilution, and affinity-purified rabbit polyclonal anti-OMA-1 Ab and anti-OMA-2 Ab (kindly provided by R. Lin, Southwestern Medical Center, Dallas, TX) were used at 1:25 dilutions. A Zeiss Axioplan 2 microscope equipped with a SPOT digital CCD camera (Diagnostic Instruments) was used to capture bright field color images. Epifluorescent images were captured with a Zeiss Axioskop microscope equipped with a Hamamatsu digital CCD camera (Hamamatsu Photonics). All images were processed with Adobe Photoshop 7.0 (Adobe).
Transgene analysis
Wild-type 5′-UTR or mutant 5′-UTR (both ATG to TTG) of gna-2 (1-141 nt of AY455927) replaced the 5′-UTR region of pPD118.20 between the ClaI and the KpnI sites (pMHL109, pMHL110, respectively). pPD118.20 has gfp driven by myo-3 promoter (Okkema et al. 1993). The extrachromosomal array lines were generated by injecting pMHL109 or pMHL110 with pRF4 and pBluescript to wild-type animals. Each array was then crossed to a smg-2 mutant background. The GFP expression of each line (two independent lines for each construct) in wild-type and smg-2 animals was compared at several stages of development.
Molecular cloning and RNA probes
Nucleotide numbering is based on the full-length gna-2 and lin-45 cDNA sequences (GenBank accession nos. are AY455927 and AY455928, respectively). yk466g5, which is nearly a full-length gna-2 cDNA (nucleotides 147-1272), was digested with HincII and SmaI, then religated resulting in nucleotides 457-1272 (pMHL51), with EcoRV and SmaI, then religated resulting in nucleotides 733-1272 (pMHL52), or with EcoRI, then religated resulting in nucleotides 1013-1272 (pMHL53). A full-length gna-2 cDNA (pMHL43) was constructed by cloning an RT-PCR product, which contains the 5′-end, into yk466g5 that was digested with SacII and HincII. The ATG-to-TTG substitution was achieved by sewing PCR using pMHL43 as a template and cloned into yk466g5 that was digested with SacII and HincII (pMHL61 with ATG [3-5] to TTG, pMHL62 with ATG [92-94] to TTG, and pMHL63 with both substitutions). yk406e8, which is nearly a full-length lin-45 cDNA (nucleotides 490-3216), was digested with SacI, then religated resulting in nucleotides 1171-3216 (pMHL47), with BamHI, then religated resulting in nucleotides 1530-3216 (pMHL48), with SmaI and HincII, then religated resulting in nucleotides 1972-3216 (pMHL49), or with NotI, then religated resulting in nucleotides 2454-3216 (pMHL50). A full-length lin-45 cDNA (pMHL58) and two other SL1 spliced lin-45 cDNAs (pMHL59 and pMHL44) were constructed by cloning an RT-PCR product, which contains the 5′-end, into yk466g5 that was digested with SmaI and ClaI.
The RNA probes used to define the GLD-1-binding region in Figure 4 are gna-2 probes: (1) SL1::1-489; (2) SL1::1-223; (3) 147-489; (4) 457-765; (5) 733-1093; (6) 1013-1272; 2M1 is the same as 2 except for an AUG (3-5) to UUG substitution; 2M2, AUG (92-94) to UUG substitution; and 2M3, both AUGs to UUGs. The lin-45 probes are (1) SL1::1-651; (2) SL1::9-651; (3) SL1::180-651; (4) 490-1212; (5) 1171-1567; (6) 1530-2033; (7) 1972-2504; and (8) 2454-3216. The wild-type tra-2 3′-UTR (tra-2 WT) probe is described in Jan et al. (1999).
Biotin-RNA pull-down assay
For gna-2 probes 1 (pMHL43), 2 (pMHL43), 3 (yk466g5), 4 (pMHL51), 5 (pMHL52), 6 (pMHL53), 2M1 (pMHL61), 2M2 (pMHL62), and 2M3 (pMHL63); for lin-45 probes 1 (pMHL58), 2 (pMHL59), 3 (pMHL44), 4 (yk357d2), 5 (pMHL47), 6 (pMHL48), 7 (pMHL49), 8 (pMHL50), and for tra-2 WT 3′-UTR probe (tra-2 WT), template DNAs for the biotin RNA synthesis were amplified by PCR using reverse primer and downstream primers, and the clones indicated inside parentheses as templates. Biotin-RNA synthesis and purification, binding reactions, and Western analysis with anti-GLD-1 Ab were performed as described (Lee and Schedl 2001).
Acknowledgments
We thank Andrew Godbey, SiQun Xu, and Ho-Yeon Kim for their excellent technical assistance; and Jens Lykke-Andersen, Patricia Kuwabara, Jim Skeath, Dave Hansen, and members of the Schedl lab for comments on the manuscript. We are grateful to Wendy Johnston and James Dennis for a gna-2::gfp strain, Rueyling Lin for oma-1 and oma-2 strains and antibodies, and Pamela Silver for anti-GFP Ab. TS NIH grant GM63310 supported this work. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1188404.
Supplemental material is available at http://www.genesdev.org.
References
- Aronoff R., Baran, R., and Hodgkin, J. 2001. Molecular identification of smg-4, required for mRNA surveillance in C. elegans. Gene 268: 153-164. [DOI] [PubMed] [Google Scholar]
- Cali B.M. and Anderson, P. 1998. mRNA surveillance mitigates genetic dominance in Caenorhabditis elegans. Mol. Gen. Genet. 260: 176-184. [DOI] [PubMed] [Google Scholar]
- Clifford R., Lee, M.H., Nayak, S., Ohmachi, M., Giorgini, F., and Schedl, T. 2000. FOG-2, a novel F-box containing protein, associates with the GLD-1 RNA binding protein and directs male sex determination in the C. elegans hermaphrodite germline. Development 127: 5265-5276. [DOI] [PubMed] [Google Scholar]
- De Moor C.T. and Richter, J.D. 2001. Translational control in vertebrate development. In Cell lineage specification and patterning of the embryo (eds. L.D. Etkin and K.W. Jeon), pp. 567-608. Academic Press, San Diego. [DOI] [PubMed]
- Detwiler M.R., Reuben, M., Li, X., Rogers, E., and Lin, R. 2001. Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev. Cell 1: 187-199. [DOI] [PubMed] [Google Scholar]
- Di Fruscio M., Chen, T., Bonyadi, S., Lasko, P., and Richard, S. 1998. The identification of two Drosophila KH domain proteins: KEP1 and SAM are members of the Sam68 family of GSG domain proteins. J. Biol. Chem. 273: 30122-30130. [DOI] [PubMed] [Google Scholar]
- Francis R., Barton, M.K., Kimble, J., and Schedl, T. 1995a. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics 139: 579-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R., Maine, E., and Schedl, T. 1995b. Analysis of multiple roles of gld-1 in germline development: Interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics 139: 607-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geballe A.P. and Sachs, M.S. 2000. Translational control by upstream open reading frames. In Translational control of gene expression (eds. N. Sorenberg et al.), pp. 595-614. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Gibert M.A., Starck, J., and Beguet, B. 1984. Role of the gonad cytoplasmic core during oogenesis of the nematode Caenorhabditis elegans. Biol. Cell 50: 77-85. [DOI] [PubMed] [Google Scholar]
- Gibson T.J., Rice, P.M., Thompson, J.D., and Heringa, J. 1993. KH domains within the FMR1 sequence suggest that fragile X syndrome stems from a defect in RNA metabolism. Trends Biochem. Sci. 18: 331-333. [DOI] [PubMed] [Google Scholar]
- Goodwin E.B., Okkema, P.G., Evans, T.C., and Kimble, J. 1993. Translational regulation of tra-2 by its 3′ untranslated region controls sexual identity in C. elegans. Cell 75: 329-339. [DOI] [PubMed] [Google Scholar]
- Grant B. and Hirsh, D. 1999. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell 10: 4311-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D., Hubbard, E.J.A., and Schedl, T. 2004a. Multi-pathway control of the proliferation versus meiotic development decision in the C. elegans germ line. Dev. Biol. 268: 342-357. [DOI] [PubMed] [Google Scholar]
- Hansen D., Wilson-Berry, L., Dang, T., and Schedl, T. 2004b. Control of the proliferation versus meiotic development decision in the C. elegans germ line through regulation of GLD-1 protein accumulation. Development 131: 93-104. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. and Martinelli, S. 1999. 1999 genetic map of Caenorhabditis elegans. Caenorhabditis Genetics Center, St. Paul, MN.
- Hodgkin J., Papp, A., Pulak, R., Ambros, V., and Anderson, P. 1989. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123: 301-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu V., Zobel, C.L., Lambie, E.J., Schedl, T., and Kornfeld, K. 2002. Caenorhabditis elegans lin-45 raf is essential for larval viability, fertility and the induction of vulval cell fates. Genetics 160: 481-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A. and Peltz, S.W. 2000. Destabilization of nonsense-containing transcripts in Saccharomyces cerevisiae. In Translational control of gene expression (eds. N. Sorenberg et al.), pp. 827-848. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Jan E., Motzny, C.K., Graves, L.E., and Goodwin, E.B. 1999. The STAR protein, GLD-1, is a translational regulator of sexual identity in C. elegans. EMBO J. 18: 258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone O. and Lasko, P. 2001. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu. Rev. Genet. 35: 365-406. [DOI] [PubMed] [Google Scholar]
- Jones A.R. and Schedl, T. 1995. Mutations in gld-1, a female germ cell-specific tumor suppressor gene in Caenorhabditis elegans, affect a conserved domain also found in Src-associated protein Sam68. Genes & Dev. 9: 1491-1504. [DOI] [PubMed] [Google Scholar]
- Jones A.R., Francis, R., and Schedl, T. 1996. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev. Biol. 180: 165-183. [DOI] [PubMed] [Google Scholar]
- Kadyk L.C. and Kimble, J. 1998. Genetic regulation of entry into meiosis in C. elegans. Development 125: 1803-1813. [DOI] [PubMed] [Google Scholar]
- Kelly W.G., Schaner, C.E., Dernburg, A.F., Lee, M.H., Kim, S.K., Villeneuve, A.M., and Reinke, V. 2002. X-Chromosome silencing in the germline of C. elegans. Development 129: 479-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer B., Crittenden, S., Gallegos, M., Moulder, G., Barstead, R., Kimble, J., and Wickens, M. 1999. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr. Biol. 9: 1009-1018. [DOI] [PubMed] [Google Scholar]
- Kuersten S. and Goodwin, E.B. 2003. The power of the 3′ UTR: Translational control and development. Nat. Rev. Genet. 4: 626-637. [DOI] [PubMed] [Google Scholar]
- Lee M.H. and Schedl, T. 2001. Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes & Dev. 15: 2408-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis H.A., Musunuru, K., Jensen, K.B., Edo, C., Chen, H., Darnell, R.B., and Burley, S.K. 2000. Sequence-specific RNA binding by a Nova KH domain: Implications for paraneoplastic disease and the fragile X syndrome. Cell 100: 323-332. [DOI] [PubMed] [Google Scholar]
- Linz B., Koloteva, N., Vasilescu, S., and McCarthy, J.E. 1997. Disruption of ribosomal scanning on the 5′-untranslated region, and not restriction of translational initiation per se, modulates the stability of nonaberrant mRNAs in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 272: 9131-9140. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu, M.D., and Steitz, J.A. 2000. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103: 1121-1131. [DOI] [PubMed] [Google Scholar]
- Maquat L.E. 2000. Nonsense-mediated RNA decay in mammalian cells: A splicing-dependent means to down-regulate the level of mRNAs that prematurely terminate translation. In Translational control of gene expression (eds. N. Sorenberg et al.), pp. 849-868. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Maquat L.E. and Carmichael, G.G. 2001. Quality control of mRNA function. Cell 104: 173-176. [DOI] [PubMed] [Google Scholar]
- Marin V.A. and Evans, T.C. 2003. Translational repression of a C. elegans Notch mRNA by the STAR/KH domain protein GLD-1. Development 130: 2623-2632. [DOI] [PubMed] [Google Scholar]
- Meijer H.A. and Thomas, A.A. 2002. Control of eukaryotic protein synthesis by upstream open reading frames in the 5′-untranslated region of an mRNA. Biochem. J. 367: 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J.T., ap Rhys, C.M., and Dietz, H.C. 2002. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science 298: 419-422. [DOI] [PubMed] [Google Scholar]
- Mitrovich Q.M. and Anderson, P. 2000. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes & Dev. 14: 2173-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris D.R. and Geballe, A.P. 2000. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 20: 8635-8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison M., Harris, K.S., and Roth, M.B. 1997. smg mutants affect the expression of alternatively spliced SR protein mRNAs in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 94: 9782-9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musco G., Stier, G., Joseph, C., Castiglione Morelli, M.A., Nilges, M., Gibson, T.J., and Pastore, A. 1996. Three-dimensional structure and stability of the KH domain: Molecular insights into the fragile X syndrome. Cell 85: 237-245. [DOI] [PubMed] [Google Scholar]
- Nabel-Rosen H., Dorevitch, N., Reuveny, A., and Volk, T. 1999. The balance between two isoforms of the Drosophila RNA-binding protein how controls tendon cell differentiation. Mol. Cell 4: 573-584. [DOI] [PubMed] [Google Scholar]
- Okkema P.G., Harrison, S.W., Plunger, V., Aryana, A., and Fire, A. 1993. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 135: 385-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page M.F., Carr, B., Anders, K.R., Grimson, A., and Anderson, P. 1999. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol. 19: 5943-5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulak R. and Anderson, P. 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes & Dev. 7: 1885-1897. [DOI] [PubMed] [Google Scholar]
- Ruiz-Echevarria M.J., Czaplinski, K., and Peltz, S.W. 1996. Making sense of nonsense in yeast. Trends Biochem. Sci. 21: 433-438. [DOI] [PubMed] [Google Scholar]
- Schedl T. 1997. Developmental genetics of the germ line. In C. Elegans II (eds. D.L. Riddle et al.), pp. 241-269. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Schisa J.A., Pitt, J.N., and Priess, J.R. 2001. Analysis of RNA associated with P granules in germ cells of C. elegans adults. Development 128: 1287-1298. [DOI] [PubMed] [Google Scholar]
- Sulston J.E. and Hodgkin, J. 1988. Methods. In The nematode Caenorhabditis elegans (ed. W.B. Wood), pp. 587-606. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Vernet C. and Artzt, K. 1997. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 13: 479-484. [DOI] [PubMed] [Google Scholar]
- Vilela C. and McCarthy, J.E. 2003. Regulation of fungal gene expression via short open reading frames in the mRNA 5′-untranslated region. Mol. Microbiol. 49: 859-867. [DOI] [PubMed] [Google Scholar]
- Vilela C., Linz, B., Rodrigues-Pousada, C., and McCarthy, J.E. 1998. The yeast transcription factor genes YAP1 and YAP2 are subject to differential control at the levels of both translation and mRNA stability. Nucleic Acids Res. 26: 1150-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. and Lykke-Andersen, J. 2002. mRNA surveillance: The perfect persist. J. Cell Sci. 115: 3033-3038. [DOI] [PubMed] [Google Scholar]
- Wickens M., Goodwin, E.B., Kimble, J., Strickland, S., and Hentze, M.W. 2000. Translational control of developmental decisions. In Translational control of gene expression (eds. N. Sorenberg et al.), pp. 295-370. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Xu L., Paulsen, J., Yoo, Y., Goodwin, E.B., and Strome, S. 2001. Caenorhabditis elegans MES-3 is a target of GLD-1 and functions epigenetically in germline development. Genetics 159: 1007-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]