Abstract
Here; we have described and tested a microarray based-method for the screening of dengue virus (DENV) serotypes. This DNA microarray assay is specific and sensitive and can detect dual infections with two dengue virus serotypes and single-serotype infections. Other methodologies may underestimate samples containing more than one serotype. This technology can be used to discriminate between the four DENV serotypes. Single-stranded DNA targets were covalently attached to glass slides and hybridised with specific labelled probes. DENV isolates and dengue samples were used to evaluate microarray performance. Our results demonstrate that the probes hybridized specifically to DENV serotypes; with no detection of unspecific signals. This finding provides evidence that specific probes can effectively identify single and double infections in DENV samples.
Keywords: dengue virus, humans, Aedes, microarrays
1. Introduction
Dengue is a mosquito-borne viral infection and a major global public health problem [1]. The most common infection produces the classical dengue fever (DF), which is characterized by a sudden onset of rash, high fever, headache, and backache. The main clinical manifestations, namely dengue haemorrhagic fever (DHF) and dengue shock syndrome (DSS), are responsible for high morbidity and mortality rates every year. Over 40% (2.5 billion) of the population in 100 tropical and subtropical countries continue to live under the threat of contracting dengue infection. It is estimated that 100 million cases of DF, 500,000 cases of DHF, and 25,000 deaths are reported annually worldwide [1]. The fatality rate due to DHF may be reduced significantly with the aid of modern supportive therapies based on early diagnosis of the specific viral infection, but in most cases, these deaths are due to the lack of an early diagnosis of DENV infection, which is caused by four distinct serotypes: DENV-1, DENV-2, DENV-3, and DENV-4. Despite extensive research, there are currently no vaccines available [2,3], although candidate dengue vaccines have recently entered phase III trials in Asia and Latin America [4,5].
DENV (genus Flavivirus, family Flaviviridae) is mosquito-borne and mainly transmitted by Aedes (Stegomyia) aegypti (L.) and Aedes (Stegomyia) albopictus (Skuse) (Diptera: Culicidae), which are infected through receptors in the midgut [6–8]. This virus has a positive-strand RNA genome of approximately 11,000 bases, which encodes three structural and seven non-structural proteins. The detection and typing of this virus, not only in the patient but also in the vector, will lead to the development of new strategies for controlling and handling DENV outbreaks. In addition, surveillance of mosquitoes infected with DENV can help monitor infection rates within vector populations harbouring specific serotypes and provide early warning of pending epidemics.
There is a growing need for rapid and reliable methods for serotype identification of DENV infections in human serum specimens and mosquito vectors. The identification of DENV serotypes is of particular importance because these infections have been associated with DHF/DSS [9–11]. Furthermore, DENV testing is required to confirm the diagnosis of DENV infection in order to differentiate it from other febrile tropical illnesses. Conventional diagnosis of DENV infections includes the detection of virus in serum by isolation in culture and detection of specific viral molecules, such as genome RNA or dengue antigens [12,13], or by detection of specific anti-dengue antibodies [14]. Isolation of DENV provides the most direct and conclusive approach, although virus isolation takes 8 to 10 days, depending on titre. Anti-dengue antibodies can be detected within the first 5 days of symptoms or the 4 to 5 days required for the immune system to produce a sufficient amount of antibodies. Moreover, antibodies produce misleading results in secondary infections due to cross-reactivities among serotype-specific antibodies and other flavivirus antibodies. Reverse transcription-polymerase chain reaction (RT-PCR) is another potential method for detection of viral RNA [15–19]. Initial studies demonstrated the need to obtain virus isolates from C6/36 cells infected with each virus sample before detection by RT-PCR [20,21]. This method is also limited by the number of species that can be detected and identified in a single test, often requiring multiple parallel reactions [22]. Real-time RT-PCR is more sensitive than RT-PCR [23] and may be used to develop a diagnostic test. Single-tube or parallel multiplex PCR assays are alternative methods that can be coupled to hybridisation using conventional microarrays or flow-thru DNA chips [22,24]. Previous studies have described the use of DNA microarrays to study respiratory viruses in hospitalised infants admitted to intensive care units as well as in paediatric units and in adult patients with influenza-like illnesses [25–27]. These authors found that global detection rates appeared to be higher with DNA microarrays than with conventional virological methods such as direct immunofluorescence, virus isolation, and RT-PCR. This is the case not only in suspected human cases but also in mosquito populations [16]. Recently, several researchers have reported the use of fully automated real-time RT-PCR assays for the detection and serotype identification of DENV in patient sera [28,29]. However, this method does not allow multiplexing for more than four targets. In contrast, microarrays are routinely used in fields where a multiplex detection approach is required [22,24,30]. Furthermore, DNA microarray technologies have found applications in disease diagnosis [30–32], gene discovery [33], genetic studies [34], pharmacogenomics [35], and toxicology [36].
Here, we demonstrate the application of this DNA microarray technology to detecting single or dual infection with two dengue virus serotypes. The system is capable of detecting DENV serotype (DENV-1, DENV-2, DENV-3, and DENV-4) during the early phase of illness or in mosquito populations. The technique provides sufficient sensitivity and reproducibility for the diagnosis of dengue in human and mosquito samples. Thus, our biosensor will facilitate early dengue diagnoses and provide a reliable estimation of dengue infection in mosquito vectors for prevention, surveillance, and control programs.
2. Experimental Section
2.1. Materials
All chemicals and solvents were purchased from Fluka-Sigma (St. Louis, MO, USA) at the highest analytical grades. Corning UltraGAPS, Aminosilane Coated Slides (Corning Incorporated, Corning, NY, USA), or Nexterion® Slide A+Aminosilane-Coated Substrate (Schott Nexterion, Jena, Germany) were used in all experiments.
2.2. Viruses
The flaviviruses used in this study included DENV-1, strain Hawaii (HA1-1944); DENV-2, strain New Guinea C (NGC-1944); DENV-3, strain Philippines (H-87); and DENV-4, strain Philippines (H-241-1946). The viruses were obtained from Dr. Duane J. Gubler (Division of Vector-borne Infectious Diseases, Centers for Disease Control, Fort Collins, CO, USA).
2.3. Serum Samples
Serum specimens were collected from 197 patients in 2000, 2001, and 2004–2006 in an epidemiologic surveillance program carried out by the Secretaria de Salud de Oaxaca and the Instituto Mexicano del Seguro Social (IMSS) of Veracruz, Mexico. DENV was evaluated by Mac-ELISA (IgM Antibody Capture Enzyme-Linked Immunosorbent Assay, MAC-ELISA; PANBIO, Brisbane, Australia) at the Secretaria de Salud [37]. This study used excess samples that remained after diagnostic testing and were stored at −70 °C. The study was reviewed and approved by the Institutional Review Board of the IMSS (Commission of Scientific Research) and the Bioethical Commission for Research in Humans of Centro de Investigación y de Estudios Avanzados del IPN (Comité de Bioética Para la Investigación en Seres Humanos, COBISH-CINVESTAV).
2.4. Mosquito Collections
Mosquitoes were collected from at least four sites in the Mexican states of Quintana Roo, Yucatan, Campeche, Tabasco, Chiapas, Veracruz, Oaxaca, Puebla, Guerrero, Morelos, Hidalgo, Mexico, Tamaulipas, Queretaro, Guanajuato, Michoacán, Colima, Jalisco, and Nayarit. Geographic locations and numbers of adult mosquitoes collected at natural sites in each city are listed in Table 1. Pools of ten adult mosquitoes were examined at each site to confirm their identities as Ae. aegypti or Ae. albopictus and then stored at −70 °C.
Table 1.
Locations, dates of collections, global positioning system coordinates and sample sizes of Ae. aegypti collections in Mexico.
| State | City | Collection Date | Coordinates (Latitude/Longitude) | Female Pools |
|---|---|---|---|---|
| Cancun | 07/08/2005 | 21°09′00.93″/86°49′39.20″ | 19 | |
| Quintana Roo | Cozumel | 08/08/2005 | 20°30′31.53″/86°56′45.42″ | 6 |
| Felipe Carrillo Puerto | 09/08/2005 | 19°35′12.24″/88°02′22.67″ | 11 | |
| Puerto Arturo | 11/08/2005 | 19°39′51.45″/89°04′57.21″ | 13 | |
| La Esperanza | 13/08/2005 | 21°02′09.70″/87°38′33.46″ | 14 | |
| Yucatan | Tinum | 15/08/2005 | 20°46′03.22″/88°23′18.78″ | 6 |
| Ticum | 17/08/2005 | 20°08′50.65″/89°12′54.87″ | 11 | |
| Merida | 19/08/2005 | 20°59′31.05″/89°38′08.56″ | 19 | |
| Hopelchen | 21/08/2005 | 19°44′37.80″/89°50′38.49″ | 9 | |
| Campeche | Calakmul | 23/08/2005 | 18°21′18.04″/89°32′15.35″ | 19 |
| Francisco Escarcega | 25/08/2005 | 18°36′00.02″/90°43′41.11″ | 13 | |
| Palizada | 27/08/2005 | 18°15′15.04″/92°05′25.22″ | 8 | |
| Tenosique | 04/09/2005 | 17°28′54.38″/91°24′59.10″ | 19 | |
| Tabasco | Jalapa | 06/09/2005 | 17°43′25.45″/92°48′44.87″ | 11 |
| Villahermosa | 08/09/2005 | 17°59′19.10″/92°55′54.18″ | 23 | |
| Paraiso | 10/09/2005 | 18°24′14.18″/93°12′17.94″ | 17 | |
| Ococingo | 12/09/2005 | 16°53′54.06″/92°06′14.24″ | 21 | |
| Chiapas | Tapachula | 14/09/2005 | 14°52′03.61″/92°16′18.06″ | 21 |
| Tuxtla Gutierrez | 16/09/2005 | 16°45′48.19″/93°06′09.98″ | 9 | |
| Arriaga | 18/09/2005 | 16°14′24.01″/93°54′34.13″ | 14 | |
| Acayucan | 02/10/2005 | 17°57′92.90″/94°55′15.11″ | 39 | |
| Veracruz | Tierra Blanca | 04/10/2005 | 18°27′33.10″/96°21′24.48″ | 18 |
| Xalapa | 06/10/2005 | 19°33′12.88″/96°54′44.00″ | 11 | |
| Tantoyuca | 08/10/2005 | 21°21′10.21″/98°13′08.71″ | 22 | |
| Tavela | 10/10/2005 | 16°39′37.79″/95°59′58.37″ | 14 | |
| Pochutla | 12/10/2005 | 15°44′52.55″/96°28′09.64″ | 37 | |
| Oaxaca I | 14/07/2004 | 17°02′34.13″/96°43′06.91″ | 22 | |
| Oaxaca | Oaxaca II | 14/10/2005 | 17°02′34.13″/96°43′06.91″ | 17 |
| Tuxtepec I | 17/07/2003 | 18°04′93.70″/96°06′78.30″ | 43 | |
| Tuxtepec II | 02/09/2004 | 18°04′93.70″/96°06′78.30″ | 45 | |
| Tuxtepec III | 16/10/2005 | 18°04′93.70″/96°06′78.30″ | 11 | |
| Nanahuatipan | 19/10/2005 | 18°07′21.42″/97°06′45.81″ | 17 | |
| Venustiano Carranza | 16/07/2005 | 20°30′31.30″/97°40′16.54″ | 11 | |
| Acateno | 17/07/2005 | 20°07′59.74″/97°12′18.12″ | 9 | |
| Puebla | Acatlan | 18/07/2005 | 18°12′00.94″/98°02′53.64″ | 2 |
| Izucar de Matamoros | 19/07/2005 | 18°35′52.52″/98°28′28.68″ | 13 | |
| Ometepec | 21/10/2005 | 16°41′04.02″/98°24′03.72″ | 4 | |
| Guerrero | Huamuxtitlan | 22/10/2005 | 17°47′50.38″/98°33′47.22″ | 22 |
| Huitzuco | 23/10/2005 | 18°18′31.09″/99°20′37.98″ | 12 | |
| Petatlan | 24/10/2005 | 17°32′12.48″/101°16′09.65″ | 17 | |
| Tepalcingo | 31/05/2005 | 18°35′57.97″/98°50′25.78″ | 8 | |
| Morelos | Yautepec | 31/05/2005 | 18°52′52.76″/99°04′02.78″ | 14 |
| Xochitepec | 30/05/2005 | 18°48′07.18″/99°14′08.09″ | 11 | |
| Puente de Ixtla | 30/05/2005 | 18°36′43.83″/99°19′58.28″ | 9 | |
| Huehuetla | 27/06/2005 | 20°31′37.64″/98°01′15.78″ | 4 | |
| Hidalgo | Jaltocan | 28/06/2005 | 21°10′23.65″/98°35′58.86″ | 2 |
| Metzitlan | 29/06/2005 | 20°35′31.25″/98°45′43.53″ | 6 | |
| Tlanalapa | 30/06/2005 | 19°49′38.55″/98°36′15.79″ | 3 | |
| Malinalco | 02/07/2005 | 18°46′49.23″/99°32′59.21″ | 14 | |
| Mexico | Nuevo Santo Tomas | 03/07/2005 | 19°10′26.34″/100°17′18.95″ | 7 |
| Tejupilco | 04/07/2005 | 18°53′23.32″/100°08′25.64″ | 9 | |
| Tlatlaya | 05/07/2005 | 18°32′17.37″/100°13′47.38″ | 9 | |
| Matamoros | 24/07/2005 | 25°50′36.99″/97°27′45.08″ | 8 | |
| Tamaulipas | Soto la Marina | 25/07/2005 | 23°46′09.07″/98°12′44.45″ | 9 |
| Ciudad Mante | 27/07/2005 | 22°45′14.20″/98°59′28.66″ | 31 | |
| Nuevo Laredo | 29/07/2005 | 27°29′06.75″/99°31′47.13″ | 3 | |
| Tancoyol | 04/06/2005 | 21°28′38.70″/99°18′53.55″ | 4 | |
| Queretaro | Landa de Matamoros | 05/06/2005 | 21°11′12.42″/99°19′13.32″ | 8 |
| Jalpan | 06/06/2005 | 21°10′17.79″/99°25′14.23″ | 3 | |
| Arroyo Seco | 07/06/2005 | 21°32′50.05″/99°41′15.58″ | 7 | |
| Guanajuato | 09/06/2005 | 21°01′12.33″/101°15′56.92″ | 7 | |
| Guanajuato | Irapuato | 11/06/2005 | 20°42′08.61″/101°22′12.88″ | 13 |
| Abasolo | 13/06/2005 | 20°27′04.59″/101°32′04.86″ | 2 | |
| Penjamo | 15/06/2005 | 20°26′13.61″/101°43′03.84″ | 6 | |
| Huetamo | 19/06/2005 | 18°37′11.89″/100°53′25.31″ | 14 | |
| Michoacan | Tacambaro | 21/06/2005 | 19°14′29.25″/101°27′13.96″ | 7 |
| Lázaro Cardenas | 23/06/2005 | 17°58′21.82″/102°13′41.12″ | 5 | |
| Apatzingan | 25/06/2005 | 19°05′21.86″/102°22′08.41″ | 9 | |
| Colima | 26/09/2005 | 19°14′43.90″/103°42′23.33″ | 21 | |
| Colima | Tecoman | 27/09/2005 | 18°54′54.16″/103°53′12.38″ | 9 |
| Minatitlan | 28/09/2005 | 19°23′16.59″/104°03′17.13″ | 4 | |
| Manzanillo | 29/09/2005 | 19°03′35.47″/104°17′18.53″ | 8 | |
| Pihuamo | 22/05/2005 | 19°15′24.83″/103°22′33.15″ | 9 | |
| Jalisco | Tequila | 24/05/2005 | 20°53′20.25″/103°50′02.26″ | 21 |
| Cihuatlan | 26/05/2005 | 19°13′58.41″/104°34′17.31″ | 4 | |
| Mismaloya | 28/05/2005 | 20°31′52.79″/105°17′09.14″ | 6 | |
| Ixtlan del Rio | 14/05/2005 | 21°01′52.83″/104°22′30.69″ | 8 | |
| Nayarit | Tepic | 16/05/2005 | 21°30′04.94″/104°53′04.03″ | 16 |
| Ixcuintla | 18/05/2005 | 21°48′04.57″/105°12′13.70″ | 6 | |
| Acaponeta | 20/05/2005 | 22°29′23.95″/105°22′29.68″ | 4 |
Aedes aegypti were collected in all cities;
Pools of Aedes albopictus collected.
2.5. Isolation of Dengue Virus
Aedes albopictus clone C6/36 cells were grown at 28 °C as described [38] to extract viral RNA from strains maintained in the laboratory and used as positive controls in our assays. After 18 h culture, the cells (2 × 106 per 100-mm plate) were infected with 0.2 mL DENV-1, -2, -3 or -4 with an input MOI of 600 PFU per plate and incubated at 28 °C for 10 days.
2.6. RNA Extraction
Total RNA was isolated from cell culture supernatants containing viruses obtained from infected cells and from mosquitoes collected and frozen in the field. RNA was obtained using Trizol LS (GIBCO BRL, Gaithersburg, MD, USA) according to the manufacturer's recommendations. Total RNA was also obtained from acute-phase plasma collected from patients in the epidemiologic surveillance program (Secretaria de Salud de Oaxaca, Mexico from 2000, 2001 and 2004–2006) using the Viral Nucleic Acid Extraction kit from Real Genomics (Real Biotech Corporation, Banqqiao, Tapei, Tiwan). The RNA was suspended in 50 μL H2O treated with diethylpyrocarbonate (DEPC, Sigma-Aldrich) and used as a template for reverse transcription-polymerase chain reaction (RT-PCR).
2.7. RT-PCR Assay
One-step RT-PCR (SuperScript One-Step RT-PCR with Platinum Taq, Invitrogen, Carlsbad, CA, USA) was performed to identify the optimal primers for DENV serotype detection. Primers targeting the C-prM [18], NS1 [17], NS3 [19,39], and NS5 [15] genes yielded products of different sizes (Table 2). Extracted RNA (5 μL) was used as a template in a 25-μL reaction volume. Assays used primers specific for DENV-1–4, but not Japanese encephalitis, Kunjin, or yellow fever viruses [19], and consensus primers common to several flaviviruses (Japanese encephalitis, Kunjin or yellow fever), including dengue viruses. Cycling conditions have been reported previously for each primer set (Table 2) [17–19,39]. Reaction mixtures were stored at −20 °C until further processing.
Table 2.
Probes and primers used for the detection of DENV serotype.
| Primer Name | Sequence | Genome Postition | Gen | Amplicon Size |
|---|---|---|---|---|
| Probes | ||||
| Texas RedDENV-1 | 5′-(Texas Red) AGTTTCTTTTCCTAAACACCTCG-3′ | 5045–5067 | NS3 | |
| 6FAMDENV-2 | 5′-(_6 FAM) CCGGTGTGCTCRGCYCTGAT-3′ | 5260–5279 | NS3 | |
| CY5DENV-3 | 5′-(_CY5) TTAGAGTYCTTAAGCGTCTCTTG-3′ | 5152–5174 | NS3 | |
| YakimaYellowDENV-4 | 5′-(_Yakima Yellow) CCTGGTTGATGACAAAAGTCTTG-3′ | 5320–5342 | NS3 | |
| Primers | ||||
| AD3 (-) [14] | 5′-CTGATTTCCAT(A, C,G,T)CC(A,G)TA-3′ | 3412–3428 | NS1 | |
| AD4 (+) | 5′-GA(C,T)ATGGG(C,G,T)TA(C,T)TGGATAGA-3′ | 3010–3029 | NS1 | 419(AD3 and AD4) |
| D1 | 5′-GAGGACCAATATCTCAG-3′ | 3135–3151 | NS1 | 265 (D1 and AD3) |
| D2 (+) | 5′-AAGCTTGAGATGGACTTT-3′ | 3235–3151 | NS1 | 194 (D2 and AD3) |
| D3 (+) | 5′-GTCTAGCTGGTCCCATT-3′ | 3150–3166 | NS1 | 271 (D3 and AD3) |
| D4 (+) | 5′-ATCATATGCGGGCCCTT-3′ | 3155–3171 | NS1 | 273 (D4 and AD3) |
| D1 [15] | 5′-TCAATATGCTGAAACGCGCGAGAAACCG-3′ | 134–161 | C-prM | |
| D2 | 5′-TTGCACCAACAGTCAATGTCTTCAGGTTC-3′ | 616–644 | C-prM | 511 (D1 and D2) |
| TS1 | 5′-CGTCTCAGTGATCCGGGGG-3′ | 568–586 | C-prM | 482 (Dl and TS1) |
| TS2 | 5′-CGCCACAAGGGCCATGAACAG-3′ | 232–252 | C-prM | 119 (Dl and TS2) |
| TS3 | 5′-TAACATCATCATGAGACAGAGC-3′ | 400–421 | C-prM | 290 (Dl and TS3) |
| TS4 | 5′-CTCTGTTGTCTTAAACAAGAGA-3′ | 506–527 | C-prM | 392 (Dl and TS4) |
| Bio-CFDJ9977a [12] | 5′- GCATGTCTTCCGTCGTCATCC-3′ | 9952–9977 | NS5 | |
| DEN1-J9243 | 5′-GCCTGAACATGCTCTATTGGCT-3′ | 9243–9264 | NS5 | 761 (J9243 and CFDJ) |
| DEN2-J9452 | 5′-TCTTCAAAAGCATTCAGCACCT-3′ | 9452–9473 | NS5 | 546 (J9452 and CFDJ) |
| DEN1-J9243 | 5′-GCCTGAACATGCTCTATTGGCT-3′ | 9243–9264 | NS5 | 761 (J9243 and CFDJ) |
| DEN2-J9452 | 5′-TCTTCAAAAGCATTCAGCACCT-3′ | 9452–9473 | NS5 | 546 (J9452 and CFDJ) |
| DEN3-9471 | 5′-CCCATCCGCTAGAGAAGAAAATTACAC-3′ | 9471–9497 | NS5 | 522 (J9471 and CFDJ) |
| DEN4-9580 | 5′- GGTTTGGCACTTCCCTCCTCTTCTTG-3′ | 9580–9605 | NS5 | 411 (9580 and CFDJ) |
| DV1 (+) [30] | 5′-GGRACKTCAGGWTCTCC-3′ | 4918–4934 | NS3 | |
| DV3 | 5′-AARTGIGCYTCRTCCAT-3′ | 5368–5384 | NS3 | 470 (DV1 and DV3) |
| DSP1 (-) [16] | 5′-AGTTTCTTTTCCTAAACACCTCG-3′ | 5067–5045 | NS3 | 169 (DV1 and DSP1) |
| DSP2 (-) | 5′-CCGGTGTGCTCRGCYCTGAT-3′ | 5279–5260 | NS3 | 362 (DV1 and DSP2 |
| DSP3 (-) | 5′-TTAGAGTYCTTAAGCGTCTCTTG-3′ | 5174–5152 | NS3 | 265 (DV1 and DSP3 |
| DSP4 (-) | 5′-CCTGGTTGATGACAAAAGTCTTG-3′ | 5342–5320 | NS3 | 426 (DV1 and DSP4) |
2.8. Probe Labelling
DENV-1, DENV-2, DENV-3, and DENV-4 specific primers (Table 2) were synthesized with the 5′ C6 amino linker by The Midland Certified Reagent Company (Midland, TX, USA). Each pair of oligonucleotides was validated by RT-PCR to verify amplification of products of the expected size.
2.9. DNA Microarray Fabrication
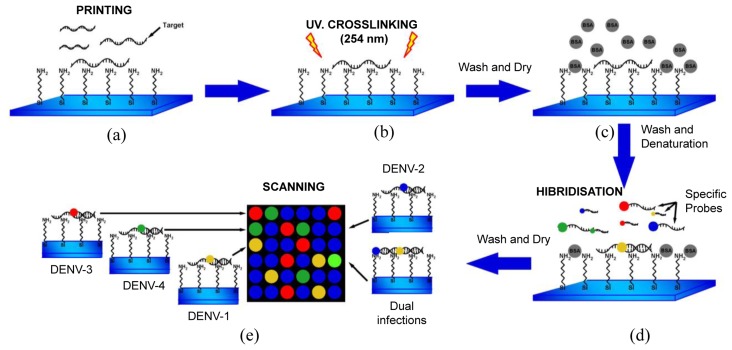
DNA microarray targets (RT-PCR product of DV1 and DV3) [19] were immobilized as follows (Figure 1). Targets (50–100 ng·μL−1) were denatured (10 min at 92 °C) and diluted in buffer [1.6 mol·mL−1 CHAPS, 1% CAPS, 50% DMSO buffer (pH 6.5)], and then printed on NEXTERION® Slide A+ aminosilane-coated slides with a Robot Spotter GeneTAC G3 (Genomic Solutions, Harvard Bioscience, Boston, MA, USA) at 55% relative humidity and a temperature of 20 °C to 25 °C (Figure 1a). Each solid pin delivered a volume of approximately 1 nL, which was designed to create a spot diameter of 300 μm. The centre-to-centre spacing of bright spots was 500 μm. The DNA was UV (254 nm)-cross linked at 250 mJ cm−2 and the slides were incubated for 20 min at 75 °C as shown in Figure 1b, followed by extensive washing with 0.1% SDS and two rinses with double-deionized water. The chips were blocked with pre-hybridisation buffer (4× SSC, 1% SDS, and 10 mg·mL−1 BSA) for 45 min at 42 °C, washed with double-deionized water for 20 s, and dried by filtered air flow (Figure 1c).
Figure 1.
(a) DNA microarray targets were printed on NEXTERION® Slide A+ aminosilane coated slides. (b) DNA was UV-cross linked at 250 mJ and slides were dried for 20 min at 75 °C. (c) Microarrays were blocked with pre-hybridisation buffer (4× SSC, 1% SDS, and 10 mg·mL−1 BSA) for 45 min at 42 °C. (d) Specific fluorescent probes (Table 2) for DENV-1, -2, -3, or -4 (5 μL) were hybridised to the DNA microarray. (e) After washing, the slides were scanned to detect Texas Red (583 nm), FAM (494 nm), Cy5 (530 nm), and Yakima yellow (646 nm).
2.10. Hybridizations, Detections and Statistical Analyses
Specific fluorescent DENV probes (Table 2) were hybridised to the immobilized DNA microarray targets. A volume of 5 μL containing specific probes for each DENV serotype (1–4) was used as shown in Figure 1d. The microarray substrates and coverslips were sealed in a humidity chamber (Bio-Rad Laboratories, Hercules, CA, USA) and incubated at 65 °C for at least 6 h. The optimal concentration of the probes for DENV-1 to -4 was 20 μM. The coverslips and supports were gently removed (4× SSC in a wash bottle) and the arrays were washed by immersion in 1× SSC, 0.1% SDS for 10 min (10 slides in 250 mL), 0.1× SSC, 0.1% SDS twice for 10 min, and 0.1× SSC twice for 10 min. Finally, the slides were blown dry by pushing the liquid away from the spots and toward the outer edges. The slides were scanned using a GeneTac LS IV Scanner (Genomic Solutions) to detect Texas Red, FAM, Cy5, and Yakima yellow fluorescence at excitation line wavelengths of 583, 494, 530, and 646 nm, respectively. Figure 1e illustrates this procedure. Fluorescence values were obtained using TM4 software [40] and statistical analyses were performed in S-Plus Statistics Software [41]. Normalization of the means was performed after background correction. The microarray data have been deposited in the NCBI Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) with GEO series accession number GPL11294-CINVESTAV-Dengue 0.6k Diagnostic Chip.
3. Results and Discussion
3.1. Selection of Probes for the Detection of DENV Serotype
To select effective primer pairs from published candidates (Table 2), we tested each PCR primer pair in a separate reaction for each serotype. Standard viral RNAs of DENV-1, strain Hawaii (HA1-1944); DENV-2, strain New Guinea C (NGC-1944); DENV-3, strain Philippines (H-87); and DENV-4, strain Philippines (H-241-1946) were assessed. Results measured by amplicon density are shown in Table 3. Optimal reproducibility was obtained with consensus primer DV1 and specific primers DSP1 to DSP4 (Table 2). DSP1–4 were labelled as described in the Experimental Section (Table 2).
Table 3.
Amplicon quantification by densitometry.
| Serotype | Primers * | Amount of DNA (ng) |
|---|---|---|
| DENV-1 | D1 and AD3 | 128 |
| D1 and TS3 | 89 | |
| DEN1-J9243 and CFDJ | 45 | |
| DV1 and DSP1 | 169 | |
| DENV-2 | D2 and AD3 | 160 |
| Dl and TS2 | 114 | |
| DEN2-J9452 and CFDJ | 39 | |
| DV1 and DSP2 | 199 | |
| DENV-3 | D3 and AD3 | 33 |
| Dl and TS3 | 92 | |
| DEN3-J9471 and CFDJ | 41 | |
| DV1 and DSP3 | 156 | |
| DENV-4 | D4 and AD3 | 25 |
| Dl and TS4 | 24 | |
| DEN4-9580 and CFDJ | 30 | |
| DV1 and DSP4 | 187 |
Table 2 lists primer sequences.
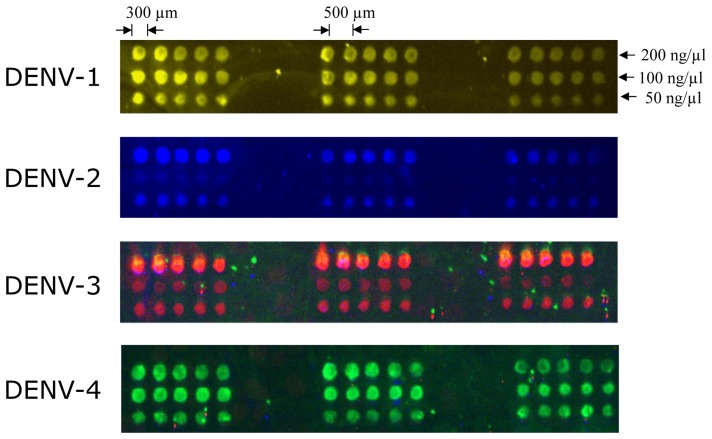
3.2. DNA Spot Concentration and Microarray Optimisation
Target density on the array surface must be optimised for effective hybridization. Several target concentrations (ranging from 50, 100, and 200 ng·μL−1) were tested as shown in Figure 2. To determine the surface density, further hybridization experiments were performed with fluorochrome-labelled probes. We found that the ideal DNA target concentration for fluorescence detection with the complementary labelled probe (20 μM) was 50 ng·μL−1. Triplicate arrays of five spots from the same sample produced virtually identical images. Probe specificity is also shown in Figure 2.
Figure 2.
Hybridisation patterns of representative DENV serotypes, indicated on the left side of each array. Vertical lanes represent different RT-PCR product concentrations (50, 100, and 200 ng·μL−1, respectively) for each DENV serotype. Specific labelled primers (Texas Red for DENV-1, FAM for DENV-2, CY5 for DENV-3 and Yakima Yellow for DENV-4) were hybridised as described in the Experimental Section and the arrays were spotted in triplicate.
3.3. Detection of Viral RNA in Clinical Serum Samples
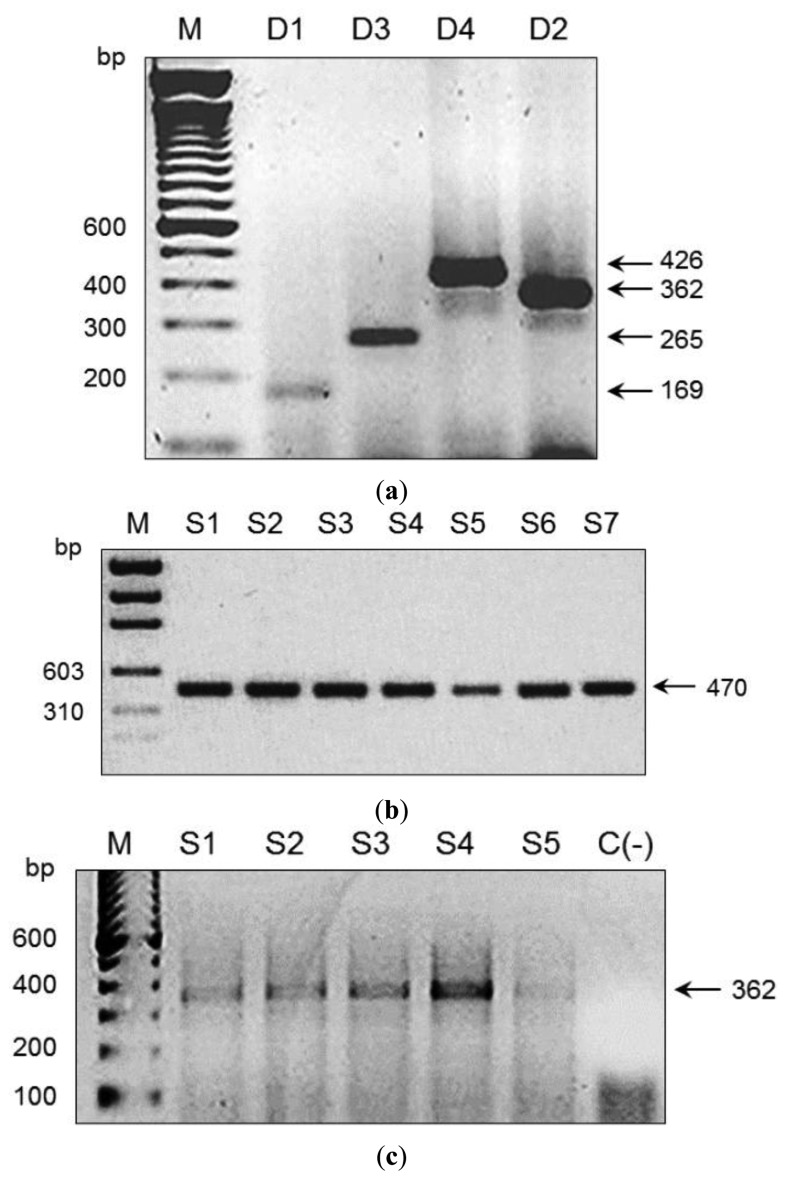
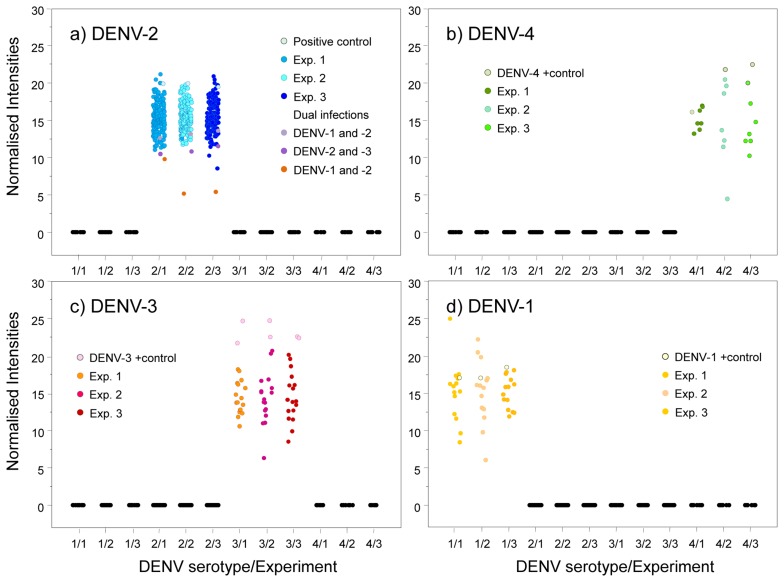
We used an amplicon derived from two consensus primers (DV1 and DV3) which contain the most conserved and specific sequences for each serotype, thus maximising the probability that all members of each viral serotype could be detected; this is a difficult and problematic task when using traditional methods. To assess the performance of our microarray in a clinical setting, the presence of DENV was evaluated in 197 human serum specimens from 12 Oaxaca cities and Veracruz. Results were validated by visual detection of the amplicon obtained with DV1 and specific primers DSP1-4 in agarose gels (Figure 3); this Figure also shows amplicons produced by consensus primers DV1 and DV3. Microarray results revealed DENV-2 in 88 samples from Oaxaca and 41 from Veracruz; DENV-3 in two samples from Oaxaca and ten from Veracruz; DENV-1 in six samples from Veracruz; and DENV-4 in seven samples from Veracruz (Figure 4). Dual infections with DENV-2 and -3 in one sample from Oaxaca and DENV-1 and -2 in a sample from Veracruz were also detected (Figure 4a). DENV serotype identity was confirmed in all samples by determining amplicon size (Figure 3). Microarray results are shown in Figure 4; two samples from the 197 analysed serum specimens that were reported as negative for DENV were also negative in this assay. Thirty-seven symptomatic patients were negative, and eleven that were reported as negative by the Mac-ELISA (IgM Antibody Capture Enzyme Linked Immunosorbent Assay) were positive by microarray analysis and confirmed by 2% agarose gel electrophoresis of the RT-PCR products. Figure 3 shows an example of these amplifications. Each assay included negative and positive controls, as well as RT-PCR products of known strains maintained in culture (isolates, VER1.1.5 for DENV-1; VER3.5.1 for DENV-2; VER1.2.1 for DENV-3, and VER1.2.7 for DENV-4). All negative controls were confirmed as negative by the microarray analysis. There were no false-negative samples, indicating the high specificity of this method. The microarray image platforms were submitted to GEO NCBI (Accession number: GPL11294, CINVESTAV, Dengue 0.6k Diagnostic Chip). These results confirmed the circulation of the four serotypes in the states of Oaxaca and Veracruz, Mexico (36). This study also showed that DENV-2 is present in serum at a higher frequency, consistent with previous assessments of the prevalence of these serotypes in the epidemics of 2000–2001 [37] and 2005–2006 [20,21].
Figure 3.
Amplification products of RT-PCR consensus and serotype-specific primer pairs. The RT-PCR products were observed in ethidium bromide-stained agarose gels (2%). Input viral RNAs were extracted from each control dengue virus strain. Results for serotypes DEN-1, DEN-2, DEN-3, and DEN-4 are shown. (a) The 169-, 362-, 265-, and 426-bp amplicons for DEN-1, DEN-2, DEN-3, and DEN-4, respectively. (b) The 470-bp amplicons expected for all four dengue serotypes using the consensus primers. (c) The 362-bp amplicons for DEN-2 obtained from five samples.
Figure 4.
Samples were evaluated for the presence of DENV-1, -2, -3, and -4. Plots represent the normalised intensity for each sample in three different experiments. Each panel represents values for DENV-1 (Texas Red), DENV-2 (FAM), DENV-3 (Cy5), or DENV-4 (Yakima yellow). (a) Fluorescence for 162 DENV-2-positive samples, 1 positive control, and 3 dual infections positive for DENV-1 and -2, DENV-2 and -3, and DENV-1 and -2. (b) Seven DENV-4-positive samples and 1 positive control. (c) Seventeen DENV-3-positive samples and 2 positive controls. (d) Fourteen DENV-1-positive samples and 1 positive control. All positive samples yielded fluorescence intensities >0.2. Each experiment is represented by spots of different colours. Each spot represents the average fluorescence intensity of triplicate spots. Analysis was performed in S-Plus software [S-PLUS: Copyright 1988, 2007 Insightful Corp. Rev Date:Wed May 02 10:07:00 2007 Buil8052. Seria#: WTB00000001. Juan Burgueño. CIMMYT, INC; 41].
3.4. Detection of Viral RNA in Field-Collected Mosquito Vectors of Dengue
Microarrays were also used to monitor and type DENV RNA in Aedes mosquitoes caught in the field (Table 1). Mosquito collection was conducted in 76 cities and 19 states with reported dengue cases (Table 1). DENV was detected in 35 cities: 28 contained DENV-2, six contained DENV-1, and three contained DENV-3 (Table 4). Mosquito pools collected in the states of Quintana Roo, Puebla, Hidalgo, Mexico, Queretaro, Guanajuato, Michoacán, and Jalisco did not contain DENV RNA. Table 1 shows the number of pools collected per city and state; Table 4 and Figure 4 illustrate serotype detection by microarray analysis. Figure 3 also shows representative examples of validation by agarose gel electrophoresis of RT-PCR products using the consensus primer DV1 and specific primers DSP1-4. All assays included positive controls for each serotype maintained in cell culture (isolates VER1.1.5 for DENV-1; VER3.5.1 for DENV-2; VER1.2.1 for DENV-3, and VER1.2.7 for DENV-4) and negative controls (42 spots) with RT-PCR products of non-infected cultured mosquito cells (Accession number: GPL11294, CINVESTAV, Dengue 0.6k Diagnostic Chip).
Table 4.
Mosquito pools tested for DENV by microarray technology and validation by RT-PCR and agarose gel electrophoresis.
| State | City | Infected Mosquito Pools | DENV Serotype * | |
|---|---|---|---|---|
|
| ||||
| Microarray | RT-PCR | |||
| Yucatan | La Esperanza | 7 | 2 | 2 |
| Tinum | 5 | 1 | 1 | |
| Ticum | 10 | 2 | 2 | |
| Mérida | 5 | 2 | 2 | |
| Campeche | Hopelchen | 9 | 2 | 2 |
| Calakmul | 11 | 2 | 2 | |
| Francisco Escarcega | 3 | 2 | 2 | |
| Palizada | 7 | 2 | 2 | |
| Tabasco | Tenosique | 14 | 3 | 3 |
| Jalapa | 4 | 2 | 2 | |
| Villahermosa | 1 | 2 | 2 | |
| Paraíso | 9 | 1 | 1 | |
| Chiapas | Ococingo | 2 | 2 | 2 |
| Tapachula ** | 7 * | 2 | 2 | |
| Tapachula | 11 | 2 | 2 | |
| Tuxtla Gutierrez | 8 | 2 | 2 | |
| Veracruz | Acayucan | 6 | 3 | 3 |
| Tierra Blanca | 7 | 2 | 2 | |
| Xalapa | 11 | 2 | 2 | |
| Tantoyuca | 2 | 2 | 2 | |
| Oaxaca | Tavela | 11 | 2 | 2 |
| 2 | 3 | 3 | ||
| 1 | 3 | 3 | ||
| Pochutla | 4 | 2 | 2 | |
| Oaxaca (I, II) | 10 | 2 | 2 | |
| Tuxtepec (I, II, III) | 31 | 2 | 2 | |
| Nanahuatipan | 12 | 2 | 2 | |
| Guerrero | Huamuxtitlan | 17 | 2 | 2 |
| Petatlan | 2 | 2 | 2 | |
| Morelos | Yautepec | 1 | 1 | ND |
| Tamaulipas | Matamoros | 8 | 2 | 2 |
| Soto la Marina | 9 | 2 | 2 | |
| Ciudad Mante | 21 | 2 | 2 | |
| 6 | 1 | 1 | ||
| 3 | 1,2 | 1,2 | ||
| Nuevo Laredo | 3 | 2 | 2 | |
| Colima | Colima | 21 | 2 | 2 |
| Tecoman | 9 | 1 | 1 | |
| Minatitlan | 4 | 2 | 2 | |
| Nayarit | Tepic | 1 | 1 | ND |
DENV serotype was determine by the microarray technology and validated by the size differences between amplicons obtained after amplification by specific primer;
Ae. Albopictus pools; ND. Not detected.
These results are crucial because these advances in microarray technology have made it possible to detect the dengue viral nucleic acid in mosquito pools from the field and in human serum specimens. This procedure was sensitive and specific for the detection of DENV up to the serotype level and dual infections (Figure 4, Table 4). Viral RNA in two of the mosquito pools (Yautepec, Morelos and Tepic, Nayarit) was not detected by RT-PCR and gel electrophoresis (Figure 3), but DENV-1 was detected in these samples by microarray analysis. This suggests that the sensitivity of the microarray may be higher than that of gel electrophoresis. We were able to detect the specific amplicon for DENV-1 by agarose gel electrophoresis using a higher concentration of cDNA. Microarray analysis also showed DENV-2 in seven pools of Ae. albopictus from Tapachula (Chiapas) and 11 pools of Ae. aegypti (Table 2); eleven pools of Ae. aegypti from Tavela (Oaxaca) contained DENV-2 and 3 DENV-3; of 30 mosquito pools of Ae. aegypti collected in Ciudad Mante (Tamaulipas), 21 contained DENV-2, six contained DENV-1, and three contained DENV-1 and -2 (Table 2).
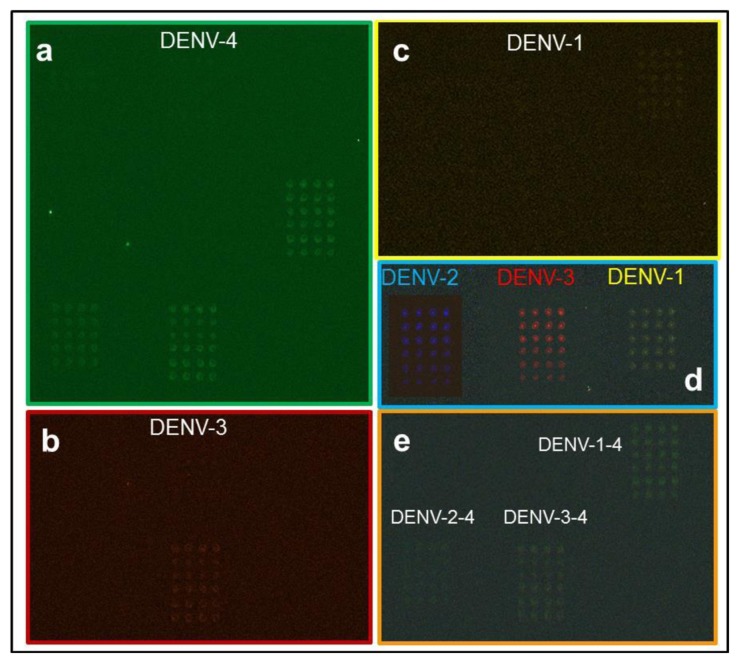
Serum and mosquito samples revealed the presence of co-incident serotypes DENV-1 and -2 and DENV-2 and -3. To test other serotype combinations, amplicons of DENV-1 and -4, DENV-2 and -4, and DENV-3 and -4 were combined and tested in the microarray assay. The results support the ability of this system to detect double infections with different serotypes (Figure 5).
Figure 5.
Samples were evaluated for the presence of dual serotypes. Panels (a)–(c) represent fluorescence for DENV-4, -3, and -1; (d) corresponds to DENV-1 (Texas Red), DENV-2 (FAM), DENV-3 (Cy5) and DENV-4 (Yakima yellow) serotype controls; and (e) shows the merged images of DENV-1 and -4, DENV-2 and -4, and DENV-3 and -4.
Although the level of detection was apparently equivalent to RT-PCR-based detection alone, microarrays have the advantage of high throughput and high sensitivity for low levels of cDNA. The detection limit of an amplicon on a standard agarose gel is approximately 20–30 ng; in the microarray, this limit is 0.05–0.2 ng cDNA per spot, derived from 1 nl of a 50–200 ng·μL−1 solution.
We also detected DENV serotype 2 in mosquito pools of Ae. albopictus, suggesting this species probably serves as a maintenance vector in rural areas of dengue-endemic countries such as Southeast Asia and the Pacific islands. This finding is also very important because this mosquito vector also has the potential to transmit DENV.
3.5. Statistical Analysis
Probe specificity was analysed by normalising the fluorescence intensities for all fluorochromes. Our results showed that only one sample yielded reproducible fluorescence signals for DENV-1 and DENV-2 (Figure 4a, yellow spots). Thus, the probes have a high specificity to diagnose the four DENV serotypes even when two different serotypes are present in the same sample.
After analysing the variability of the fluorescence-normalised values, we found that the variability between samples was higher than the variability between triplicate samples, with the exception of the Yakima yellow results in experiment 2 and the Cy5 results in experiment 1 (Table 5).
Table 5.
Variability coefficient (%) of sample fluorescence in the microarray.
| Label | Samples * | Experiment | ||
|---|---|---|---|---|
|
| ||||
| 1 | 2 | 3 | ||
| Fluorescence-Normalised Value (%) | ||||
| FAM | DENV-2 | 13.5 | 12.6 | 14.5 |
| 7.4 | 7.5 | 5.0 | ||
| Yellow | DENV-4 | 9.2 | 33.2 | 27.1 |
| 3.3 | 33.6 | 10.6 | ||
| DENV-3 | 19.4 | 26.1 | 24.3 | |
| 21.3 | 20.9 | 16.7 | ||
| DENV-1 | 25.1 | 25.8 | 12.8 | |
| 8.8 | 15.8 | 12.7 | ||
Variability between analysed samples (bold font); Variability between replicates (normal font).
The primary advantages of this microarray technique include the speed at which high numbers of specimens can be screened for the presence of DENV with high sensitivity and specificity for single and dual serotype infections. The speed of specimen detection can be improved by using a microfluidic microarray system [42,43]. Fluorescence variability was very low in our assays, suggesting this technology will be very useful for DENV diagnoses. Detection of two cases of dual infections (DENV-2 and -1; and DENV-2 and -3) confirm the feasibility of using this microarray assay to diagnose dual infections (Figure 4) [20,21,44].
This and previous reports [45–47] have demonstrated the great potential of microarray technology for viral detection and identification. These studies pave the way to designing a universal viral signature chip that can be used to determine the presence of DENV, as well as the specific serotype, in a given sample. The experience gained in this study can be applied to any method that relies on the formation of hybrids between sample-derived nucleic acids and short oligonucleotide probes. Additional work is needed to test in silico-designed primers for detecting additional DENV variants on the same chip, thereby supporting studies of DENV evolution across different geographic areas.
We have developed a chip that may be used for diagnosis for a large number of human serum specimens or specimens from mosquitoes and have demonstrated its utility for the identification and surveillance of DENV serotypes. The chip can be used to detect other viruses, since the target amplicon was obtained with consensus primers (DV1 and DV3) that recognize other flaviviruses such as Japanese encephalitis, Kunjin, and yellow fever. This level of sensitivity is very important because these viruses induce symptoms clinically indistinguishable from those of true DENV. We plan to develop such a chip with primers specific to these additional flaviviruses.
Robotic microarray techniques and laser-based image analysis have been applied to the design and development of DNA microarrays to analyse transcriptional responses of cells and microorganisms [33,43]. The use of defined oligonucleotide probes is especially attractive for the synthesis of specific microarrays for viral pathogens circulating in large human populations [47]. The first reported chip assay for a human pathogen (human herpes virus) was highly effective in a global assessment of human cytomegalovirus [48] and for more recent evaluations of genetic variability in West Nile virus [32]. We used a similar approach to synthesise a chip specifically for DENV serotype determination. The high specificity of the probes described in this study can also be adapted to the recently described DNA Biosensor base on a nanoporous alumina membrane [49] while maintaining specificity for each DENV serotype.
4. Conclusions/Outlook
We have developed a microarray for accurate typing of DENV serotypes in human serum or mosquitoes; the system can distinguish serotypes 1, 2, 3, and 4 as well as dual infections with different serotypes. This microarray may also detect other human animal flaviviruses, since the consensus primers DV1 and DV3 target RNA from Japanese encephalitis, Kunjin, and yellow fever viruses. This diagnostic tool has many applications in the study of viral pathogenesis and, perhaps equally importantly, may facilitate viral identification and infections caused by two different DENV serotypes.
Acknowledgments
This work was supported by CONACYT grant CB-2005-01-50603. We thank all members of the Multidisciplinary Network of Microarrays RIIMA for helpful discussion.
Conflicts of Interest
No competing financial interests exist.
References
- 1.World Health Organization Dengue haemorrhagic fever: early recognition, diagnosis and hospital management—An audiovisual guide for health care workers responding to outbreaks. Wkly Epidemiol. Rec. 2006;81:362–363. [PubMed] [Google Scholar]
- 2.Lee H.C., Yen Y.T., Chen W.Y., Wu-Hsieh B.A., Wu S.C. Dengue type 4 live-attenuated vaccine viruses passaged in Vero cells affect genetic stability and dengue-induced hemorrhaging in mice. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osorio J.E., Huang C.Y.H., Kinney R.M., Stinchcomb D.T. Development of DENVax: A chimeric dengue-2 PDK-53-based tetravalent vaccine for protection against dengue fever. Vaccine. 2011;29:7251–7260. doi: 10.1016/j.vaccine.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guy B., Barrere B., Malinowski C., Saville M., Teyssou R., Lang J. From research to phase III: Preclinical, industrial and clinical development of the Sanofi Pasteur tetravalent dengue vaccine. Vaccine. 2011;29:7229–7241. doi: 10.1016/j.vaccine.2011.06.094. [DOI] [PubMed] [Google Scholar]
- 5.Barban V., Munoz-Jordan J.L., Santiago G.A., Mantel N., Girerd Y., Gulia S., Claude J.B., Lang J. Broad neutralization of wild-type dengue virus isolates following immunization in monkeys with a tetravalent dengue vaccine based on chimeric Yellow Fever 17D/Dengue viruses. Virology. 2012;429:91–98. doi: 10.1016/j.virol.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Munoz M.L., Cisneros A., Cruz J., Das P., Tovar R., Ortega A. Putative dengue virus receptors from mosquito cells. FEMS Microbiol. Lett. 1998;168:251–258. doi: 10.1111/j.1574-6968.1998.tb13281.x. [DOI] [PubMed] [Google Scholar]
- 7.Mercado-Curiel R.F., Esquinca-Aviles H.A., Tovar R., Diaz-Badillo A., Camacho-Nuez M., Muñoz M.L. The four serotypes of dengue recognize the same putative receptors in Aedes aegypti midgut and Ae. albopictus cells. BMC Microbiol. 2006;6:85–94. doi: 10.1186/1471-2180-6-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mercado-Curiel R.F., Black W.C., Muñoz M.L. A dengue receptor as possible genetic marker of vector competence in Aedes aegypti. BMC Microbiol. 2008;8:118–132. doi: 10.1186/1471-2180-8-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guzmán M.G., Kouri G.P., Bravo J., Soler M., Vazquez S., Morier L. Dengue hemorrhagic fever in Cuba, 1981: A retrospective seroepidemiologic study. Am. J. Trop. Med. Hyg. 1990;42:179–184. doi: 10.4269/ajtmh.1990.42.179. [DOI] [PubMed] [Google Scholar]
- 10.Nisalak A., Endy T.P., Nimmannitya S., Kalayanarooj S., Thisayakorn U., Scott R.M., Burke D.S., Hoke C.H., Innis B.L., Vaughn D.W. Serotype-specific dengue virus circulation and dengue disease in Bangkok, Thailand from 1973 to 1999. Am. J. Trop. Med. Hyg. 2003;68:191–202. [PubMed] [Google Scholar]
- 11.Alvarez M., Rodriguez-Roche R., Bernardo L., Vazquez S., Morier L., Gonzalez D., Castro O., Kouri G., Halstead S.B., Guzman M.G. Dengue hemorrhagic Fever caused by sequential dengue 1-3 virus infections over a long time interval: Havana epidemic, 2001-2002. Am. J. Trop. Med. Hyg. 2006;75:1113–1117. [PubMed] [Google Scholar]
- 12.Koraka P., Burghoorn-Maas C.P., Falconar A., Setiati T.E., Djamiatun K., Groen J., Osterhaus A.D. Detection of immune-complex-dissociated nonstructural-1 antigen in patients with acute dengue virus infections. J. Clin. Microbiol. 2003;41:4154–4159. doi: 10.1128/JCM.41.9.4154-4159.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliveira DePaula S., Malta Lima D., Clotteau M., Pires Neto R.J., Lopes da Fonseca B.A. Improved detection of dengue-1 virus from IgM-positive serum samples using C6/36 cell cultures in association with RT-PCR. Intervirology. 2003;46:227–231. doi: 10.1159/000072432. [DOI] [PubMed] [Google Scholar]
- 14.Innis B.L., Nisalak A., Nimmannitya S., Kusalerdchariya S., Chongswasdi V., Suntayakorn S., Puttisri P., Hoke C.H. An enzyme-linked immunosorbent assay to characterize dengue infections where dengue and Japanese encephalitis co-circulate. Am. J. Trop. Med. Hyg. 1989;40:418–427. doi: 10.4269/ajtmh.1989.40.418. [DOI] [PubMed] [Google Scholar]
- 15.Chang G.J., Trent D.W., Vorndam A.V., Vergne E., Kinney R.M., Mitchell C.J. An integrated target sequence and signal amplification assay, reverse transcriptase-PCR-enzyme-linked immunosorbent assay, to detect and characterize flaviviruses. J. Clin. Microbiol. 1994;32:477–483. doi: 10.1128/jcm.32.2.477-483.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris E., Roberts T.G., Smith L., Selle J., Kramer L.D., Valle S., Sandoval E., Balmaseda A. Typing of dengue viruses in clinical specimens and mosquitoes by single-tube multiplex reverse transcriptase PCR. J. Clin. Microbiol. 1998;36:2634–2639. doi: 10.1128/jcm.36.9.2634-2639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henchal E.A., Polo S.L., Vorndam V., Yaemsiri C., Innis B.L., Hoke C.H. Sensitivity and specificity of a universal primer set for the rapid diagnosis of dengue virus infections by polymerase chain reaction and nucleic acid hybridization. Am. J. Trop. Med. Hyg. 1991;45:418–428. doi: 10.4269/ajtmh.1991.45.418. [DOI] [PubMed] [Google Scholar]
- 18.Lanciotti R.S., Calisher C.H., Gubler D.J., Chang G.J., Vorndam A.V. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction of the cited article. J. Clin. Microbiol. 1992;3:545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seah C.L.K., Chow V.T.K., Tan H.C., Chan Y.C. Rapid, single-step RT-PCR typing of dengue viruses using five NS3 gene primers of the cited article. J. Virol. Methods. 1995;51:193–200. doi: 10.1016/0166-0934(94)00104-o. [DOI] [PubMed] [Google Scholar]
- 20.Gardella-Garcia C.E., Perez-Ramirez G., Navarrete-Espinosa J., Cisneros A., Jimenez-Rojas F., Ramírez-Palacios L.R., Rosado-Leon R., Camacho-Nuez M., Munoz M.L. Specific genetic markers for detecting subtypes of dengue virus serotype-2 in isolates from the states of Oaxaca and Veracruz, Mexico. BMC Microbiol. 2008;8 doi: 10.1186/1471-2180-8-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cisneros A., Díaz-Badillo A., Cruz-Martínez G., Tovar R., Ramírez-Palacios L.R., Jiménez-Rojas F., Beaty B., Black IV W.C., Munoz M.L. Dengue 2 genotypes in the state of Oaxaca, Mexico. Arch. Virol. 2006;151:113–125. doi: 10.1007/s00705-005-0595-9. [DOI] [PubMed] [Google Scholar]
- 22.Dobnik D., Morisset D., Gruden K. NAIMA as a solution for future GMO diagnostics challenges. Anal. Bioanal. Chem. 2010;396:2229–2233. doi: 10.1007/s00216-009-3197-7. [DOI] [PubMed] [Google Scholar]
- 23.Raymond F., Carbonneau J., Boucher N., Robitaille L., Boisvert S., Wu W.K., de Serres G., Boivin G., Corbeil J. Comparison of automated microarray detection with real-time PCR assays for detection of respiratory viruses in specimens obtained from children. J. Clin. Microbiol. 2009;47:743–750. doi: 10.1128/JCM.01297-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiu C.Y., Alizadeh A.A., Rouskin S., Merker J.D., Yeh E., Yagi S., Schnurr D., Patterson B.K., Ganem D., DeRisi J.L. Diagnosis of a critical respiratory illness caused by human metapneumovirus by use of a pan-virus microarray. J. Clin. Microbiol. 2007;45:2340–2343. doi: 10.1128/JCM.00364-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frobert E., Escuret V., Javouhey E., Casalegno J.S., Bouscambert-Duchamp M., Moulinier C., Gillet Y., Lina B., Floret D., Morfin F. Respiratory viruses in children admitted to hospital inten- sive care units: Evaluating the CLART1 Pneumovir DNA array. J. Med. Virol. 2011;83:150–155. doi: 10.1002/jmv.21932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Renois F., Talmud D., Huguenin A., Moutte L., Strady C., Cousson J., Lévêque N., Andréoletti; L. Rapid detection of respiratory tract viral infections and co-infections in patients with influenza-like illnesses by use of RT-PCR DNA microarray Systems. J. Clin. Microbiol. 2010;48:3836–3842. doi: 10.1128/JCM.00733-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huguenin A., Moutte L., Renois F., Leveque N., Talmud D., Abely M., Nguyen Y., Carrat F., Andreoletti L. Broad Respiratory Virus Detection in Infants Hospitalized for Bronchiolitis by Use of a Multiplex RT-PCR DNA Microarray System. J. Med. Virol. 2012;84:979–985. doi: 10.1002/jmv.23272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shu P.Y., Chang S.F., Kuo Y.C., Yueh Y.Y., Chien L.J., Sue C.L., Lin T.H., Huang J.H. Development of group- and serotype-specific one-step SYBR green I-based real-time reverse transcription-PCR assay for dengue virus. J. Clin. Microbiol. 2003;41:2408–2416. doi: 10.1128/JCM.41.6.2408-2416.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paudel D., Jarman R., Limkittikul K., Klungthong C., Chamnanchanunt S., Nisalak A., Gibbons R., Chokejindachai W. Comparison of real-time SYBR green dengue assay with real-time taqman RT-PCR dengue assay and the conventional nested PCR for diagnosis of primary and secondary dengue infection. N. Am. J. Med. Sci. 2011;3:478–485. doi: 10.4297/najms.2011.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryabinin V.A., Kostina E.V., Maksakova G.A., Neverov A.A., Chumakov K.M., Sinyakov A.N. Universal oligonucleotide microarray for sub-typing of influenza a virus. PLoS One. 2011;6 doi: 10.1371/journal.pone.0017529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gauthier M., Bonnaud B., Arsac M., Lavocat F., Maisetti J., Kay A., Simon F., Zoulim F., Vernet G. Microarray for hepatitis B virus genotyping and detection of 994 mutations along the genome. J. Clin. Microbiol. 2010;48:4207–4215. doi: 10.1128/JCM.00344-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grinev A., Lu Z., Chizhikov V., Rios M. Application of a full-genome microarray-based assay for the study of genetic variability of West Nile virus. J. Virol. Methods. 2012;183:219–223. doi: 10.1016/j.jviromet.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 33.Zhang S.N., Sun H.H., Jin Y.M., Piao L.Z., Jin D.H., Lin Z.H., Shen X.H. Identification of differentially expressed genes in gastric cancer by high density cDNA microarray. Cancer Genet. 2012;205:147–155. doi: 10.1016/j.cancergen.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Yang H.C., Liang Y.J., Huang M.C., Li L.H., Lin C.H., Wu J.Y., Chen Y.T., Fann C.S.J. A genome-wide study of preferential amplification/hybridization in microarray-based pooled DNA experiments. Nucl. Acids Res. 2006;34:e106.. doi: 10.1093/nar/gkl446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burmester J.K., Sedova M., Shapero M.H., Mansfield E. NAIMA as a Solution for Future GMO Diagnostics Challenges. In: Chittur S.V., editor. Microarray Methods for Drug Discovery. 1st ed. Volume 632. Human Press; New York, NY, USA: 2010. pp. 99–124. [Google Scholar]
- 36.Lee M.Y., Kumar R.A., Sukumaran S.M., Hogg M.G., Clark D.S., Dordick J.S. Three-dimensional cellular microarray for high-throughput toxicology assays. Proc. Natl. Acad. Sci. USA. 2008;105:59–63. doi: 10.1073/pnas.0708756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cisneros-Solano A., Moreno-Altamirano M.M.B., Martínez-Soriano U., Jiménez-Rojas F., Diaz-Badillo A., Muñoz M.L. Sero-epidemiological and virological investigation of dengue infection in Oaxaca, México, during 2000–2001. Dengue Bull. 2004;28:28–34. [Google Scholar]
- 38.Igarashi A. Isolation of a Singh's Aedes albopictus cell clone sensitive to Dengue and Chikungunya viruses. J. Gen. Virol. 1978;40:531–544. doi: 10.1099/0022-1317-40-3-531. [DOI] [PubMed] [Google Scholar]
- 39.Chow V.T.K., Seah C.L.K., Chan Y.C. Use of NS3 consensus primers for the polymerase chain reaction amplification and sequencing of dengue viruses and other flaviviruses. Arch. Virol. 1993;133:157–170. doi: 10.1007/BF01309751. [DOI] [PubMed] [Google Scholar]
- 40.Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 41.MathSoft S-PLUS User's Manual, Version 8.0. Data Analysis Products Division. Insightful Management. Insightful Corporation; Seattle, WA,USA: 2007. [Google Scholar]
- 42.Peytavi R., Raymond F.R., Gagné D., Picard F.J., Jia G., Zoval J., Madou M., Boissinot K., Boissinot M., Bissonnette L., et al. Microfluidic device for rapid (<15 min) automated microarray hybridization. Clin. Chem. 2005;51:1836–1844. doi: 10.1373/clinchem.2005.052845. [DOI] [PubMed] [Google Scholar]
- 43.Wang L., Li P.C.H. Optimization of a microfluidic microarray device for the fast discrimination of fungal pathogenic DNA. Anal. Biochem. 2010;400:282–288. doi: 10.1016/j.ab.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 44.Gubler D.J., Kuno G., Sather G.E., Waterman S.H. A case of natural concurrent human infection with two dengue viruses. Am. J. Trop. Med. Hyg. 1985;34:170–173. doi: 10.4269/ajtmh.1985.34.170. [DOI] [PubMed] [Google Scholar]
- 45.Townsend M.B., Dawson E.D., Mehlmann M., Smagala J.A., Dankbar D.M., Moore C.L., Smith C.B., Cox N.J., Kuchta R.D., Rowlen K.L. Experimental evaluation of the FluChip diagnostic microarray for influenza virus surveillance. J. Clin. Microbiol. 2006;44:2863–2871. doi: 10.1128/JCM.00134-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang R.F., Beggs M.L., Robertson L.H., Cerniglia C.E. Design and evaluation of oligonucleotide-microarray method for the detection of human intestinal bacteria in fecal samples. FEMS Microbiol. Lett. 2002;213:175–182. doi: 10.1111/j.1574-6968.2002.tb11302.x. [DOI] [PubMed] [Google Scholar]
- 47.Chou C.C., Lee T.T., Chen C.H., Hsiao H.Y., Lin Y.L., Ho M.S., Yang P.C., Peck K. Design of microarray probes for virus identification and detection of emerging viruses at the genus level. BMC Bioinform. 2006;7 doi: 10.1186/1471-2105-7-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chambers J., Angulo A., Amaratunga D., Guo H., Jiang Y., Wan J.S., Bittner A., Frueh K., Jackson M.R., Peterson P.A., et al. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J. Virol. 1999;73:5757–5766. doi: 10.1128/jvi.73.7.5757-5766.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng J., Toh C.S. Impedimetric DNA biosensor based on a nanoporous alumina membrane for the detection of the specific oligonucleotide sequence of dengue virus. Sensors. 2013;13:7774–7785. doi: 10.3390/s130607774. [DOI] [PMC free article] [PubMed] [Google Scholar]