Abstract
Chondrogenesis is a multistep process that is essential for endochondral bone formation. Previous results have indicated a role for β-catenin and Wnt signaling in this pathway. Here we show the existence of physical and functional interactions between β-catenin and Sox9, a transcription factor that is required in successive steps of chondrogenesis. In vivo, either overexpression of Sox9 or inactivation of β-catenin in chondrocytes of mouse embryos produces a similar phenotype of dwarfism with decreased chondrocyte proliferation, delayed hypertrophic chondrocyte differentiation, and endochondral bone formation. Furthermore, either inactivation of Sox9 or stabilization of β-catenin in chondrocytes also produces a similar phenotype of severe chondrodysplasia. Sox9 markedly inhibits activation of β-catenin-dependent promoters and stimulates degradation of β-catenin by the ubiquitination/proteasome pathway. Likewise, Sox9 inhibits β-catenin-mediated secondary axis induction in Xenopus embryos. β-Catenin physically interacts through its Armadillo repeats with the C-terminal transactivation domain of Sox9. We hypothesize that the inhibitory activity of Sox9 is caused by its ability to compete with Tcf/Lef for binding to β-catenin, followed by degradation of β-catenin. Our results strongly suggest that chondrogenesis is controlled by interactions between Sox9 and the Wnt/β-catenin signaling pathway.
Keywords: Chondrocyte differentiation, Sox9, β-catenin
Chondrogenesis, an obligatory process in endochondral bone formation, starts with the recruitment of chondrogenic mesenchymal cells into condensations. This is followed by the differentiation of these cells into chondrocytes, which produce cartilage-specific extracellular matrix (ECM) proteins including type II collagen and the proteoglycan aggrecan. Chondrocytes then undergo a unidirectional proliferation to form orderly parallel columns, exit the cell cycle, become prehypertrophic, and then hypertrophic. Sox9, a high-mobility-group (HMG-box) transcription factor, is required at sequential steps in this pathway (Bi et al. 1999, 2001; Akiyama et al. 2002).
Both the human disease campomelic dysplasia, which is caused by heterozygous mutations in the Sox9 gene and is due to Sox9 haploinsufficiency, as well as Sox9 heterozygous mutant mice are characterized by a general hypoplasia of endochondral bones (Foster et al. 1994; Wagner et al. 1994). Inactivation of Sox9 in limb buds using the Cre recombinase/loxP recombination system before chondrogenic mesenchymal condensations results in the complete absence of mesenchymal condensations and of subsequent cartilage and bone formation, indicating that Sox9 is needed for an early step in chondrogenesis, that of mesenchymal condensations (Akiyama et al. 2002). A similar conclusion was also reached by analysis of mouse embryo chimeras derived from homozygous Sox9 mutant embryonic stem (ES) cells (Bi et al. 1999). That Sox9 is needed at sequential steps is shown by the severe generalized chondrodysplasia of mouse embryos in which Sox9 is deleted after chondrogenic mesenchymal condensations (Akiyama et al. 2002). The severe reduction of cartilage-specific matrix production and lack of proliferating chondrocytes in these mutants is in large part attributable to the absence of Sox5 and Sox6, which themselves are needed for overt chondrocyte differentiation but not mesenchymal condensations (Smits et al. 2001). Indeed, Sox9 is required for expression of Sox5 and Sox6 in all endochondral skeletal elements (Akiyama et al. 2002). Moreover, the presence of hypertrophic chondrocytes in embryos, in which Sox9 was inactivated after mesenchymal condensations, suggested the hypothesis that Sox9 inhibits the transition of proliferating chondrocytes into hypertrophic chondrocytes in the growth plate (Akiyama et al. 2002). This hypothesis is further supported by the existence of an enlarged zone of hypertrophic chondrocytes and premature mineralization of endochondral bones in heterozygous Sox9 mouse mutants (Bi et al. 2001). Previous in vitro studies have shown that Sox9 binds to and activates chondrocyte-specific enhancer elements in Col2a1, Col11a2, Aggrecan, and CD-RAP genes (Lefebvre et al. 1997; Bridgewater et al. 1998; Xie et al. 1999; Sekiya et al. 2000) and that L-Sox5 and Sox6 cooperate with Sox9 to activate the Col2a1and aggrecan genes (Lefebvre et al. 1998). However, how Sox9 regulates chondrocyte proliferation and differentiation remains unknown.
β-Catenin is a cytoplasmic protein that functions in cell-cell adhesion by interacting with cadherins. N-Cadherin is believed to have a major role in cell-cell interactions during chondrogenic mesenchymal condensations, participating in the assembly of a cadherin-catenin-actin adhesion complex (Oberlender and Tuan 1994; Delise and Tuan 2002). β-Catenin is also a downstream intracellular signaling molecule in the canonical Wnt pathway, which controls cell differentiation, cell proliferation, and cell migration in many aspects of embryonic development (Huelsken and Birchmeier 2001; Moon et al. 2002). In the absence of Wnt signals, cytoplasmic β-catenin is constitutively phosphorylated at its N terminus and efficiently degraded by the ubiquitination/26S proteasome pathway. Canonical Wnt signaling inhibits phosphorylation of β-catenin, resulting in its stabilization, cytoplasmic accumulation, and translocation into the nucleus, where it interacts with members of the T-cell-factor (Tcf)/lymphoid-enhancer-factor (Lef) HMG-box family to activate target genes. Recent studies have shown that forced expression of β-catenin in prechondrogenic cells in vitro inhibits overt chondrocyte differentiation, whereas its overexpression in chick limb buds accelerates hypertrophic conversion of proliferating chondrocytes (Hartmann and Tabin 2000; Ryu et al. 2002). These results, that is, inhibition of overt chondrocyte differentiation and premature hypertrophic chondrocyte differentiation, are analogous to the phenotype of mouse embryos in which Sox9 is inactivated during and after mesenchymal condensations (Akiyama et al. 2002). In addition, a previous study showed that Sox3 and Sox17 bind to β-catenin and inhibit Tcf/Lef-mediated signaling activity of β-catenin (Zorn et al. 1999). Thus, these results raise the possibility of functional interactions between Sox9 and β-catenin during chondrocyte differentiation.
To clarify the roles of Sox9 during chondrocyte differentiation, we generated mutant mice in which Sox9 is overexpressed in chondrocytes. This overexpression results in a generalized chondrodysplasia characterized by inhibition of cell proliferation and differentiation of chondrocytes. These phenotypic changes are similar to those we found in mice in which β-catenin is inactivated in chondrocytes. Conversely, activation of a dominant stable β-catenin mutation in chondrocytes results in a similar phenotype as that of embryos in which the Sox9 alleles are inactivated in chondrocytes (Akiyama et al. 2002). Hence, the physical and functional interactions between Sox9 and β-catenin that we identified provide evidence that chondrocyte differentiation, proliferation, and maturation to hypertrophy are controlled by crosstalk between Sox9 and the canonical Wnt pathway.
Results
Generation of mutant mice overexpressing Sox9
Type II collagen is highly expressed in all chondrocytes. The Col2a1-null mutant mice exhibit a severe chondrodysplasia and die perinatally, whereas the Col2a1 heterozygous mutant mice have essentially no skeletal phenotype (Li et al. 1995). To analyze the function of Sox9 during chondrocyte differentiation in vivo, we inserted a 3xHA-tagged Sox9 cDNA into one Col2a1 allele and generated mice in which all chondrocytes overexpressed Sox9. Using the mating scheme described in the Materials and Methods section, we produced animals that were heterozygous for Col2a1/Sox9 (Fig. 1A). Because we used ES cells harboring Protamine 1-Cre transgenes (O'Gorman et al. 1997), the floxed Pgk-neo-bpA cassette was removed in the germ cells of chimeras (Fig. 1A-C). Indeed, more than 95% of newborn heterozygous mutant mice no longer carried the Pgk-neo-bpA cassette.
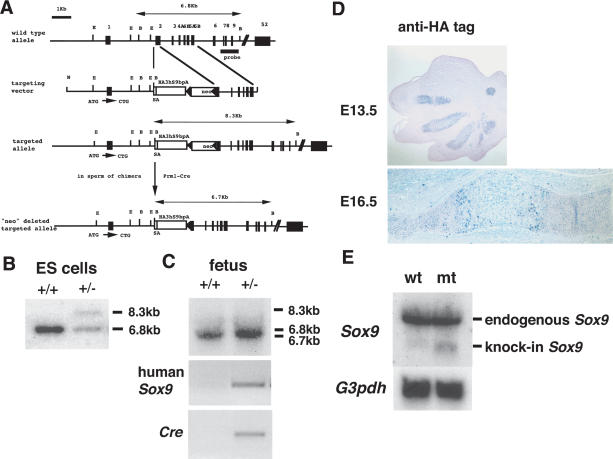
Figure 1.
Targeting strategy for insertion of Sox9 cDNA in the Col2a1 locus. (A) Structure of the genomic Col2a1 locus, targeting vector, and targeted allele. Exons are depicted as closed boxes, and intronic sequences are shown as solid lines. DNA fragments revealed in Southern blot analysis are indicated by double arrows. (B) BamHI; (E) EcoRI; (N) NotI. (B,C) Southern blot analysis of ES cells and fetal genomic DNA. Genomic DNA isolated from ES cells or the skin of wild-type and Col2a1/Sox9 knock-in mutants is digested with BamHI and then hybridized with the 3′ probe. The wild-type, mutant, and mutant alleles without neo are detected as 6.8-kb, 8.3-kb, and 6.7-kb fragments, respectively. (D) Expression of 3xHA-tagged Sox9 protein in limb buds of E13.5 and E16.5 Col2a1/Sox9 knock-in mutants. (E) Expression of 3xHA-tagged Sox9 mRNA in cartilage of wild-type and Col2a1/Sox9 knock-in mutant newborn mice using a mouse Sox9 cDNA probe. The same result occurs with a human Sox9 cDNA probe (data not shown).
We assessed the onset and levels of expression of knocked-in Sox9 by immunohistochemistry with antibody to HA epitope tag. The expression of knocked-in Sox9 was detectable, although at a low level, in limb bud mesenchymal cells before chondrogenic mesenchymal condensations appeared in embryonic day 11.5 (E11.5; data not shown). Expression of 3xHA-tagged Sox9 was detected in condensed mesenchymal cells and in differentiated chondrocytes except hypertrophic chondrocytes (Fig. 1D). Northern blot analysis using total RNA purified from rib cartilages of newborn mice revealed that the level of knocked-in Sox9 gene expression was ∼20% of that of endogenous Sox9 gene expression (Fig. 1E).
Delay in endochondral bone formation in Sox9 knock-in heterozygous mutants
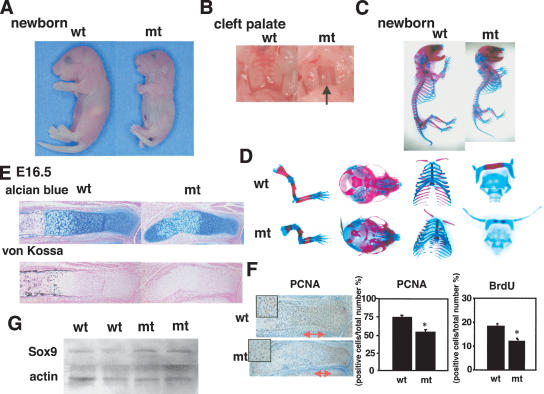
Of the Sox9 knock-in heterozygous mutant mice, 80% died in the immediate postnatal period from respiratory distress, probably caused by a cleft secondary palate and a small rib cage (Fig. 2A-C). Only 20% were viable and fertile, perhaps because of a much smaller cleft secondary palate in these animals, but their growth was retarded (Fig. 5A, below).
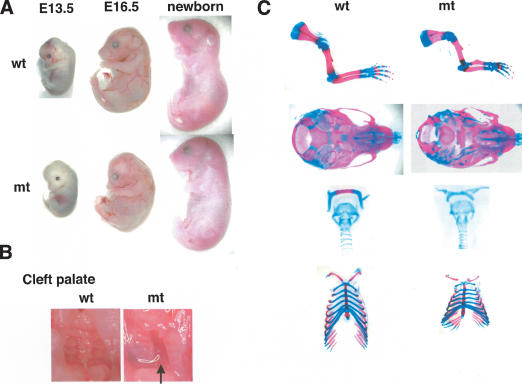
Figure 2.
Analysis of skeletal phenotypes in Col2a1/Sox9 knock-in mutant embryos. (A) Gross appearance of E13.5 and E16.5 embryos and newborn mice. (B) A cleft secondary palate in mutant newborn mice (the arrow). (C) Skeletons of newborn mice stained by alcian blue followed by alizarin red. Cartilage and endochondral bones in the mutant are hypoplastic.
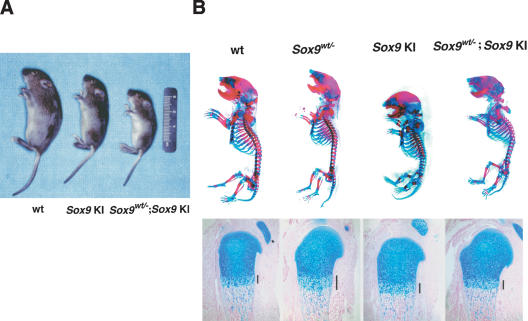
Figure 5.
Rescue of Sox9 heterozygous mice with Col2a1/Sox9 knock-in mutant mice. (A) Appearance of 14-day-old mice. (B) Skeletons of newborn mice stained with alcian blue followed by alizarin red. In Sox9wt/-; Sox9 knock-in (KI) double mutants, the chondrodysplasia is partially corrected. Alcian blue staining of humerus shows the reduction of width of the hypertrophic zone (the bars) in Sox9wt/-; Sox9 knock-in (KI) double mutants, compared with that in Sox9wt/-.
During embryogenesis, we could not detect any difference between E13.5 wild-type and mutant embryos (Fig. 2A). However, from E16.5 on, mutant embryos exhibited craniofacial deformities characterized by a domed skull and a short snout, as well as short limbs (Fig. 2A). A few newborn mutants displayed a straight back, lacking the physiological curvature of the spine (Fig. 2A). Staining of skeletal preparations from newborn mutant mice with alcian blue and alizarin red revealed that all skeletal elements derived by endochondral bone formation were shorter (Figs. 2C, 5B [below]). Especially, the radius was markedly hypoplastic and angulated, and the smallness of the rib cage was obvious. In severely affected animals, the interlaminar joints of the cervical and thoracic vertebrae were fused to each other (data not shown). In the mutants, the cartilages of the skull base and the hyoids were formed normally but showed markedly delayed maturation of the cartilage and subsequent ossification (Fig. 2C).
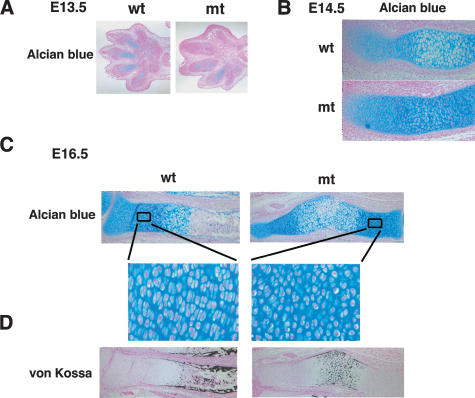
Histological analysis showed that chondrogenic mesenchymal condensations in the mutants were comparable with those in wild-type embryos (Fig. 3A). In the radius of wild-type embryos, chondrocytes matured and became hypertrophied in the center of cartilage anlagen in E14.5 (Fig. 3B). The bone collars were formed and calcified cartilages were replaced by bones in E16.5 (Fig. 3C). In contrast, in the radius of the mutants, chondrocytes had not become hypertrophic in E14.5 (Fig. 3B). In E16.5, the size of the radius was much reduced, compared with that in wild-type embryos (Fig. 3C). Hypertrophic chondrocytes were formed, but bone collar formation and cartilage replacement by bones were markedly delayed (Fig. 3C,D). Furthermore, in contrast to wild-type embryos, proliferating chondrocytes were small and round, and the orderly parallel columnar structures were not observed in the growth plates of the mutants (Fig. 3C).
Figure 3.
Histological analysis of limbs in Col2a1/Sox9 knock-in mutant embryos. (A) Alcian blue staining of limb buds in E13.5 embryos. Chondrogenic mesenchymal condensations normally occur in the mutants. (B) Alcian blue staining of radius in E14.5 embryos. No hypertrophic chondrocytes are seen in mutant embryos. (C) Alcian blue staining of radius in E16.5 embryos shows delay of endochondral bone formation in the mutants. Boxed regions are shown at a higher magnification. (D) Staining by von Kossa's method visualizes mineral deposition of radius in E16.5 wild-type and mutant embryos.
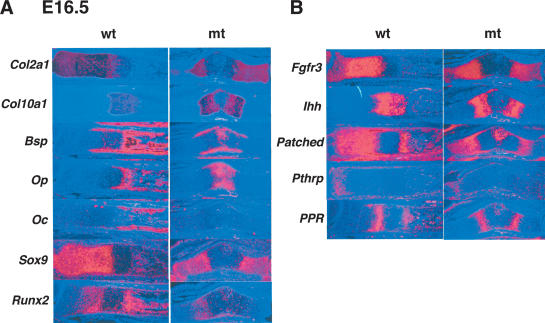
To understand the delay of endochondral bone formation, we analyzed the expression of several marker genes of chondrogenesis and osteogenesis by in situ hybridization (Fig. 4A). In E16.5 mutant embryos, Col2a1 and Col10a1, markers of chondrocytes and hypertrophic chondrocytes, respectively, were expressed normally. Runx2 was expressed in chondrocytes and bone collar, and Bone sialoprotein (Bsp) was expressed in bone collar of the mutants. However, Osteopontin (Op) and Osteocalcin (Oc) were not expressed in the bone collar of the mutants at this stage, indicating that osteoblast differentiation occurred during endochondral bone formation but was delayed.
Figure 4.
Expression of chondrocyte and osteoblast markers and signaling molecules in Col2a1/Sox9 knock-in mutant embryos. Sections of limb buds of E16.5 wild-type and mutant embryos are hybridized with Col2a1, Col10a1, Bsp, Op, Oc, Sox9, and Runx2 probes (A); and Fgfr3, Ihh, Patched, Pthrp, and PPR probes (B).
No change in expression of several signaling molecules in Sox9 knock-in mutant embryos
Indian hedgehog (Ihh) and Parathyroid hormone-related peptide (Pthrp) as well as Fibroblast growth factor receptor (Fgfr) 3 signaling control the rate of chondrocyte proliferation and the transition of proliferating chondrocytes to hypertrophy (Vortkamp et al. 1996; Ornitz and Marie 2002). Ihh also coordinates these processes with the onset of osteogenesis in bone collars (St-Jacques et al. 1999). Figure 4B shows that in E16.5 mutant radius, Ihh, Pthrp, Pthrp receptor (PPR), Patched, and Fgfr3 were each expressed normally. Thus, the delay of endochondral bone formation in the mutant embryos was not caused by the alteration of expression of these signaling molecules.
Phenotypic rescue of the Sox9 heterozygous mutant mice by Sox9 knock-in mutant mice
Sox9 heterozygous mutants die shortly after birth from respiratory distress (Bi et al. 2001). These mutants exhibit hypoplasia of all skeletal elements derived by endochondral bone formation, which is caused by haploinsufficiency of Sox9 (Fig. 5B). We crossed Sox9 heterozygous mutant male chimeras, which we previously reported (Bi et al. 1999), and Sox9 knock-in mutant female mice that survived after birth and then generated Sox9 heterozygous mutant mice carrying the Col2a1/Sox9 knock-in gene. Only 30% of Sox9 heterozygous mutant mice carrying the Col2a1/Sox9 knock-in gene survived after birth. At 14 d after birth, these rescued mutant mice were very small, compared with wild-type littermates (Fig. 5A), but, unlike the Sox9 heterozygous mutants, did not have a cleft secondary palate (data not shown). Staining of skeletal preparations from rescued newborn mutant mice with alcian blue and alizarin red revealed that, although they still exhibited a mild hypoplasia of skeletal elements, the bending of long bones, deformity of scapula and pelvis, and small rib cage were corrected (Fig. 5B). In Sox9 heterozygous mutant mice, the zone of hypertrophic chondrocytes was wider than in wild-type embryos (Bi et al. 2001). Histological analysis showed a normal width of the hypertrophic zone in the rescued mice (Fig. 5B). These results indicate that the additional small amounts of Sox9 can partially rescue the chondrodysplasia, characteristics of Sox9 heterozygous mutants.
Reduced chondrocyte proliferation and Cyclin D1 expression in Sox9 knock-in mutant embryos
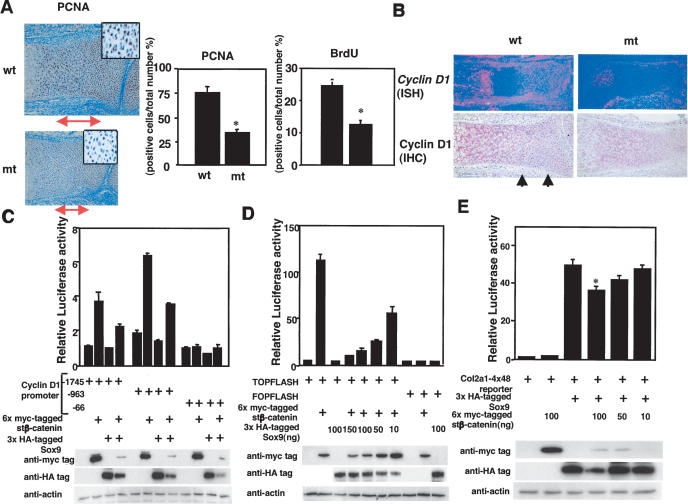
The delay of endochondral bone formation in Sox9 knock-in mutant embryos could have been caused by a reduction of chondrocyte proliferation. To test this hypothesis, we performed staining of proliferative cell nuclear antigen (PCNA), a good marker for proliferating chondrocytes, in the radius of E16.5 (Fig. 6A). PCNA was abundantly positive in columnar chondrocytes of wild-type growth plates. In contrast, in the mutant growth plates, the number of PCNA-positive chondrocytes was reduced to 50% of that in wild-type growth plates. BrdU in vivo pulse-labeling also revealed a marked reduction (50% reduction) in cell proliferation of proliferating chondrocytes in mutant embryos compared with their wild-type siblings (Fig. 6A), indicating that proliferation of mutant chondrocytes was inhibited.
Figure 6.
Inhibition of cell proliferation and Cyclin D1 expression in Col2a1/Sox9 knock-in mutant embryos, and negative functional interactions between Sox9 and β-catenin in vitro. (A) PCNA staining shows a decrease in PCNA-positive cells (brown nuclei) in the radius of E16.5 mutant embryos. The double arrows indicate the zone of proliferating chondrocytes. Boxed regions show a higher magnification of proliferating chondrocytes. BrdU incorporation also decreases in the radius of E16.5 mutant embryos. Statistical significance is assessed by one-way analysis of variance and unpaired Student's t-test. (*) Statistically significant difference between wild-type and mutant embryos at p < 0.001. (B) Expression of Cyclin D1 mRNA (in situ hybridization; ISH) and protein (immunohistochemistry; IHC) in the radius of E16.5 wild-type and mutant embryos. The arrows indicate Cyclin D1-positive proliferating chondrocytes. (C) Activation of the Cyclin D1 promoter by stβ-catenin is inhibited by Sox9 in 293 cells. Cells are cotransfected with 6x myc-tagged stβ-catenin and 3xHA-tagged Sox9. Activity of the Cyclin D1 promoter is measured by luciferase assay. Cell extracts are fractionated by electrophoresis and blotted with anti-myc and anti-HA antibodies. Cotransfection of Sox9 and β-catenin results in a reduction of the levels of both proteins. (D) Activation of TOPFLASH by stβ-catenin is inhibited by Sox9. TOPFLASH and FOPFLASH activities are measured by luciferase assay. Cotransfection of 6x myc-tagged stβ-catenin and 3xHA-tagged Sox9 in 293 cells results in a reduction of the levels of these proteins in a dose-dependent manner. (E) Activation of the Col2a1/4 × 48-bp enhancer reporter construct was inhibited by stβ-catenin in C3H10T1/2 cells. Cotransfection of 6x myc-tagged stβ-catenin and 3xHA-tagged Sox9 results in a reduction of the levels of these proteins in a dose-dependent manner. Statistical significance is assessed by one-way analysis of variance and unpaired Student's t-test. (*) Statistically significant difference between Sox9 transfection and Sox9 and stβ-catenin cotransfection at p < 0.001.
Cyclin D1 is expressed specifically in proliferating chondrocytes of growth plates and is required for their normal proliferation (Beier et al. 2001; Long et al. 2001). In the growth plates of E16.5 Sox9 knock-in mutants, the expression of Cyclin D1 mRNA and protein was severely reduced (Fig. 6B), indicating that reduction of chondrocyte proliferation in Sox9 knock-in mutant embryos was partly caused by the inhibition of Cyclin D1 expression.
Sox9 represses β-catenin-mediated transcriptional activity and β-catenin inhibits Sox9 transcriptional activity
β-Catenin interacts with Tcf/Lef family transcription factors, which are sequence-specific DNA-binding proteins that contain a single HMG box (Clevers and van de Wetering 1997). The β-catenin/Tcf complex binds the Tcf/Lef consensus site in the Cyclin D1 promoter region and transactivates the Cyclin D1 gene (Tetsu and McCormick 1999). To determine whether Sox9 modulates the transcription of Cyclin D1 by the β-catenin/Tcf complex, we transfected luciferase reporter genes into human embryonic kidney 293 cells, which endogenously express Lef1. The Cyclin D1 promoter plasmids, -1745CD1 and -963CD1, contain five potential Tcf-binding sites, whereas -66CD1 has no Tcf-binding site (Tetsu and McCormick 1999). Figure 6C shows that both -1745CD1 and -963CD1 but not -66CD1 reporters were strongly activated in response to stβ-catenin and that Sox9 inhibited this activity. We also transfected an artificial luciferase reporter construct, TOPFLASH, which contains multimeric Tcf/Lef1-binding sites upstream of the c-fos promoter, versus the negative control FOPFLASH, which harbors mutated Tcf-binding sites (Korinek et al. 1997). stβ-Catenin strongly induced the transcription of TOPFLASH, not of FOPFLASH, and Sox9 inhibited the transcriptional activity of β-catenin in a dose-dependent manner (Fig. 6D).
The inhibition of β-catenin activity by Sox9 raises the possibility that β-catenin could modulate the activity of Sox9. To test this hypothesis, we transfected a Col2a1 promoter/4 × 48-bp enhancer reporter construct into mouse embryonic C3H10T1/2 fibroblast cells. Sox9 activated the chondrocyte-specific Col2a1 promoter/4 × 48-bp enhancer reporter construct (Fig. 6E) as well as a Col2a1 promoter/2 × 231-bp enhancer reporter construct (data not shown). β-Catenin significantly inhibited this activity of Sox9 in a dose-dependent manner (Fig. 6E).
Sox9 does not bind a Tcf consensus DNA-binding site, and Tcf does not bind a chondrocyte-specific enhancer
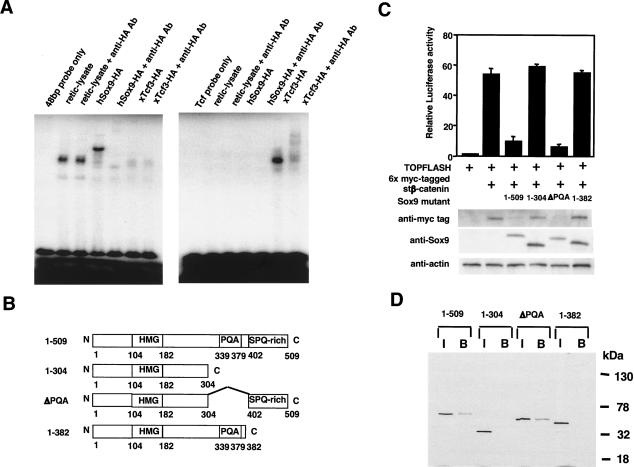
The Sox and Tcf/Lef protein families contain well-conserved DNA-binding HMG domains that recognize similar consensus sequences (Clevers and van de Wetering 1997). To determine if the inhibition by Sox9 of β-catenin transcriptional activity was mediated through competition between Sox9 and Tcf/Lef proteins for binding to Tcf/Lef DNA-binding sites, we examined the binding specificity of Sox9 and Tcf/Lef by EMSA (Fig. 7A). 3xHA-tagged xTcf3 bound to a DNA probe containing a Tcf/Lef consensus sequence, and antibodies directed against HA tag supershifted xTcf3-probe complexes. In contrast, Sox9 did not bind to the Tcf/Lef probe. 3xHA-tagged Sox9 bound a consensus sequence within the chondrocyte-specific 48-bp enhancer of the Col2a1 gene, and anti-HA antibodies supershifted the complex. xTcf3 did not bind to this probe. Our data indicate that Sox9 inhibited transcriptional activities of β-catenin and that this inhibition did not result from competition by Sox9 with Tcf/Lef for Tcf/Lef DNA-binding sites.
Figure 7.
Functional and structural analysis of the interactions between Sox9 and β-catenin. (A) EMSAs performed using in vitro translated 3xHA-tagged Sox9 and 3xHA-tagged xTcf-3 proteins show that Sox9 does not bind to Tcf/Lef DNA-binding sites and that Tcf does not bind to Sox9 consensus DNA-binding sites. (B) Schematic representation of the Sox9 deletion mutants. (C) Activation of TOPFLASH by stβ-catenin is inhibited by Sox9 mutants containing the C-terminal transactivation domain. Cotransfection of 6x myc-tagged stβ-catenin and Sox9 mutants containing the C-terminal transactivation domain results in a reduction of the levels of these proteins. (D) In vitro binding of Sox9 deletion mutants to 6xHis-tagged β-catenin bound to a nickel-resin. (I) Input; (B) bound to resin containing 6xHis-tagged β-catenin.
The C-terminal transactivation domain of Sox9 binds Armadillo repeats of β-catenin
To further investigate how Sox9 inhibits β-catenin/Tcf activity, we mapped the site of Sox9 necessary for inhibition of β-catenin transcriptional activity. Reporter analyses using cotransfection of the TOPFLASH reporter gene, stβ-catenin, and Sox9 deletion constructs (Fig. 7B) revealed that the C-terminal transactivation domain of Sox9 was needed for its inhibitory activity on β-catenin (Fig. 7C). In vitro binding assays showed that deletion mutants of Sox9 that lacked the C-terminal transactivation domain failed to interact with β-catenin (Fig. 7D). Thus, the C-terminal transactivation domain of Sox9 bound β-catenin, the same region required for inhibition of β-catenin activity.
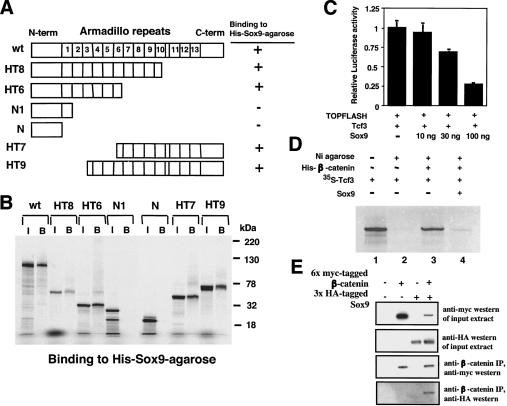
Next, we mapped the Sox9-binding site in β-catenin. Truncated 35S-labeled β-catenin proteins (Fig. 8A; Fagotto et al. 1996; Martin et al. 2002) were incubated with nickel-agarose coupled with 6xHis-tagged Sox9 recombinant protein. We found that Sox9 was able to bind within the segment of Armadillo repeats 4-10 of β-catenin. This segment overlaps with the Tcf-binding site in β-catenin (Fig. 8B; Behrens et al. 1996; Bienz and Clevers 2003). Because Sox9 inhibited the activity of Tcf using the TOPFLASH reporter in a dose-dependent manner (Fig. 8C), and because Sox9 also inhibited binding of Tcf to β-catenin in vitro (Fig. 8D), our results suggest that Sox9 and Tcf/Lef compete for overlapping sites in β-catenin.
Figure 8.
Functional and structural analysis of the interactions between Sox9 and β-catenin. (A) Schematic representation of the β-catenin deletion mutants. (B) In vitro binding of β-catenin deletion mutants to 6xHis-tagged Sox9 bound to a nickel-resin. (I) Input; (B) bound to resin containing 6xHis-tagged Sox9. (C) Activation of TOPFLASH in C3H10T1/2 cells by Tcf3 is inhibited by Sox9 in a dose-dependent manner. (D) Sox9 inhibits binding of Tcf3 to β-catenin in vitro. Sox9 and 35S-labeled Tcf3 proteins are incubated with 200 ng of 6xHis-tagged β-catenin immobilized to a nickel-resin. Of the input Tcf3 protein (lane 1) and bound Tcf3 proteins (lanes 2-4), 10% are resolved on SDS-PAGE. (E) Binding between Sox9 and β-catenin in Cos-7 cells. Cos-7 cells are cotransfected with 6x myc-tagged stβ-catenin and 3xHA-tagged Sox9 for 24 h. Cell lysates are incubated with anti-β-catenin antibody and protein G-agarose beads under gentle agitation for 3 h at 4°C. The beads are washed three times with buffer (50 mM Tris at pH 8.0, 150 mM NaCl, 1% Triton X-100), resuspended in 10 μL of SDS-polyacrylamide gel electrophoresis sample loading buffer, and boiled for 2 min. Western immunoblotting is then performed. Anti-HA:HRP monoclonal antibody (Roche) and anti-myc:HRP antibody (Invitrogen) are diluted 1:1000 and 1:5000, respectively.
To confirm the interaction between Sox9 and β-catenin in intact cells, coimmunoprecipitation experiments were performed with 6x myc-tagged stβ-catenin and 3xHA-tagged Sox9. Cos-7 cells were transfected with 3xHA-tagged Sox9 and 6x myc-tagged stβ-catenin expression vectors. β-Catenin was immunoprecipitated from the cell lysates, and the immunoprecipitates were then subjected to Western blot analysis with anti-HA antibody. As shown in Figure 8E, 3xHA-tagged Sox9 co-precipitated with 6x myc-tagged stβ-catenin, indicating that Sox9 and β-catenin, indeed, formed a physical complex.
Sox9 inhibits β-catenin-induced secondary axis formation in Xenopus embryos
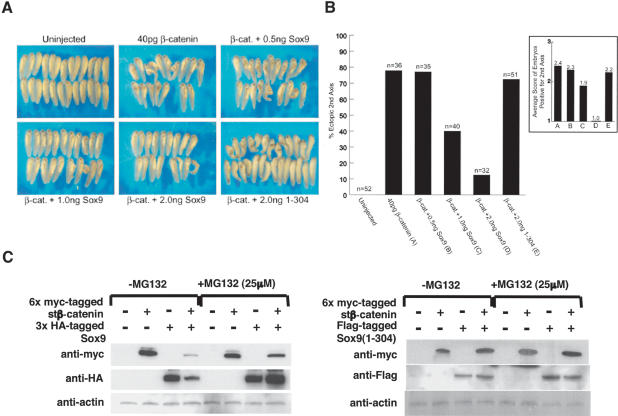
To obtain direct evidence of inhibitory effects of Sox9 on β-catenin activity in vivo, we performed the secondary axis assay in Xenopus laevis embryos. As expected, microinjection of 40 pg of wild-type β-catenin RNA alone into a single ventral-vegetal blastomere (four-cell stage) produced secondary axes in a large percentage of injected embryos (77.8%; Fig. 9A). Coinjection of Sox9 with β-catenin RNA significantly suppressed β-catenin-mediated secondary axis formation in a dose-dependent manner. Whereas the coinjection of 0.5 ng of Sox9 with β-catenin did not significantly suppress secondary-axis formation (77.1%), 1.0 ng of Sox9 inhibited secondary-axis formation to a notable extent (40.0%), and 2.0 ng of Sox9 resulted in a near complete suppression of duplicate axes (12.5%). In contrast, 2.0 ng of Sox9 (1-304) mutant, which lacks the transactivation domain, did not inhibit secondary-axis formation (72.5%). To further characterize the extent of secondary-axis formation, a value of 1 = weak, 2 = moderate, or 3 = strong was assigned to each embryo positive for a second axis phenotype (Fig. 9B). This semiquantitative analysis revealed that Sox9, but not Sox9 (1-304), suppressed the strength of β-catenin-mediated secondary-axis formation in a dose-dependent manner.
Figure 9.
(A,B) Sox9 inhibits β-catenin-mediated secondary-axis formation in Xenopus embryos. (A) Representative tail bud stage Xenopus embryos injected with the indicated RNAs into a single ventral-vegetal blastomere at the four-cell stage. (B) Summary of second axis assays from two separate experiments. Injection of mRNA encoding wild-type β-catenin (40 pg) results in a high frequency (77.8%) of embryos with secondary axes. Coinjection of increasing levels of Sox9 mRNA (0.5, 1.0, and 2.0 ng) with β-catenin (40 pg) results in a dose-dependent inhibition of β-catenin-induced secondary-axis formation. Sox9 (1-304) mRNA (2.0 ng) does not inhibit β-catenin-induced secondary-axis formation. Semiquantitative analysis of each embryo positive for a secondary axis (1 = weak, 2 = moderate, 3 = strong) further indicates that β-catenin-mediated secondary-axis formation is inhibited by increasing levels of Sox9, but not by Sox9 (1-304). (C) Proteasome inhibitor, MG132, restores the levels of β-catenin and Sox9 in 293 cells transfected with 6x myc-tagged stβ-catenin and 3xHA-tagged Sox9. Mutant Sox9 (1-304) does not decrease the levels of β-catenin, and MG132 has no effect on the levels of either β-catenin or Sox9 in these experiments.
Sox9 and β-catenin induce degradation of β-catenin and Sox9, respectively
To determine how Sox9 inhibits β-catenin signaling activity, we examined the levels of expression of 3xHA-tagged Sox9 and 6x myc-tagged stβ-catenin proteins by Western blot analysis. In cotransfection experiments with 3xHA-tagged Sox9 and 6x myc-tagged stβ-catenin, the levels of both proteins was markedly reduced (Figs. 6C-E, 9C). The reduction in β-catenin levels could be restored by treatment for 4 h with the proteasome inhibitor, MG132, 24 h after cotransfection (Fig. 9C). In contrast, deletion mutants of Sox9 that lacked the C-terminal transactivation domain did not reduce the level of β-catenin (Figs. 7C, 9C), and MG132 had no effects on this level (Fig. 9C). The level of this Sox9 mutant was also unchanged in cotransfection experiments with β-catenin, and MG132 had no effects on the level of the Sox9 mutant (Fig. 9C). These results strongly suggest that Sox9:β-catenin complex formation results in their mutual destruction by the ubiquitination/26S proteasome pathway.
Sox9 knock-in mutants and β-cateninflox/flox; Col2a1-Cre have similar phenotypes
To support our hypothesis that Sox9 and β-catenin inhibit their activities mutually in chondrocytes, we generated conditional β-catenin-null mutant mice using Col2a1-Cre transgenes to delete β-catenin genes in chondrocytes and compared the phenotypes of these mice with Sox9 knock-in mutants. β-cateninflox/flox; Col2a1-Cre mutants died at birth from respiratory distress caused by a large cleft secondary palate and a small rib cage (Fig. 10A,B). The mutant newborns exhibited craniofacial deformities characterized by a domed skull and a short snout, as well as short limbs (Fig. 10A). Staining of skeletal preparations from newborn mutant mice with alcian blue and alizarin red revealed that all skeletal elements derived by endochondral bone formation were shorter and very hypoplastic (Fig. 10C). In the mutants, the cartilages of the skull base, limbs, rib cage, and the hyoids showed severely delayed endochondral bone formation (Fig. 10D).
Figure 10.
Analysis of skeletal phenotypes in conditional β-catenin-null mutants with the Col2a1-Cre transgene. (A) Gross appearance of newborn mice. (B) A cleft secondary palate in mutant newborn mice (the arrow). (C,D) Skeletons of newborn mice stained by alcian blue followed by alizarin red. (E) Alcian blue staining of the radius in E16.5 mice shows delay of endochondral bone formation in the mutant embryos. Staining by von Kossa's method shows no mineral deposition of the radius in E16.5 mutant embryos. (F) PCNA staining shows a decrease in PCNA-positive cells (brown nuclei) of the radius in E16.5 mutant embryos. The double arrows indicate the zone of proliferating chondrocytes. Boxed regions show a higher magnification of proliferating chondrocytes. BrdU incorporation also decreases in the radius in E16.5 mutant embryos. Statistical significance is assessed by one-way analysis of variance and unpaired Student's t-test. (*) Statistically significant difference between wild-type and mutant embryos at p < 0.001. (G) Rib cartilages of E19.0 wild-type or β-cateninflox/flox; Col2a1-Cre mutant embryos are lysed in RIPA buffer, and the Sox9 proteins are analyzed by Western blot using polyclonal anti-Sox9 antibody.
Whole-mount alcian blue staining showed that chondrogenic mesenchymal condensations in E13.5 mutants were comparable with those in wild-type embryos (data not shown). Histological analysis showed that in the radius of mutants, chondrocytes had not become hypertrophic in E14.5 (data not shown). In E16.5 mutants, the size of the radius was much reduced, compared with that in wild-type embryos (Fig. 10E). Hypertrophic chondrocytes were formed, but bone collar formation and cartilage replacement by bones had not occurred yet (Fig. 10E). PCNA staining showed that in E16.5 mutant growth plates, the number of PCNA-positive chondrocytes was reduced to 70% of that in wild-type growth plates (Fig. 10F). BrdU in vivo pulse-labeling also revealed a marked reduction (34% reduction) in proliferation of proliferating chondrocytes in mutant embryos compared with their wild-type siblings (Fig. 10F), indicating that proliferation of mutant chondrocytes was inhibited. Thus, these lines of evidence indicate that Sox9 knock-in mutants and β-cateninflox/flox; Col2a1-Cre mutants have very similar phenotypes. In addition, Western analysis showed detectable increase of the level of Sox9 in chondrocytes of β-cateninflox/flox; Col2a1-Cre mutants (Fig. 10G), which may contribute to the skeletal phenotype of β-cateninflox/flox; Col2a1-Cre mutants.
Activation of a β-catenin dominant stable mutation in chondrocytes and inactivation of Sox9 in chondrocytes result in similar phenotypes in vivo
As increased expression of Sox9 in chondrocytes results in a similar phenotype to that produced by inactivation of β-catenin in chondrocytes, we asked whether increased β-catenin signals in chondrocytes may produce a similar phenotype to inactivation of Sox9 in chondrocytes (Akiyama et al. 2002). Constitutively active β-catenin can be produced in mice by activating a gain-of-function mutation in the β-catenin gene by using the Cre/loxP system (Harada et al. 1999). This conditional mutation of β-catenin was activated in chondrocytes by using Col2a1-Cre transgenic mice.
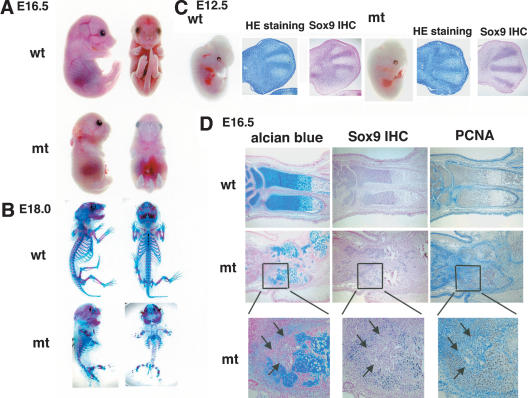
The stable β-catenin mutants (β-catenin+/flox(ex3); Col2a1-Cre) died around E18.0-E18.5. The mutant embryos were characterized by a very severe and generalized chondrodysplasia, very similar to that in Sox9flox/flox; Col2a1-Cre (Fig. 11A; Akiyama et al. 2002). Alcian blue and alizarin red staining of skeletal preparations of E18.0 mutant embryos indicated that all the skeletal elements formed by endochondral bone formation, including ribs, limbs, and vertebrae, were extremely small (Fig. 11B). Alcian blue staining was severely reduced, indicating that cartilage formation was markedly defective.
Figure 11.
Severe, generalized chondrodysplasia in β-catenin+/flox(ex3); Col2a1-Cre embryos. (A) Gross appearance of E16.5 embryos. (B) Skeletons of E18.0 embryos stained by alcian blue followed by alizarin red. Mutant embryos are characterized by a very severe and generalized chondrodysplasia. (C) Gross appearance of E12.5 mutant embryos is comparable to that of wild-type littermates. Histological analysis of limb buds stained by hematoxylin and Treosin at 12.5 dpc. Both wild-type and mutant limb buds have discernible chondrogenic mesenchymal condensations. Immunohistochemistry shows comparable expression of Sox9 protein in condensed mesenchymal cells in E12.5 wild-type and mutant embryos. (D) Alcian blue and nuclear fast-red staining, PCNA staining, and immunohistochemistry of Sox9 protein in ulna of E16.5 wild-type and mutant embryos, respectively. Only cells surrounded by alcian blue-stainable matrix are PCNA-positive and express Sox9. Small round cells (arrows) lack expression of Sox9.
Histological analyses showed that in E12.5 mutants chondrogenic mesenchymal condensations took place normally and that expression of Sox9 was comparable to that in condensations of wild-type littermates (Fig. 11C). Figure 11D shows that in E16.5 mutants, the size of the ulna was much reduced, compared with wild-type ulna. Most cells did not undergo overt differentiation into chondrocytes. Furthermore, in contrast to wild-type skeletal elements, the orderly parallel columnar structures of proliferating chondrocytes were not observed. Occasionally small clusters of cells resembling proliferating chondrocytes were detected, probably caused by mosaicism in the recombination of loxP sites in the β-catenin gene. Immunohistochemistry revealed that many cells in the mutant growth plates were negative for Sox9. These cells were also PCNA-negative. Cells that are surrounded by alcian blue-stainable matrix were both PCNA-positive and positive for Sox9. Hence, cell proliferation was severely inhibited in cells in which Sox9 could no longer be detected. Overall, this phenotype presents striking similarities with that of Sox9flox/flox; Col2a1-Cre in which Sox9 was inactivated in chondrocytes (Akiyama et al. 2002). These experiments indicate that constitutively active β-catenin strongly reduces Sox9 levels.
Discussion
Functional interactions between Sox9 and the Wnt/β-catenin signaling pathway in cartilage differentiation
The Wnt family of secreted glycoproteins are signaling molecules that play important roles in controlling a wide range of developmental processes, including tissue patterning, cell fate, and cell proliferation (Cadigan and Nusse 1997; Wodarz and Nusse 1998). Wnt signaling is transduced through distinct signaling pathways, both canonical and noncanonical (Korswagen 2002; Pandur et al. 2002; Veeman et al. 2003). In the canonical Wnt pathway, binding of secreted Wnts to the Frizzled family of cell surface receptors inactivates Gsk3-β, resulting in stabilization and nuclear translocation of β-catenin and activation of Wnt target genes. The noncanonical pathways also signal through the Frizzled receptors. The planar cell polarity pathway activates the rho family of GTPases and the Jun N-terminal kinase, and modifies cytoskeletal organization and epithelial cell polarization. The Wnt/Ca2+ pathway stimulates the intracellular increase of Ca2+ through activation of protein kinase C and calmodulin-dependent kinase II.
In vitro studies of chondrogenic mesenchymal cells in micromass cultures show that LiCl, which inhibits Gsk3-β and mimics canonical Wnt pathway activation, or a proteasome inhibitor, which stabilizes β-catenin, as well as overexpression of stβ-catenin, markedly inhibit expression of the major chondrocyte marker Col2a1, suggesting that β-catenin inhibits overt chondrocyte differentiation (Ryu et al. 2002). Misexpression of stβ-catenin in developing chick limb buds produces a shortening of the cartilage elements with accelerated maturation of chondrocytes into hypertrophy and increased expression of type X collagen (Hartmann and Tabin 2000). Together the two sets of experiments suggest that β-catenin and the canonical Wnt pathway both inhibit chondrocyte differentiation of chondrocyte precursors and promote progression of differentiated chondrocytes to hypertrophy.
In this study, we show that mouse embryo mutants overexpressing Sox9 in chondrocytes display a phenotype of dwarfism with inhibition of chondrocyte proliferation and a marked delay in the transition of proliferating chondrocytes to hypertrophy. This phenotype is very similar to that of loss-of-function mutants in which the β-catenin alleles are inactivated in chondrocytes. These mutants show a modest but detectable increase in Sox9 levels in chondrocytes. Conversely, expression of a stabilized β-catenin in chondrocytes produces a phenotype of severe chondrodysplasia, with many cells showing a loss of Sox9 and a marked inhibition of overt chondrocyte differentiation. This phenotype is very similar to that observed when Sox9 is inactivated in chondrocytes. It is possible that, as in these Sox9 mutants, the absence of proliferation in cells lacking Sox9 in embryos, in which stabilized β-catenin is activated, might be caused by the virtual absence of ECM around these cells (Akiyama et al. 2002). Thus, increased expression of Sox9 in chondrocytes resembles the phenotype produced by loss of function of β-catenin, and loss of expression of Sox9 in chondrocytes resembles the phenotype produced by constitutive activity of β-catenin in these cells.
Expression of the Cyclin D1 gene and the activity of its promoter are increased by the β-catenin/Tcf-Lef complex. Sox9 overexpression in chondrocytes in vivo induces a marked down-regulation of Cyclin D1. Sox9 also inhibits the β-catenin-mediated increase in Cyclin D1 promoter activity as well as the β-catenin-dependent activity of the synthetic TOPFLASH promoter. These lines of evidence strongly suggest negative functional interactions between β-catenin and Sox9. Indeed, Sox9 inhibits Xenopus secondary-axis formation by β-catenin in vivo. There are two possible models to explain how Sox9 represses β-catenin/Tcf-Lef complex activities. Our in vitro binding assays indicate that Sox9 can bind to Armadillo repeats that overlap the Tcf/Lef-binding site within β-catenin. We also show that Sox9 inhibits binding of Tcf to β-catenin and activation by Tcf of a TOPFLASH reporter. Thus, it is likely that Sox9 binds to β-catenin and excludes Tcf/Lef from interacting with β-catenin. This inhibitory model of Sox9 is a feature common to two other Sox proteins, xSox17α/β and xSox3 (Zorn et al. 1999). In in vitro cotransfection assays, Sox9 also causes β-catenin degradation, contributing to a reduction of β-catenin transcriptional activity. Hence, Sox9 plays a role in modulating the responsiveness of chondrocytes to canonical Wnt signaling. We also speculate that Wnt/β-catenin signaling inhibits Sox9 activities through binding of β-catenin to the transactivation domain of Sox9. One possible mechanism is the induction of Sox9 degradation by β-catenin as suggested by data in this study. Another may be inhibition of Sox9 transcriptional complex formation through the binding of β-catenin to the Sox9 transactivation domain.
β-Catenin is highly expressed in mesenchymal cells committed to the chondrocyte lineage (Ryu et al. 2002). At the stage of chondrogenic mesenchymal condensation, its expression is down-regulated and can no longer be clearly detected by immunohistochemistry in the nucleus of overtly differentiated chondrocytes (data not shown). β-Catenin, however, has an obvious role in chondrocyte differentiation, as demonstrated by the abnormal phenotype of the growth plate in mutants in which the β-catenin alleles have been inactivated in chondrocytes.
Recent studies showed that several Wnts regulate skeletal development (Yamaguchi et al. 1999; Hartmann and Tabin 2000; Church et al. 2002; Yang et al. 2003), although their precise functions in chondrocyte differentiation still need to be clarified. In the growth plate, Wnt4 is expressed in developing joints, Wnt5a at the boundary of proliferating and prehypertrophic chondrocytes and in perichondrium/periosteum, and Wnt5b in prehypertrophic chondrocytes (Hartmann and Tabin 2000; Church et al. 2002; Yang et al. 2003). Wnt1 misexpression in the developing chick limbs and craniofacial region severely inhibits chondrogenesis (Rudnicki and Brown 1997), strongly suggesting that the canonical Wnt pathway inhibits overt chondrocyte differentiation. The effect of Wnt4 misexpression in the developing chick limbs, which is mediated by canonical Wnt signals, is to accelerate hypertrophic chondrocyte differentiation, similar to the phenotype resulting from misexpression of stβ-catenin (Hartmann and Tabin 2000). In contrast, Wnt5a misexpression in the developing chick limbs delays the maturation of chondrocytes (Hartmann and Tabin 2000; Church et al. 2002). Mice ectopically expressing Wnt5a in chondrocytes exhibit delayed chondrocyte differentiation and hypertrophy, partly mediated by inhibition of Cyclin D1 expression in differentiated chondrocytes (Yang et al. 2003). A recent study showed that Wnt5a inhibits the canonical Wnt pathway by promoting β-catenin degradation and that canonical Wnt activity in Wnt5a-null mutant limb buds is up-regulated (Topol et al. 2003). These experiments are in agreement with the findings that overexpression of β-catenin inhibits the differentiation of chondroprogenitors and accelerates hypertrophic chondrocyte differentiation. However, the interpretation of the phenotypes of Wnt5a-null mutants and of transgenic mice ectopically expressing either Wnt5a or Wnt5b is more complex (Yang et al. 2003). One possible explanation is that several Wnts and their Frizzled receptors coordinate canonical and noncanonical Wnt signaling in vivo and that the phenotypes of mutants and transgenic mice that are observed are the result of a mixed activation of both canonical and non-canonical pathways. Wnts could also regulate other signaling molecules involved in endochondral bone formation, including the Ihh/Pthrp loop and FGF signaling, resulting in indirectly affecting chondrocyte proliferation and differentiation.
Overexpression of Sox9 in committed chondrogenic mesenchymal cells did not affect cell viability (data not shown) as well as the timing and the overall size of mesenchymal condensations, indicating that although Sox9 is required for chondrogenic mesenchymal condensations, a small amount of additional Sox9 has no effect on chondroprogenitors in vivo. Moreover, in Sox9-overexpressing embryos, overt differentiation into chondrocytes of cells present in mesenchymal condensations occurred normally. In addition, expression of genes for a series of signaling molecules, including Fgfr3, Ihh, Pthrp, and PPR, was comparable to that in wild-type embryos. These data indicate that inhibition of cell proliferation and differentiation is not caused by changes in the levels of signaling molecules known to control chondrocyte proliferation and differentiation.
Cyclin D1 controls progression through the G1 phase of the cell cycle, and its expression is strictly regulated by transcriptional mechanisms (Beier et al. 1999; Tetsu and McCormick 1999). In growth plates, Cyclin D1 is specifically expressed in proliferating chondrocytes (Long et al. 2001), and Cyclin D1-null mutant mice display a smaller zone of proliferating chondrocytes (Fantl et al. 1995; Sicinski et al. 1995; Beier et al. 2001), confirming the requirement for Cyclin D1 for normal chondrocyte proliferation. Our in vivo data showing that Sox9 overexpression inhibited BrdU incorporation, and PCNA and Cyclin D1 expression indicate that Sox9 regulates chondrocyte proliferation and that this effect is at least in part mediated by Cyclin D1. This conclusion is consistent with the previous in vitro finding that in CFK2 cells stably transfected with Sox9, an increased proportion of cells was observed in the G0/G1 phase compared with empty vector-transfected cells (Panda et al. 2001). Thus, in addition to the role of Sox9 in the activation of Col2a1, Col11a2, Aggrecan, and presumably many other genes for cartilage ECM components (Bi et al. 1999), Sox9 also controls chondrocyte proliferation by inhibition of Cyclin D1 expression.
Level of Sox9 expression is tightly regulated in chondrocytes
In humans, mutations in a single allele of Sox9 result in a severe skeletal malformation syndrome called campomelic dysplasia due to haploinsufficiency of Sox9 (Foster et al. 1994; Wagner et al. 1994). Sox9 heterozygous mutant mice exhibit similar skeletal anomalies as those observed in campomelic dysplasia patients (Bi et al. 2001). When Sox9 was deleted in chondrocytes through the Cre recombinase/loxP recombination system, the Sox9-null mutant chondrogenic cells were unable to proliferate and to produce a cartilage-specific ECM (Akiyama et al. 2002). On the other hand, Sox9 knock-in heterozygous mutants show an increase in Sox9 expression in chondrocytes to a level of ∼120% of that in chondrocytes of wild-type embryos. This small additional amount of Sox9 is sufficient to produce a very abnormal skeletal phenotype. Overexpression of Sox9 also partially rescues the skeletal phenotype of Sox9 heterozygous mice. The relatively small increase in Sox9 levels in these double mutants is presumably sufficient to correct some of the skeletal defects caused by a 50% reduction in Sox9. Given that both half the levels of Sox9 and a modest increase above the normal levels of Sox9 produce severe skeletal anomalies, the level of Sox9 expression in chondrocytes in vivo must be tightly regulated.
In conclusion, Sox9 is a transcription factor required for the proper progression of the chondrocyte differentiation pathway during endochondral bone formation. We provide evidence that Sox9 and β-catenin physically and functionally interact with each other (Fig. 12). The homologies between the phenotypes of embryos overexpressing Sox9 in chondrocytes and those in which the β-catenin alleles are inactivated in chondrocytes, and conversely the similarities of phenotypes of embryos, in which the Sox9 alleles are inactivated in chondrocytes, and of those expressing a dominant stable mutation of the β-catenin gene in chondrocytes, suggest that the interactions between Sox9 and β-catenin regulate the activity of the canonical Wnt signaling pathway in chondrocytes and, thereby, mediate, at least in part, chondrocyte proliferation and differentiation.
Figure 12.
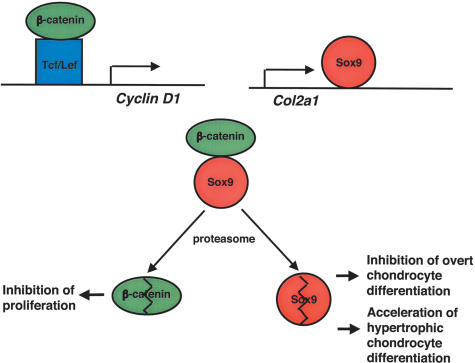
Model describing functional and physical interactions between Sox9 and β-catenin in chondrocytes. β-Catenin activates the Cyclin D1 gene and regulates chondrocyte proliferation. Sox9 activates ECM genes including Col2a1 and regulates chondrocyte differentiation. Sox9 inhibits β-catenin/Tcf-Lef activity by competing with the binding sites of Tcf/Lef within β-catenin. Sox9 also promotes degradation of β-catenin by the ubiquitination/26S proteasome pathway through formation of a Sox9:β-catenin complex. This results in inhibition of proliferation, and in delayed hypertrophic chondrocyte differentiation. In addition, formation of a Sox9:β-catenin complex also causes degradation of Sox9, inhibiting overt chondrocyte differentiation and accelerating hypertrophic chondrocyte differentiation. The model predicts that the relative levels of Sox9 and β-catenin control chondrocyte differentiation.
Materials and methods
Generation of mutant mice
To construct the targeting vector, a 6.0-kb fragment of the Col2a1 gene, covering 3 kb of 5′-flanking exon 1 with a mutation of ATG to CTG for the initial methionine and 3 kb of intron 1 containing a chondrocyte-specific enhancer region, was inserted upstream of a 3xHA-tagged human Sox9 cDNA-bpA and floxed Pgk-neo-bpA cassette, which contained a splicing acceptor signal (SA) at the 5′-end. For the short vector arm, a 2.4-kb fragment covering from a part of exon 2 to exon 5 was inserted downstream from this cassette (Fig. 1A). 129SvEv ES clones harboring Protamine 1-Cre transgenes (O'Gorman et al. 1997) with a Col2a1/Sox9 heterozygous mutation were screened by Southern blot analysis with a Col2a1 probe located outside of the homology regions used for gene recombination. Mouse chimeras were generated by C57BL/6 host blastocyst injection of mutant ES cell clones, and chimeras obtained were bred with 129SvEv mice to Sox9 knock-in heterozygous mice in which a Pgk-neo-bpA cassette was deleted in germ cells of chimeras (Fig. 1A-C).
β-Catenin floxed mice were obtained from The Jackson Lab. In a first cross, Col2a1-Cre transgenic mice (Ovchinnikov et al. 2000) were mated with mice heterozygous for the β-catenin floxed allele. The offspring inheriting Col2a1-Cre and a floxed allele were then mated with β-catenin floxed heterozygous mice to obtain embryos harboring the Col2a1-Cre transgene together with two β-catenin floxed alleles. Routine mouse genotyping was performed by PCR as described previously (Brault et al. 2001).
Mice carrying an allele of β-catenin that contains loxP sites flanking exon 3 (Harada et al. 1999) were crossed with Col2a1-Cre transgenic mice to obtain β-catenin gain-of-function mutations.
Histological analysis
Whole-mount alcian blue staining of mouse embryos and alcian blue and alizarin red staining of skeleton were performed as described previously (Hogan et al. 1994). For the histological analysis, embryos were fixed with 4% paraformaldehyde and embedded in paraffin. Sections of 7 μm were stained with alcian blue and nuclear fast red, or with the von Kossa reaction and nuclear fast red. Immunohistochemical staining was performed by using peroxidase chromogens (Zymed)/TrueBlue substrate (KPL). The following antibodies were used: goat polyclonal anti-Sox9 (1:100, Santa Cruz); rabbit polyclonal anti-Sox9 (1:100); rabbit polyclonal anti-HA (1:500, Covance); rabbit polyclonal anti-Cyclin D1 (1:100, Santa Cruz); and mouse monoclonal anti-β-catenin (1:100, BD Transduction Lab). Cell proliferation analysis was performed on paraffin-embedded sections using the PCNA Staining Kit (Zymed) following the manufacturer's protocol. Cell proliferation was also evaluated by BrdU pulse-labeling. BrdU was injected intraperitoneally into pregnant mice 4 h before death and was detected by the Zymed BrdU staining kit following the manufacturer's protocol. RNA in situ hybridization analysis was carried out as previously described (Conlon and Rossant 1992; Albrecht et al. 1997). Pictures of hybridization signals were taken with a red filter and superimposed with blue fluorescence images of cell nuclei stained with Hoechst 33258 dye.
Cell culture and transient transfections
C3H10T1/2 cells, Cos-7 cells, and 293 cells were cultured and transfected by Fugene 6 (Roche) according to the manufacturer's instruction. For reporter assays, 450 ng of reporters and 150 ng of pcDNA-LacZ, used as an internal control for transfection efficiency, were cotransfected with the indicated expression plasmids. Transfection was done in triplicate. Cells were harvested after 24 h, and luciferase and β-galactosidase activities were assayed as described previously (Lefebvre et al. 1997).
Electrophoretic mobility shift assays (EMSAs)
Sox9 and Tcf3 proteins were synthesized by TNT in vitro transcription/translation systems (Promega), and protein products were confirmed by Western blot analysis (data not shown). DNA-binding reactions and EMSAs were performed as described previously (Lefebvre et al. 1997).
In vitro protein-binding assays
Recombinant human Sox9 protein and recombinant Xenopus stabilized (st) β-catenin protein, in which glycogen synthase kinase 3 (Gsk3) serine and threonine phosphorylation sites are converted to alanine, were expressed in bacteria and purified by nickel-coupled agarose resin. 35S-labeled stβ-catenin proteins, 35S-labeled Tcf-3 protein, Sox9, and deleted Sox9 proteins were produced by TNT in vitro transcription/translation systems (Promega). Ten microliters of translation reactions was incubated with 1 μg of recombinant His-tagged protein coupled to agarose resin at 4°C in 400 μL of binding buffer (20 mM Tris at pH 8.0, 150 mM NaCl, 0.5% NP-40). Nickel-agarose pellets were washed five times in binding buffer, and bound proteins were resolved on SDS-PAGE and visualized by autoradiography.
Xenopus laevis secondary-axis assays
Xenopus β-catenin (McCrea et al. 1991) and human Sox9 cDNAs were subcloned into the pCS2+ vector. Plasmids were linearized with NotI and transcribed in vitro into capped mRNAs using the SP6 mMACHINE Kit (Ambion). Unincorporated nucleotides were removed by filtration through Quick Spin Sephadex G-50 columns (Boehringer Mannheim). The concentration and integrity of the mRNAs were evaluated by measuring the optical density (OD260/280) and mobility on a 1% RNA formaldehyde agarose gel.
Xenopus females were injected with 800 units of human chorionic gonadotropin 10-16 h prior to manual squeezing to isolate eggs. The eggs were placed in 0.1× MMR (pH 7.4) and fertilized in vitro. Prior to the first cleavage, the embryonic jelly coat was removed by incubation in a 2% cysteine HCl (pH 8.0) solution. Borosilicate glass microinjection pipettes (Sutter Instrument Co.) were pulled using a P-30 pulling instrument (Sutter), beveled with a K.T. Brown Type Micropipette beveller (Sutter), and microinjections were performed using the NA-1 oil-driven microinjector (Sutter). Embryos were microinjected at the four-cell cleavage stage into the equatorial region of a single vegetal-ventral blastomere and placed in a solution of 5% Ficoll in 1× MMR for 60-90 min prior to transfer to 0.1× MMR. mRNAs were injected at a volume of 10 nL per blastomere. Embryonic phenotypes were evaluated using a standard binocular dissecting microscope (Nikon SMZ-U) and captured as digital images (Nikon CoolPix 995) at the tadpole stage.
Acknowledgments
We are grateful to Phyllis LuValle, Veronique Lefebvre, Andrew P. McMahon, Viktor Wixler, Francois Fagotto, and Henry M. Kronenberg for plasmids and probes. We thank Steve O'Gorman for PC3 ES cells. We also thank Janie Finch for editorial assistance. This work was funded by NIH grant PO1 AR42919 and a grant from The G. Harold & Leila Y. Mathers Charitable Foundation to B.d.C. and National Institutes of Health Training Grant GM-5-T32-HD07325 and American Legion Auxiliary Fellowship to J.P.L. We also acknowledge NIH grant CA16672 for DNA sequence analysis and veterinary services.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1171104.
Corresponding authors.
References
- Akiyama H., Chaboissier, M.C., Martin, J.F., Schedl, A., and de Crombrugghe, B. 2002. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes & Dev. 16: 2813-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht U., Eichele, G., Helms, J.A., and Lu, H.C. 1997. Visualization of gene expression patterns by in situ hybridization. In Molecular and cellular methods in developmental toxicology (ed. G.P. Daston), pp. 23-48. CRC Press, Boca Raton.
- Behrens J., von Kries, J.P., Kuhl, M., Bruhn, L., Wedlich, D., Grosschedl, R., and Birchmeier, W. 1996. Functional interaction of β-catenin with the transcription factor LEF-1. Nature 382: 638-642. [DOI] [PubMed] [Google Scholar]
- Beier F., Lee, R.J., Taylor, A.C., Pestell, R.G., and LuValle, P. 1999. Identification of the cyclin D1 gene as a target of activating transcription factor 2 in chondrocytes. Proc. Natl. Acad. Sci. 96: 1433-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier F., Ali, Z., Mok, D., Taylor, A.C., Leask, T., Albanese, C., Pestell, R.G., and LuValle, P. 2001. TGFβ and PTHrP control chondrocyte proliferation by activating cyclin D1 expression. Mol. Biol. Cell 12: 3852-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W., Deng, J.M., Zhang, Z., Behringer, R.R., and de Crombrugghe, B. 1999. Sox9 is required for cartilage formation. Nat. Genet. 22: 85-89. [DOI] [PubMed] [Google Scholar]
- Bi W., Huang, W., Whitworth, D.J., Deng, J.M., Zhang, Z., Behringer, R.R., and de Crombrugghe, B. 2001. Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc. Natl. Acad. Sci. 98: 6698-6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz M. and Clevers, H. 2003. Armadillo/β-catenin signals in the nucleus—Proof beyond a reasonable doubt? Nat. Cell Biol. 5: 179-182. [DOI] [PubMed] [Google Scholar]
- Brault V., Moore, R., Kutsch, S., Ishibashi, M., Rowitch, D.H., McMahon, A.P., Sommer, L., Boussadia, O., and Kemler, R. 2001. Inactivation of the β-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128: 1253-1264. [DOI] [PubMed] [Google Scholar]
- Bridgewater L.C., Lefebvre, V., and de Crombrugghe, B. 1998. Chondrocyte-specific enhancer elements in the Col11a2 gene resemble the Col2a1 tissue-specific enhancer. J. Biol. Chem. 273: 14998-15006. [DOI] [PubMed] [Google Scholar]
- Cadigan K.M. and Nusse, R. 1997. Wnt signaling: A common theme in animal development. Genes & Dev. 11: 3286-3305. [DOI] [PubMed] [Google Scholar]
- Church V., Nohno, T., Linker, C., Marcelle, C., and Francis-West, P. 2002. Wnt regulation of chondrocyte differentiation. J. Cell Sci. 115: 4809-4818. [DOI] [PubMed] [Google Scholar]
- Clevers H. and van de Wetering, M. 1997. TCF/LEF factor earn their wings. Trends Genet. 13: 485-489. [DOI] [PubMed] [Google Scholar]
- Conlon R.A. and Rossant, J. 1992. Exogenous retinoic acid rapidly induces anterior ectopic expression of murine Hox-2 genes in vivo. Development 116: 357-368. [DOI] [PubMed] [Google Scholar]
- Delise A.M. and Tuan, R.S. 2002. Analysis of N-cadherin function in limb mesenchymal chondrogenesis in vitro. Dev. Dyn. 225: 195-204. [DOI] [PubMed] [Google Scholar]
- Fagotto F., Funayama, N., Gluck, U., and Gumbiner, B.M. 1996. Binding to cadherins antagonizes the signaling activity of β-catenin during axis formation in Xenopus. J. Cell Biol. 132: 1105-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantl V., Stamp, G., Andrews, A., Rosewell, I., and Dickson, C. 1995. Mice lacking cyclin D1 are small and show defects in eye and mammary gland development. Genes & Dev. 9: 2364-2372. [DOI] [PubMed] [Google Scholar]
- Foster J.W., Dominguez-Steglich, M.A., Guioli, S., Kowk, G., Weller, P.A., Stevanovic, M., Weissenbach, J., Mansour, S., Young, I.D., Goodfellow, P.N., et al. 1994. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature 372: 525-530. [DOI] [PubMed] [Google Scholar]
- Harada N., Tamai, Y., Ishikawa, T., Sauer, B., Takaku, K., Oshima, M., and Taketo, M.M. 1999. Intestinal polyposis in mice with a dominant stable mutation of the β-catenin gene. EMBO J. 18: 5931-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C. and Tabin, C.J. 2000. Dual roles of Wnt signaling during chondrogenesis in the chicken limb. Development 127: 3141-3159. [DOI] [PubMed] [Google Scholar]
- Hogan B., Beddington, R., Costantini, F., and Lacey, E. 1994. Manipulating the mouse embryo: A laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Huelsken J. and Birchmeier, W. 2001. New aspects of Wnt signaling pathways in higher vertebrates. Curr. Opin. Genet. Dev. 11: 547-553. [DOI] [PubMed] [Google Scholar]
- Korinek V., Barker, N., Morin, P.J., van Wichen, D., de Weger, R., Kinzler, K.W., Vogelstein, B., and Clevers, H. 1997. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC-/- colon carcinoma. Science 275: 1784-1787. [DOI] [PubMed] [Google Scholar]
- Korswagen H.C. 2002. Canonical and non-canonical Wnt signaling pathways in Caenorhabditis elegans: Variations on a common signaling theme. Bioessays 24: 801-810. [DOI] [PubMed] [Google Scholar]
- Lefebvre V., Huang, W., Harley, V.R., Goodfellow, P.N., and de Crombrugghe, B. 1997. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro α1(II) collagen gene. Mol. Cell. Biol. 17: 2336-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V., Li, P., and de Crombrugghe, B. 1998. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 17: 5718-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.W., Prockop, D.J., Helminen, H., Fassler, R., Lapvetelainen, T., Kiraly, K., Peltarri, A., Arokoski, J., Lui, H., Arita, M., et al. 1995. Transgenic mice with targeted inactivation of the Col2 α 1 gene for collagen II develop a skeleton with membranous and periosteal bone but no endochondral bone. Genes & Dev. 9: 2821-2830. [DOI] [PubMed] [Google Scholar]
- Long F., Schipani, E., Asahara, H., Kronenberg, H., and Montminy, M. 2001. The CREB family of activators is required for endochondral bone development. Development 128: 541-550. [DOI] [PubMed] [Google Scholar]
- Martin B., Schneider, R., Janetzky, S., Waibler, Z., Pandur, P., Kuhl, M., Behrens, J., von der Mark, K., Starzinski-Powitz, A., and Wixler, V. 2002. The LIM-only protein FHL2 interacts with β-catenin and promotes differentiation of mouse myoblasts. J. Cell Biol. 159: 113-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrea P.D., Turck, C.W., and Gumbiner, B. 1991. A homolog of the armadillo protein in Drosophila (plakoglobin) associated with E-cadherin. Science 254: 1359-1361. [DOI] [PubMed] [Google Scholar]
- Moon R.T., Bowerman, B., Boutros, M., and Perrimon, N. 2002. The promise and perils of Wnt signaling through β-catenin. Science 296: 1644-1646. [DOI] [PubMed] [Google Scholar]
- Oberlender S.A. and Tuan, R.S. 1994. Expression and functional involvement of N-cadherin in embryonic limb chondrogenesis. Development 120: 177-187. [DOI] [PubMed] [Google Scholar]
- O'Gorman S., Dagenais, N.A., Qian, M., and Marchuk, Y. 1997. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl. Acad. Sci. 94: 14602-14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz D.M. and Marie, P.J. 2002. FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes & Dev. 16: 1446-1465. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov D.A., Deng, J.M., Ogunrinu, G., and Behringer, R.R. 2000. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis 26: 145-146. [PubMed] [Google Scholar]
- Panda D.K., Miao, D., Lefebvre, V., Hendy, G.N., and Goltzman, D. 2001. The transcription factor SOX9 regulates cell cycle and differentiation genes in chondrocytic CFK2 cells. J. Biol. Chem. 276: 41229-41236. [DOI] [PubMed] [Google Scholar]
- Pandur P., Maurus, D., and Kuhl, M. 2002. Increasingly complex: New players enter the Wnt signaling network. Bioessays 24: 881-884. [DOI] [PubMed] [Google Scholar]
- Rudnicki J.A. and Brown, A.M. 1997. Inhibition of chondrogenesis by Wnt gene expression in vivo and in vitro. Dev. Biol. 185: 104-118. [DOI] [PubMed] [Google Scholar]
- Ryu J.H., Kim, S.J., Kim, S.H., Oh, C.D., Hwang, S.G., Chun, C.H., Oh, S.H., Seong, J.K., Huh, T.L., and Chun, J.S. 2002. Regulation of the chondrocyte phenotype by β-catenin. Development 129: 5541-5550. [DOI] [PubMed] [Google Scholar]
- Sekiya I., Tsuji, K., Koopman, P., Watanabe, H., Yamada, Y., Shinomiya, K., Nifuji, A., and Noda, M. 2000. SOX9 enhances aggrecan gene promoter/enhancer activity and is up-regulated by retinoic acid in a cartilage-derived cell line, TC6. J. Biol. Chem. 275: 10738-10744. [DOI] [PubMed] [Google Scholar]
- Smits P., Li, P., Mandel, J., Zhang, Z., Deng, J.M., Behringer, R.R., de Croumbrugghe, B., and Lefebvre, V. 2001. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev. Cell 1: 277-290. [DOI] [PubMed] [Google Scholar]
- Sicinski P., Donaher, J.L., Parker, S.B., Li, T., Fazeli, A., Gardner, H., Haslam, S.Z., Bronson, R.T., Elledge, S.J., and Weinberg, R.A. 1995. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell 82: 621-630. [DOI] [PubMed] [Google Scholar]
- St-Jacques B., Hammerschmidt, M., and McMahon, A.P. 1999. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes & Dev. 13: 2072-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetsu O. and McCormick, F. 1999. β-Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398: 422-426. [DOI] [PubMed] [Google Scholar]
- Topol L., Jiang, X., Choi, H., Garrett-Beal, L., Carolan, P.J., and Yang, Y. 2003. Wnt-5a inhibits the canonical Wnt pathway by promoting GSK-3-independent β-catenin degradation. J. Cell Biol. 162: 899-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman M.T., Axelrod, J.D., and Moon, R.T. 2003. A second canon. Functions and mechanisms of β-catenin-independent Wnt signaling. Dev. Cell 5: 367-377. [DOI] [PubMed] [Google Scholar]
- Vortkamp A., Lee, K., Lanske, B., Segre, G.V., Kronenberg, H.M., and Tabin, C.J. 1996. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science 273: 613-622. [DOI] [PubMed] [Google Scholar]
- Wagner T., Wirth, J., Meyer, J., Zabel, B., Held, M., Zimmer, J., Pasantes, J., Bricarelli, F.D., Keutel, J., Hustert, E., et al. 1994. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 79: 1111-1120. [DOI] [PubMed] [Google Scholar]
- Wodarz A. and Nusse, R. 1998. Mechanisms of Wnt signaling in development. Annu. Rev. Cell Dev. Biol. 14: 59-88. [DOI] [PubMed] [Google Scholar]
- Xie W.F., Zhang, X., Sakano, S., Lefebvre, V., and Sandell, L.J. 1999. Trans-activation of the mouse cartilage-derived retinoic acid-sensitive protein gene by Sox9. J. Bone Miner. Res. 14: 757-763. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T.P., Bradley, A., McMahon, A.P., and Jones, S. 1999. A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development 126: 1211-1223. [DOI] [PubMed] [Google Scholar]
- Yang Y., Topol, L., Lee, H., and Wu, J. 2003. Wnt5a and Wnt5b exhibit distinct activities in coordinating chondrocyte proliferation and differentiation. Development 130: 1003-1015. [DOI] [PubMed] [Google Scholar]
- Zorn A.M., Barish, G.D., Williams, B.O., Lavender, P., Klymkowsky, M.W., and Varmus, H.E. 1999. Regulation of Wnt signaling by Sox proteins: XSox17 α/β and XSox3 physically interact with β-catenin. Mol. Cell 4: 487-498. [DOI] [PubMed] [Google Scholar]