Abstract
Robust development of the early embryo may benefit from mechanisms that ensure that not all pluripotent cells differentiate at exactly the same time: such mechanisms would build flexibility into the process of lineage allocation. This idea is supported by the observation that pluripotent stem cells differentiate at different rates in vitro. We use a clonal commitment assay to confirm that pluripotent cells commit to differentiate asynchronously even under uniform differentiation conditions. Stochastic variability in expression of the Notch target gene Hes1 has previously been reported to influence neural versus mesodermal differentiation through modulation of Notch activity. Here we report that Hes1 also has an earlier role to delay exit from the pluripotent state into all lineages. The early function of Hes1 to delay differentiation can be explained by an ability of Hes1 to amplify STAT3 responsiveness in a cell-autonomous manner. Variability in Hes1 expression therefore helps to explain why STAT3 responsiveness varies between individual ES cells, and this in turn helps to explain why pluripotent cells commit to differentiate asynchronously. Stem Cells 2013;31:1511–1522
Keywords: Embryonic Stem Cells, LIF, Stat3, Hes1, Notch
Introduction
The earliest stages of amniote development are remarkably robust: cells can be removed from the early embryo with no significant impact on subsequent formation of the embryo [1, 2]. To facilitate this level of flexibility it would seem advantageous for cells in the early epiblast to commit to differentiation asynchronously. It is not easy to test whether commitment to differentiation of pluripotent cells is synchronous in vivo. However, in cultures of embryonic stem (ES) cells differentiation can be readily monitored on a cell-by cell basis. From experiments of this type it is clear that populations of ES cells differentiate at different rates even when exposed to a uniform environment [3, 4].
We have previously reported that that mesoderm differentiation can be reduced and neural differentiation can be enhanced from ES cells by activation of Notch [3]. Others have reported that ES cells readily differentiate into mesoderm upon suppression of Notch [5, 6]. Notch is, however, not an absolute requirement for differentiation of pluripotent cells into any lineage but rather influences the probability that a cell will respond to prevailing differentiation conditions [3, 7, 8]. Notch signaling therefore provides a flexible mechanism through which cells could adapt to changes in the composition of the surrounding tissue.
Flexibility in the differentiation response of pluripotent cells can further be explained by the finding that transcription factors including Nanog, Rex1, Hex, and Hes1 are expressed in a heterogeneous and dynamic expression pattern in the early epiblast [4, 9-11]. Hes1 is also expressed in a mosaic distribution in the early embryo [12] and is both a direct target [13] and an upstream modulator [11] of the Notch pathway.
Hes1 acts in multiple tissues to influence both the overall timing of differentiation and the final fate of differentiating cells. For example, Hes1 initially delays the differentiation of neural stem cells, keeping them in a self-renewing state, and later favors their differentiation into astrocytes at the expense of neurons [14]. Here we report that Hes1 acts in a similar manner in ES cells. Hes1 has already been reported to bias the direction of differentiation through modulation of Notch activity [11]. We now find that Hes1 also has an earlier role to delay commitment to all lineages. This earlier role cannot be explained by the ability of Hes1 to modulate Notch activity so we set out to find out what other mechanisms mediate this effect of Hes1.
In other cell types, Hes1 acts primarily as a transcriptional repressor [14], but it can also act though nontranscriptional mechanisms. For example, in neuroepithelial cells Hes1 binds and helps activate signal transducer and activator of transcription 3 (STAT3), a component of the leukemia inhibitory factor (LIF) signaling pathway [15]. LIF/STAT3 signaling maintains pluripotency of ES cells [16] and is sufficient for conversion of epiblast stem cells (EpiSC) to ES cells under permissive culture conditions [17]. LIF acts at least in part by maintaining expression of Klfs [18, 19]. Differentiation of ES cells is regulated not only by availability of LIF but also by the ability of cells to respond to it. Most notably, the transition of naive ES cells to differentiation primed-epiblast is accompanied by loss of Stat3 responsiveness which seems to be due in part to downregulation of Stat3 [17, 20].
Here we report that Hes1 can amplify STAT3 activity in pluripotent cells. This explains how Hes1 imposes a delay on ES cell differentiation. It also helps to explain why STAT3 activity is heterogeneous even in the face of uniform high levels of LIF. These findings shed some light on the observation that a minor subpopulation of pluripotent cells spontaneously differentiate in the presence of LIF. It also provides an example of how differentiation depends not only on exposure to prevailing cues but also on responsiveness to those cues.
Materials and Methods
Experiments were performed in triplicate and statistical significance was calculated using a paired Student's t test.
Commitment Assay
ES cells were plated at 104 cells per centimeter square in differentiation media on gelatinized dishes for 1, 2, 3, or 4 days (challenge phase) then trypsinized, counted, and replated at 103 cells per centimeter square under ES cell self-renewal conditions (LIF + FCS) for 5 days and then stained for alkaline phosphatase (AP) activity. Cells that had failed to commit to differentiate during the challenge phase would form AP positive ES cell colonies. These colonies were counted and expressed relative to the number of colonies on control dishes. The control dishes contained cells from the same starting population that had undergone the same process but were grown under ES cell self-renewal conditions (LIF + FCS) during the challenge phase.
Western Blot Analysis
Cells were lysed for Western blots in RIPA buffer (Sigma) supplemented with 0.5 mM Pefabloc (Fluka) and complete protease inhibitors (Roche, Basel, Switzerland, http://www.roche-applied-science.com). For coimmunoprecipitation experiments, cells were lysed and subject to immunoprecipitation following the procedure described in [15].
Luciferase Assays
Luciferase activity was quantified using the Dual-Luciferase kit (Promega, Madison, WI, http://www.promega.com) using SV40-renilla for normalization. The APRE-luciferase reporter construct for quantifying STAT3 activity was reported previously [21]. The 12xCSL-luciferase Notch reporter [13] was a gift from U. Lendahl.
Mouse ES Cell Culture
ES cells were maintained in glasgow minimum essential medium (GMEM) supplemented with 2-mercaptoethanol, nonessential amino acids, glutamine pyruvate, 10% fetal calf serum (FCS), and 100 units/mL LIF on gelatinized tissue culture flasks [22]. 2i culture [23] and EpiSC culture [24, 25] were carried out as described in the cited publications. Gamma secretase inhibitor (GSI) was obtained from Calbiochem (San Diego, CA, http://www.emdbiosciences.com; cat. 565771) and used at a concentration of 4 μM unless otherwise stated.
Cell Lines
Unless otherwise stated below, all cell lines used in this study are derivatives of E14-TG2a.IV (129/Ola) ES cells. The doxycycline (dox) inducible Hes1 expression cell line was made by first modifying the ROSA locus in mouse E14-TG2a.IV ES cells by homologous recombination with a gene targeting vector pAW2 designed to result in constitutive expression of a reverse tetracycline transactivator (rtTA2S-M2) [26] and to enable subsequent recombinase-mediated cassette exchange (RMCE) at the ROSA locus. This targeting vector contained the coding sequence of the rtTA2S-M2 joined to a splice acceptor such that following integration by homologous recombination the rtTA2S-M2 coding sequence is expressed by transcription from the endogenous ROSA gene promoter. The vector contained a RMCE acceptor cassette (frt/hprt-Δ5′/loxP/MC1neopA/lox511), which was made by modification of a previously described chromosome engineering cassette [27]. After electroporation of linearized pAW2 targeting vector into the mouse E14-TG2a.IV (129/Ola strain) cell line, which is hypoxanthine phosphoribosyl transferase deficient (HPRT−), G418 resistant clones were screened for correct integration by homologous recombination at the ROSA locus by Southern blotting with a flanking external probe to derive the cell line AW2. A plasmid, pAW5, was constructed containing a RMCE donor cassette loxP/hprt−Δ5′/frt/lox511, also made by modification of a previously described chromosome engineering cassette [27]. The pAW5 plasmid can be used to deliver into the RMCE acceptor cassette in the AW2 ROSA locus any desired cDNA joined to a minimal CMV promoter with tet operator sites, termed the tetracycline responsive element (TRE), by first inserting this at a unique AscI restriction site in pAW5. Coelectroporation of the plasmid pAW5 (modified with insertion of TRE-cDNA sequence at the AscI site) with a Cre expression plasmid pCAGGS-Cre-IRESpuro into AW2 cells results in a Cre-mediated RMCE reaction (supporting information Fig. 3A) that reconstructs a functional hprt minigene in the ROSA locus to give HPRT+ cells with concomitant deletion of the neo gene and integration of the TRE-cDNA. Subsequently electroporation with a FLPe recombinase expressing plasmid pCAGGS-FLPe-IRESpuro deletes the hprt minigene via flanking frt sites to leave a selection marker-free modified ROSA locus with an rtTA transgene directly followed by the integrated TRE-cDNA. For the study described herein a DNA restriction fragment comprising a TRE joined to Hes1 cDNA (with an N terminal FLAG tag) linked to an IRES sequence joined to cDNA encoding a truncated human CD2 protein and followed by a poly adenylation addition signal sequence was ligated into the AscI site of pAW5 plasmid. 50 μg of pAW5-TRE Hes1 cDNA-IREShCD2 plasmid + 25 μg pCAGGS-Cre-IRESpuro plasmid were coelectroporated into 5 × 107 AW2 cells and these selected in medium containing hypoxanthine, aminopterin, and thymidine. HPRT+ cells were recovered at a frequency of 10−5 to 10−6. Correct RMCE was verified by demonstrating loss of G418 resistance in HPRT+ cells surviving the selection. In the final modification step, 5 × 106 HPRT+ cells were electroporated with 25 μg pCAGGS-FLPe-IRESpuro and selected in medium containing 10 μM 6-thioguanine. HPRT− cells were recovered at a frequency of approximately 10−2.
Figure 3.
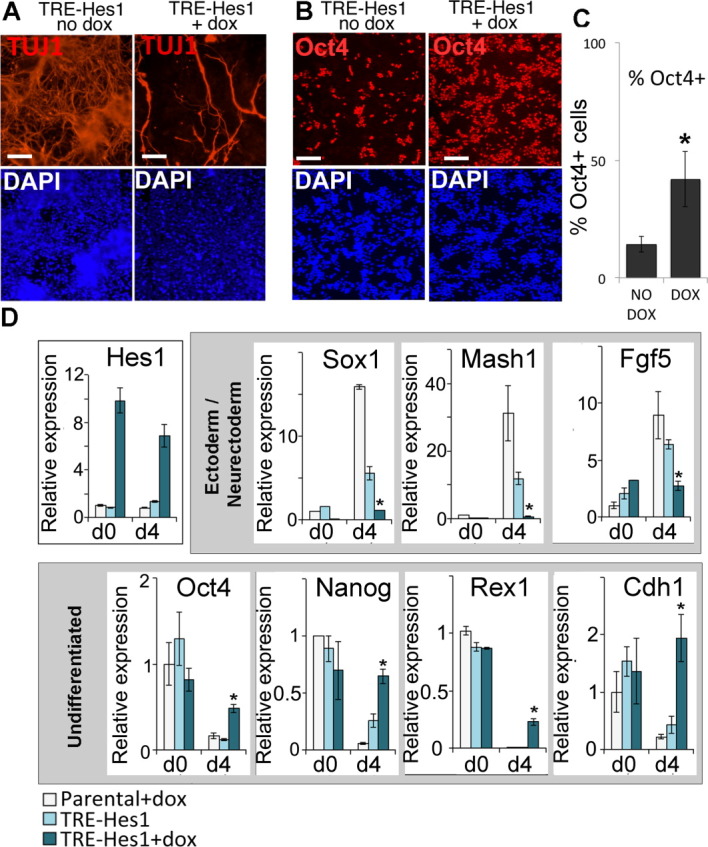
Hes1 delays neural induction. (A): TRE-Hes1 embryonic stem (ES) cells in the absence of dox or induced for 24 hours with 0.3 μg/mL of dox before undergoing neuronal differentiation for 6 days in the continued presence or absence of dox. Cells were replated onto laminin for the final 2 days of the protocol. (B, C): TRE-Hes1 ES cells in the absence of dox or induced for 24 hours with 0.3 μg/mL of dox before undergoing neuronal differentiation for 6 days in the continued presence or absence of dox. Cells were replated onto laminin for the final 2 days of the protocol and then replated onto laminin for the final 4 hours to permit quantification of Oct4 positive cells. 10,000 cells were scored per condition in triplicate. (D): Quantitative polymerase chain reaction analysis of TRE-Hes1 ES cells in the absence of dox or induced for 24 hours with 0.3 μg/mL of dox before undergoing neural differentiation in the continued presence or absence of dox alongside dox-treated parental control ES cells. Scale bars = 200 μM. All data are represented as mean ± SD. *Indicates p < .05 relative to TRE-Hes1 no-dox control. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; TRE, tetracycline responsive element.
Hes1 homozygous null cells [11] (TT2: C57BL/6/CBA) were kindly provided by R. Kageyama. To generate Hes1-rescue ES cell lines the coding sequence for Hes1 was fused to the FKPB12 Destablization domain [28], placed downstream of the CAGS promoter in the plasmid pPyCAGIP [29]. This plasmid was linearized and electroporated into Hes1 homozygous null ES cells [11]. Cells were selected in hygromycin to recover clones with random chromosomal integrations of the plasmid.
Differentiation Protocols
Monolayer neural differentiation is described in detail in [30]. Briefly, ES cells were washed to remove all traces of serum and then plated on gelatin-coated tissue culture plastic in N2B27 serum-free medium. N2B27 consists of a 1:1 ratio of Dulbecco's modified Eagle's medium/F12 and Neurobasal media supplemented with 0.5% modified N2 (made in house as described in [30]), 0.5% B27 (Gibco, Grand Island, NY, http://www.invitrogen.com) and 2-mercaptoethanol. Medium was changed every second day. Mesoderm differentiation was carried out as previously described [31]. For differentiation induced by LIF withdrawal, cells were plated in ES cell propagation media (GMEM+10% FCS) without addition of exogenous LIF. Primers used for quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) are described in supporting information Table 1.
Immunofluorescence and Fluorescence Activated Cell Sorting
Primary antibodies were obtained from the following sources: neuronal β-III tubulin (TUJ1: Covance), Hes1 (gift of T. Sudo [32]), Oct4 (C10; Santa Cruz Biotechnology, Santa Cruz, CA, http://www.scbt.com), Flag (clone M2; Sigma), and Stat3-P (Cell Signaling Technologies, Beverly, MA, http://www.cellsignal.com). The Nanog antibody has been described previously [4, 33]. Fluorescence activated cell sorting (FACS) analysis was performed using a Becton Dickinson FACS Calibur flow cytometer.
Quantification of Nuclear Immunostaining
In order to quantify the fluorescence signal in each individual cell, we generated an automated pipeline for image analysis. Briefly, RGB pictures were registered and preprocessed using our own plugin in ImageJ (http://rsbweb.nih.gov/ij/). The preprocessing step consisted of a background subtraction in each channel as well as a gamma correction of the blue channel to reveal low intensity nuclei. Then, to detect single cell nuclei, the blue channel (4′,6-diamidino-2-phenylindole (DAPI)) was segmented using a previously published algorithm [34] with the following parameter values: for ES cells, sigma = 0.15, minimum nucleus size = 350 pixels, and fusion threshold = 4, for cells at day 3 of differentiation, sigma = 0.15, minimum nucleus size = 350, and fusion threshold = 1. This algorithm provides a picture within which each nucleus is labeled with a unique color in the image. Using a homemade java application that we developed with eclipse (www.eclipse.org), the signal in the red and green channels of the preprocessed RGB picture was measured in the superimposed area of each nucleus to calculate the average intensity. Applications developed for this analysis can be downloaded at http://www.crm.ed.ac.uk/research/group/embryonic-stem-cell-differentiation.
Results
Notch Amplifies STAT3 Activity in ES Cells
Our initial aim was to identify the mechanism by which Notch signaling influences differentiation of pluripotent cells. We focused on signaling pathways that are associated with regulation of pluripotency. LIF/STAT3 signaling and Notch both tend to inhibit non-neural differentiation [3, 5, 6, 35]. We therefore tested whether Notch was able to amplify LIF/STAT3 signaling in ES cells. To this end we made use of ES cells constitutively expressing an activated form of Notch under the control of the Rosa26 locus (R26NotchIC) [3]. We found that R26NotchIC cells express increased levels of phosphorylated STAT3 during steady state culture (Fig. 1A) and of the STAT3 target gene Socs3 after acute LIF stimulation [19] (Fig. 1B) when compared to clonally related control cells [3].
Figure 1.
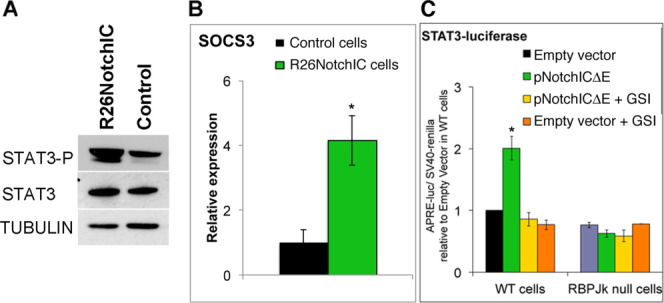
NotchIC amplifies STAT3 activity in embryonic stem (ES) cells. (A): Western blot analysis for phospho-Stat3 and total Stat3 in R26NotchIC cells or clonally related parental control cells cultured in LIF plus fetal calf serum. (B): Quantitative polymerase chain reaction analysis for Socs3 in R26NotchIC cells or clonally related parental control cells plated in N2B27 serum-free media overnight and then stimulated with LIF for 2 hours. (C): Normalized luciferase activity in D3 parental cells or RBPJk homozygous null ES cells after transfection with a STAT3-luciferase plasmid and an SV40-renilla plasmid together with a γ-secretase-dependent form of NotchIC (pNotchICΔE) or empty vector plasmid, either in the presence of absence of 4 μM GSI. All data are represented as mean ± SD. *Indicates p < .05 relative to parental or empty vector controls. Abbreviation: GSI, gamma secretase inhibitor; WT, wild type.
STAT3 luciferase reporter assays [36] confirmed this result: Stat3 activity was increased in wild-type E14-TG2a.IV cells transfected with a plasmid driving constitutive expression of a γ-secretase-dependent form of activated Notch (NotchICΔE [37]) and could be reduced to levels lower than control cells by application of a GSI (Fig. 1C). In RBPJκ homozygous null ES cells, which are unable to activate Notch target genes [38], NotchICΔE had no impact on STAT3 activity (Fig. 1C). The ability of Notch to amplify STAT3 therefore depends on transcription of Notch target genes. This raises the question of which Notch target gene is responsible for amplifying STAT3 activity in ES cells.
Mosaic Expression of Hes1 Is Independent of Notch Activity in ES Cells
Notch target genes include members of the Hes family of transcription factors [14]. We focused on Hes1 because this is a direct Notch target gene [13] that is expressed in a subpopulation of ES cells [11]. We confirmed that Hes1 is upregulated in R26NotchIC cells compared to clonally related controls (supporting information Fig. 1A) and downregulated in wild-type E14-TG2a.IV ES cells upon acute inhibition of the Notch pathway using GSIs (supporting information Fig. 1B). Hes1 expression is, however, not absolutely dependent on Notch in ES cells: Notch inhibitors were only able to reduce Hes1 expression by only around 50% (supporting information Fig. 1B), even though these inhibitors were shown to be highly effective as measured using luciferase reporters of Notch activity (supporting information Fig. 1C). This is in keeping with the observation that Hes1 can be expressed independently of Notch in vivo [39].
Antibody staining confirmed that Hes1 has a mosaic expression in populations of ES cells, as previously reported [11] (Fig. 2A). Uniform expression of activated Notch under the control of the ubiquitous Rosa26 promoter [3, 40] (supporting information Fig. 1E), or inhibition of Notch activity through ablation of RBPJκ [6] (supporting information Fig. 1G), changed the intensity of Hes1 staining but did not eliminate its heterogeneous distribution, suggesting that this heterogeneity is unlikely to arise by Notch-mediated lateral inhibition and perhaps instead reflects autonomous oscillations of Hes1 expression [41] or regulation by another heterogeneously expressed transcription factor.
Figure 2.
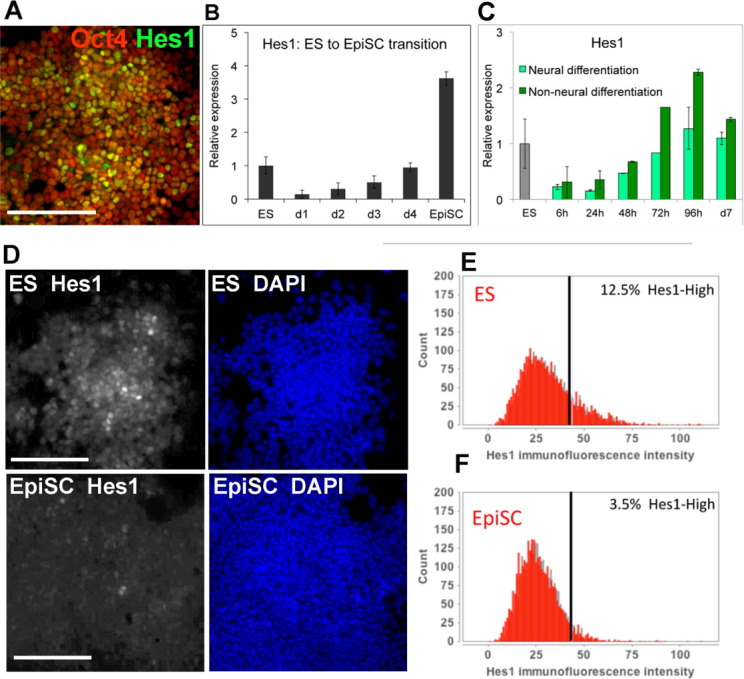
Hes1 is transiently downregulated upon LIF withdrawal. (A): Antibody staining for Hes1 and Oct4 in wild-type E14-TG2a.IV ES cells. (B): Quantitative polymerase chain reaction (qPCR) for Hes1 in E14-TG2a.IV ES cells plated under EpiSC conditions for the stated times or in established cultures of EpiSC. (C): qPCR for Hes1 in E14-TG2a.IV ES cells undergoing neural differentiation in serum-free monolayer or undergoing non-neural differentiation in serum without LIF. (D): Antibody staining for Hes1 in E14-TG2a.IV ES cells or EpiSC. (E, F): Quantification of Hes1 immunofluorescence in E14-TG2a.IV ES cells (E) or EpiSC (F). Scale bars = 200 μM. All data are represented as mean ± SD. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; ES, embryonic stem; EpiSC, epiblast stem cell.
Mosaic Expression of Hes1 Is Independent of Nanog in ES Cells
Nanog is a regulator of ES cell differentiation that is expressed in a mosaic distribution within pluripotent cells [4]. Hes1 has two Nanog binding sites within its promoter [42] and it has been suggested that Hes1 may positively correlate with Nanog in ES cells, at least at the transcript level [11]. We decided to further investigate the relationship between expression of Hes1 and Nanog in ES cells.
We made use of Nanog reporter cell lines in which green fluorescent protein (GFP) has been shown to faithfully report expression of Nanog [4]. We did not observe any clear correlation between NanogGFP and Hes1 protein (supporting information Fig. 2A, 2B). Furthermore, Hes1 is still expressed in a heterogeneous distribution in Nanog homozygous null ES cells [4] (supporting information Fig. 2C). We conclude that Hes1 does not demarcate the same subpopulation as Nanog and that heterogeneity in Hes1 expression is not a consequence of heterogeneity in Nanog.
Hes1 Is Transiently Downregulated at the Onset of Differentiation
Having observed heterogeneity in Hes1 expression within populations of ES cells (Fig. 2A), we next asked whether Hes1 expression changes as ES cells move toward differentiation. We found that Hes1 is transiently downregulated as ES cells move through the transition toward EpiSC (Fig. 2B) or toward differentiation into neural or non-neural lineages (Fig. 2C). This dip in Hes1 expression during differentiation corresponds to the time that cells are transiting to an epiblast-like state [43] and losing LIF responsiveness [17].
In order to compare the distribution of Hes1 protein within purified cultures of purified ES cells and EpiSC, we made use of a cell line, Oct4GIP, in which a puromycin resistance gene is knocked into the Oct4 locus so that any differentiated cells can be eliminated through puromycin selection [44]. In puromycin-selected Oct4GIP ES cells there is, as expected, considerable variation in Hes1 protein expression between individual cells (Fig. 2D, 2E). In contrast, in established cultures of Oct4-GIP-purifed EpiSC Hes1 protein becomes more uniform, with only a very minor subpopulation expressing high levels of Hes1 protein (Fig. 2F).
Hes1 Inhibits Differentiation of ES Cells
Hes1 has been reported to favor mesoderm and suppress neural differentiation under neural conditions [11]. In other cell types, Hes1 influences the timing of differentiation as well as the final differentiated fate [14]. We asked how Hes1 affects the kinetics with which pluripotent cells differentiate.
In order to test this, we generated a cell line expressing Hes-1 under tight control of the dox inducible reverse tet transcriptional activator (rtTA2S-M2) system [26]. To achieve this, we first modified mouse ES cells at the ubiquitously expressed ROSA locus by homologous recombination (supporting information Fig. 3A(i)) using a gene targeting vector designed to express the rtTA2S-M2 coding sequence from the endogenous ROSA gene promoter. This gene-targeting vector also simultaneously integrated site-specific recombination sites (lox and frt sites) and selection markers to enable subsequent RMCE at the ROSA locus. The resulting rtTA2S-M2 expressing cell line, called AW2, was then used for integration of a Hes1 cDNA expression cassette by genetically selectable RMCE. This Hes1 cDNA expression cassette consisted of a TRE joined to a N-terminal FLAG tagged Hes1 cDNA linked to an internal ribosome entry site (IRES) sequence joined to hCD2 sequence and a polyadenylation addition signal sequence. In a two step process involving Cre and FLPe recombination (supporting information Fig. 3A(ii)) a cell line was therefore generated with a selection marker-free modified ROSA locus for optimal inducible expression of FLAG tagged Hes1, induction of which could also be measured by hCD2 expression. TRE-Hes1 cells created in this way were demonstrated to exhibit a dose-dependent upregulation of Hes1 in response to dox (supporting information Fig. 3B, 3C). This system allows for control over both the time and the amount of Hes1 overexpression. We selected a dose of dox (0.3 μg/mL) that gave moderate but sustained expression at close to physiological levels.
Hes1 suppresses differentiation of neurons from neural progenitors [14]. In order to validate the activity of Hes1 transgene in this system we therefore made use of an ES cell-based neurogenesis assay. We confirmed that application of dox dramatically reduced the number of neurons that emerge from ES cells during this assay (Fig. 3A). Dox-treated cultures contained a higher proportion of Oct4+ cells compared with control-treated cultures (Fig. 3B, 3C) suggesting that Hes1 inhibits neural induction as well as subsequent neuronal differentiation. Indeed, Hes1 has been reported to suppress not only neuronal differentiation from neural progenitors but also neural induction of ES cells, an effect that is mediated through downregulation of Notch ligands [11]. We confirmed that sustained expression of Hes1 suppresses neural differentiation in a directed neural monolayer differentiation protocol [30] (Fig. 3D).
We then tested the effect of sustained Hes1 expression during a directed mesoderm monolayer differentiation protocol in the presence of collagen IV and serum, designed to drive cells predominantly into a mesodermal fate [31]. To our surprise, we found that Hes1 prevented the upregulation of the mesoderm marker platelet-derived growth factor receptor alpha (PDGFRα) and instead sustained expression of the ES cell marker stage-specific embryonic antigen 1 (SSEA1) (Fig. 4A–4C). Upon undirected differentiation induced by LIF withdrawal in the continued presence of serum for 4 days, dox-treated cultures contain mixtures of flag-Hes1-positive cells and Flag-Hes1-negative cells due to heterogeneity in activation of the transgene after differentiation. Under these conditions, flag-Hes1 appears to act cell autonomously to maintain Oct4 expression and to suppress the characteristic changes in morphology that accompany ES cell differentiation (Fig. 4D).
Figure 4.
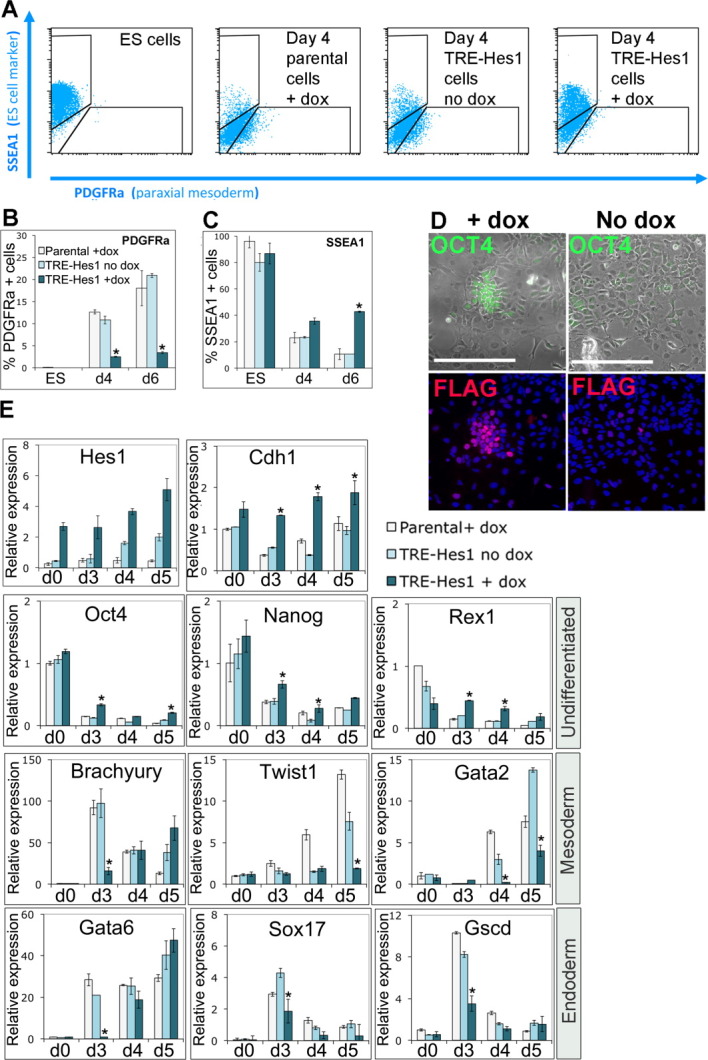
Hes1 delays mesoderm and endoderm differentiation. (A): Fluorescence activated cell sorting analysis of TRE-Hes1 ES cells in the absence of dox or induced for 24 hours with 0.3 μg/mL of dox before undergoing directed mesoderm differentiation in the continued presence or absence of dox alongside dox-treated parental control ES cells. (B, C): Quantification of data shown in (A). (D): Antibody staining of TRE-Hes1 ES cells 4 days after withdrawal of LIF in the continued presence of fetal calf serum, treated with 0.3 μg/mL of dox for the preceding 4 days or in the absence of dox. (E): Quantitative polymerase chain reaction analysis of TRE-Hes1 ES cells in the absence of dox or induced for 24 hours with 0.3 μg/mL of dox before undergoing embryoid body differentiation in the continued presence or absence of dox alongside dox-treated parental control ES cells. Scale bars = 200 μM. All data are represented as mean ± SD. *Indicates p < .1 relative to TRE-Hes1 no-dox control. Abbreviations: dox, doxycycline; ES, embryonic stem; PDGFRα, platelet-derived growth factor receptor alpha; SSEA-1, stage-specific embryonic antigen 1; TRE, tetracycline responsive element.
We went on to test the effect of Hes1 on a population level during embryoid body culture. Under these conditions, activation of Hes1 by dox resulted in a delay in differentiation into both mesoderm and endoderm (Fig. 4E). We conclude that Hes1 not only inhibits differentiation of ES cells into the neural linage but also delays mesoderm and endoderm differentiation.
Ablation of Hes1 Accelerates Differentiation of ES Cells
Hes1 homozygous null ES cells differentiate more homogenously into neural progenitors compared with wild-type controls [11]. We found that Hes1 homozygous null cells downregulate pluripotency markers more rapidly both under neural differentiation conditions (Fig. 5A) and under non-neural differentiation conditions (Fig. 5B) when compared with wild-type control cells, consistent with the observation that Hes1 delays differentiation into all three germ layers (Fig. 5B). Early lineage-specific differentiation markers were prematurely upregulated in Hes1 homozygous null cells, appearing after only 3 days under both differentiation conditions (Fig. 5C, 5D). Choice of lineage was determined by the extrinsic culture conditions, with early neural markers Sox1 and Zfp521 appearing after 3 days under neural differentiation conditions (Fig. 5C), and early mesoderm/endoderm markers T, Gscd, Wnt3a, and Pax7 appearing after 3 days under non-neural differentiation conditions (Fig. 5D). Therefore, Hes1 is required to suppress premature onset of differentiation markers associated with all three germ layers.
Figure 5.
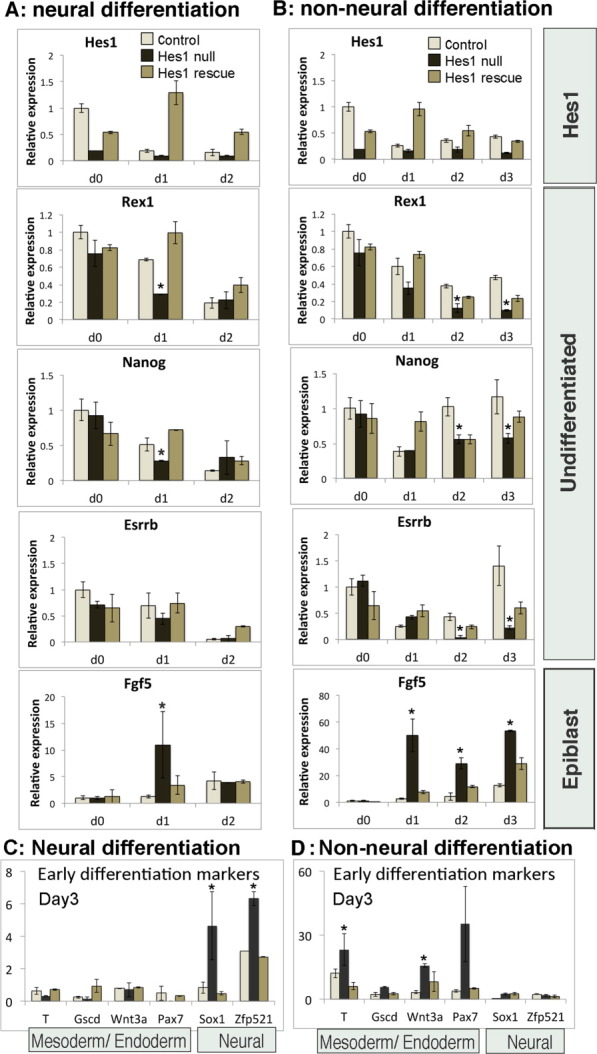
Hes1 homozygous null embryonic stem (ES) cells differentiate more rapidly into both neural and non-neural lineages. (A–D): Quantitative polymerase chain reaction analysis of control wild-type ES cells, Hes1 homozygous null ES cells, or Hes1 homozygous null cells rescued with a constitutively expressed Hes1 transgene, undergoing neural differentiation in serum-free monolayer (A, C) or undergoing non-neural differentiation in serum without LIF (B, D). All data are represented as mean ± SD. *Indicates p < .1 relative to wild-type control cells.
We next asked whether we could rescue this phenotype by delivering a Hes1 transgene to the Hes1 homozygous null ES cells under a constitutive CAGS promoter [45]. Initial attempts were unsuccessful because the high levels of Hes1 driven by this promoter were toxic to ES cells. In order to overcome this problem we generated a destabilized form of Hes1 by fusing it to the FKBP12-L106 destablilization domain [28] and generated stable cell lines expressing the Hes1-FKBP12 fusion protein under the CAGS promoter. Fusion to FKBP12 was not sufficient to completely degrade Hes1 protein, but rather it reduced the levels to a range within which cells were able to tolerate the exogenous Hes1 and expand normally under self-renewal conditions.
After restoring Hes1 to Hes1 homozygous null cells using this strategy, the kinetics of differentiation were restored to a rate similar to parental wild-type cells (Fig. 5A–5D). Thus, the accelerated rate of differentiation of Hes1 homozygous null ES cells can be directly attributed to the absence of Hes1.
Hes1 Delays Commitment
Results described above indicate that Hes is both necessary and sufficient to impose a delay on differentiation of ES cells into derivatives of all three germ layers. This suggests that there may be an early role for Hes1 to modulate initial exit from the pluripotent state, independent from the later, previously reported, role to bias the direction of differentiation.
These results are based on quantification of molecular markers of pluripotency and differentiation in pooled populations. We next went on to test the ability of Hes1 to delay the loss of pluripotency markers on a cell-by-cell basis.
We placed ES cells under differentiation conditions for 3 days and then counted the number of cells that remained positive for the pluripotency marker Nanog. A significantly higher proportion of cells remained Nanog positive after 3 days in dox-treated cultures compared with control-treated cultures (supporting information Fig. 4). The majority of cells did however lose Nanog expression within this time frame, even in dox-treated cultures. Immunofluorescence for the flag epitope that is fused to the exogenous Hes1 protein revealed that the majority of cells had lost the expression of the Hes1 transgene during the course of differentiation. The clusters of cells that remained Nanog positive were, however, almost always associated with clusters of cells that retained the flag-Hes1 transgene (supporting information Fig. 4). This further supports the idea that sustained expression of Hes1 maintains pluripotency markers during ES cell differentiation.
We went on to test these findings using a functional assay of the commitment status of individual cells (Fig. 6A). In this assay, cells are challenged with uniform differentiation conditions in order to give cells the opportunity to commit to differentiation if they are so inclined. Cells are then assayed for their commitment status: any cell that resists differentiation during the “challenge” phase should be able to form an AP-positive colony when replated at clonal density under ES cell self-renewal culture conditions (LIF + FCS). This assay confirms that wild-type ES cells commit asynchronously to differentiation under uniform neural differentiation conditions (serum-free N2B27 media: (Fig. 6B)).
Figure 6.
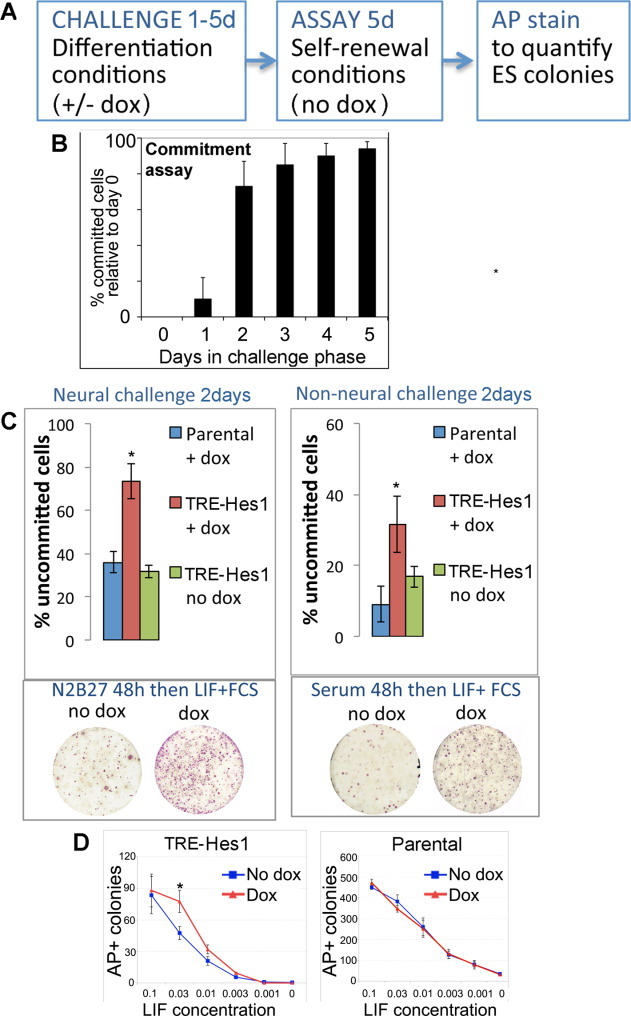
Hes1 delays exit from an uncommitted pluripotent state. (A): Schematic illustration of commitment assay. (B): Commitment assay showing the proportion of cells that lose the ability to self-renew after placing in serum-free N2B27 media for the stated number of days, expressed relative to cells retained in LIF + FCS throughout. (C): Quantification of % uncommitted cells after a 48-hour commitment assay under neural or non-neural challenge conditions. (D): Quantification of AP positive colonies forming in GMEM+FCS with stated dose of LIF, expressed as a fraction of the normal working concentration. Cells under test are TRE-Hes1 ES cells or Parental control cells treated with or without 0.3 μg/mL of dox. All data are represented as mean ± SD. *Indicates p < .1 relative to TRE-Hes1 no-dox control. Abbreviations: AP, alkaline phosphatase; dox, doxycycline; FCS, fetal calf serum; LIF, leukemia inhibitory factor; TRE, tetracycline responsive element.
In order to test the effect of Hes1 on commitment, dox was applied to dox-inducible Hes1 cell only during the “challenge” phase of the commitment assay. Dox is not added once cells have been moved to LIF + FCS to assay for self-renewal (see schematic illustration in Fig. 6A).
Preliminary tests indicated that the majority of control unmanipulated cells undergo commitment, that is, lost the ability to form a self-renewing colony, within 48 hours of exposure to either neural or non-neural differentiation conditions. We therefore used a 48-hour challenge period in order to test whether exogenous Hes1 is able to delay commitment to differentiation. We found that dox-treated cultures contain more than twice as many uncommitted cells compared with control cultures after 48-hour exposure to either neural or non-neural differentiation conditions (Fig. 6C). The effect of exogenous Hes1 was to delay rather than absolutely block differentiation: induction of Hes1 using dox is not sufficient to sustain self-renewal for 5 days in the absence of LIF (Fig. 6D). However, self-renewal over this time period could be sustained at a lower dose of LIF in dox-treated cells compared with control-treated cells (Fig. 6D).
Hes1 Cell Autonomously Amplifies STAT3 Activity
Results described above suggest that Hes1 is operating by more than one mechanism: (a) to downregulate Notch ligands and thus suppress neural differentiation, as previously reported [11], and (b) to act via an unknown mechanism to delay exit from the pluripotent state.
We speculated that this second mechanism could operate though amplification of STAT3 activity. This idea is based on the ability of Hes1 to bind and activate STAT3 [15], on our observation that Notch can amplify both Hes1 expression and STAT3 activity in ES cells (Fig. 1; supporting information Fig. 1) and on the observation that exogenous Hes1 reduces the threshold level of LIF for self-renewal of ES cells (Fig. 6D).
Making use of the dox-inducible Hes1 ES cells, we confirmed that moderate sustained expression of Hes1 can amplify STAT3 activity in ES cells as read out by qPCR for Socs3 (Fig. 7A), luciferase assays for STAT3 activity (Fig. 7B), or levels of phosphorylated STAT3 (Fig. 7C).
Figure 7.
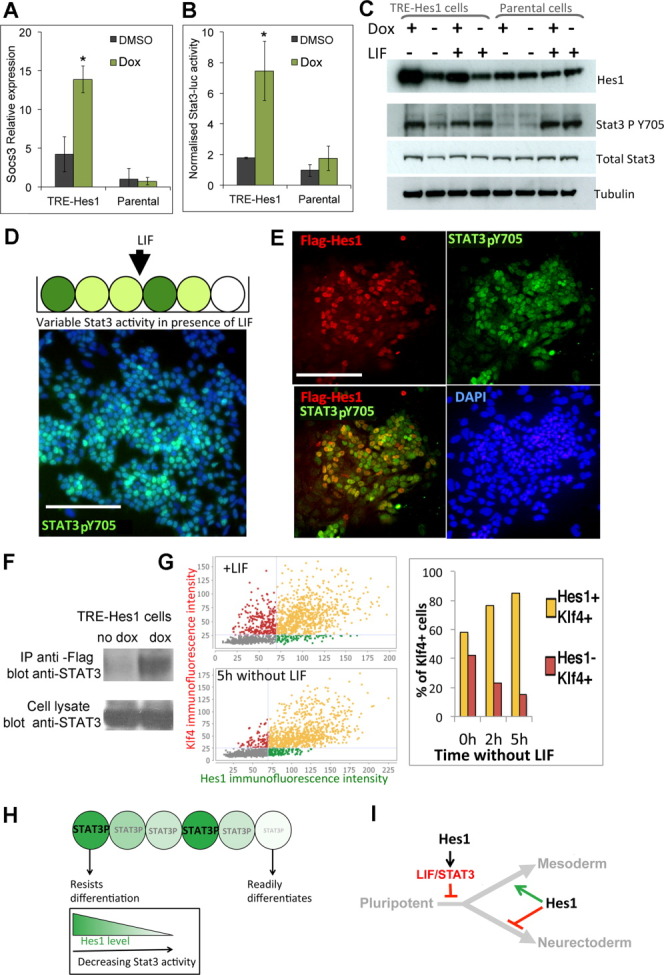
Hes1 cell-autonomously maintains Stat3 activity. (A): Quantitative polymerase chain reaction analysis for Socs3 in TRE-Hes1 embryonic stem (ES) cells or parental control cells in the absence of dox or induced with 0.3 μg/mL of dox for 48 hours. (B): Normalized luciferase activity in TRE-Hes1 ES cells or parental control cells or R26-NotchIC cells after transfection with a STAT3-luciferase plasmid and an SV40-renilla plasmid. (C): Western blot for indicated protein expression in TRE-Hes1 ES cells or parental control cells cultured in the absence of dox or induced with 0.3 μg/mL of dox for 48 hours in the continued presence of LIF or 24 hours after LIF withdrawal in the presence of fetal calf serum. (D): Antibody staining for Stat3-pY705 and DAPI (blue) in E14-TG2a.IV cells cultured in LIF + FCS. (E): Antibody costaining for pSTAT3 and for the flag epitope in TRE-Hes1 ES cells induced for 48 hours with 0.1 μg/mL of dox. (F): Hes1 proteins coprecipitate STAT3 in ES cells. TRE-Hes1 ES cells were treated with dox for 48 hours to induce flag-Hes1 or were left untreated. The cells were lysed and subjected to IP with antibodies to Flag followed by Western blotting for Stat3. (G): Quantitative immunofluorescence for Klf4 and Hes1 in E14-TG2a.IV cells cultured in LIF + FCS or after LIF withdrawal for the stated times. (H, I): Model. We propose that there is an early role for Hes1 to delay the transition toward differentiation through maintenance of STAT3 activity (G, H). Hes1 subsequently modulates lineage choice by regulating expression of Notch ligands (H). Scale bars = 200 μM. All data are represented as mean ± SD. *Indicates p < .05 relative to TRE-Hes1 no-dox control. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; DMSO, dimethyl sulfoxide; dox, doxycycline; IP, immunoprecipitation; LIF, leukemia inhibitory factor; STAT3, signal transducer and activator of transcription 3; TRE, tetracycline responsive element.
This suggests that one mechanism by which Hes1 can bias differentiation is through a cell-autonomous increase in the antidifferentiation signal mediated by LIF. Heterogeneity in Hes1 would therefore result in heterogeneity in STAT3 activity, which in turn would result in a variable response to differentiation cues. We used phospho-STAT3 antibodies to confirm that STAT3 activity was indeed very variable in ES cells, even in the presence of saturating concentrations of exogenous LIF (Fig. 7D). Furthermore, when low levels of dox were used to upregulate flag-tagged Hes1 in a subpopulation of cells, we saw a consistent cell-autonomous maintenance of STAT3-P in cells that expressed the Flag-Hes1 transgene (Fig. 7E). We performed coimmunoprecipitation experiments to confirm that Hes1 directly interacts with STAT3 in ES cells (Fig. 7F), as previously reported for other cell types [15].
Klf4 is also part of a network of pluripotency-associated transcription factors and marks a subpopulation of ES cells that is associated with expression of the core pluripotency regulators. In addition to being regulated by the pluripotency transcriptional network, Klf4 expression is activated by Stat3 signaling. Under self-renewal conditions, we found no particular association between expression of Klf4 and expression of Hes1 (Fig. 7G), in keeping with our finding that Hes1 does not correlate with Nanog in self-renewing ES cell cultures. However, upon removal of LIF we find that Klf4 is selectively maintained in the Hes1-positive subpopulation (Fig. 7G). This is consistent with our finding that Hes1 maintains Stat3 activity after removal of exogenous LIF. These data support a model in which Hes1 acts cell-autonomously to amplify STAT3 activity (Fig. 7H) and consequently to delay differentiation of pluripotent cells (Fig. 7I).
Discussion
We propose that Hes1 acts as a buffer against differentiation though amplification of STAT3. Hes1 therefore plays two roles during the differentiation of pluripotent cells: an early role, reported here, to delay the transition toward differentiation through maintenance of STAT3 activity, and a later role to modulate lineage choice by regulating expression of Notch ligands [11]. This is reminiscent of the dual role played by Hes1 in the nervous system: first to delay differentiation into all neural cell types, and subsequently to favor astrocyte differentiation at the expense of neurogenesis [14].
Our findings highlight the idea that differentiation depends not only on exposure to exogenous cues but also on the ability to readily respond to those cues. Indeed, it has been reported previously that Stat3 activity varies between individual ES cells both in the presence of saturating LIF and in the absence of exogenous LIF [46] [47]. We propose that heterogeneous expression of Hes1 provides one explanation for the source of this variability. Oscillations of Hes1 and Stat3 oscillations are linked in fibroblasts [41], and Hes1 modulates Stat3 activity in neuroepithelial cells [15], but this is the first report to our knowledge demonstrating that Hes1 modulates Stat3 in pluripotent cells.
The transcription factor Nanog is a core component of the transcriptional network that maintains pluripotency [48]. Nanog, like Hes1, is expressed in a mosaic distribution in ES cells [4] and in the inner cell mass of the preimplantation blastocyst [49]. Heterogeneity in the expression of Nanog is an important factor in explaining why pluripotent cells differentiate asynchronously. We observe no correlation between the expression of Nanog and Hes1 in ES cells and find that Hes1 is still detectable in a mosaic distribution in Nanog- homozygous null ES cells. Hes1 is therefore not merely a downstream readout of Nanog heterogeneity but could be operating in parallel to confer variability in the differentiation response.
How, then, does Hes1 come to be heterogeneously expressed? Hes1 is a direct target gene of the Notch pathway [13]. However, Hes1 does not mimic the effect of Notch on the differentiation decisions of ES cells [3, 11]: effects of Notch in ES cells seem likely to mediated by another target gene Hes3 [50]. Furthermore, mosaic expression of Hes1 protein is still seen, albeit at slightly reduced levels, in the absence of Notch activity. It therefore seems unlikely that variability in Hes1 is dependent on Notch signaling: rather, it likely results from autonomous oscillations driven by negative feedback [14] We speculate that this type of nondeterministic mechanism for desynchronizing differentiation may be the best way to confer robustness to changes in the composition of the surrounding tissue.
Hes1 mutants proceed successfully through gastrulation: defects do not become apparent until after E8.5. This is consistent with the fact that Hes1 is neither absolutely necessary nor sufficient to instruct differentiation in vitro but rather confers flexibility in the differentiation response by modulating the threshold with which cells will respond to differentiation environments. Furthermore, any requirement for Hes1 for robust development is likely to be masked by redundancy with Id genes, which are coexpressed with Hes1 in early development and upregulated in Hes1 mutants [51]. Furthermore, Id1, like Hes1, delays the exit from the pluripotent state and subsequently favors mesodermal over neural lineages [35, 52].
Conclusion
In summary, we propose that robustness and flexibility in lineage allocation may be achieved through transiently delaying commitment of a subset of cells by making them more responsive to threshold levels of anticommitment cues, while other cells are deaf to these cues. This highlights the idea that mechanisms designed to confer robustness and flexibility to the developmental process in vivo are likely to confound attempts to achieve reliable and predictable differentiation in culture. Unpicking these mechanisms is the first step toward gaining a better command over the differentiation of pluripotent cells
Acknowledgments
We thank R. Kagayama for providing Hes1 homozygous null ES cells, D. Vassilopoulou for help with Stat3 assays, U. Lendahl for the GSI-dependent NotchIC construct and Notch reporter constructs, T. Schroeder for RBPJκ- homozygous null ES cells, T. Sudo for the Hes1 antibody, and T. Wandless and L. Banaszynski for the FKBP12-L106 construct. S.L. is a Wellcome Trust Career Development Fellow. This work was supported by the Welcome Trust (WT082232AIA) and the BBSRC (BB/I006680/1 and G15381). I.C. and R.O. were supported by the Medical Research Council of the UK and by a scholarship from CONACYT.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.Stern CD. Evolution of the mechanisms that establish the embryonic axes. Curr Opin Genet Dev. 2006;16:413–418. doi: 10.1016/j.gde.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Nagy A, Gertsenstein M, Vintersten K, et al. Manipulating the Mouse Embryo: A Laboratory Manual. 3rd ed. Cold Spring Harbor Laboratory Press. New York: 2002. [Google Scholar]
- 3.Lowell S, Benchoua A, Heavey B, et al. Notch promotes neural lineage entry by pluripotent embryonic stem cells. PLoS Biol. 2006;4:e121. doi: 10.1371/journal.pbio.0040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chambers I, Silva J, Colby D, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 5.Nemir M. Induction of cardiogenesis in embryonic stem cells via downregulation of Notch1 signaling. Circ Res. 2006;98:1471–1478. doi: 10.1161/01.RES.0000226497.52052.2a. [DOI] [PubMed] [Google Scholar]
- 6.Schroeder T, Meier-Stiegen F, Schwanbeck R, et al. Activated Notch1 alters differentiation of embryonic stem cells into mesodermal cell lineages at multiple stages of development. Mech Dev. 2006;123:570–579. doi: 10.1016/j.mod.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Andersson ER, Sandberg R, Lendahl U. Notch signaling: Simplicity in design, versatility in function. Development (Cambridge, England. 2011;138:3593–3612. doi: 10.1242/dev.063610. [DOI] [PubMed] [Google Scholar]
- 8.Muñoz-Descalzo S, de Navascues J, Arias AM. Wnt-Notch signalling: An integrated mechanism regulating transitions between cell states. Bioessays. 2012;34:110–118. doi: 10.1002/bies.201100102. [DOI] [PubMed] [Google Scholar]
- 9.Toyooka Y, Shimosato D, Murakami K, et al. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development (Cambridge, England) 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- 10.Canham MA, Sharov AA, Ko MSH, et al. Functional heterogeneity of embryonic stem cells revealed through translational amplification of an early endodermal transcript. PLoS Biol. 2010;8:e1000379–9. doi: 10.1371/journal.pbio.1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi T, Mizuno H, Imayoshi I, et al. The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev. 2009;23:1870–1875. doi: 10.1101/gad.1823109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatakeyama J, Kageyama R. Notch1 expression is spatiotemporally correlated with neurogenesis and negatively regulated by Notch1-independent Hes genes in the developing nervous system. Cereb Cortex. 2006;16(suppl 1):i132–i137. doi: 10.1093/cercor/bhj166. [DOI] [PubMed] [Google Scholar]
- 13.Jarriault S, Brou C, Logeat F, et al. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 14.Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: Repressors and oscillators that orchestrate embryogenesis. Development (Cambridge, England. 2007;134:1243–1251. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- 15.Kamakura S, Oishi K, Yoshimatsu T, et al. Hes binding to STAT3 mediates crosstalk between Notch and JAK–STAT signalling. Nat Cell Biol. 2004;6:547–554. doi: 10.1038/ncb1138. [DOI] [PubMed] [Google Scholar]
- 16.Smith AG, Heath JK, Donaldson DD, et al. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 17.Yang J, van Oosten AL. Theunissen TW et al. Stat3 activation is limiting for reprogramming to ground state pluripotency. Stem Cells. 2010;7:319–328. doi: 10.1016/j.stem.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niwa H, Ogawa K, Shimosato D, et al. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- 19.Hall J, Guo G. Wray J et al. Oct4 and LIF/Stat3 additively induce Krüppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell. 2009;5:597–609. doi: 10.1016/j.stem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Aiba K, Sharov AA, Carter MG, et al. Defining a developmental path to neural fate by global expression profiling of mouse embryonic stem cells and adult neural stem/progenitor cells. Stem Cells. 2006;24:889–895. doi: 10.1634/stemcells.2005-0332. [DOI] [PubMed] [Google Scholar]
- 21.Morii E, Tsujimura T, Jippo T, et al. Regulation of mouse mast cell protease 6 gene expression by transcription factor encoded by the mi locus. Blood. 1996;88:2488–2494. [PubMed] [Google Scholar]
- 22.Smith AG. Culture and differentiation of embryonic stem cells. J Tiss Cult Meth. 1991;13:89–94. [Google Scholar]
- 23.Ying Q-L, Wray J, Nichols J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tesar PJ, Chenoweth JG, Brook FA, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 25.Brons IG, Smithers LE, Trotter MW, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 26.Urlinger S, Baron U, Thellmann M, et al. Exploring the sequence space for tetracycline-dependent transcriptional activators: Novel mutations yield expanded range and sensitivity. Proc Natl Acad Sci USA. 2000;97:7963–7968. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallace HAC, Marques-Kranc F, Richardson M, et al. Manipulating the mouse genome to engineer precise functional syntenic replacements with human sequence. Cell. 2007;128:197–209. doi: 10.1016/j.cell.2006.11.044. [DOI] [PubMed] [Google Scholar]
- 28.Banaszynski LA, Chen LC, Maynard-Smith LA, et al. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chambers I, Colby D, Robertson M, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 30.Pollard SM, Benchoua A, Lowell S. Conversion of embryonic stem cells to neural stem cells, neurons and glia. Methods Enzymol. 2006;418:151–169. doi: 10.1016/S0076-6879(06)18010-6. [DOI] [PubMed] [Google Scholar]
- 31.Nishikawa SI, Nishikawa S, Hirashima M, et al. Progressive lineage analysis by cell sorting and culture identifies FLK1+VE-cadherin+ cells at a diverging point of endothelial and hemopoietic lineages. Development (Cambridge, England. 1998;125:1747–1757. doi: 10.1242/dev.125.9.1747. [DOI] [PubMed] [Google Scholar]
- 32.Ito T, Udaka N, Yazawa T, et al. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development (Cambridge, England) 2000;127:3913–3921. doi: 10.1242/dev.127.18.3913. [DOI] [PubMed] [Google Scholar]
- 33.Yates A, Chambers I. The homeodomain protein Nanog and pluripotency in mouse embryonic stem cells. Biochem Soc Trans. 2005;33(pt 6):1518–1521. doi: 10.1042/BST0331518. [DOI] [PubMed] [Google Scholar]
- 34.Li G, Liu T. Tarokh A et al. 3D cell nuclei segmentation based on gradient flow tracking. BMC Cell Biol. 2007;8:40. doi: 10.1186/1471-2121-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ying QL, Nichols J, Chambers I, et al. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 36.Schumann RR, Kirschning CJ, Unbehaun A, et al. The lipopolysaccharide-binding protein is a secretory class 1 acute-phase protein whose gene is transcriptionally activated by APRF/STAT/3 and other cytokine-inducible nuclear proteins. Mol Cell Biol. 1996;16:3490–3503. doi: 10.1128/mcb.16.7.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kopan R, Schroeter EH, Weintraub H, et al. Signal transduction by activated mNotch: Importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci USA. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schroeder T, Fraser ST, Ogawa M, et al. Recombination signal sequence-binding protein Jkappa alters mesodermal cell fate decisions by suppressing cardiomyogenesis. Proc Natl Acad Sci USA. 2003;100:4018–4023. doi: 10.1073/pnas.0438008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.la Pompa de JL, Wakeham A, Correia KM, et al. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development (Cambridge, England. 1997;124:1139–1148. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- 40.Zambrowicz BP, Imamoto A, Fiering S, et al. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci USA. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshiura S, Ohtsuka T, Takenaka Y, et al. Ultradian oscillations of Stat, Smad, and Hes1 expression in response to serum. Proc Natl Acad Sci USA. 2007;104:11292–11297. doi: 10.1073/pnas.0701837104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katoh M, Katoh M. Integrative genomic analyses on HES/HEY family: Notch-independent HES1, HES3 transcription in undifferentiated ES cells, and Notch-dependent HES1, HES5, HEY1, HEY2, HEYL transcription in fetal tissues, adult tissues, or cancer. Int J Oncol. 2007;31:461–466. [PubMed] [Google Scholar]
- 43.Sterneckert J, Stehling M, Bernemann C, et al. Neural induction intermediates exhibit distinct roles of Fgf signaling. Stem Cells. 2010;28:1772–1781. doi: 10.1002/stem.498. [DOI] [PubMed] [Google Scholar]
- 44.Ying QL, Nichols J, Evans EP, et al. Changing potency by spontaneous fusion. Nature. 2002;416:545–548. doi: 10.1038/nature729. [DOI] [PubMed] [Google Scholar]
- 45.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 46.Davey RE, Zandstra PW. Spatial organization of embryonic stem cell responsiveness to autocrine gp130 ligands reveals an autoregulatory stem cell niche. Stem Cells. 2006;24:2538–2548. doi: 10.1634/stemcells.2006-0216. [DOI] [PubMed] [Google Scholar]
- 47.Peerani R, Onishi K, Mahdavi A, et al. Manipulation of signaling thresholds in “engineered stem cell niches” identifies design criteria for pluripotent stem cell screens. Plos One. 2009;4:e6438. doi: 10.1371/journal.pone.0006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chambers I, Smith A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene. 2004;23:7150–7160. doi: 10.1038/sj.onc.1207930. [DOI] [PubMed] [Google Scholar]
- 49.Tang F, Barbacioru C, Bao S, et al. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6:468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Androutsellis-Theotokis A, Leker RR, Soldner F, et al. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442:823–826. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- 51.Ishibashi M, Ang SL, Shiota K, et al. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- 52.Zhang K, Li L, Huang C, et al. Distinct functions of BMP4 during different stages of mouse ES cell neural commitment. Development (Cambridge, England. 2010;137:2095–3105. doi: 10.1242/dev.049494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


