Summary
Centrosomes duplicate only once per cell cycle, but the controls that govern this process are largely unknown. We have identified Cep76, a centriolar protein that interacts with CP110. Cep76 is expressed at low levels in G1 and is induced in S and G2 phase, during which point centrioles have already commenced duplication. Interestingly, depletion of Cep76 drives the accumulation of centriolar intermediates in certain types of cancer cells. Enforced Cep76 expression specifically inhibits centriole amplification in cells undergoing multiple rounds of duplication without preventing the formation of extra procentrioles from a parental template. Furthermore, elevated levels of Cep76 do not affect normal centriole duplication. Thus, Cep76 specifically prevents centriole re-duplication by limiting duplication to once per cell cycle. Our findings also point to mechanistic differences between normal duplication and aberrant centriole amplification as well as distinctions between diverse modes of amplification.
Keywords: Cep76, CP110, centriole duplication, centriole re-duplication, centriole amplification
Introduction
Although the recent identification of key centrosome components has increased our understanding of this organelle, many mechanistic aspects of the centrosome cycle in mammalian cells remain shrouded in mystery. It is known that the centrosome is composed of several hundred polypeptides, the majority of which have no assigned function (Andersen et al., 2003). It is also known that centrosomes duplicate once per cell cycle in a highly choreographed manner. During mitosis, each daughter cell inherits a single centrosome containing two centrioles surrounded by a proteinaceous cloud of pericentriolar matrix (PCM). The two centrioles, termed the mother and daughter centrioles (or parental centrioles), undergo disengagement from one another and become primed for the next round of duplication by early G1 (Tsou and Stearns, 2006). In late G1 phase, centrioles further separate from one another, initiating a semi-conservative duplication process in which a single procentriole emerges from each parental centriole (Hinchcliffe and Sluder, 2001b). Procentriole elongation and maturation occur during S and G2 phases, and during prophase, centrosomes begin to separate, subsequently establishing the mitotic spindle.
The controls that govern the strict timing and singularity of events during centrosome duplication are largely unknown, although it has been established that centrosome duplication is tied to the periodic activity of cyclin-dependent kinases (CDKs) (Hinchcliffe and Sluder, 2001a). Several candidate proteins have emerged as CDK targets that could profoundly affect centrosome cycle events. Among them, nucleophosmin (NPM1), Mps1, and CP110 are the best characterized (Chen et al., 2002; Fisk and Winey, 2001; Okuda et al., 2000). NPM1 localizes to centrosomes in G1 phase cells prior to centrosome duplication, and subsequent phosphorylation by cyclin E/CDK2 at the G1/S transition triggers its dissociation from centrosomes, allowing duplication to occur (Okuda et al., 2000). Mps1, like NPM1, is a centrosomal protein implicated in regulating centrosome duplication (Fisk and Winey, 2001; Kasbek et al., 2007). Phosphorylation of Mps1 by CDK2 is necessary to prevent Mps1 from proteasome-mediated degradation, thus allowing Mps1 to remain at centrosomes and participate in duplication events (Fisk and Winey, 2001; Kasbek et al., 2007). CP110, a protein first identified as a centriolar CDK2 substrate, has also emerged as an important player in centrosome function (Chen et al., 2002). Ablation of CP110 CDK2 phospho-acceptor sites gives rise to unscheduled centrosome separation and polyploidy, and depletion of this protein prevents centrosome amplification during prolonged S phase arrest, leads to premature centrosome separation, and causes cytokinesis defects (Chen et al., 2002; Tsang et al., 2006). Ultrastructural studies show that CP110 plays an early role in centriole biogenesis, during which it associates with the distal tip of growing procentrioles (Kleylein-Sohn et al., 2007). Biochemical studies have shown that CP110 associates with multiple proteins, forming complexes varying in size between several hundred kDa and 3 MDa (Tsang et al., 2006). We have characterized a subset of these complexes, and using immuno-affinity purification and protein-protein interaction traps, we identified several CP110 interacting partners, including calmodulin (CaM) and centrin, two small calcium binding proteins, Cep97, and Cep290 (Spektor et al., 2007; Tsang et al., 2008; Tsang et al., 2006). Interestingly, depletion of either CP110 or Cep97 gives rise to abnormally long centriolar filaments and aberrant assembly of primary cilia, suggesting that these proteins could function in part by limiting centriolar length and by blocking the ciliogenic branch of centriole function.
The mechanisms by which centrosomes duplicate only once per cell cycle are not fully understood. This “once per cell cycle” control has been postulated to involve two mechanistically distinct “rules”: a cell cycle control and a copy number control (Nigg, 2007). Cell cycle control refers to the mechanisms that limit the number of duplication events to one per cell cycle (Nigg, 2007). In addition to the role of separase in licensing parental centrioles for a new round of duplication (Tsou and Stearns, 2006), controls must also exist to prevent re-duplication after a normal round of duplication has taken place. Experiments based on cell fusion assays have led to the idea that centrosome re-duplication can be blocked by an unknown organelle-intrinsic mechanism that is not dependent on the centrosome-to-nucleus ratio (Wong and Stearns, 2003). Consistent with this notion, laser ablation of procentrioles induces re-duplication of the parental centriole in S phase-arrested cells (Loncarek et al., 2008), suggesting that the re-duplication block requires the physical presence of centrioles. Copy number control refers to the formation of only one procentriole per parental centriole during normal duplication (Nigg, 2007). Both control mechanisms must be tightly coordinated for faithful duplication, but very little is known about the molecular pathways or players that regulate these two processes.
In an effort to dissect the mechanisms underlying the duplication of centrosomes, we have attempted to identify proteins associated with CP110. Here, we have begun to characterize an interacting protein, Cep76. We show that Cep76 associates with CP110 and a second CP110-interacting protein, Cep97. The protein localizes to centrosomes and is expressed in a cell cycle-dependent manner, peaking in S and G2 phase cells. Significantly, depletion of Cep76 leads to the accumulation of centriolar intermediates, a subset of which morphologically resembles centrioles. This phenotype is not universal, as it is restricted to certain types of cancer cells, suggesting that additional factors act in concert with Cep76 to restrict centriolar duplication. Conversely, Cep76 over-expression suppresses centriole amplification generated through multiple rounds of duplication in hydroxyurea (HU)-treated, prolonged S-phase arrested cells. In striking contrast, enforced expression does not prevent centriole amplification mediated by Polo-like kinase 4 (Plk4), wherein parental centrioles give rise to multiple adjoining procentrioles that simultaneously form within a single cell cycle (Kleylein-Sohn et al., 2007). In addition, ectopic Cep76 expression does not affect normal centrosome duplication. Taken together, our data suggest that Cep76 is required to specifically block centrosome re-duplication. We propose that Cep76 could represent a centrosome-intrinsic factor that limits reproduction of centrioles to once per cell cycle.
Results
Identification of Cep76 as a CP110-interacting protein
In a recent proteomic screen for CP110-interacting proteins, we identified several proteins including Cep97 (Spektor et al., 2007) and a previously uncharacterized protein, Cep76. Cep76 appeared to be a valid centrosomal target in light of a previous mass spectrometric analysis of purified centrosomes in which this protein was identified (Andersen et al., 2003). The protein has no recognizable motifs except for a partial WD-40 repeat and a calcium/lipid-binding, or CaLB, domain. Cep76 appears to have been highly conserved from mammals to zebrafish and frogs, although obvious homologs were not identified in flies or yeast. To test whether CP110 and Cep76 interact in cells, we Flag epitope-tagged Cep76, transfected 293T cells, and performed anti-Flag immunoprecipitations. Both proteins were co-immunoprecipitated, and conversely, we detected Flag-Cep76 after immunoprecipitating CP110 (Fig. S1A). Next, we performed immunofluorescence experiments to detect epitope-tagged Cep76 and found that the protein localized to centrosomes (Fig. S1B).
In light of these localization and interaction data, we raised antibodies specific for Cep76 to detect the endogenous protein. In agreement with our enforced expression studies, we showed that native Cep76 interacted with CP110 (Fig. 1A). Our previous work indicated that CP110 associates with a cadre of proteins involved in the centrosome cycle or ciliogenesis, and we investigated potential interactions with known CP110-associated proteins. We found that Cep76 also interacted with Cep97, although we did not observe interactions between Cep76 and centrin or CaM (Fig. 1A and data not shown). We performed domain-mapping studies to identify the region of CP110 responsible for its interactions with Cep76 (Fig. S1C). A portion of CP110 encompassing residues 200–565 robustly interacted with Cep76, and the interaction domain most likely maps more finely between residues 350 and 565. We have shown previously that amino-terminal residues 1–223 of CP110 were required to interact with Cep97, and carboxy-terminal residues were required to interact with CaM, suggesting that non-competitive binding by CP110 to each of these proteins and Cep76 could occur. In total, these studies indicate that Cep76 interacts with a subset of centriolar proteins that likely form distinct CP110-Cep97-Cep76 assemblies separable from CP110-centrin complexes.
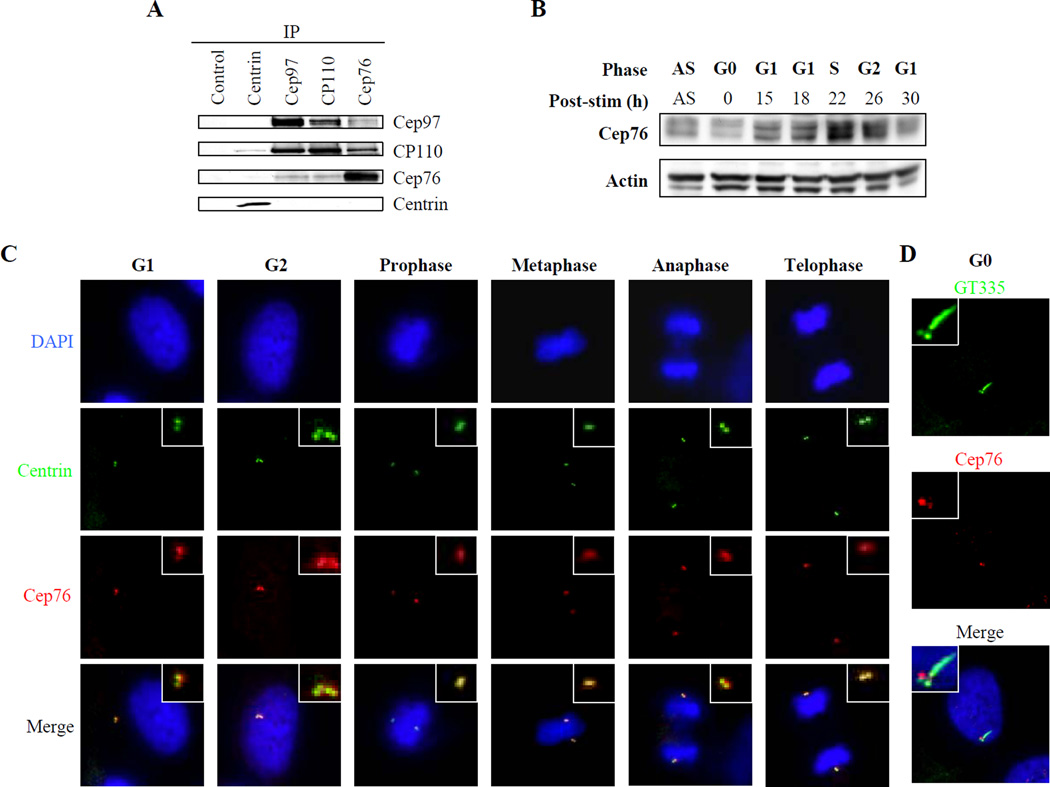
Figure 1.
Cep76 interacts with CP110 and is periodically expressed during the cell cycle. (A) Western blotting of endogenous Cep97, CP110, Cep76, and centrin after immunoprecipitation with anti-centrin, anti-Cep97, anti-CP110, anti-Cep76, or control (anti-Flag) antibody from 293T cell extracts. (B) T98G cells were synchronized by serum starvation, stimulated to initiate cell cycle re-entry, and lysates from different cell cycle stages were collected. Western blots show endogenous Cep76 protein levels. Actin was used as a loading control. “AS,” asynchronous population. (C) RPE-1 cells in diverse stages of the cell cycle were processed for immunofluorescence with anti-Cep76 (red) and anti-centrin (green) antibodies. DNA was stained with DAPI (blue). (D) Quiescent RPE-1 cells were processed for immunofluorescence with anti-Cep76 antibody (red) and anti-glutamylated tubulin antibody (GT335, green). DNA was stained with DAPI (blue).
Cep76 localization and protein levels in quiescent and cycling cells
Next, we assessed Cep76 levels as a function of cell cycle progression. We synchronized human T98G cells by mitogen deprivation, released cells back into the cycle, and determined Cep76 levels in each population. We found that Cep76 levels were relatively low in quiescent cells, and they increased as cells progressed through G1 (Figs. 1B and S1D). Protein levels rose by about two-fold, peaked during S and G2 phase, and decreased by roughly the same amount as cells re-entered the subsequent G1 phase. Similar cell cycle-dependent fluctuations in Cep76 levels were observed in U2OS and Saos-2 cells (Figs. S2A–D). Thus, in this setting, Cep76 protein levels oscillate and peak at a time subsequent to initiation of centriole duplication. We used immunofluorescence microscopy to detect Cep76 localization in U2OS cells and at different stages of the cell cycle in diploid human retinal pigment epithelial (RPE-1) cells. Cep76 localized to centrioles as determined by its extensive overlap with the centriolar marker, centrin (Fig. 1C). Identical results were obtained with a second antibody raised against a different portion of Cep76 (data not shown). Moreover, we found that Cep76 staining was enhanced in G2 as compared to G1 phase (Fig. 1C). The Cep76 signal appeared to diminish as cells progressed through mitosis (Fig. 1C), suggesting possible post-translational regulation, akin to what we have observed for CP110. We also analyzed Cep76 localization during the process of ciliogenesis. RPE-1 cells were rendered quiescent, a setting in which the majority of cells develop primary cilia. We detected cilia assembly using antibodies against glutamylated tubulin, which visualizes both primary cilia and daughter centrioles. We found that Cep76 was substantially reduced on the ciliated mother centriole (basal body) relative to the daughter centriole (Fig. 1D). Furthermore, Cep76 staining was never observed along the ciliary axoneme (Fig. 1D). This pattern is reminiscent of, but not identical to, the localization of both CP110 and Cep97 in ciliated cells (Spektor et al., 2007) and suggests that Cep76 levels and/or localization are tightly regulated in G0/G1 cells.
Depletion of Cep76 leads to accumulation of centriolar intermediates
We investigated the functional impact of depleting Cep76 using RNA interference-mediated gene silencing. We transfected either pools of four small interfering RNAs (siRNAs), two individual siRNAs, or two distinct microRNAs (miRNAs), and significantly reduced Cep76 levels in human U2OS cells, resulting in depletion of 80–90% of Cep76 at the RNA and protein levels (Figs. 2A, S3A and S3D and data not shown). We microscopically confirmed the near-complete removal of Cep76 from centrosomes in siRNA-treated cells (Fig. 2B). Depletion of Cep76 did not affect the levels or localization of either CP110 or Cep97 at centrioles (Figs. 2D, S3D and data not shown), suggesting that CP110 and Cep97 are recruited to centrosomes independently of Cep76. Likewise, loss of CP110 or Cep97 does not trigger loss of Cep76 from centrosomes (Fig. S3B). Of note, suppression of Cep76 expression had no obvious impact on cell cycle progression (Fig. S3C).
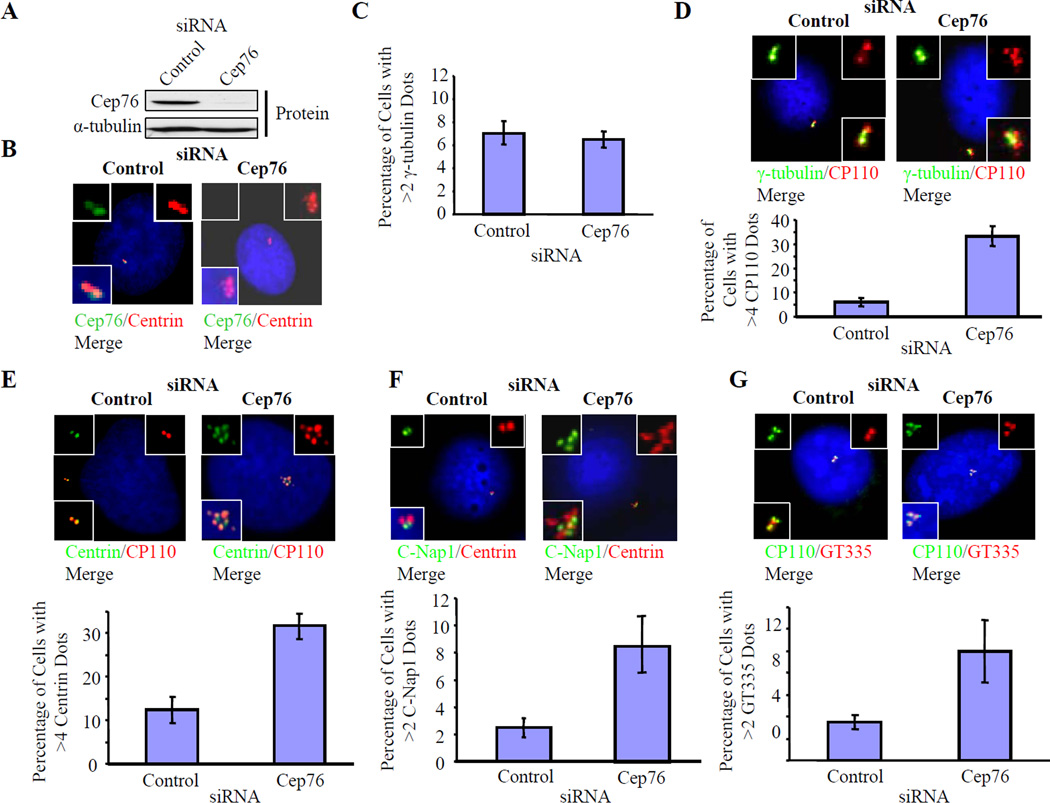
Figure 2.
Depletion of Cep76 induces amplification of centriole markers in U2OS cells. (A) Cep76 protein levels in cells treated with control or Cep76 siRNAs as indicated. α-tubulin was used as a loading control. (B, left) Co-localization of endogenous Cep76 with centrin in cells treated with control siRNA. (B, right) Cep76 is essentially abolished after transfection of an siRNA targeting Cep76. (C–G) The percentages of cells with (C) more than two γ-tubulin dots, (D) greater than four CP110 dots, (E) greater than four centrin dots, (F) more than two C-Nap1 dots, and (G) more than two poly-glutamylated tubulin dots (GT335) in control or Cep76-depleted cells were determined. Representative pictures are shown in D–G. In C–G, average data obtained from three independent experiments are shown. About 100 cells for each siRNA transfection were scored each time. Error bars represent +/− S.D.
Next, we examined the impact of Cep76 depletion on centrosome duplication in U2OS cells. We monitored asynchronously growing, Cep76-depleted cells by counting dots stained with antibodies against γ-tubulin, which primarily localizes to the PCM, and detected no abnormalities in the number of dots (Figs. 2C–D). Similar observations were made with the use of another PCM marker, pericentrin (data not shown). Remarkably, however, when we performed immunofluorescence using antibodies against known centriolar markers, including centrin and CP110, we found that Cep76 depletion provoked the accumulation of an abnormal complement of dots (>4) containing both proteins (Figs. 2D–E). Indeed, loss of Cep76 led to a 3–6-fold increase in the number of cells with amplified dots. This phenotype was also observed with a pool of siRNAs or two individual siRNAs derived from this pool, and two unrelated sequences incorporated into miRNAs gave identical results (Fig. S3E).
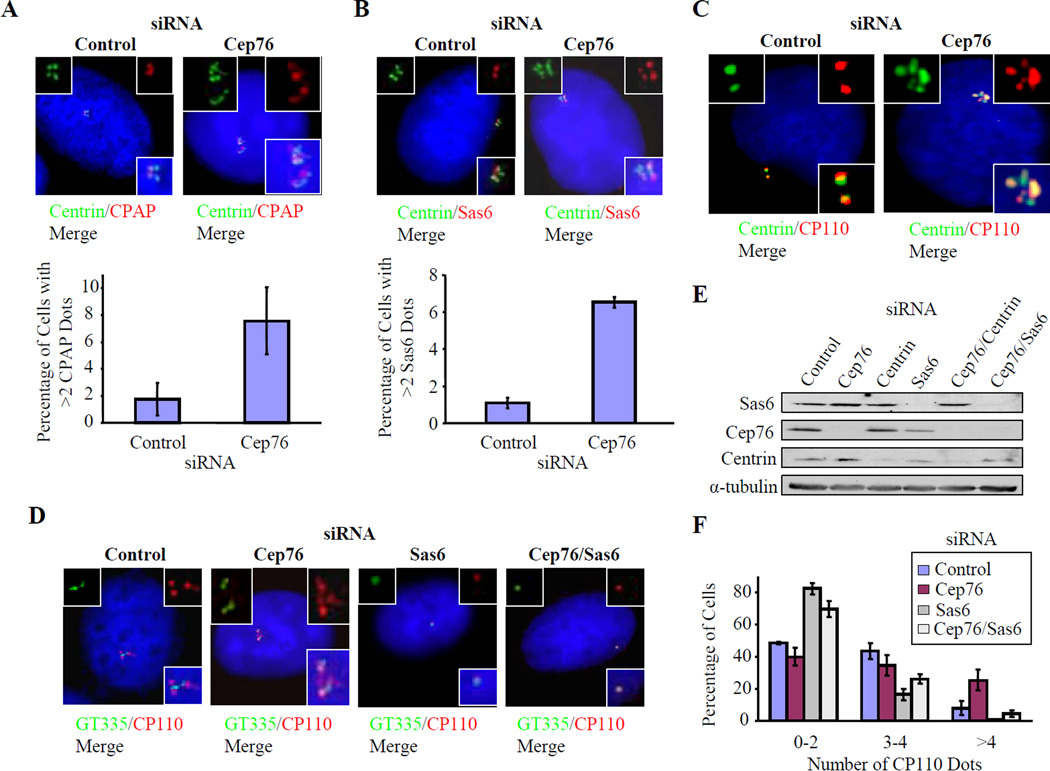
Several lines of evidence indicate that these supernumerary dots may represent ectopic centriolar intermediates and centrioles. First, we observed amplification using antibodies against several centriolar markers, including the distal centriolar proteins centrin and CP110 (Figs. 2D–E), proximal centriolar markers, including C-Nap1, CPAP, and Sas6, and a marker of stabilized centriolar microtubules, glutamylated tubulin (Figs. 2F–G, 3A–B). Second, these supernumerary dots are resistant to the microtubule de-polymerizing drug nocodazole, consistent with the notion that they represent intrinsic, stable components of centrosomes (Fig. 3C). Third, combined suppression of Cep76 and Sas6, a protein required for centriole duplication and whose depletion leads to a reduction in centriole numbers (Strnad et al., 2007), reversed the Cep76 knock-down phenotype and mimicked Sas6 ablation (Figs. 3D–F). These results suggest that Sas6 is required for the formation of supernumerary dots and that Sas6 and Cep76 could function antagonistically. Fourth, the dots resulting from Cep76 depletion were morphologically homogeneous and tightly clustered. Furthermore, they did not co-localize with PCM-1 at centriolar satellites (Fig. 4A), which, by definition, are smaller in size, more numerous, scattered throughout the cytoplasm, and sensitive to nocodazole treatment (Kubo et al., 1999).
Figure 3.
Depletion of Cep76 induces amplification of centriole markers that is dependent on Sas6. U2OS cells were treated with control or Cep76 siRNA. (A, B) The percentages of cells with (A) more than two CPAP dots and (B) more than two Sas6 dots were determined. Representative pictures showing co-localization of CPAP or Sas6 (red) with centrin (green) are shown. (C) Control and Cep76-siRNA depleted cells were treated with 10 µM nocodazole for 1 hour to induce microtubule de-polymerization. (D–F) U2OS cells were transfected with the indicated siRNAs. (D) Cells were processed for immunofluorescence with anti-glutamylated tubulin (GT335, green) and anti-CP110 antibodies (red). DNA was stained with DAPI (blue). (E) Western blotting of Cep76, Sas6, and centrin after transfection of indicated siRNAs. α-tubulin was used as a loading control. (F) The percentages of cells with the indicated number of CP110 dots were determined. In A–B and F, average data obtained from two independent experiments are shown. At least 100 cells for each siRNA transfection were scored each time. Error bars represent +/− S.D.
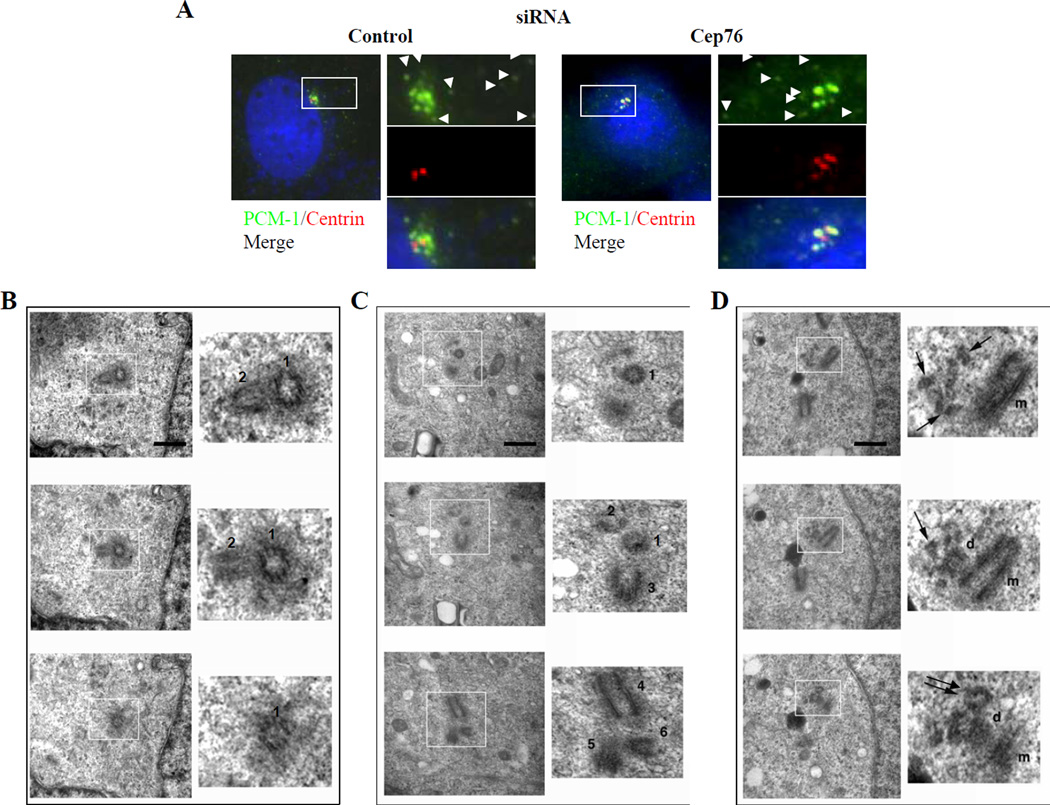
Figure 4.
Depletion of Cep76 induces the formation of centriolar intermediates that do not co-localize with centriolar satellites. (A) Co-staining of centrin and PCM-1 in cells treated with control or Cep76 siRNA. Arrowheads indicate centriolar satellites. (B–D) U2OS cells transfected with control or Cep76 siRNA were fixed and analyzed by serial section EM. (B, C) Selected sections spanning a control (B) and a Cep76-depleted cell (C), showing a total of 2 and 6 centrioles (indicated with numbers), respectively. (D) In this particular Cep76-depleted cell, a parental (m)-procentriole (d) pair possessing an extra centriole (indicated with double arrows) and abnormal microtubular structures (indicated with single arrows) were observed. Scale bar, 500 nm.
To determine whether the supernumerary dots represent ectopic centriolar intermediates and centrioles, we examined control and Cep76-depleted cells by ultra-thin serial section electron microscopy (EM). We found that Cep76-depleted cells (3/18 examined) exhibited a striking centriolar phenotype in comparison with control siRNA-transfected cells (0/11 examined)(Figs. 4B–D), in accordance with immunofluorescence experiments performed in parallel (supernumerary CP110 dots were observed in 5% and 22% of control and Cep76-depleted cells, respectively). Of the three Cep76-depleted cells examined by EM, one cell possessed ectopic, recognizable centrioles (Fig. 4C), and the other two displayed aberrant microtubular structures, a subset of which morphologically resembled centrioles (Fig. 4D and data not shown). These aberrant microtubular structures 1) were primarily located in proximity to, but were shorter than, procentrioles; 2) were morphologically similar to “granddaughter” centrioles described previously (Vidwans et al., 2003); and 3) were distinguishable from centriolar satellites (or fibrous granules), which appear as small (~50 nm), densely staining granules by EM (Kubo et al., 1999; Prosser et al., 2009). Our studies further rule out the possibility that our supernumerary dots are centrin aggregates, as these assemblies, observed under conditions of stress, are not known to associate with any other centrosomal components and are not visible by EM (Loncarek et al., 2008). Indeed, extra CP110 dots were detected in Cep76-depleted cells even when centrin was co-depleted (Figs. 3E, S4A–B). Taken together, our data indicate that depletion of Cep76 results in aberrant accumulation of centriole intermediates and centriole assembly at or near existing procentrioles. However, we note that the number of γ-tubulin or pericentrin dots did not increase upon Cep76 knock-down (Fig. 2C). This observation could indicate that pericentriolar material has not been recruited to the centriolar intermediates or that a common cloud of PCM envelopes the tightly clustered centrioles and centriolar intermediates. We cannot currently discriminate between these possibilities, and because all of these structures may not represent fully “mature” centrioles (see below), we will refer to these structures as centriolar intermediates.
We next examined mitotic cells that had been treated with Cep76 siRNAs to complement our studies in interphase cells. Cep76 depletion did not lead to a significant increase in the mitotic index or in the percentage of abnormal mitotic spindles relative to a control (Fig. S5A). To investigate this further, we transfected cells with Cep76 siRNAs, synchronized cells in S phase, released cells back into the cycle, and stained them for centriolar markers at different times after release. While we observed significant amplification of centrin dots in interphase and prophase cells, we did not detect a corresponding increase in centrin dots in metaphase, anaphase, and telophase cells depleted of Cep76 (Figs. S5B–C). Together, these data suggest that amplification of centrin dots arising from Cep76 depletion does not lead to mitotic catastrophe and raise the possibility that the additional centriolar intermediates, by virtue of their immaturity, tendency to cluster, and possible inability to accumulate PCM, cannot organize extra or abnormal mitotic spindles. As a consequence, they may be inactivated or destroyed sometime between prophase and metaphase.
The Cep76 depletion phenotype is cancer cell-specific
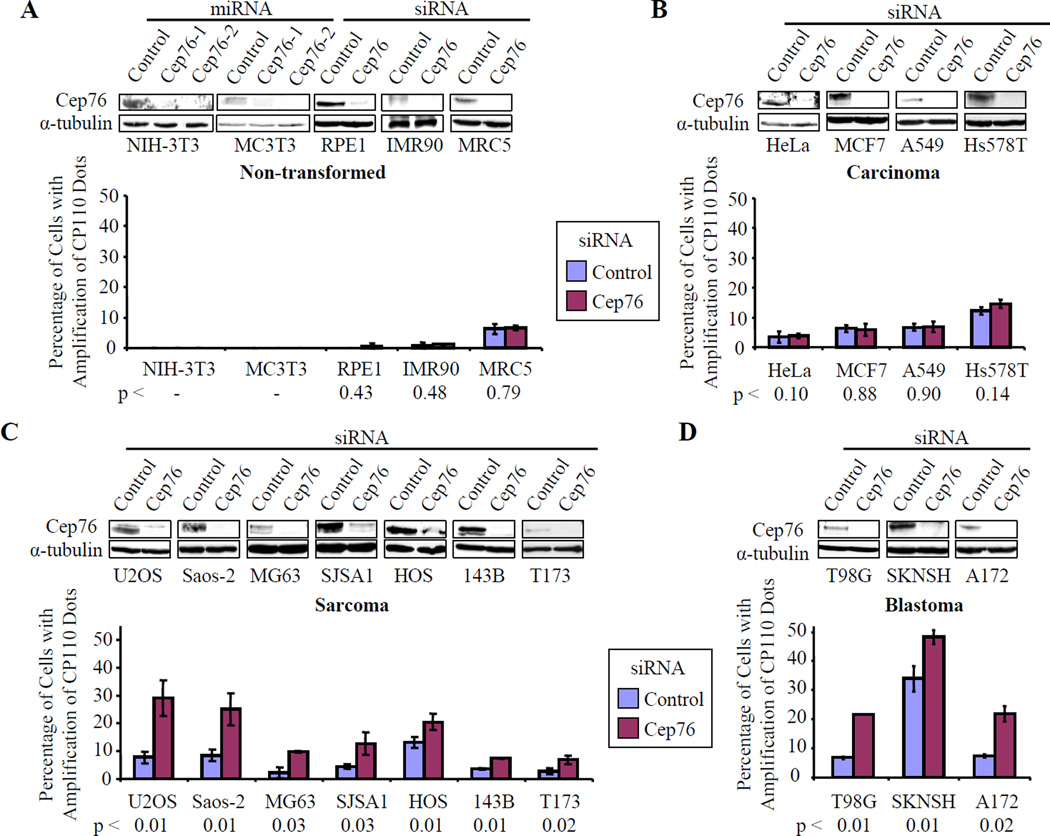
To rule out the possibility that our observations were restricted to a single cell line, we performed analogous experiments in a second cell line, Saos-2. In agreement with our previous experiments, we observed amplification of CP110, centrin, and C-Nap1 dots upon ablating Cep76 in this cell line (Figs. S4C–F). U20S and Saos-2 cells are derived from patients with osteosarcoma, and like U2OS, we show that Saos-2 cells also undergo centriole amplification during prolonged HU treatment (Balczon et al., 1995; and data not shown). Next, we sought to determine whether the Cep76 phenotype is restricted to osteosarcomas or if it is also observed in non-transformed, diploid cells and other types of transformed and cancer cell lines. Furthermore, we asked whether all cell lines that exhibit the Cep76 depletion phenotype also display centriole amplification after prolonged HU treatment. We successfully depleted Cep76 in five different non-transformed, diploid (3T3, mouse embryonic fibroblast; MC3T3, mouse osteoblast; RPE1, retinal pigment epithelial cells; IMR90, lung fibroblast; MRC5, lung fibroblast) cell lines derived from tissues of diverse origin with either siRNAs or miRNAs, and showed that none of them exhibited amplification of CP110 dots (Fig. 5A). We then carried out additional Cep76 ablation experiments in a variety of cancer cell lines to determine whether specific types of tumors were susceptible to loss of this protein (Figs. 5B–D). Cep76 loss did not induce accumulation of an abnormal complement of CP110 dots in four different carcinoma cell lines (HeLa and MCF7, cervical and breast adenocarcinomas respectively; A549, lung carcinoma; Hs578T, invasive ductal carcinoma)(Fig. 5B) or two virally transformed (HEK293 and IMCD3, mouse inner medullary collecting duct cells) cell lines (data not shown). In striking contrast, depletion of Cep76 led to amplification of CP110 dots in all seven osteosarcoma cell lines (U2OS, Saos-2, MG63, SJSA1, HOS, 143B, T173) (Fig. 5C) and all three blastomas (T98G and A172, glioblastomas; SKNSH, neuroblastoma) tested, although the degree of amplification was cell-line dependent (Fig. 5D). Together, these results indicate that non-transformed and transformed cells respond differently to Cep76 depletion and that the biological function of Cep76 is more obvious in certain types of cancer cells.
Figure 5.
Depletion of Cep76 induces amplification of CP110 dots in osteosarcomas and blastomas. (A) Non-transformed or diploid (mouse 3T3, mouse MC3T3, RPE-1, IMR90, MRC5), (B) carcinoma (HeLa, MCF7, A549, Hs578T), (C) osteosarcoma (U2OS, Saos-2, MG63, SJSA1, HOS, 143B, T173), and (D) blastoma (T98G, SKNSH, A172) cell lines were treated with control, Cep76 siRNAs, or Cep76 miRNAs, as indicated. Western blot detection of endogenous Cep76 is shown for each cell line, with α-tubulin used as a loading control. The percentages of cells showing amplification of CP110 dots were determined. Average data obtained from two to three independent experiments are shown. For 3T3 and MC3T3 cells, experiments were carried out once with two independent miRNAs (with similar results), so data were not averaged. At least 100 cells for each siRNA or miRNA transfection were scored each time. Error bars represent +/− S.D. p values comparing control and Cep76 siRNA transfected cells are indicated below.
We observed that only a subset of osteosarcomas (U2OS, Saos-2 and HOS) displayed centriole over-duplication during prolonged HU treatment (data not shown), although each of the osteosarcoma lines displayed sensitivity to Cep76 loss. Thus, there does not appear to be a simple one-to-one correlation between the ability of a cell line to over-duplicate its centrioles during prolonged HU treatment and accumulation of centriolar intermediates upon Cep76 ablation. Furthermore, Cep76 levels were comparable in both HU arrested and proliferating cells (Fig. S4G). These data suggest that the mechanisms underlying amplification induced by HU treatment or Cep76 loss may not be identical, although we cannot rule out the possibility of partially overlapping pathways.
The Cep76 depletion phenotype is dependent on CP110 and Cep97
One prediction from our studies is that Cep76 has an inhibitory effect on the ability of CP110 to promote centriole assembly, and therefore, that it might lie “upstream” of CP110 in the series of events that culminates in centriolar assembly. If true, then we would expect loss of CP110 to function epistatically to suppress accumulation of centriolar intermediates induced by Cep76 loss. This is indeed what we observed when we simultaneously suppressed both Cep76 and CP110 (Figs. S6A–B). Similar results were obtained when both Cep76 and Cep97 were depleted simultaneously (Fig. S6C), and this could be due to the disappearance of CP110 upon Cep97 loss (Spektor et al., 2007) or to a requirement for Cep97. These results are significant because they reinforce the notion that Cep76 functions in concert with its interacting partners, CP110 and Cep97. Furthermore, our results are strikingly reminiscent of other studies in which amplification of centrioles mediated by either prolonged S phase arrest (HU treatment) or Plk4 could be suppressed by ablation of CP110 (Chen et al., 2002; Kleylein-Sohn et al., 2007). Taken together, our studies suggest that depletion of Cep76 results in amplification of centriolar intermediates and that this process depends on CP110 and Cep97.
Cep76 actively suppresses one type of centriole amplification
There are at least two settings in which centriole amplification can occur. First, during prolonged S phase arrest provoked by HU treatment, centrosomes escape the re-duplication block, leading to multiple rounds of centriole duplication and disengagement (Balczon et al., 1995; Kuriyama et al., 2007; Loncarek et al., 2008). Under these conditions, procentriole formation is thought to be highly synchronous in the first round of duplication. Subsequent rounds of duplication are progressively less synchronous, with the second round of duplication taking place at approximately 24 hours after the first round of duplication (Loncarek et al., 2008). Centriole amplification is also observed upon ectopically expressing Plk4. Expression of Plk4 leads to concurrent production of multiple procentrioles adjoining a single parental centriole within S phase of one cell cycle, resulting in a “flower” or “rosette”-like pattern that is morphologically distinct from the pattern observed during S phase arrest (Kleylein-Sohn et al., 2007). These “flower”-like centrioles persist and subsequently disengage during passage through mitosis (Kleylein-Sohn et al., 2007). Recent observations suggest that HU-induced and Plk4-induced centriole amplification are mechanistically different from one another, since “flowers” are rarely observed during prolonged S phase arrest (Loncarek et al., 2008). We speculated that if Cep76 were a rate-limiting suppressor of centriole amplification, then enforced expression of this protein would negate the appearance of supernumerary centrioles generated through one or both pathways.
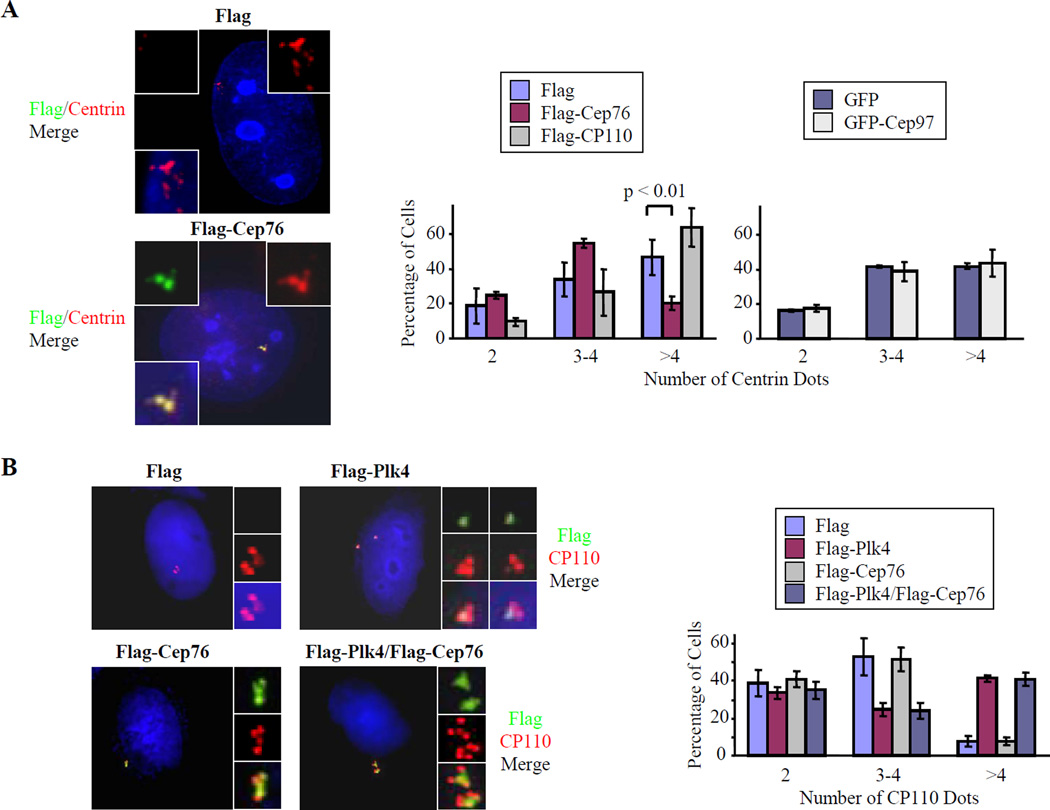
To test this idea, we ectopically expressed Flag-Cep76 in U2OS cells and treated them with HU to induce amplification. Interestingly, we found that elevated levels of Cep76 markedly suppressed HU-mediated centriole amplification (Fig. 6A). In contrast, expression of a second CP110-interacting protein, Cep97, had no effect in the presence of HU. Furthermore, over-expression of CP110 itself led to a modest increase in centriole amplification (Fig. 6A), consistent with our previous finding that this protein is required to permit amplification in HU-arrested cells (Chen et al., 2002).
Figure 6.
Ectopic expression of Cep76 prevents HU-induced but not Plk4-induced centriole amplification. (A) U2OS cells were transiently transfected with plasmids expressing Flag, Flag-Cep76, Flag-CP110, GFP, or GFP-Cep97 and treated with 2 mM HU for 48 hours to allow centriole amplification. (Left panels) Transfected cells were stained with antibodies to Flag (green), centrin (red), and DAPI (blue). (Right panels) The number of centrin dots in transfected cells was counted. (B) U2OS cells were transiently transfected with plasmids expressing Flag, Flag-Plk4, Flag-Cep76, or Flag-Plk4 and Flag-Cep76. (Left panels) Transfected cells were stained with antibodies to Flag (green), CP110 (red), and with DAPI (blue). (Right panels) The number of CP110 dots in transfected cells was counted. In A–B, about 100 transfected cells were scored for each condition, and averaged data obtained from two to three independent experiments are shown. Error bars represent +/− S.D. Under our experimental conditions, “flowers” were observed in about 40–50% of asynchronously growing U2OS cells and generally contain 3–4 “petals” or procentrioles.
Next, we asked whether centriole amplification mediated by ectopic Plk4 could also be suppressed by enforced expression of Cep76. We transfected U2OS cells with Flag-Cep76 and Flag-Plk4, individually or in combination, and scored cells for the presence of centriolar “rosettes.” Expression of Cep76 was unable to suppress Plk-4-mediated centriole amplification (Fig. 6B), suggesting that Cep76 function is specifically associated with suppression of amplification generated through multiple rounds of duplication. Similar results were obtained upon enforced expression of myc-tagged Plk4 (data not shown). Our data also strengthen the notion that HU- and Plk4-mediated amplification are mechanistically separable and pinpoint Cep76 as a protein that distinguishes the two processes.
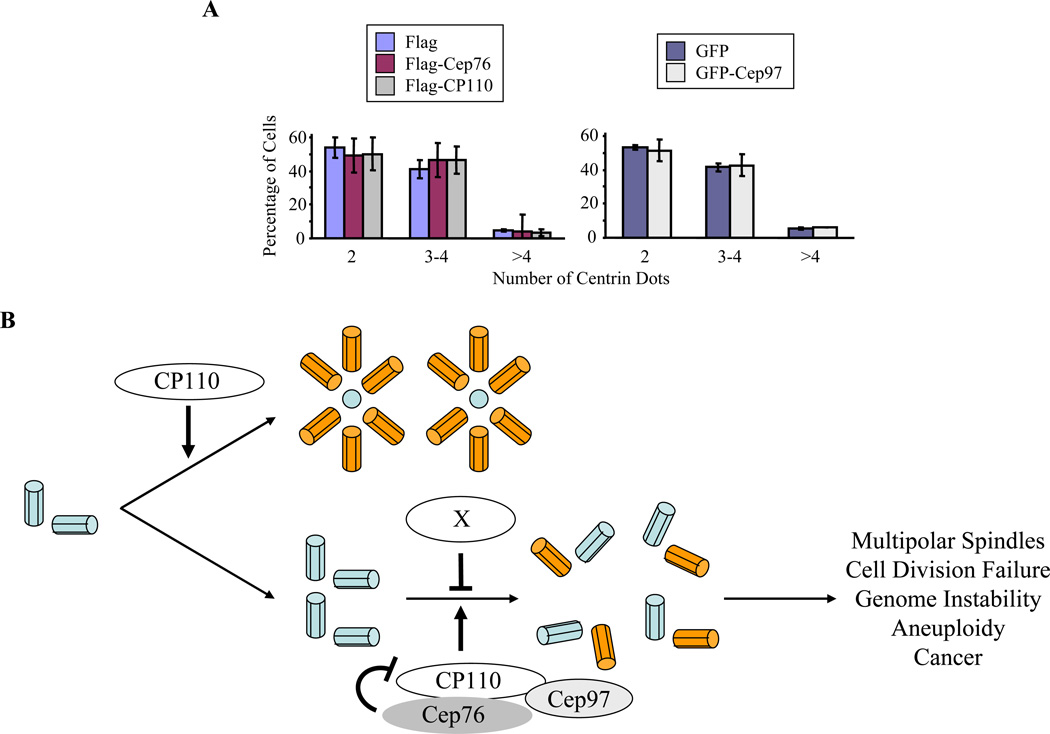
Cep76 does not suppress normal centriole duplication
The foregoing results suggested that Cep76 expression could inhibit events associated with centriole duplication. To distinguish whether the protein played a role in normal centriole duplication, we performed analogous experiments in which we expressed Cep76 in U20S cells that were otherwise untreated and counted centrin dots. We found that, in contrast to HU-treated cells, there was no apparent impact on the number of centrioles (Fig. 7A). Similar results were obtained after counting centriole numbers in untreated cells expressing either Cep97 or CP110 as controls (Fig. 7A). Taken together, our data strongly suggest that Cep76 specifically plays a role in inhibiting centriole re-duplication rather than normal duplication. Our data further imply that Cep76-dependent pathways could distinguish events associated with normal versus abnormal centriole duplication.
Figure 7.
Ectopic expression of Cep76 does not prevent normal centriole duplication. (A) U2OS cells were transiently transfected with plasmids expressing Flag, Flag-Cep76, Flag-CP110, GFP, or GFP-Cep97 and grown for 72 hours. The number of centrin dots in transfected cells was counted. About 100 transfected cells were scored for each condition, and averaged data obtained from three independent experiments are shown. Error bars represent +/− S.D. (B) Model depicting the role of Cep76 in suppressing amplification of centrioles. Cep76 interacts with CP110 and Cep97 to specifically restrain centriole re-duplication. In non-transformed, diploid cells or transformed cells that originate from certain cell types, the mechanisms suppressing centriole amplification could involve additional unknown factor(s)(protein X). Centriole amplification, in turn, could contribute to mitotic failure, genome instability, aneuploidy, and cancer.
Discussion
Recently, a number of groups have begun dissecting the controls regulating centriole duplication and ciliogenesis. Our work has focused on the role of CP110, which has been shown to play an integral role in centriole biogenesis, cytokinesis, and ciliogenesis (Chen et al., 2002; Kleylein-Sohn et al., 2007; Spektor et al., 2007; Tsang et al., 2008; Tsang et al., 2006). Given the multitude of diverse CP110 complexes ranging in size from several hundred kDa to a few MDa (Tsang et al., 2008; Tsang et al., 2006), it will be essential to elucidate the individual activities of each of these complexes. Based on studies presented here and elsewhere (Spektor et al., 2007; Tsang et al., 2008; Tsang et al., 2006), it is clear that CP110 exhibits diverse roles, since certain CP110 complexes play a role in centriole duplication and other centrosome cycle events, and others play an inhibitory role through control of centriole length and ciliogenesis. Here, we have identified an uncharacterized protein, Cep76, which provides an additional layer of control involved in regulating the process of centriole duplication.
A model for suppression of centriole re-duplication
Based on our findings, we postulate that depletion of Cep76 causes cells to become permissive to multiple rounds of centriole duplication (Fig. 7B). This permissiveness could be exquisitely sensitive to Cep76 levels. Thus, our model predicts that mitotic and G1 phase cells, wherein Cep76 is much less abundant, would be able to replicate their centrioles, whereas cells that have entered S phase have exited the permissive stage, at least in part through the up-regulation of Cep76. Since enforced Cep76 expression specifically attenuates HU-induced amplification but has no impact on Plk4-induced centriole amplification or normal centriole duplication, it is possible that Cep76 functions to specifically prevent the production of supernumerary centrioles through multiple rounds of duplication rather than through a single round of amplification. Consistent with the notion that centriole amplification induced by Plk4 is mechanistically different from that of Cep76, the supernumerary centrioles and intermediates produced following Cep76 ablation generally do not resemble a “flower” or “rosette” pattern (data not shown), as is frequently seen following Plk4 expression. In light of previous findings that both Plk4- and HU-mediated centriole amplification are dependent on CP110 (Chen et al., 2002; Kleylein-Sohn et al., 2007), it is intriguing that Cep76 specifically affects one pathway. In other words, according to the two “rules” of control proposed for centrosome duplication (Nigg, 2007), Cep76 could specifically exert cell cycle control by providing a centrosome-intrinsic block to re-duplication after normal duplication has occurred (Wong and Stearns, 2003), thereby restricting the number of duplication events to one per cycle. At present, we do not know precisely how Cep76 suppresses centriole re-duplication at a molecular level, and experiments are currently underway to identify the compendium of proteins with which Cep76 and CP110 interact. We note that Cep76 contains a calcium-dependent lipid-binding (CaLB) domain, and given the pivotal role that calcium-CaM signaling plays in the cell cycle, cytokinesis, and centrosome function, it will be interesting to determine whether calcium signaling also plays a role in Cep76 function and suppression of centriole over-duplication.
Consequences of centriole amplification
Centrosome amplification is a common hallmark in cancer cells and correlates with multipolar spindles, genome instability and aneuploidy (Brinkley, 2001; Lingle et al., 2002; Lingle et al., 1998). Whether centrosome amplification is the cause or consequence of cancer is not yet clear. Recent evidence indicates that centrosome amplification does not necessarily lead to genomic instability and cancer and that cells possess mechanisms to compensate for the presence of extra centrioles, thereby suppressing multipolar mitoses. Supernumerary centrioles are known to coalescence or cluster, resulting in the formation of a normal bipolar spindle (Basto et al., 2008; Kwon et al., 2008; Quintyne et al., 2005). In addition, extra centrioles can prolong mitosis by delaying satisfaction of the spindle assembly checkpoint (Basto et al., 2008; Yang et al., 2008). This mechanism is thought to allow cells, especially cancer cells, extra time to reorganize their chromatin and to divide with biopolar spindles. We speculate that a different mechanism could allow cells to cope with extra centrioles: centrioles that have not yet elongated to full length and matured to the point that they have accumulated pericentriolar material and become functional microtubule-organizing centers (i.e. centriolar intermediates) are selectively destroyed or inactivated during mitosis. Identification of mechanisms controlling the formation, degradation, or inactivation of centriolar intermediates will permit further characterization of the potential functionality of these transient structures. It is interesting to note that centriole inactivation has been previously reported in Drosophila cell lines stably expressing Plk4 (Basto et al., 2008); however, it is uncertain as to whether all the Plk4-induced supernumerary centrioles have reached full “maturity”. It is possible that centrioles that have elongated to full length were retained and formed clusters at the spindles, whereas others that have not reached full length were inactivated in mitosis, potentially explaining why some centrioles clustered whereas others were destroyed in this model system.
Cep76 in normal versus cancer cells and in cancer cells derived from tissues of diverse origin
The amplification phenotype associated with Cep76 loss was observed in osteosarcomas and blastomas, but not in non-transformed, diploid human or mouse cell lines or in carcinomas. We believe that these results have potential implications for human cancer, and may offer clues about how centrosomes are regulated in different tissues. Since carcinomas arise from cells of epithelial origin, one intriguing possibility is that centriole amplification, which can potentially lead to cell division failure, genomic instability, aneuploidy and cancer, may play a more prominent role during cellular transformation in tissues of non-epithelial origin. Alternatively, it is possible that non-transformed cells or transformed cells of epithelial origin possess additional mechanisms for suppressing centriole amplification. This mechanism may involve the participation of multiple gene products at the centrosome (Cep76 and factor X; Fig. 7B), such that a defect in one gene (Cep76 or factor X) would not be sufficient to induce the production of supernumerary centrioles. In other words, a guardian role for Cep76 is only uncovered after cells have been “pushed” into a less stable position through an antecedent event, namely, loss of another, undefined regulatory protein (factor X). Given that the phenotype of cells depleted of Cep76 is observed in certain cancers, it will be important to determine whether this gene is rearranged or mutated in human cancer and whether there are additional genes whose loss predisposes cells to centriole amplification in the absence of Cep76. In any event, identification of Cep76 could facilitate the identification of additional proteins necessary for the strict maintenance of appropriate centriole numbers in normal cells.
Experimental Procedures
Cell culture and plasmids
RPE-1 hTERT (RPE-1) cells were kindly provided by A. Khodjakov. NIH-3T3, MC3T3, IMR90, MRC5, HeLa, MCF7, A549, Hs578T, U2OS, Saos-2, MG63, SJSA1, HOS, 143B, T173, T98G, SKNSH, A172, and 293T cells were obtained from ATCC. All cells were cultured in DMEM supplemented with 10% FBS at 37°C in a humidified 5% CO2 atmosphere. Plasmids expressing recombinant Flag-CP110 and GFP-Cep97 proteins were described previously (Spektor et al., 2007; Tsang et al., 2006). Plasmids expressing recombinant epitope-tagged Plk4 were obtained from N.K. Soung, K. Lee, and E. Nigg.
Identification of Cep76
The identification of CP110-interacting proteins using immuno-affinity purification and mass spectrometric sequencing was described previously (Spektor et al., 2007). Two Cep76 peptides were identified using these methods.
Antibodies
To generate rabbit anti-Cep76 antibodies, a glutathione-S-transferase (GST) fusion protein containing residues 1–143 and 315–451 of Cep76 was expressed in E. coli and purified to homogeneity. Antibodies against Cep76 were purified by affinity chromatography. Other antibodies used included polyclonal rabbit anti-CP110 (Chen et al., 2002), anti-Cep97 (Spektor et al., 2007), anti-centrin mouse monoclonal 20H5 (J. Salisbury), anti-C-Nap-1 (E. Nigg), anti-Sas6 (P. Gonczy), anti-CPAP (T. Tang), anti-PCM-1 (A. Merdes), anti-α-tubulin, anti-Flag, and anti-γ-tubulin (all from Sigma-Aldrich), anti-phospho-Histone H3(Ser10) (Upstate), and anti-glutamylated tubulin GT335 (C. Janke).
Cell cycle synchronization and FACS analysis
T98G cells were synchronized by serum withdrawal and re-stimulation as described (Tsang et al., 2007). U2OS and Saos-2 cells were synchronized with 0.4 mM mimosine (G1), 2 mM HU (G1/S), or 40 ng/ml nocodazole (G2/M) for 24 hours. Cells released from the HU block for 6–7, 10, and 12–14 hours progressed into S/G2, G2, and M phase, respectively. Propidium iodide staining and FACS analysis were performed as described previously (Tsang et al., 2007).
RT-PCR
Extraction of total RNA was performed using TRIzol reagent (Invitrogen). cDNA synthesis was performed using SuperScript First-Strand synthesis kit (Invitrogen), and resulting cDNAs were amplified by PCR using gene-specific primers for Cep76 and actin. Two independent reactions were performed for each set of primers. Linear amplification was ensured in each case. Primer sequences are available upon request.
Immunoprecipitation, immunoblotting, and immunofluorescence microscopy
Cells were lysed with buffer containing 50 mM Hepes pH 7, 250 mM NaCl, 5 mM EDTA/pH 8, 0.1% NP-40, 1 mM DTT, 0.5 mM PMSF, 2 µg/ml leupeptin, 2 µg aprotinin, 10 mM NaF, 50 mM β-glycerophosphate, and 10% glycerol at 4°C for 30 minutes. For most experiments, 2 mg of extract was immunoprecipitated, and precipitated polypeptides were analyzed by SDS-PAGE and immunoblotting. Typically, 50–100 µg of lysate were loaded in the input (IN) lane. For expression and mapping studies, 293T cells were transfected with Flag-tagged constructs using calcium phosphate. Cells were harvested 48–72 hours after transfection. Indirect immunofluorescence was performed as described (Chen et al., 2002). Briefly, cells were grown on glass coverslips and fixed with cold methanol for 2 minutes. Cells were then permeabilized with 1% Triton X-100/PBS for 5 minutes. Slides were blocked with 3% BSA in 0.1% Triton X-100/PBS prior to incubation with primary antibodies. Secondary antibodies used were Cy3- or FITC-conjugated donkey anti-mouse or anti-rabbit IgG (Jackson Immunolabs). Cells were also stained with DAPI. Image acquisition was performed on a Zeiss Axiovert 200M microscope (63X objective lens, N.A. 1.4, 1.63 Optovar) equipped with a cooled Retiga 2000R CCD (QImaging) and Metamorph Software (Molecular Devices).
Electron Microscopy
Transmission electron microscopy was performed as described previously (Duensing et al., 2007).
RNA interference
Synthetic siRNA oligonucleotides were purchased from Dharmacon. Transfection of siRNAs was performed using Oligofectamine (Invitrogen) or sImporter (Upstate) per manufacturer’s instructions. The 21-nucleotide siRNA sequence for the non-specific control was 5’-AATTCTCCGAACGTGTCACGT-3’. The 21-nucleotide siRNA sequences for Cep76 were: 5’-GATAGACAATCGACATTAATT-3’, 5’-CAACACGGAGGTATCTTTATT-3’, 5’-GAGCGTACAACAAGTATATTT-3’, and 5’-GCAGAGAAAGAGCGATTATTT-3’. The siRNAs for CP110 and Cep97 silencing were described previously (Spektor et al., 2007). The miRNA sequences for Cep76 used were 5’-GATCATGTTTGCTTGTAAA-3’ and 5’-CTGATTCAACAACAATGTT-3’. The miRNAs sequences were cloned into the Mir30 miRNA expression system (Open Biosystems), and the resultant plasmids were transfected into 293T Phoenix cells to generate retroviruses. Cells were infected three times over a period of 24 hours and selected in 2 µg/ml puromycin for three days to generate stable lines.
Statistical Analysis
The statistical significance of the difference between two means was determined using a two-tailed Student’s t-test. Differences were considered significant when p<0.05.
Acknowledgements
We are grateful to William Lane (Harvard Microchemical Facility) for his assistance with mass spectrometric identification of CP110-interacting proteins, and to Joseph Suhan for invaluable help with electron microscopy. We thank D. Khoo for his initial involvement in this work, which suggested a role for Cep76 in centriole amplification. G. Hsu is also acknowledged for his assistance. We thank all members of the Dynlacht laboratory for constructive advice and encouragement. We thank J. Salisbury, P. Gonczy, T. Tang, A. Merdes, E. Nigg, and C. Janke for providing antibodies, and N.K. Soung, K. Lee, and E. Nigg for providing epitope-tagged Plk4 plasmids. B.D.D. was supported in part by an Irma T. Hirschl Career Scientist Award and March of Dimes research grant, for which he expresses his gratitude. S. D. was supported by NIH grant R01 CA112598 and a Research Scholar grant from the American Cancer Society. A.S. and W.Y.T. were supported by a DOD Prostate Cancer pre-doctoral fellowship and an Alberta Heritage Foundation for Medical Research full-time postdoctoral fellowship, respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Balczon R, Bao L, Zimmer WE, Brown K, Zinkowski RP, Brinkley BR. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J Cell Biol. 1995;130:105–115. doi: 10.1083/jcb.130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, Raff JW. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkley BR. Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol. 2001;11:18–21. doi: 10.1016/s0962-8924(00)01872-9. [DOI] [PubMed] [Google Scholar]
- Chen Z, Indjeian VB, McManus M, Wang L, Dynlacht BD. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev Cell. 2002;3:339–350. doi: 10.1016/s1534-5807(02)00258-7. [DOI] [PubMed] [Google Scholar]
- Duensing A, Liu Y, Perdreau SA, Kleylein-Sohn J, Nigg EA, Duensing S. Centriole overduplication through the concurrent formation of multiple daughter centrioles at single maternal templates. Oncogene. 2007;26:6280–6288. doi: 10.1038/sj.onc.1210456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk HA, Winey M. The mouse Mps1p-like kinase regulates centrosome duplication. Cell. 2001;106:95–104. doi: 10.1016/s0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH, Sluder G. Centrosome duplication: three kinases come up a winner! Curr Biol. 2001a;11:R698–R701. doi: 10.1016/s0960-9822(01)00412-2. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH, Sluder G. "It takes two to tango": understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev. 2001b;15:1167–1181. doi: 10.1101/gad.894001. [DOI] [PubMed] [Google Scholar]
- Kasbek C, Yang CH, Yusof AM, Chapman HM, Winey M, Fisk HA. Preventing the degradation of mps1 at centrosomes is sufficient to cause centrosome reduplication in human cells. Mol Biol Cell. 2007;18:4457–4469. doi: 10.1091/mbc.E07-03-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Kubo A, Sasaki H, Yuba-Kubo A, Tsukita S, Shiina N. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J Cell Biol. 1999;147:969–980. doi: 10.1083/jcb.147.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama R, Terada Y, Lee KS, Wang CL. Centrosome replication in hydroxyurea-arrested CHO cells expressing GFP-tagged centrin2. J Cell Sci. 2007;120:2444–2453. doi: 10.1242/jcs.008938. [DOI] [PubMed] [Google Scholar]
- Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, Pellman D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle WL, Barrett SL, Negron VC, D'Assoro AB, Boeneman K, Liu W, Whitehead CM, Reynolds C, Salisbury JL. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci U S A. 2002;99:1978–1983. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle WL, Lutz WH, Ingle JN, Maihle NJ, Salisbury JL. Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc Natl Acad Sci U S A. 1998;95:2950–2955. doi: 10.1073/pnas.95.6.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J, Hergert P, Magidson V, Khodjakov A. Control of daughter centriole formation by the pericentriolar material. Nat Cell Biol. 2008;10:322–328. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. Centrosome duplication: of rules and licenses. Trends Cell Biol. 2007;17:215–221. doi: 10.1016/j.tcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Okuda M, Horn HF, Tarapore P, Tokuyama Y, Smulian AG, Chan PK, Knudsen ES, Hofmann IA, Snyder JD, Bove KE, Fukasawa K. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 2000;103:127–140. doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- Prosser SL, Straatman KR, Fry AM. Molecular dissection of the centrosome overduplication pathway in S-phase arrested cells. Mol Cell Biol. 2009 doi: 10.1128/MCB.01124-08. (accepted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–129. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and CP110 suppress a cilia assembly program. Cell. 2007;130:678–690. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gonczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WY, Bossard C, Khanna H, Peranen J, Swaroop A, Malhotra V, Dynlacht BD. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev Cell. 2008;15:187–197. doi: 10.1016/j.devcel.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WY, Spektor A, Luciano DJ, Indjeian VB, Chen Z, Salisbury JL, Sanchez I, Dynlacht BD. CP110 cooperates with two calcium-binding proteins to regulate cytokinesis and genome stability. Mol Biol Cell. 2006;17:3423–3434. doi: 10.1091/mbc.E06-04-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WY, Wang L, Chen Z, Sanchez I, Dynlacht BD. SCAPER, a novel cyclin A-interacting protein that regulates cell cycle progression. J Cell Biol. 2007;178:621–633. doi: 10.1083/jcb.200701166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- Wong C, Stearns T. Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nat Cell Biol. 2003;5:539–544. doi: 10.1038/ncb993. [DOI] [PubMed] [Google Scholar]
- Yang Z, Loncarek J, Khodjakov A, Rieder CL. Extra centrosomes and/or chromosomes prolong mitosis in human cells. Nat Cell Biol. 2008;10:748–751. doi: 10.1038/ncb1738. [DOI] [PMC free article] [PubMed] [Google Scholar]