Abstract
Cryo-electron microscopy is an increasingly popular tool for studying the structure and dynamics of biological macromolecules at high resolution. A crucial step in automating single-particle reconstruction of a biological sample is the selection of particle images from a micrograph. We present a novel algorithm for selecting particle images in low-contrast conditions; it proves more effective than the human eye on close-to-focus micrographs, yielding improved or comparable resolution in reconstructions of two macromolecular complexes.
Keywords: particle selection, machine-learning, automation, high-resolution, cryo-EM
Introduction
The term single-particle reconstruction refers to the reconstruction of a macromolecule from multiple projections, each presenting a single, freestanding copy of the macromolecule. These projections are obtained by cryo-electron microscopy (cryo-EM). The plunge-freeze procedure traps the molecules in a thin layer of vitreous ice. A low-dose electron beam captures a low-contrast, two-dimensional projection image (referred to as a micrograph) containing a collection of the molecules trapped in random orientations. The images of the molecules are then subjected to a computational workflow commonly referred to as single-particle analysis, which results in a 3D density map of the macromolecule.
A single high-resolution reconstruction of a 3D macromolecular complex requires the collection of thousands of micrographs, which typically yield hundreds of thousands of particle images. In cases where contrast is extremely low (e.g. with low electron exposures and low defocus settings), a researcher currently spends a substantial amount of time picking particle images from the micrographs. From an image-processing standpoint, the particle-picking problem can be broken down into two steps. First, candidate particle images must be selected from the micrograph; this step historically has been referred to as particle selection. Second, the "true" particles (i.e., those representing biological molecules) must be identified among those candidates that may contain falsely discovered non-particles such as contaminants or noise; this step is commonly referred to as particle verification. This effort is often compounded by specimen heterogeneity, i.e. multiple conformational states coexisting within the same sample. This problem makes it necessary to collect a larger dataset to ensure there is sufficient relevant data left, after classification, to build a high-resolution map of the structure of interest. Hence, particle picking, especially the second step of particle verification, represents a significant barrier to a completely automated, reproducible single-particle analysis workflow.
Considerable effort has been made to develop algorithms that aid the human eye in selecting good particle images in these extremely low-contrast micrographs (Glaeser, 2004; Langlois et al., 2011; Zhu et al., 2004). A strategy often used is to employ a cross-correlation search over the micrograph in identifying data windows containing candidate particles and then manually verify each window (Rath and Frank, 2004; Roseman, 2003). Another approach to limit the false discovery rate (Langlois and Frank, 2011) is to use hand-tuned thresholds, which can be applied on a micrograph-by-micrograph basis (Chen and Grigorieff, 2007; Tang et al., 2007) or over the entire set (Voss et al., 2009). The elements of subjectivity can be reduced using a machine-learning algorithm referred to as a classifier, a supervised learning tool which requires the user to define an initial selection comprising several hundred examples of “good” and “bad” windows (Arbeláez et al., 2011; Langlois et al., 2011; Zhao et al., 2013). Alternatively, candidate particle images identified can be aligned in 2D and then clustered into classes based on intrinsic information; this enables the user to look at the average of each class and either verify or reject the entire class, or further inspect individual particles within that class (Arbeláez et al., 2011; Shaikh et al., 2008). Nevertheless, current methods still require significant effort by the user to verify particles.
We envision a new type of tool that uses unsupervised learning to select particles from the micrograph with minimal user intervention. The user is only required to provide the approximate size of the macromolecule. Unsupervised learning leverages the observation that images of physical objects have limited complexity, and thus, can be described by a compact representation. We seek to further reduce this compact representation by exploiting the fact that the views of the macromolecules are linked by rigid-body transformations: azimuthal rotation and translation.
In the present study, we introduce a two-step automated particle-picking procedure. The first step is a modified template-matching procedure, termed AutoPicker, which identifies a set of candidate particle images from a collection of micrographs and rejects high-contrast contamination and noise using an unsupervised learning procedure. The second step employs an unsupervised one-class classifier, termed View Classifier or ViCer, which exploits the similarity among aligned true particles to reject outliers. To assess the quality of the final particle selection, we have applied the algorithm to identify and verify particles from two independent datasets recorded under low-contrast conditions: one of micrographs containing 70S ribosomes from E. coli and the second containing molecules of the V/A-ATPase from Thermus thermophilus. The density maps obtained using the automatically selected particle images were compared to maps derived from manually selected particle images, which led to high-quality structures. We demonstrate that the particle images selected from of the AutoPicker/ViCer workflow lead to density maps with comparable, if not better, resolved features, and find that this outcome is in part a consequence of AutoPicker/ViCer’s ability to identify additional true particles in close-to-focus micrographs.
Methods
Proposed Particle-Picking Algorithm
The proposed automated particle-picking algorithm naturally reduces to two steps: 1) identification as well as an initial verification of potential particles with AutoPicker and 2) further verification using outlier rejection with ViCer.
AutoPicker
The AutoPicker algorithm, as outlined in Supplemental Figure 1a, uses template matching to identify windows that contain candidate particle images in a micrograph and classification by unsupervised learning to reject both high contrast contaminants and noise windows. Template matching alone provides an excellent ranking of low-contrast, noisy particle (SNR ~0.06) windows over noise, yet provides no means for selecting the optimal threshold to distinguish these two groups. In addition, a micrograph may contain high-contrast contaminants such as ice crystals and bubbles in the ice after radiation damage of the specimen; depending on their size, windows containing contaminants are ranked, according to the cross-correlation score between each window and a template, higher than, or at the same level as, those containing particles. The unsupervised learning algorithm introduced by AutoPicker handles both of these limitations.
First, AutoPicker employs principal component analysis (PCA) over the power spectra of the extracted image windows, reducing each image to a single principal component. Then, assuming a Gaussian distribution, it rejects windows that fall in the tail, i.e. more than 4 standard deviations from the mean. While this cutoff might seem extreme, in practice only the noise windows follow a Gaussian distribution, whereas contaminants tend to follow a more skewed distribution on the tail. This cutoff targets only a specific type contaminant that proves deleterious to the next step. AutoPicker then repeats this procedure over the background surrounding the particle as defined by a ring around the particle; the size of the ring is defined as the particle radius multiplied by the exclusion multiplier and the width is the exclusion distance. Large contaminants and aggregation violate this ring of exclusion, and consequently, become outliers. This step eliminates the most obvious high-contrast contaminants.
Second, AutoPicker applies Otsu’s algorithm (Otsu, 1979) on the cross-correlation scores of the remaining windows with the template in order to determine the optimal threshold that separates candidate particles from noise. Note that the order of these two steps is important because high-contrast contaminants tend to skew the cross-correlation histogram, causing Otsu’s method to find a suboptimal threshold. In this work, the template was chosen as a disk with a radius corresponding to the particle size and its edges softened by application of a kernel with a Gaussian falloff.
ViCer 2.0
For relatively clean micrographs lacking ice crystals and other artifacts, the AutoPicker algorithm is sufficient to ensure good particle selection. However, many contingencies can contrive to produce less than ideal micrographs and in such cases additional contaminant removal proves necessary. The View Classifier (ViCer) can then be used to further clean the candidate particles of contaminants.
The original ViCer outlier rejection algorithm (Langlois et al., 2012), as outlined in Supplemental Figure 1b, works by maximizing the similarity between true particles and, as a byproduct, is able to recognize contaminants as outliers. ViCer requires that the particle images have been aligned and grouped into views; it then uses the translation-invariant bispectral transforms of the particle images to further increase the similarity among true particles. Next, principal component analysis (PCA) is used to represent the bispectral transforms in a two-dimensional feature space. Visual inspection of this space revealed that the true projections tend to form a single cluster, surrounded by outlier contaminants.
The new ViCer algorithm includes two substantial improvements over the original algorithm. First, the PCA is replaced with an outlier robust version of PCA called DHR-PCA (Feng et al., 2012). This robust PCA prevents corruption of the covariance matrix by contaminants, and as a consequence, yields principal components that better separate contaminants from true particles. Second, the Mahalanobis distance score (a multivariate z-score) replaces the ad hoc multivariate extension of the median absolute deviation (MAD) score (Hoaglin et al., 1983) to define the decision boundary between true particles and outlier contaminants. The Mahalanobis distance is defined as follows:
where x is a vector comprising the first two components from PCA, μ is a vector representing the mean for each coordinate in x and S is the covariance matrix estimated from the data in factor space.
In addition, the covariance matrix used to estimate the Mahalanobis distance is estimated using the robust minimum covariance determinant algorithm (Rousseeuw, 1984). The decision boundary cutoff is then determined by a chi-squared distribution (Pearson, 1900), with two degrees of freedom (corresponding to the two principal components) and a probability of 0.97. This probability value is a meaningful parameter, as it defines an outlier as any image with a probability less than 0.03. Note that while this parameter value can be adjusted based on the needs of the experimentalist, the current value performs consistently well in empirical trials.
Characterization of the Datasets
70S Ribosome from E. coli
Micrographs of 70S E. coli ribosomes were imaged with an FEI (Portland, OR, USA) Tecnai F30 Polara electron microscope equipped with a field emission gun operating at 300 kV. The data were recorded under low-dose conditions (~20e−/Å2) at a temperature of −180° C and captured on Kodak (Rochester, NY) SO-163 film. The objective aperture in the microscope was 100 µm, and the magnification was set to 59,000. Defocus ranged from 1.5 to 5 µm. A Zeiss-Imaging scanner (Intergraph, Huntsville, Alabama, USA) was used to digitize the micrographs with a step size of 7 µm; for more details see (Pallesen et al., 2013).
V/A-ATPase from T. thermophilus
Micrographs of V/A-ATPase from T. thermophilus HB8 were obtained for a previously described study (Lau and Rubinstein, 2012). Specimens were imaged with an FEI Tecnai F20 electron microscope equipped with a field emission gun and operated at 200 kV. The data were recorded under low-dose conditions (~18–20 e−/Å2) at a temperature of −180° C and captured on Kodak SO-163 film at a magnification of 50,000 with a defocus range of 2.5 to 4.5 µm. The micrographs were digitized with a Zeiss-Imaging scanner with a 7µm step size.
Manually Verified Benchmarks
This work applies the AutoPicker/ViCer particle-picking software to two independent datasets that had already led to high-quality structures (Lau and Rubinstein, 2012; Pallesen et al., 2013) by employing manual particle verification. The manual picking for each dataset was performed using different protocols that are standard to each lab. Note that the selections obtained by manual picking represent a substantial investment of time and effort by each lab and that it would be arduous to create a more consistent benchmark. Each protocol is subsequently described in detail.
SPIDER LFCPick (70S Ribosome)
This work includes two manually verified subsets of particle windows for the ribosome dataset, which are used as benchmarks for the described algorithm. The particle windows for the first benchmark were selected using SPIDER’s LFCPick (Adiga et al., 2005) and then manually verified by one of the authors (J.P.). The particle windows for the second benchmark were selected using DoGLFC with AffinityRank (Langlois et al., 2011) and then manually verified by another coworker (Gyanesh Sharma, G.S.).
MRC Ximdisp (ATP synthase)
This work also includes a benchmark derived from a single set of manually verified particle windows from a T. thermophilus V/A-ATPase dataset. The particles images were interactively selected with Ximdisp (Crowther et al., 1996).
Single-particle Reconstruction
The orientation parameters are derived from references as described in the previous subsections. Both complexes were subjected to “gold standard” refinement for spatial frequencies higher than 1/(40 Å) and the angular search was oversampled by two orders (Scheres, 2012a). The final density maps were filtered to the target resolution and amplitude enhanced using the program bfactor (http://emlab.rose2.brandeis.edu/grigorieff/download_b.html). CTF correction was performed using the defocus values as estimated by standard SPIDER procedures for the 70S ribosome and CTFFFIND (Mindell and Grigorieff, 2003) for the V/A-ATPase.
Results and Discussion
The primary concern when replacing manual with automated particle picking is whether the procedure employing the automated algorithm can perform comparably to the manual particle picking. Such a procedure will have several significant advantages including speed, consistency and reproducibility. Therefore, we chose two different asymmetric complexes as test cases, ATP synthase (a rod-like protein complex embedded in an amorphous detergent micelle) and the 70S ribosome (a globular protein/RNA complex), in order to determine whether the AutoPicker/ViCer workflow could reliably detect particle images in two widely different cases of sample type and experimental conditions. The structures of both complexes were previously determined using manual particle picking leading to high-quality structures (Lau and Rubinstein, 2012; Pallesen et al., 2013). Thus these two datasets represent interesting, real-life cases where particle picking presents a challenge due to low contrast in the image and imperfect experimental conditions that give rise to contaminants or aggregated particles.
Earlier studies have measured the performance of an automated particle-picking algorithm against a "gold standard" benchmark data set created by an expert manually picking the particles within the micrograph (Langlois and Frank, 2011; Zhu et al., 2004). The often-used dataset in the field (Zhu et al., 2004) is derived from a high-contrast data collection of keyhole limpet hemocyanin (KLH) molecules where the micrographs contain minimal contamination. This is, however, not a dataset typical for current high-resolution studies where micrographs are usually collected higher voltage and have less contrast. Under these conditions, it is hard to define a "good" particle or "true positive" with a sufficient level of confidence or agreement among experts (Zhao et al., 2013). In addition, this task comes with the superfluous and onerous stipulation that one of the two prominent views should be discarded, requiring the algorithm to perform view classification in addition to particle selection. We conclude that, for low-contrast datasets, benchmarking the automated picking directly against manual picking will not result in a meaningful comparison given both the ambiguity in assigning true positives and in the lack of a meaningful benchmark.
The lack of a meaningful comparison is a known issue in the field and efforts are currently underway to remedy this situation. One notable effort is the 3DEM benchmark1, which ambitiously aims to provide a high-quality benchmark for every step in single-particle reconstruction. This project is bound to have a large impact on methods development for the field, but it is currently still a work in progress.
Instead, we compare automated to manual selection/verification using the end product of the single-particle analysis workflow: the density map. As outlined in Methods, we apply the same workflow to process both datasets using RELION (Scheres, 2012b) for automated angular refinement, which measures the resolution using the Fourier Shell Correlation (FSC) at 0.143 (Rosenthal and Henderson, 2003). During map refinement with RELION, the dataset is divided into two halves and each half is refined independently in order to prevent an overly optimistic resolution estimate from the FSC resulting from overfitted noise within the data. This approach provides an unbiased means to compare automated to manual selection/verification while preventing the influence of most subjective decisions by the user, which could lead to a misleading comparison of the maps. Subjectivity could be introduced through either a custom mask or tailored parameter settings; we avoid this problem by not including a mask and by using the same parameter settings for both angular refinements.
A concern with the application of the FSC to judge the quality of candidate particles selected either manually or automatically is that the quantity and not the quality of the particles might inflate the resolution estimate. For example, in the case were one processes all the data together and then divides the data into half-sets for resolution estimation. However, the use of "gold-standard" refinement measures the resolution from two independent datasets, avoiding inflation of the resolution due to overfitting of the data.
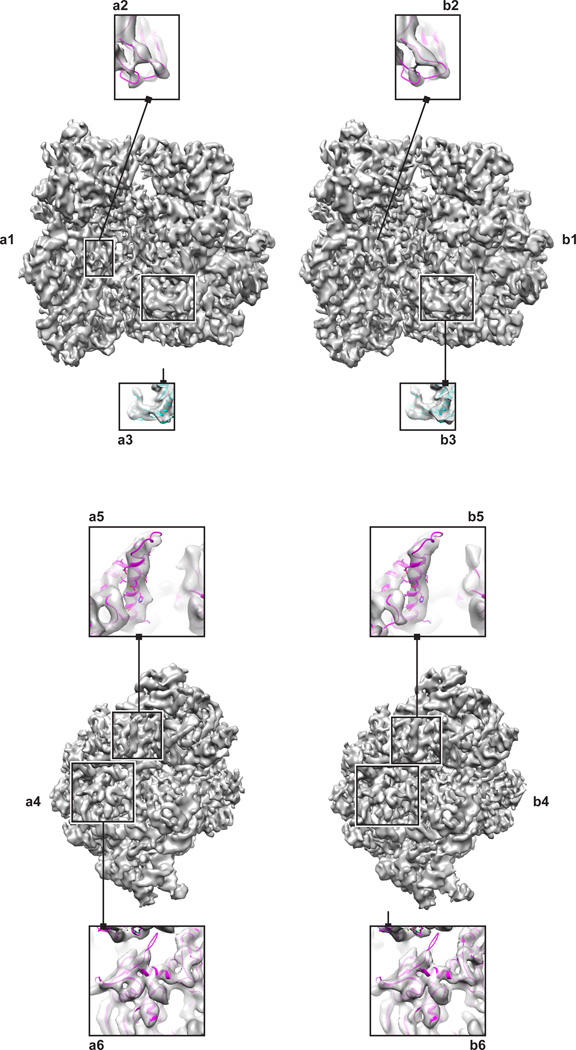
In the case of the 70S ribosome dataset, angular refinement of the automated AutoPicker/ViCer selection/verification produced a higher-resolution map, at 7.1 Å, compared to two manual verifications of the particle images found by template matching, at 7.4 Å (J.P) and 8.5 Å (G.S.). A visual inspection of the density maps obtained by AutoPicker/ViCer versus the manual verification (as performed by J.P.) reveals that the automated method produces a better-quality map than the better of the two manual verifications (as performed by J.P.). That is, the map derived from the automated method (Figure 1a) contains numerous features that are better resolved when compared to the map derived from manual verification (Figure 1b). Consider as an example the loop region (residues 79–81) between strands of a β-sheet in protein S12 of the 30S subunit where the map derived from our automated particle-picking workflow contains sufficient density to accommodate the entire loop (Figure 1a2), whereas the density mass in the same region of the map derived from manual particle verification is fragmented (Figure 1b2). The density mass encompassing a β-sheet in the protein L6 of the 50S subunit (residues 88–121) exhibits some separation between the strands in the map from AutoPicker/ViCer selection/verification (Figure 1a3), whereas the corresponding density in the map from manual verification exhibits no such separation (Figure 1b3). Further inspection of an α-helix in the protein S8 of the 30S subunit (residues 29–51) reveals density in the AutoPicker/ViCer map corresponding to a large aromatic side chain (Figure 1a5), which is entirely missing in the manual map (Figure 1b5). Finally, the density enveloping two neighboring secondary structure elements in protein S2 of the 30S subunit (residues 154–163) resolves the separation between the two elements in the automated map (Figure 1a6) but not in the manually verified map (Figure 1b6).
Figure 1.
70S Ribosome reconstructed from a) selections made by AutoPicker/ViCer b) manual verification performed by J.P. The left panel (a1, b1) shows the ribosome in the classic view with the 30S subunit on the left and the 50S on the right. Within this view, specific features highlight differences in resolvability of a loop (a2,b2) and a β-sheet (a3,b3). The right panel (a4,b4) shows an end on view of the 30S subunit.. Within this view, specific features highlight differences in resolvability of an α-helix (a5,b5) and the separation of secondary structure elements (a6,b6).
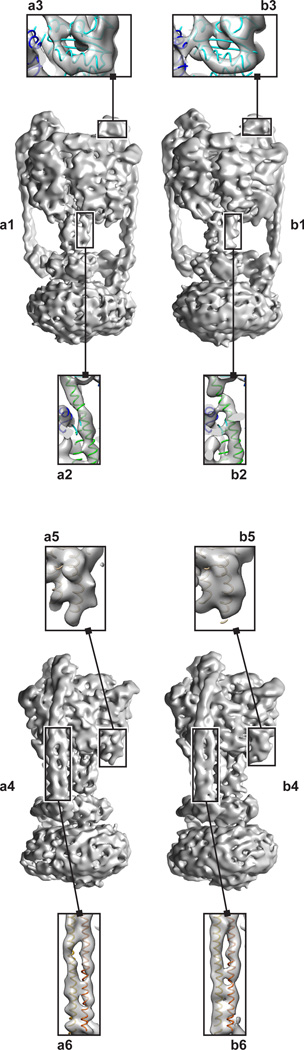
Similarly, a map calculated by RELION for the V/A-ATPase from the automatically picked particle images was at least as good as a map calculated from manually selected particle images. One of the most striking features in the published map of the T. thermophilus V/A-ATPase (Lau and Rubinstein, 2012) was the ability to resolve some of the α-helices in soluble and membrane regions of the complex. The membrane region of the complex contains a ring of helical-hairpin subunits known as subunit L, which is equivalent to subunit c in eukaryotic V- and F-type ATPases. The L-ring is comprised of two concentric rings of α-helices, with each subunit contributes one α-helix to the inner ring and one α-helix to the outer ring. The maps produced by RELION from both the manually and automatically selected particle images resolve the α-helices of the outer ring. Furthermore, the two extended peripheral stalk structures in the soluble region of the complex consist of a right-handed coiled coil of elongated α-helices from the E and G subunits. The α-helices of the E and G subunits were also partially resolved in both of the maps calculated here (Fig. 2). Other short α-helices in the structure could also be resolved (Fig. 2, inset).
Figure 2.
3D map of the V/A-ATPase from T. thermophilus. A map reconstructed from a) automatically and b) manually picked particle images. The left panel (a1,b1) shows the V/A-ATPase from the front. Within this view, specific features highlight the resolvability of an α-helix from the central core, residues 1–37 of the D subunit, (a2,b2) versus a peripheral α-helix, residues 195–214 of the A subunit, (a3,b3). The right panel (a4,b4) shows the V/A-ATPase from the side. Within this view, specific features that highlight resolvability of a helix-turn-helix motif, residues 538–576 of chain A, (a5,b5) as compared to an α-helix in the stalk, residues 3–57 of the B subunit and residues 25–79 of the E subunit, (a6,b6).
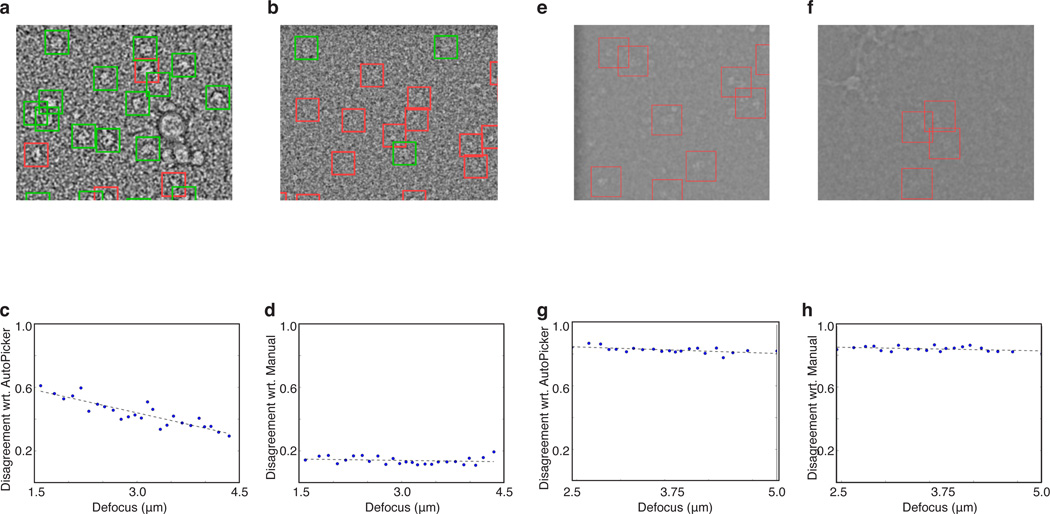
We observed that the number of particles found by the AutoPicker/ViCer workflow was the same irrespective of changes in defocus, whereas for manual picking this number decreased on micrographs captured closer to focus, e.g. fewer particles are typically selected manually from the micrographs shown in Figure 3a,e compared to those shown in Figure 3 b,f. This trend is quantified in Figure 3c,g, which plots, for each defocus group (x-axis), the fraction of particles (y-axis) found by AutoPicker/ViCer but missed by manual selection/verification (ATP-synthase) or only verification (70S ribosome) with respect to the total number of particles found by AutoPicker/ViCer; this fraction is denoted as disagreement with respect to AutoPicker. From these values we note that the level of disagreement increases as the micrograph is captured closer to focus. As a control, we also plot the level of disagreement with respect to manual verification (Figure 3d) and manual selection/verification (Figure 3h), which measures, for each defocus group (x-axis), the fraction of particles (y-axis) found by manual verification but missed by AutoPicker/ViCer with respect to the total number of particles found by manual verification. In this case, the level of disagreement is virtually the same irrespective of changes to defocus, demonstrating that the fraction of manually verified particles missed by AutoPicker/ViCer does not change with defocus.
Figure 3.
A comparison of manual selection versus AutoPicker/ViCer selection at close and far from focus. Two example micrographs from the 70S ribosome dataset (a) far from focus (4.1 µm) and (b) close to focus (1.8 µm) followed by two example micrographs from the V/A-ATPase dataset (e) far from focus (4.1 µm) and (f) close to focus (2.6 µm). The green windows in panels a and b coincide with those manually verified by J.P. The windows marked in red contain unverified particles. Note that due to the low amount of overlap in the V/A-ATPase dataset, no green windows appear in right most micrographs (e,f). All micrographs have been preprocessed in the same manner: Gaussian band-pass filter, outlier pixel removal and down-sampled by a factor of four. Plots comparing AutoPicker/ViCer to manual selection over the 70S dataset (c,d) and ATP synthase dataset (g, h) showing disagreement with respect to (wrt) AutoPicker (c,g) and wrt manual verification (d, h) on the y-axis and defocus on the x-axis; the data was partitioned into 25 defocus groups (blue dots). The black dashed line is a linear trend line fit to the data.
This observed trend, where the proficiency of manual verification decreased with decreasing defocus, proved stronger for the 70S ribosome dataset (Figure 3c) than the V/A ATPase dataset (Figure 3g). This difference can be attributed to the difference in contrast of the particles arising from differences in the composition of the molecule and the conditions unique to each data collection. That is, the 70S ribosome dataset was imaged at 300 kV with the defocus ranging from 1.5 to 5 µm whereas the V/A-ATPase dataset at 200 kV, with the defocus ranging from 2.5 to 5 µm.
The automated particle-picking algorithm has already been employed to calculate density maps for several other biomolecular complexes. For example, two publication-quality density maps of the 40S subunit in complex with the protein DHX29 (Hashem et al., 2013b) and the HCV IRES (Hashem et al., 2013a) have been obtained recently in a short period of time, thanks in part to the proficiency of the new automated particle-picking algorithm. This algorithm fills a gap in automated data processing making it possible to perform a fully automated single-particle reconstruction while simultaneously collecting data. In other words, the experimentalist can now view a preliminary structure before the data collection has even finished and, as a consequence, judge whether the quality of the data is sufficient for further image processing.
While the AutoPicker/ViCer workflow will significantly advance high-throughput processing of images captured by cryo-electron microscopy, it has certain inherent restraints. First, it relies on template matching to perform a fast initial search of the micrograph so that when there is a significant amount of contamination or aggregation the current peak exclusion misses good particles, as seen with the V/A-ATPase dataset. This is still an open problem and will be the focus of future work. Second, as a prerequisite for being completely unsupervised in the steps following template matching, this approach makes assumptions about the distribution of data within the micrograph, i.e. that particles representing biomolecules will constitute the largest fraction of objects in the micrograph. This assumption does not present a major problem, however, because in almost all cases, the user screens both grids and micrographs to ensure a minimum level of sample quality.
The rapid advances in the technology underlying data collection in cryo-EM necessitate on-going development of the image-processing workflow. To this end, the AutoPicker/ViCer algorithms have been released as part of Arachnid, a new open-source image-processing package aimed toward images collected by cryo-EM (http://www.arachnid.us).
In sum, we demonstrate that our automated particle-picking algorithm, AutoPicker/ViCer, can accurately identify true particles for very different macromolecular complexes imaged under imperfect experimental conditions: low contrast with significant levels of contamination. The two samples differ in two important ways. First, the samples are composed of different types of macromolecular assembly, i.e. the 70S ribosome is composed of both RNA and protein while the V/A-ATPase is composed of protein with a detergent micelle. Second, the samples present very different shapes, with the 70S ribosome being globular and the V/A-ATPase rod-like in shape. Despite these differences, the selections made by the AutoPicker/ViCer workflow yielded high-quality density maps without manual intervention, saving substantial costs in time and labor and preventing the results from being influenced by subjective decisions.
Supplementary Material
Acknowledgments
This work was supported by the Howard Hughes Medical Institute and NIH grants R37 GM 29169 and R01 GM 55440 (to J. Frank). We would like the thank Melissa Thomas for assistance with the preparation of the illustrations and Gyanesh Sharma for performing a second manual verification. We would also like to thank early adopters of AutoPicker in our group, namely BoChen, Bingxin Shen, Ming Sun, Nam Ho, Amedee DesGeorges, Yaser Hashem and Sanchaita Das, for their feedback.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
None
References
- Adiga U, Baxter WT, Hall RJ, Rockel B, Rath BK, et al. Particle picking by segmentation: A comparative study with SPIDER-based manual particle picking. Journal of Structural Biology. 2005;152:211–220. doi: 10.1016/j.jsb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Arbeláez P, Han B-G, Typke D, Lim J, Glaeser RM, et al. Experimental evaluation of support vector machine-based and correlation-based approaches to automatic particle selection. Journal of Structural Biology. 2011;175:319–328. doi: 10.1016/j.jsb.2011.05.017. [DOI] [PubMed] [Google Scholar]
- Chen JZ, Grigorieff N. SIGNATURE: A single-particle selection system for molecular electron microscopy. Journal of Structural Biology. 2007;157:168–173. doi: 10.1016/j.jsb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Crowther RA, Henderson R, Smith JM. MRC Image Processing Programs. Journal of Structural Biology. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- Feng J, Xu H, Yan S. Robust PCA in High-dimension: A Deterministic Approach. International Conference on Machine Learning; Edinburgh, Scotland. 2012. [Google Scholar]
- Glaeser RM. Historical background: why is it important to improve automated particle selection methods? Journal of Structural Biology. 2004;145:15–18. doi: 10.1016/j.jsb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, et al. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature. 2013a;503:539–543. doi: 10.1038/nature12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, et al. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell. 2013b;153:1108–1119. doi: 10.1016/j.cell.2013.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoaglin DC, Mosteller F, Tukey JW. Understanding Robust and Exploratory Data Analysis. John Wiley & Sons.; 1983. [Google Scholar]
- Langlois R, Frank J. A Clarification of the Terms Used in Comparing Semi-automated Particle Selection Algorithms in Cryo-EM. Structural Biology. 2011;175:348–352. doi: 10.1016/j.jsb.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois R, Pallesen J, Frank J. Reference-free particle selection enhanced with semi-supervised machine learning for cryo-electron microscopy. Journal of Structural Biology. 2011;175:353–361. doi: 10.1016/j.jsb.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois R, Ash JT, Pallesen J, Frank J. Fully Automated Particle Selection and Verification in Single-Particle Cryo-EM. In: Frank J, Herman G, editors. Minisymposium on Computational Methods in Three-Dimensional Microscopy Reconstruction; New York, NY. 2012. [Google Scholar]
- Lau WCY, Rubinstein JL. Subnanometre-resolution structure of the intact Thermus thermophilus H+-driven ATP synthase. Nature. 2012;481:214–218. doi: 10.1038/nature10699. [DOI] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of Structural Biology. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Otsu N. A Threshold Selection Method from Gray-Level Histograms. IEEE Transactions on Systems, Man and Cybernetics. 1979;9:62–66. [Google Scholar]
- Pallesen J, Hashem Y, Korkmaz Gr, Koripella R, Huang C, et al. Cryo-EM visualization of the ribosome in termination complex with apo-RF3 and RF1. 2013 doi: 10.7554/eLife.00411. eLife 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson K. On the criterion that a given system of deviations from the probable in the case of a correlated system of variables is such that it can be reasonably supposed to have arisen from random sampling. Philosophical Magazine Series 5. 1900;Vol. 50:157–175. [Google Scholar]
- Rath BK, Frank J. Fast automatic particle picking from cryo-electron micrographs using a locally normalized cross-correlation function: a case study. Journal of Structural Biology. 2004;145:84–90. doi: 10.1016/j.jsb.2003.11.015. [DOI] [PubMed] [Google Scholar]
- Roseman AM. Particle finding in electron micrographs using a fast local correlation algorithm. Ultramicroscopy. 2003;94:225–236. doi: 10.1016/s0304-3991(02)00333-9. [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal Determination of Particle Orientation, Absolute Hand, and Contrast Loss in Single-particle Electron Cryomicroscopy. Journal of Molecular Biology. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Rousseeuw PJ. Least median of squares regression. Journal of the American Statistical Association. 1984;79:871–880. [Google Scholar]
- Scheres SHW. A Bayesian View on Cryo-EM Structure Determination. Journal of Molecular Biology. 2012a;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. Journal of Structural Biology. 2012b;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TR, Trujillo R, LeBarron JS, Baxter WT, Frank J. Particle-verification for single-particle, reference-based reconstruction using multivariate data analysis and classification. Journal of Structural Biology. 2008;164:41–48. doi: 10.1016/j.jsb.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, et al. EMAN2: An extensible image processing suite for electron microscopy. Journal of Structural Biology. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy. Journal of Structural Biology. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Brubaker MA, Rubinstein JL. TMaCS: A hybrid template matching and classification system for partially-automated particle selection. Journal of Structural Biology. 2013;181:234–242. doi: 10.1016/j.jsb.2012.12.010. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Carragher B, Glaeser RM, Fellmann D, Bajaj C, et al. Automatic particle selection: results of a comparative study. Journal of Structural Biology. 2004;145:3–14. doi: 10.1016/j.jsb.2003.09.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.