Abstract
Activation by glycosaminoglycans (GAGs) is an emerging trend among extracellular proteases important in disease. ProMMP-7, the zymogen of a matrix metalloproteinase secreted by mucosal epithelial and tumor cells, is activated at their surfaces by sulfated GAGs, but how? ProMMP-7 is activated in trans by representative heparin oligosaccharides in a length-dependent manner, with a large jump in activation at lengths of 16 monosaccharides. Imaging by atomic force microscopy visualized small complexes of proMMP-7 molecules linked by 8-mer lengths of heparinoids and extended assembles formed with 16-mer lengths of heparin. Complexes of proMMP-7 with polydisperse heparin or heparan sulfate were more diverse. Heparinoids evidently accelerate activation by tethering multiple proMMP-7 molecules together for proteolytic attack among neighbors. Removal of either the prodomain or C-terminal peptide sequence of KRSNSRKK from MMP-7 prevents formation of the long arrays induced by heparin 16-mers or heparan sulfate. The role of the C-terminus in activation assays suggests it contributes to remote, allosteric binding of GAGs. Enhancement of proteolytic velocity of MMP-by GAGs indicates them to be effectors of V-type allostery. GAGs from proteoglycans appear to assemble proMMP-7 molecules for activation, an event preceding its tumorigenic or anti-bacterial proteolytic activities at cell surfaces.
Keywords: zymogen activation, glycosaminoglycans, protein-carbohydrate interactions, atomic force microscopy, allostery, enzyme kinetics
INTRODUCTION
Zymogens from four major classes of proteases were found to be regulated by sulfated glycosaminoglycans (GAGs) which physiologically radiate from the core proteins of proteoglycans on and outside of animal cells. GAGs accelerate the activation of procathepsins B, L and S (lysozomal cysteine proteases),1–3 the serine protease tryptase,4 and aspartic proteases such as β-secretase.5, 6 GAGs promote autolytic activation of proMMP-2 and -7,7, 8 plus transactivation of proMMP-2 by proteases.9–11 The zymogens of MMPs comprise a prodomain, a catalytic domain (Figure 1A), and usually a hemopexin-like domain.12, 13 GAGs can position the protease to attack substrate, e.g. heparin proteoglycan linking chymase to substrate14 or GAG chains recruiting MMP-7 to proHB-EGF at surfaces of mucosal epithelial cells in female reproductive organs15 or to pro-α-defensins to activate bactericidal activity.8, 16 GAGs also regulate some active proteases. For example, heparin accelerates antithrombin inhibition of thrombin17 and chondroitin-4-sulfate (C4S) makes cathepsin K collagenolytic.18 The work herein asks by what mechanism(s) does GAG-dependent activation of proMMP-7 proceed? Greater understanding should offer perspective on GAG activation of other pathophysiologically important proteases as well.
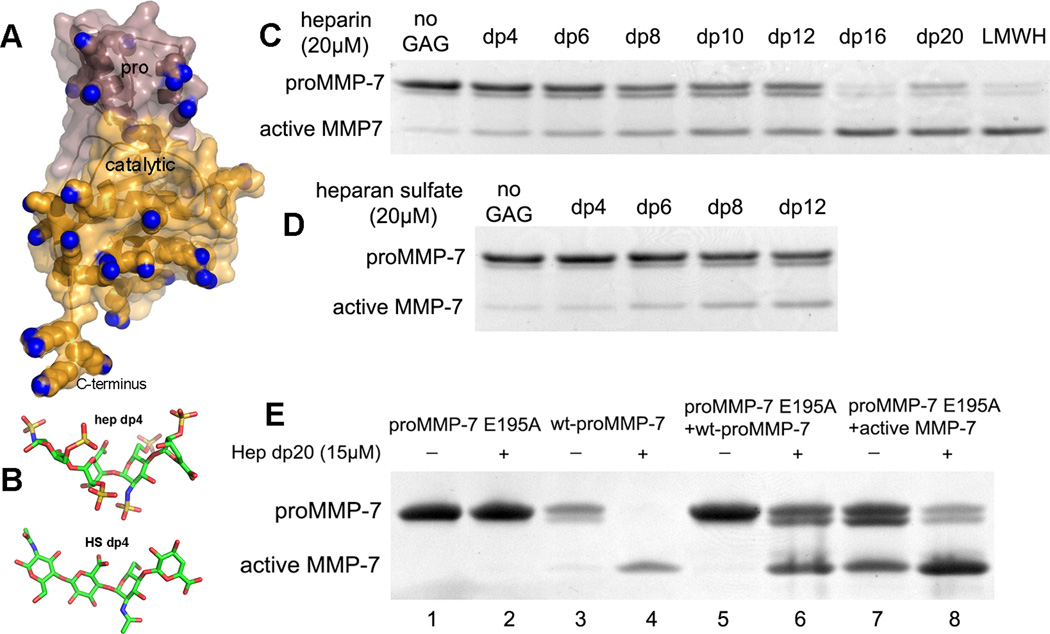
Figure 1. GAGs mediate intermolecular activation of proMMP-7.
(A) The positive charges of arginine and lysines are colored blue on the domains in the homology model of proMMP-7.43 (B) Crystallographic coordinates are plotted for tetrasaccharides from heparin and from HS (PDB codes 1BFB, 3E7J). (C, D) Heparin and HS oligosaccharides of defined length accelerate proMMP-7 activation in a length-dependent manner. Wild-type proMMP-7 (5.5 µM, 27.5 kDa) was incubated with 20 µM (C) heparin oligosaccharides from four to 20 saccharides units long (dp4 to dp20), low molecular weight heparin (LMWH) (C), or (D) HS oligosaccharides four to 12 saccharide units long at 37°C for 2 h before stopping reactions. (E) In the presence of heparin fragments, proMMP-7 is activated through intermolecular proteolysis. ProMMP-7-E195A (7.3 µM) was incubated with wt proMMP-7 (1.8 µM, lanes 5, 6) or active wt-MMP-7 (2.6 µM, lanes 7, 8) with or without Hep dp20 (15 µM, even lanes) at 37°C for 3 h before stopping the reactions.
GAG-stimulated activation of proMMP-7 initiates its role in innate immunity8 and very possibly in tumorigenesis.15, 19 Heparan sulfate (HS) appears to dock proMMP-7 to epithelial cells.20, 21 Proteolytic activity of MMP-7 in mucosal epithelia promotes antibacterial defense in the small intestine,16 wound closure in the lung,22 tumor cell survival (by decreasing FasL-triggered apoptosis),23, 24 and mammary tumor cell proliferation (via release of a domain from the ErbB4 receptor).25 High MMP-7 expression is associated with early stage breast cancer,19 its metastasis to bone,26 as well as invasion, metastasis and poor outcome of colorectal cancer.27
The activation of proMMPs can be grouped under mechanisms of stepwise proteolysis and allosteric displacement of the prodomain not requiring proteolysis.12 Stepwise proteolysis can be triggered by destabilizing agents such as proteases and thiol-reactive agents.12 Allosteric activation without proteolysis has resulted from protein-protein interactions with the pro-forms of proMMP-9, -2, and -3.12, 28, 29 Heparin and chondroitin-4,6-sulfate (CSE) activate proMMP-7.8, 30 One hypothesized mechanism for this activation is an allosteric bridging of the GAG chain between pro- and catalytic domains to expose the active site.13 Such bridging in a 1:1 complex that unmasks the active site is the working hypothesis for autoactivation of procathepsin B.1 It was speculated that heparin might enhance activity of MMP-7 through conformational change.30 Another possibility would be GAGs linking proMMP-7 molecules, analogous to GAGs of proteoglycans bridging MMP-7 to substrates of pro-α-defensin8, 13 or pro-HB-EGF in complex with its ErbB4 receptor.15 Simultaneous binding of serine protease inhibitors (serpins) and serine proteases to heparin results in inhibition.31, 32 Considering such alternative hypotheses and elucidating the mechanism of GAG activation of proMMP-7 is the purpose of this work.
The heparinoids HS and heparin are extended, undulating polymers of disaccharides of uronic acid α(1–4)-linked to D-glucosamine with a higher level of sulfation in heparin and more N-acetyl groups in HS.33, 34 A tetrasaccharide therein forms one period of the sinusoidal structure (Figure 1B).35 The composition and length of HS especially varies among tissue and cell types.33 Heparin from bovine and porcine mucosa is widely used in research but polydisperse. Heparin oligosaccharides of defined length have served as a simplifying tool for investigation of GAG-protein interactions and in this study of the activation of proMMP-7. AFM recently imaged carbohydrate-lectin complexes36 and studied forces in an enzyme-GAG association.37 The novel AFM images of GAG-enzyme interactions reveal structural attributes of GAG-induced assemblies with proMMP-7. Heparin fragments appear to activate proMMP-7 primarily by bridging the protease molecules into extended complexes. Formation of these complexes that are efficient at activation requires heparin oligosaccharides of sufficient length, the prodomain, and the C-terminal basic peptide. GAGs then act on mature enzyme to enhance proteolytic velocity, suggesting allosteric transmission across the catalytic domain.
RESULTS
GAGs associating with proMMP-7
The ability of physiological GAGs to interact with proMMP-7 was surveyed by their competition with surface-tethered heparin, using surface plasmon resonance. ProMMP-7 was made stable by the E195A substitution of the general base, which inactivates mature MMP-7 by 1900-fold.38 Heparin, chondroitin sulfate E, and dermatan disulfate (DS) competed well for proMMP-7-E195A and contain two or more sulfate groups per disaccharide repeated in their structures (SI Figures S1, S2). HS, DS, and chondroitin sulfates A and C (averaging one sulfate per disaccharide) and chondroitin sulfate D (with two sulfates per disaccharide) failed to outcompete heparin (SI Figure S2). The lesser affinity of its physiological partner HS for proMMP-7 sufficed for efficient, saturable activation (SI Figure S1B). This suggests that HS may not only recruit proMMP-7 into cell surface complexes,15 but also accelerate its maturation to the active form.
ProMMP-7 is activated in trans
We asked if GAGs act on individual or pairs of proMMP-7 molecules to induce activation. We exploited oligosaccharides of HS and heparin in assays of proMMP-7 activation for two reasons: (i) to distinguish GAG effects on a single proenzyme from linking of proenzyme molecules and (ii) to introduce homogeneity in size. HS oligosaccharides were available in lengths up to 12 saccharide units (HS dp12). Heparin oligosaccharides are available up to 20 saccharide units (Hep dp20). Heparin oligosaccharides are experimentally well-behaved and may represent physiological partners HS and CSE. Heparin shares similar disaccharide structure with HS and shares high sulfation with CSE (SI Figure S1C). Incubation of wt proMMP-7 with Hep or HS dp4 to dp12 increased autolytic activation moderately, with appearance of active catalytic domain increasing with oligosaccharide length (Figure 1C,D). The activation was much more complete using Hep dp16, Hep dp20, or low molecular weight heparin. The dramatic boost in activation at lengths of 16 monosaccharides or greater suggests the need for this length for tethering and positioning of the proenzymes together.
We tested the hypothesis of heparin-dependent colocalization promoting activation. E195A-inactivated proMMP-7 is incompetent for the Hep dp20-induced activation that readily proceeds with dilute wt proMMP-7 (Figure 1E, lanes 1–4). Addition of wt proMMP-7 activated part of the proMMP-7-E195A once Hep dp20 was added (Figure 1E, lanes 5–6). Addition of 0.36 equivalents of active wt MMP-7 activated part of proMMP-7-E195A without Hep dp20 and much more with it (Figure 1E, lanes 7–8). Thus, longer heparin chains appear to draw (pro)MMP-7 molecules together for activation to proceed in trans.
Hydrodynamics of assembly with heparin fragments
We investigated the average size of proMMP-7 complexes formed in solution as a function of the length of the heparin oligosaccharide, using hydrodynamics. Dynamic light scattering (DLS) established proMMP-7-E195A to be monomeric with estimated MW of 27.6 kDa (and diameter of 2.5 nm; SI Table S1) matching calculated MW of 27.7 kDa. After adding a 10-fold molar excess of Hep dp8, DLS estimated the MW to be 30 kDa and diameter at 2.6 nm, suggesting a single proenzyme in complexes. Addition of Hep dp16 to 5-fold molar excess introduced excessive light scattering dominated by a component appearing to be 6 µm in diameter (SI Table S1), suggesting aggregation, instead of a 2:1 complex hypothesized initially.
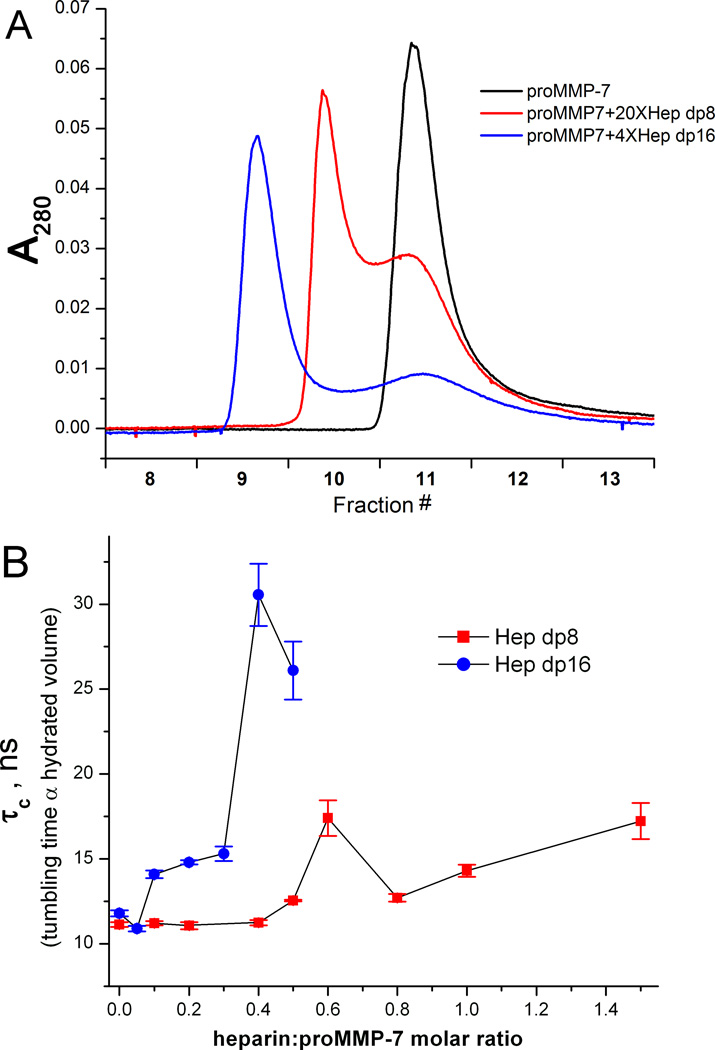
ProMMP-7-E195A eluted as a sharp peak from an analytical gel filtration column (Figure 2A). Addition of Hep dp8 to 10-fold molar excess gave rise to a similar but slightly broader peak consistent with DLS evidence of a single proenzyme in complexes. Addition of the Hep dp8 to 20-fold molar excess added a faster migrating peak at fraction 10, suggesting lower affinity association into a higher MW complex. Addition of 4-fold molar excess of Hep dp16 introduced an even larger complex (eluting at fraction 9), while a peak still eluting at fraction 11 likely contained a single proenzyme (Figure 2A). After correcting for retardation in migration of free proMMP-7-E195A in 150 mM NaCl (from 0.67 to 0.81 column volumes), the proMMP-7 with Hep dp8 may have eluted around 2.2-fold the apparent MW of free proMMP-7, consistent with complexes containing two proenzyme molecules. ProMMP-7-E195A with Hep dp16 may have eluted with apparent MW around 4.1-fold that of free proMMP-7.
Figure 2. ProMMP-7 and heparin oligosaccharides form complexes containing two or more proenzyme molecules.
(A) ProMMP-7-E195A (50 µM) and its mixtures with Hep dp8 or dp16 oligosaccharides (at molar excesses indicated) were run on an analytical gel permeation column. (B) NMR-detected titrations of proMMP-7-E195A (15N-labeled, 300 µM) with Hep dp8 or dp16 were monitored with a proxy for apparent hydrodynamic volume, the rotational correlation time (τC) from 15N NMR relaxation.
As a surrogate for apparent MW, rotational correlation times (τC of diffusional tumbling) were measured throughout Hep dp8 and dp16 titrations of 15N-labeled proMMP-7-E195A using 15N NMR relaxation (detecting most enzyme states except aggregates). τC is proportional to the hydrated volume of a globular protein complex. Addition of 0.6:1 Hep dp8 to proMMP-7-E195A increased τC by 1.6-fold to almost 18 ns (Figure 2B). This ratio and the slowed tumbling might suggest formation of assemblies containing possibly two proenzymes per chain of Hep dp8. The drop in τC upon addition of the dp8 to 0.8 to 1 equivalents parallels the “template effect” typified by heparin bridging between serpins and serine proteases.17, 32, 39 This suggests that the 0.8 to 1.0 equivalents of dp8 competed some proMMP-7-E195A away from some ternary complexes (one dp8 per two proenzymes). Addition of Hep dp16 at 0.4:1 to proMMP-7-E195A slowed tumbling by almost 3-fold to ~31 ns. Both the slowing and the ratio suggest formation of complexes that include cases of two to three (or more) proenzyme molecules bound per Hep dp16. NMR line broadening became severe at 0.5:1 Hep dp16:proenzyme and prohibitive at higher concentrations, consistent with high MW.
Oligomerization and oligosaccharide length
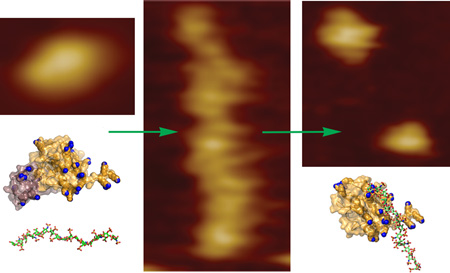
Atomic force microscopy (AFM) enabled us to ask further how do strongly activating complexes with Hep dp16 oligosaccharides differ in structure from weakly activating complexes with dp8? The maximum heights of proMMP-7-E195A appear to peak around 2.5 nm for the free state and complexes with Hep dp16, and just above 2 nm in the Hep dp8 complexes (Figure 3E). These heights are consistent with the diameter of the catalytic domain in the crystal structure (PDB code 1MMQ). ProMMP-7-E195A alone appears as monodisperse monomers, some of which look pear-shaped (Figure 3A). Considering the dimensions of MMP structures containing pro- and catalytic domains, the smaller (sometimes faint) lobe in the AFM-observed pear shape may represent the prodomain and the larger lobe the catalytic domain.
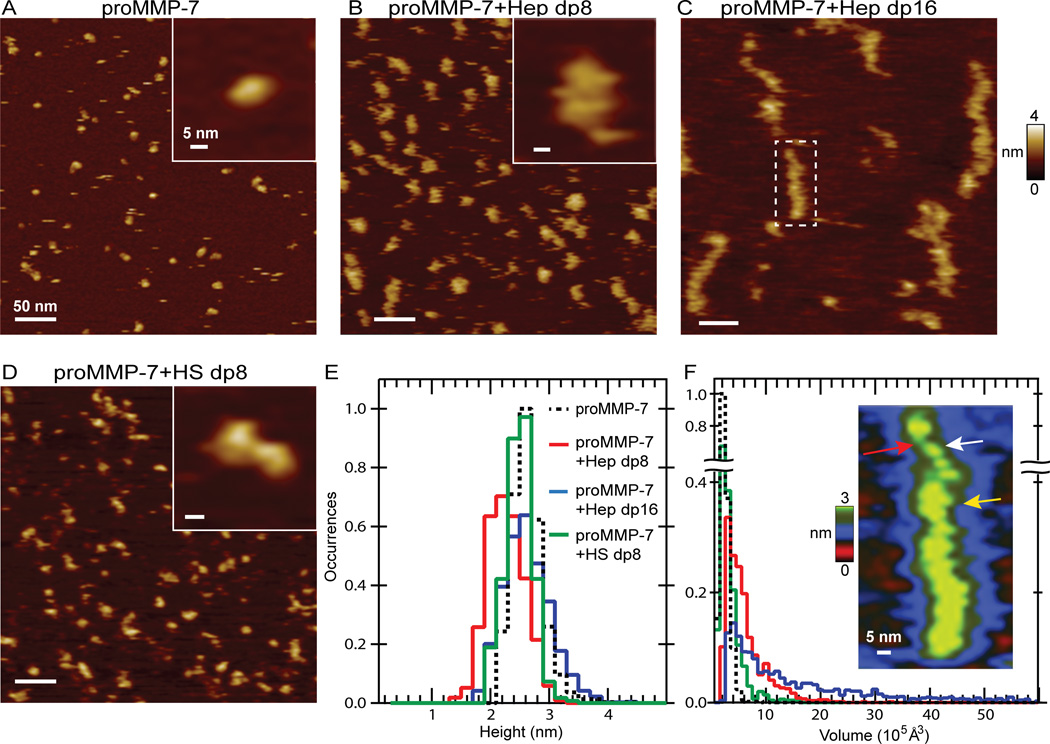
Figure 3. Atomic force microscopy (AFM) reveals tethering of proMMP-7 molecules by heparin oligosaccharides that becomes extensive with lengths of 16 monosaccharide units.
400×400 nm AFM images are shown for (A) proMMP7-E195A (100 nM) only or (B) 50 nM zymogen with 100 nM Hep dp8, (C) 100 nM zymogen with 100 nM Hep dp16, or (D) 50 nM zymogen with 100 nM HS dp8. Insets show enlargements. (E) The maximum heights of proMMP-7-E195A in the free state or with Hep dp8 or dp16 oligosaccharides present are tabulated as histograms with bins of 2 Å. In E and F, the black dashed line represents proMMP-7-E195A only (N = 903 features), red the mixture with Hep dp8 (N = 980), blue with Hep dp16 (N = 1379), and green with HS dp8 (N = 826 features). (F) Volumes of the same features are tallied into bins of 1×105 Å3 in the histogram. The inset of (F) expands the boxed feature from panel C. The red arrow marks an apparent Hep dp16 chain. The white arrow marks a width of one enzyme while the yellow arrow marks a width of two enzymes.
What do apparent volumes from AFM suggest about size of complexes formed? While proMMP-7-E195A is sharply defined in volume, its assemblies with Hep dp8 present have a broad distribution of volumes, and those with Hep dp16 a very broad distribution of volumes. The apparent volumes of the free monomers of proMMP-7-E195A are well-defined around 1.4×105 Å3, with most between 0.8 and 2.5×105 Å3 (Figure 3F). In the mixture with Hep dp8 with proMMP-7-E195A, the features are more diffuse with biggest population around 2.0 to 5.5×105 Å3 (Figure 3F). This being about twice the monomer volume suggests two proenzymes to be present in these particles. This is consistent with NMR τC evidence for two proenzymes bound per Hep dp8 chain near the 0.6 dp8 per proMMP-7 point in the NMR titration (Figure 2B). AFM images suggest mixtures with HS dp8 are also enriched in apparent tandem dimers, but have a higher proportion of particles containing a single proenzyme and a lower proportion containing multiple proenzyme molecules (Figure 3D,F). Lesser but significant populations with Hep dp8 or HS dp8 appeared around 3.4×105, 4.0×105, and 5.4×105 Å3 (Figure 3F), suggestive of complexes containing three, four, or five proenzyme molecules, respectively. Less common features with Hep dp8 exhibit an apparent volume of 9.4×105 Å3 (Figure 3F), suggesting larger complexes containing perhaps eight to ten proMMP-7 molecules.
These AFM data do not account for tip convolution. Deconvolution to minimize the contribution of tip geometry reduced the volume of proMMP-7-E195A and its complexes to around 70% (SI Figure S3); quoted volumes are thus small overestimates.
Striking extended, chain-like arrays with links of diffuse objects formed upon mixing of Hep dp16 with proMMP-7-E195A (Figure 3C). Most of these, extended arrays have apparent volumes of 6 to 30×105 Å3, though some range up to 60×105 Å3 (Figure 3F). Such large features are likely to contain tens of proMMP-7 molecules. Nonetheless, with Hep dp16 the most abundant apparent volumes are near 2.8×105 and 4×105 Å3, suggesting two to four proenzyme molecules in the complexes. Hep dp16 minimizes the number of single enzyme particles present on the mica surface, in contrast to images of dp8-containing mixtures. –This paucity and the extended arrays suggest that Hep dp16 draws smaller complexes together into the extended arrays.
Assemblies with heparinoids
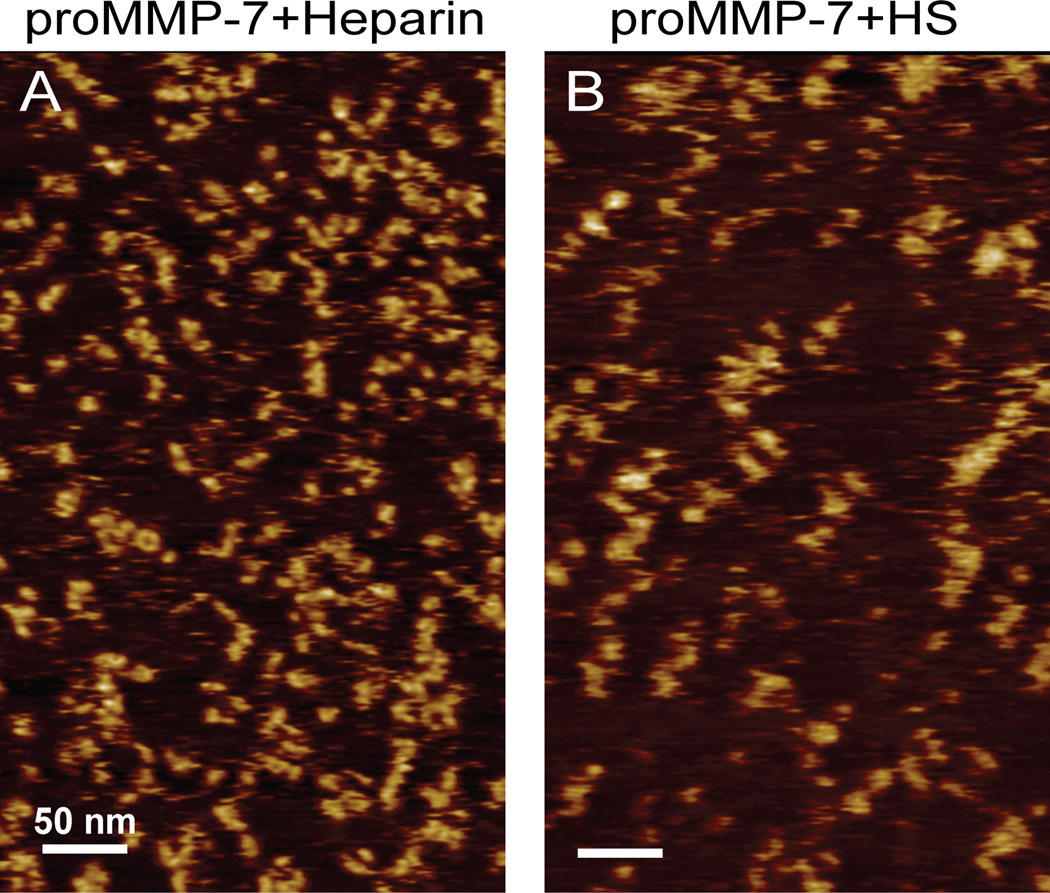
Whether these trends extend to heparin and HS was considered. Bovine heparin or HS, each polydisperse with chain lengths spanning 5 to 50 kDa, was mixed with proMMP-7-E195A and monitored by AFM (Figure 4). In both cases, the features ranged across the size range observed with Hep dp8 to some as large as observed with Hep dp16, but with more variability in morphology. Extended complexes are visible and partly resemble those induced by Hep dp16, both in images with HS and in larger numbers with heparin (Figure 4). Apparent volumes of the AFM features with heparin, HS, Hep dp8 appear, however, to be similar within experimental uncertainties (SI Figure S4). (Numerous smaller particles are present in the polydisperse mixtures, despite the human eye focusing on the larger particles). There is greater spreading of the larger features with HS and heparin. Lower resolution of imaged features seems to accompany longer GAG chains, which might allow more freedom of movement on the surface.
Figure 4. AFM images of proMMP-7-E195A mixtures with heparin (A) and heparan sulfate (B) suggest similarities in the heterogeneous tandem assemblies.
300×500 nm images are plotted for mixtures of (A) 100 nM proMMP-7-E195A with 2.5 µg/ml bovine lung heparin or (B) 50 nM proMMP7-E195A with 2.5 ug/ml bovine heparan sulfate. Note that bovine heparin and HS are polydisperse, with chain lengths from 5 to 50 kDa. The vertical scale spans 0 to 4 nm for both panels.
Activation relies on favorable electrostatics
Favorable electrostatic interactions between polyanionic Hep dp16 and the extensive positive charge of proMMP-7 (Figure 1A,B) were tested for importance in activation by adding NaCl up to 1 M in activation assays. 100 mM NaCl decreased activation to about half and 500 mM NaCl to about 10% (SI Figure S5). (Salt had no effect on proteolytic activity of the catalytic domain.)
Prodomain required for assembly
We tested whether the positively charged prodomain (Figure 1A) participates in oligomerization. Without the prodomain, no extended arrays form. Mixing active wt MMP-7 with Hep dp8, Hep dp16, or HS preserved its monodisperse AFM images, heights, and volumes (SI Figure S6), suggesting that the particles mainly contain a single enzyme in each case. Thus, the prodomain appears necessary for heparinoid-induced formation of oligomers.
C-terminal roles in activation and assembly
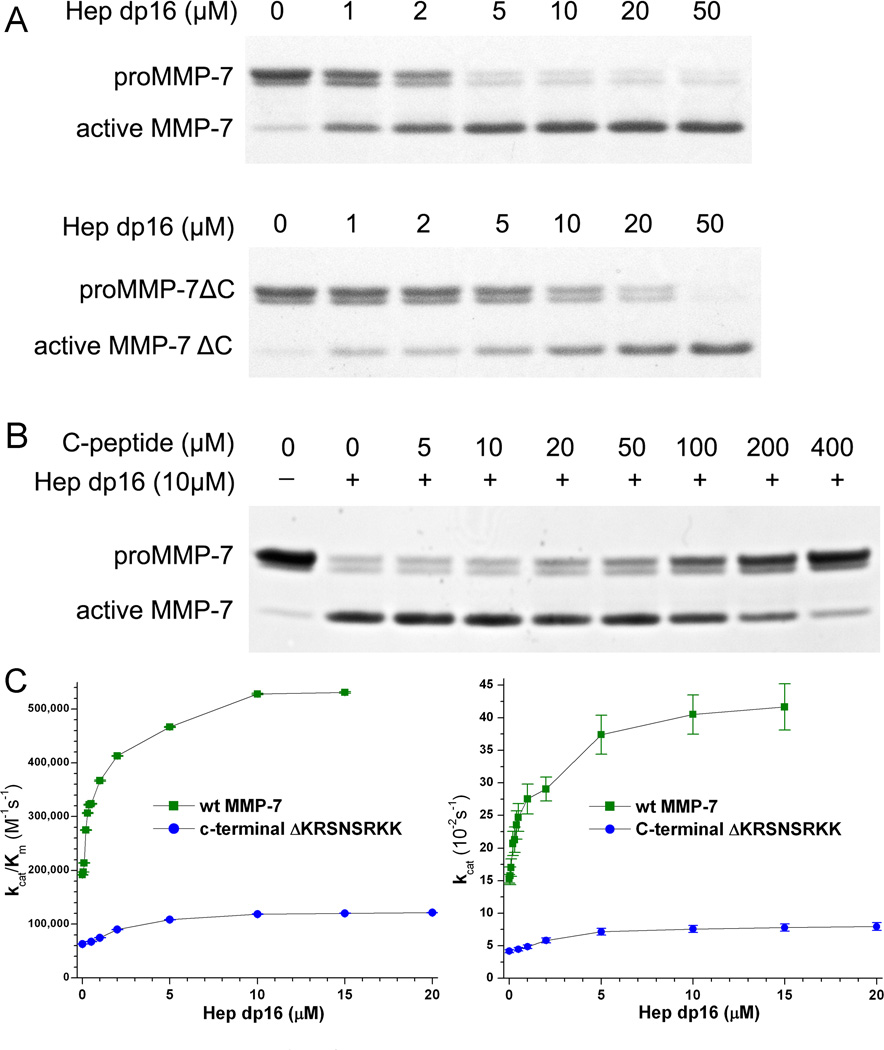
Since a synthetic C-terminal decapeptide from rat MMP-7 binds heparin,20 we tested whether the corresponding basic peptide from human MMP-7, KRSNSRKK, affects heparin-induced activation of proMMP-7. Deletion of this C-terminus impaired Hep dp16-induced activation, requiring at least 4-fold as much dp16 for as much activation as wt enzyme (Figure 5A). Moreover, synthetic KRSNSRKK peptide competed with Hep dp16-induced activation of wt proMMP-7, as 400 µM peptide nearly abolished it (Figure 5B). The C-terminus must then bind heparin in the activation assay.
Figure 5. The basic C-terminus is a remote site of allosteric, activating binding by Hep dp16 oligosaccharides.
(A) ProMMP-7 (5.5 µM) with wt sequence or deletion of KRSNSRKK was incubated with increasing concentrations of Hep dp16 at 37 °C for 2 h until each reaction was stopped. (B) The synthetic peptide with the KRSNSRKK sequence from the C-terminus interferes in Hep dp16-induced activation of proMMP-7. (C) Hep dp16 effects on catalytic efficiency (kcat/KM) and proteolytic turnover (kcat) of soluble Knight's peptide were monitored by FRET (25 °C, pH 7.5) are compared between active wt-MMP-7 and MMP-7ΔC.
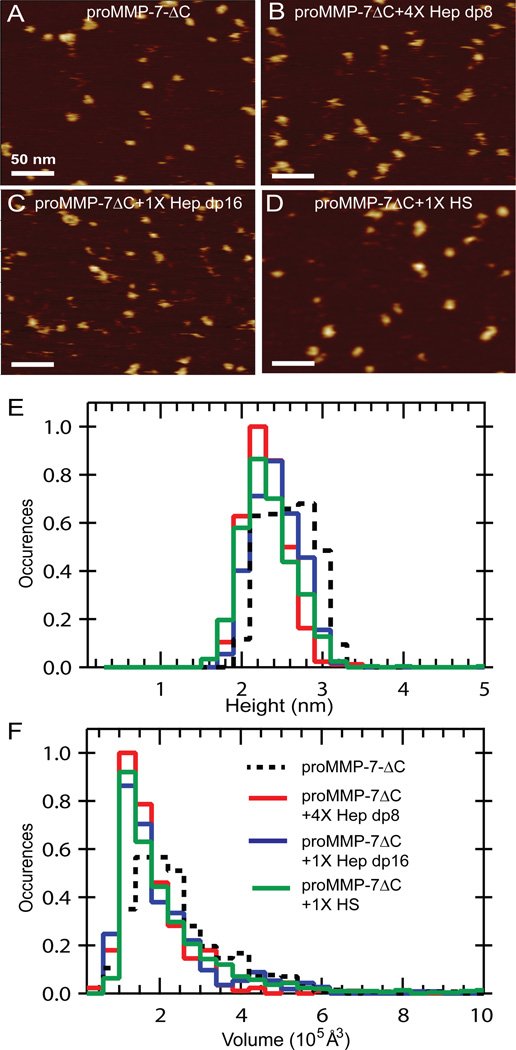
A role for the C-terminus in oligomerization was tested by AFM imaging. Hep dp8, dp16 or HS did not enlarge appearance, height or volume of proMMP-7ΔC particles (Figure 6) Hep dp16 mixed 1:1 with proMMP-7ΔC was dominated by volumes (~1×105 Å3; Figure 6C,F) equaling those of monomeric proMMP-7 (Figure 3A,F). The C-terminus is thus important in oligomerization needed for efficient activation.
Figure 6. Deletion of the basic C-terminus abrogates formation of extended complexes in the presence of heparin oligosaccharides.
400×400 nm AFM images are shown for (A) 50 nM proMMP-7ΔC alone, (B) 50 nM proMMP-7ΔC mixed with 200 nM Hep dp8, (C) 50 nM proMMP-7ΔC mixed with 50 nM Hep dp16, or (D) 100 nM proMMP-7ΔC mixed with 5 µg/ml HS. The number of features tabulated for the histograms are N = 456 for the free state, N = 284 for the mixture with Hep dp8, N = 362 for the 1:1 mixture with Hep dp16, and and N = 957 for the mixture with HS. (B) The height histograms use bins of 2 Å. (C) The volume histograms use bins of 0.4×105 Å3.
GAGs enhance velocity by allostery
We investigated whether GAGs modulate activity of mature MMP-7. HS, DS, and low molecular weight heparin, in a concentration-dependent manner, enhanced MMP-7 catalytic efficiency and velocity of proteolysis of Knight’s soluble peptide substrate40 of MMPs (SI Table S2). Fitting of progress curves41 found the enhanced catalytic efficiency (kcat/KM) mainly results from increased velocity Vmax or catalytic turnover kcat. The increased velocity and binding outside the active site categorize this as V-type allostery. DS boosted the catalytic efficiency the most of the intact GAGs, i.e by 1.7-fold, along with a 2.1-fold boost of kcat (SI Table S2). HS was the most efficient in needing only 0.1 mg/ml to achieve enhancement of kcat.
We asked whether structural features important in activation of the zymogen are important in enhancing mature MMP-7 activity. Proteolytic velocity increased with length of heparin fragment (SI Table S3). Hep dp16 delivered a 2.8-fold increase in both kcat/KM and kcat, which was superior to enhancements by Hep dp8, dp12 or low molecular weight heparin (Figure 5C, SI Tables S3, S2). Moreover, enhancement by Hep dp16 saturated at the much lower concentration of 10 µM (Figure 5C).
We checked if the C-terminus participates in this allosteric enhancement. Hep dp16 enhanced the catalytic efficiency and kcat of MMP-7ΔC by 1.9-fold (Figure 5C), implicating its binding after loss of the C-terminus. MMP-7ΔC retained only one-third of wild-type catalytic efficiency (Figure 5C). This impairment is surprising because this C-terminus is remote from the active site, outside the fold of this enzyme, and disordered, judging from sharp, random coil NMR peaks. Nonetheless, the C-terminus acts akin to an internal allosteric activator.
DISCUSSION
Trans-activation and assembly via GAG binding
We investigated the question of how proMMP-7 is activated by heparin and HS. The principles may extend to CSE and DS because of their analogous charge complementarity to proMMP-7. Heparin oligosaccharides induce trans-activation of proMMP-7 molecules they bridge (Figure 1). The significantly greater activation by heparin chains 16 or more saccharides long suggests side-by-side binding of zymogens, which is corroborated by AFM images (Figures 3, 4). HS and CSE are probable physiological partners for proMMP-7 that are linked to proteoglycans in vivo.8, 13, 15 Tandem assembly and trans-activation of basic proMMP-7 along acidic chains of HS or CSE emanating from proteoglycans can be predicted of mucosal epithelial cell surfaces.
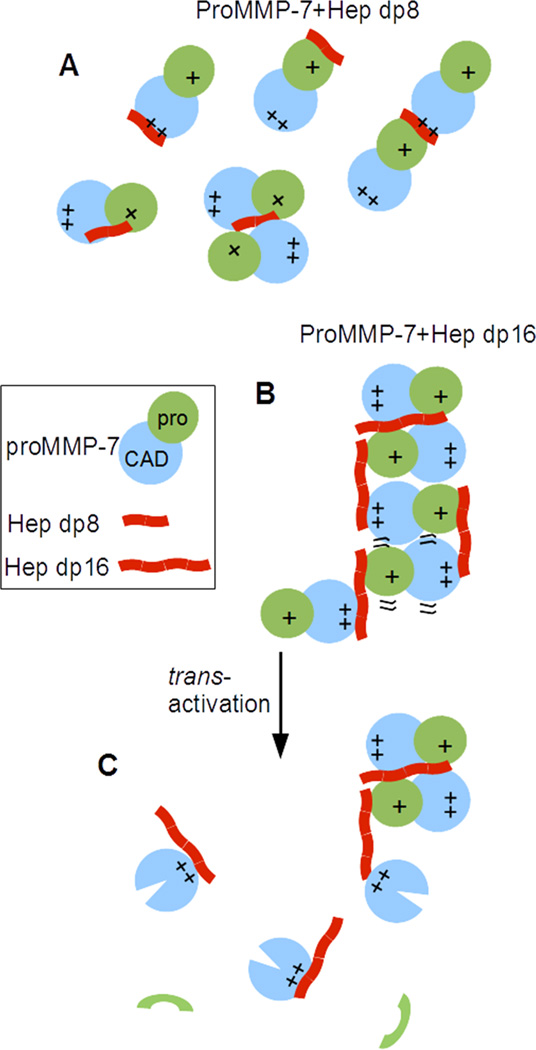
The importance of heparin-binding by the prodomain and basic C-terminus in normal assembly processes is evident from their deletion preventing formation of the arrays induced by Hep dp16 or HS (SI Figure S4, Figure 6). Mixtures of Hep or HS dp8 with proMMP-7 are likely to be enriched in dimers (Figure 3) sandwiched around an 8-saccharide chain bound, as well as 1:1 complexes (Figure 7A). The prodomain and C-terminus of proMMP-7 working together with Hep dp16 to create long tandem arrangements is symbolized in Figure 7B.
Figure 7. Speculative and simplified models for assembly of proMMP-7 induced by short and long heparin oligosaccharides.
(A) 8-monosaccharide lengths (dp8) of heparin are likely to encourage formation of sandwich dimers, in addition to forming 1:1 complexes with proenzymes. (B) 16-monosaccharide lengths (dp16) of heparin trigger formation of extended tandem arrangements that promote proteolysis of neighbors. (C) Removal of the prodomain renders the mature MMP-7 unlikely to remain in the arrays.
A long GAG chain may provide enough freedom for one proMMP-7 to attack the vulnerable sequence linking pro- and catalytic domains of its neighbor on the GAG chain. As activation minimally needs three molecules to interact (two proMMP-7 and GAG chain to link them), the velocity of activation must depend strongly on their concentrations. Extended complexes on longer heparinoid chains (Figures 3C, 4) have locally high concentrations that promote activation. Multiple proenzymes in the oligomers increase the probability that a transiently active proenzyme will contact and hydrolyze a neighboring zymogen.
For covalently intact proMMP-7 to be transiently an activating protease, the pro- and catalytic domains must separate enough to allow a loop of another proMMP-7 to enter its active site. The precedent of prodomain being spatially separated from the active site while covalently joined has been inferred from allosteric activations of proMMP-9, -2, and -3 by protein-protein interactions12, 28, 29 and from GAG activation of procathepsin B.1 ProMMP-9 activated in this fashion was inhibited by TIMP-1, confirming separation of the connected pro- and catalytic domains.28
Allostery
The hypothesis of GAG activation of proMMP-7 being allosteric was deduced from the need for pro- and catalytic domains to separate and for the GAG to bind away from active site.13 All GAG constructs tested accelerated proteolytic velocity of MMP-7 (SI Tables S2, S3), establishing them as allosteric activators. The complexes of catalytic domain with Hep dp16 or HS containing only one enzyme molecule in over 90% of particles (SI Figure S4) has two implications. First, proteolytic activation of proMMP-7 is likely to free the catalytic domain from GAG-induced oligomers, even while the catalytic domain can continue to associate with GAG chains (Figure 7C). Second, the allosteric enhancement of its proteolytic velocity by the GAG bound is probably transmitted across the catalytic domain.
The need for C-terminus and prodomain in heparinoid-induced assembly and activation (Figures 3, 5, 6, S4) identify them as remote GAG binding sites. The C-terminus must be a key binding site because its deletion seems to diminish the higher affinity component of acceleration by Hep dp16 (Figure 5C). However, the remaining lower affinity component of 1.9-fold acceleration induced by Hep dp16 (Figure 5C and SI Table S4) means that heparin oligosaccharides also bind elsewhere in the catalytic domain to allosterically enhance velocity. Indeed, several other positive charges on the back of the catalytic domain (Figure 1A) were hypothesized to tether rat MMP-7 to the HS of uterine epithelial cells and basement membranes.20 The C-terminus is needed not only for binding heparin but also for full enzyme activity. This suggests a network of communication between C-terminus and active site.
Regulatory bridges of GAGs to proteases
Parallels can be seen between GAG-dependent activations of proMMP-7 and proteases from two other classes. Favorable electrostatics are important in the GAG activations of proMMP-7, procathepsin B, L and S,1–3 β-secretase and procathepsin D.5, 6 GAG chains bind both pro- and catalytic domains of procathepsin B and proMMP-7. GAG activation of procathepsin D increases with length of heparin oligosaccharide,6 quite like trans-activation of proMMP-7 (Figure 1). Cathepsin K binding to C4S (CSA) introduces collagenase activity and “beads-on-a-string” oligomeric complexes.18 However, their regular repetition in crystal lattices42 contrasts the irregularity of proMMP-7 upon Hep dp16, heparin, or HS on solution-covered surfaces (Figures 3, 4).
Tethering between active protease and substrate is analogous to tethering between protease molecules. GAG chains were proposed to promote proMMP-2 activation by tethering it to active MMP-2 7 or thrombin.11 Heparin proteoglycans may draw chymase together with its substrate of thrombin.14 GAG chains link MMP-7 to pro-α-defensin for its maturation to a bactericidal peptide.8 HS proteoglycan positions MMP-7 to process HB-EGF to its mature form to bind its ErbB4 receptor and support cell survival.15
Joint, ionic binding to heparin, 16 monosaccharides or longer, brings serine proteases close to serpins for inhibition,17, 32, 39 similar to 16-mers and longer bridging among proMMP-7 molecules (Figures 1, 3, 4). In contrast to crystallized ternary complexes of serpin-heparin-thrombin,32 multiple copies of proMMP-7 assemble with 16-residue fragments of heparin up to full length heparin and HS (Figures 2, 3, 4). The use of two binding sites by proMMP-7 appears to support its repeating bridging by heparin and HS chains.
CONCLUSIONS
The initial hypotheses that GAG activation of proMMP-7 depends on either (i) 1:1 GAG binding with bridging and separation of pro- from catalytic domain13 or (ii) formation of ternary complexes with two proMMP-7 bound side-by-side to a GAG chain8 (like serpin-heparin-thrombin complexes) must be modified. Heparin-induced activation proceeds mainly in trans in assemblies initially containing multiple zymogens, rather than the simple 2:1 complexes expected. (There is precedent for extended complexes of another GAG-dependent protease.42) Heparinoid binding that is highly productive for activation of proMMP-7 bridges it to neighboring proMMP-7 molecules by binding prodomains and C-termini. This tethering together must increase likelihood of proteolytic attack among neighbors. Subsequent to the activation, binding of heparinoid chains to mature MMP-7 (at C-terminus and elsewhere) allosterically enhances its proteolytic velocity. The idea of preventing activation of proMMP-7 as a novel cancer therapeutic strategy with fewer side effects13 can now consider intervening in C-terminal and prodomain roles in activation.
METHODS
Surface plasmon resonance
Porcine intestinal heparin (Celsus) was immobilized on a streptavidin chip (GE Healthcare). For competition analysis, 500 nM of proMMP7 pre-mixed with 1000 nM of the GAG in HBS-P buffer (GE Healthcare) was flowed over the heparin chip at 30 µl/min at 25°C, followed by 2 min of flow of HBS-P buffer for dissociation, and surface regeneration using 2 M NaCl.
GAG activation of proMMP-7
ProMMP-7 (wt, E195A-inactivated, or with C-terminal KRSNSRKK deletion) was incubated with bovine HS (Sigma), low molecular weight heparin from porcine intestinal mucosa (Sigma), or the heparin oligosaccharides. The reaction mixtures were incubated for 2 or 3 h at 37°C, stopped using LDS sample buffer (Novex), separated on Bis-tris 4–12% SDS-PAGE gels (Novex), and stained with Coomassie Blue. The protein bands were scanned and quantified by Quantity One software (Biorad).
Hydrodynamics
Analytical gel filtration chromatography used an analytical TSK-GEL G3000SW column (Tosoh) at 22°C. The column and samples were equilibrated with gel filtration buffer listed in SI. 50 µM proMMP-7 was mixed with Hep dp8 at 20-fold molar excess or Hep dp16 at 4-fold molar excess and filtered through a 0.22 µm centrifugal filter (Millipore). 80 µL of each mixture was injected, eluted at 0.8 mL/min, and 1 mL fractions collected.
Rotational correlation times τC were measured with a Bruker 800 MHz Avance III NMR system from 15N relaxation (see SI), at each point of titrations of proMMP-7-E195A (300 µM) with heparin oligosaccharides.
Atomic force microscopy
ProMMP-7-E195A and truncated forms were prepared at 50 or 100 nM in imaging buffer (10 mM CaCl2, pH 7.5; see SI). Mixtures with Hep dp8 or dp16, heparin, HS, or HS dp8 were examined. Each solution was deposited immediately after mixing on a freshly cleaved muscovite mica surface (V-1 grade, Spi Supplies) and incubated for ~5 min. The surface was then rinsed three times with ~100 µL of imaging buffer. AFM images were acquired in imaging buffer in tapping mode using a commercial instrument (Cypher, Asylum Research). SNL tips (sharp nitride levers, SNL-10, Bruker) with measured spring constants of ~0.2 N/m were used. Images were recorded at ~30°C with an estimated tip-sample force < 100 pN, deduced by comparing the free space tapping amplitude (~5 nm) to the imaging set point amplitude. Under such conditions, minimal protein distortion is expected. Image analysis is detailed in SI.
Kinetics of proteolysis
Digestion of soluble Knight's peptide40 (FS-6; EMD) was monitored by FRET with excitation at 328 nM and emission at 393 nM. Progress curves of the digestions at 25°C in proteolysis buffer (see SI) were monitored with a BioTek Synergy MX plate reader. kcat/Km, Km and kcat were obtained by fitting of a few progress curves as described.41
Supplementary Material
Acknowledgements
We thank W. Parks for pointing out the need for mechanistic information on GAG activation of proMMP-7. This work was supported by NIH grant R01GM057289. G. King acknowledges support from the Burroughs Wellcome Fund Career Award at the Scientific Interface. The University of Missouri and NIH grant RR022341 supported acquisition of the 800 MHz NMR equipment.
Abbreviations
- AFM
atomic force microscopy
- CAD
catalytic domain
- CSA or C4S
chondroitin-4-sulfate
- CSE
chondroitin sulfate E (chondroitin-4,6-sulfate)
- dp
degree of polymerization
- DS
dermatan sulfate
- DLS
dynamic light scattering
- FRET
Förster resonance energy transfer
- GAG
glycosaminoglycan
- HB-EGF
heparin-binding epidermal growth factor
- Hep
heparin
- HS
heparan sulfate
- MMP
matrix metalloproteinase
- serpin
serine protease inhibitor
- SPR
surface plasmon resonance
- wt
wild-type
Footnotes
ASSOCIATED CONTENT
Supporting Information
Supporting information includes supplementary figures, tables, sources of materials, preparative methods, buffers, and methods details. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Caglič D, Pungerčar JR, Pejler G, Turk V, Turk B. Glycosaminoglycans Facilitate Procathepsin B Activation through Disruption of Propeptide-Mature Enzyme Interactions. J. Biol. Chem. 2007;282:33076–33085. doi: 10.1074/jbc.M705761200. [DOI] [PubMed] [Google Scholar]
- 2.Fairhead M, Kelly SM, van der Walle CF. A heparin binding motif on the pro-domain of human procathepsin L mediates zymogen destabilization and activation. Biochem. Biophys. Res. Commun. 2008;366:862–867. doi: 10.1016/j.bbrc.2007.12.062. [DOI] [PubMed] [Google Scholar]
- 3.Vasiljeva O, Dolinar M, Pungerčar JR, Turk V, Turk B. Recombinant human procathepsin S is capable of autocatalytic processing at neutral pH in the presence of glycosaminoglycans. FEBS Lett. 2005;579:1285–1290. doi: 10.1016/j.febslet.2004.12.093. [DOI] [PubMed] [Google Scholar]
- 4.Hallgren J, Karlson U, Poorafshar M, Hellman L, Pejler G. Mechanism for activation of mouse mast cell tryptase: dependence on heparin and acidic pH for formation of active tetramers of mouse mast cell protease 6. Biochemistry. 2000;39:13068–13077. doi: 10.1021/bi000973b. [DOI] [PubMed] [Google Scholar]
- 5.Beckman M, Holsinger RMD, Small DH. Heparin Activates β-Secretase (BACE1) of Alzheimer's Disease and Increases Autocatalysis of the Enzyme. Biochemistry. 2006;45:6703–6714. doi: 10.1021/bi052498t. [DOI] [PubMed] [Google Scholar]
- 6.Beckman M, Freeman C, Parish CR, Small DH. Activation of cathepsin D by glycosaminoglycans. FEBS J. 2009;276:7343–7352. doi: 10.1111/j.1742-4658.2009.07444.x. [DOI] [PubMed] [Google Scholar]
- 7.Crabbe T, Ioannou C, Docherty AJ. Human progelatinase A can be activated by autolysis at a rate that is concentration-dependent and enhanced by heparin bound to the C-terminal domain. EurJBiochem. 1993;218:431–438. doi: 10.1111/j.1432-1033.1993.tb18393.x. [DOI] [PubMed] [Google Scholar]
- 8.Ra HJ, Harju-Baker S, Zhang F, Linhardt RJ, Wilson CL, Parks WC. Control of promatrilysin (MMP7) activation and substrate-specific activity by sulfated glycosaminoglycans. J. Biol. Chem. 2009;284:27924–27932. doi: 10.1074/jbc.M109.035147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butler GS, Butler MJ, Atkinson SJ, Will H, Tamura T, Schade van Westrum S, Crabbe T, Clements J, d'Ortho MP, Murphy G. The TIMP2 membrane type 1 metalloproteinase "receptor" regulates the concentration and efficient activation of progelatinase A. A kinetic study. J. Biol. Chem. 1998;273:871–880. doi: 10.1074/jbc.273.2.871. [DOI] [PubMed] [Google Scholar]
- 10.Iida J, Wilhelmson KL, Ng J, Lee P, Morrison C, Tam E, Overall CM, McCarthy JB. Cell surface chondroitin sulfate glycosaminoglycan in melanoma: role in the activation of pro-MMP-2 (pro-gelatinase A) Biochem. J. 2007;403:553–563. doi: 10.1042/BJ20061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koo B-H, Han JH, Yeom YI, Kim D-S. Thrombin-dependent MMP-2 Activity Is Regulated by Heparan Sulfate. J. Biol. Chem. 2010;285:41270–41279. doi: 10.1074/jbc.M110.171595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hadler-Olsen E, Fadnes B, Sylte I, Uhlin-Hansen L, Winberg J-O. Regulation of matrix metalloproteinase activity in health and disease. FEBS J. 2011;278:28–45. doi: 10.1111/j.1742-4658.2010.07920.x. [DOI] [PubMed] [Google Scholar]
- 13.Tocchi A, Parks WC. Functional interactions between matrix metalloproteinases and glycosaminoglycans. FEBS J. 2013;280:2332–2341. doi: 10.1111/febs.12198. [DOI] [PubMed] [Google Scholar]
- 14.Pejler G, Sadler JE. Mechanism by Which Heparin Proteoglycan Modulates Mast Cell Chymase Activity. Biochemistry. 1999;38:12187–12195. doi: 10.1021/bi991046b. [DOI] [PubMed] [Google Scholar]
- 15.Yu WH, Woessner JF, Jr, McNeish JD, Stamenkovic I. CD44 anchors the assembly of matrilysin/MMP-7 with heparin-binding epidermal growth factor precursor and ErbB4 and regulates female reproductive organ remodeling. Genes Dev. 2002;16:307–323. doi: 10.1101/gad.925702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, Lopez-Boado YS, Stratman JL, Hultgren SJ, Matrisian LM, Parks WC. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science. 1999;286:113–117. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]
- 17.Griffith MJ. Kinetics of the heparin-enhanced antithrombin III/thrombin reaction. Evidence for a template model for the mechanism of action of heparin. J. Biol. Chem. 1982;257:7360–7365. [PubMed] [Google Scholar]
- 18.Li Z, Yasuda Y, Li W, Bogyo M, Katz N, Gordon RE, Fields GB, Brömme D. Regulation of Collagenase Activities of Human Cathepsins by Glycosaminoglycans. J. Biol. Chem. 2004;279:5470–5479. doi: 10.1074/jbc.M310349200. [DOI] [PubMed] [Google Scholar]
- 19.Lynch CC, Vargo-Gogola T, Martin MD, Fingleton B, Crawford HC, Matrisian LM. Matrix metalloproteinase 7 mediates mammary epithelial cell tumorigenesis through the ErbB4 receptor. Cancer Res. 2007;67:6760–6767. doi: 10.1158/0008-5472.CAN-07-0026. [DOI] [PubMed] [Google Scholar]
- 20.Yu WH, Woessner JF., Jr Heparan sulfate proteoglycans as extracellular docking molecules for matrilysin (matrix metalloproteinase 7) J. Biol. Chem. 2000;275:4183–4191. doi: 10.1074/jbc.275.6.4183. [DOI] [PubMed] [Google Scholar]
- 21.Berton A, Selvais C, Lemoine P, Henriet P, Courtoy PJ, Marbaix E, Emonard H. Binding of matrilysin-1 to human epithelial cells promotes its activity. Cell. Mol. Life Sci. 2007;64:610–620. doi: 10.1007/s00018-007-6415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen P, Abacherli LE, Nadler ST, Wang Y, Li Q, Parks WC. MMP7 Shedding of Syndecan-1 Facilitates Re-Epithelialization by Affecting α2β1 Integrin Activation. PLoS ONE. 2009;4:e6565. doi: 10.1371/journal.pone.0006565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitsiades N, Yu W-h, Poulaki V, Tsokos M, Stamenkovic I. Matrix Metalloproteinase-7-mediated Cleavage of Fas Ligand Protects Tumor Cells from Chemotherapeutic Drug Cytotoxicity. Cancer Res. 2001;61:577–581. [PubMed] [Google Scholar]
- 24.Vargo-Gogola T, Fingleton B, Crawford HC, Matrisian LM. Matrilysin (Matrix Metalloproteinase-7) Selects for Apoptosis-resistant Mammary Cells in Vivo. Cancer Res. 2002;62:5559–5563. [PubMed] [Google Scholar]
- 25.Lynch CC, Vargo-Gogola T, Martin MD, Fingleton B, Crawford HC, Matrisian LM. Matrix Metalloproteinase 7 Mediates Mammary Epithelial Cell Tumorigenesis through the ErbB4 Receptor. Cancer Res. 2007;67:6760–6767. doi: 10.1158/0008-5472.CAN-07-0026. [DOI] [PubMed] [Google Scholar]
- 26.Voorzanger-Rousselot N, Juillet F, Mareau E, Zimmermann J, Kalebic T, Garnero P. Association of 12 serum biochemical markers of angiogenesis, tumour invasion and bone turnover with bone metastases from breast cancer: a crossectional and longitudinal evaluation. BrJCancer. 2006;95:506–514. doi: 10.1038/sj.bjc.6603285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang W-S, Chen P-M, Wang H-S, Liang W-Y, Su Y. Matrix metalloproteinase-7 increases resistance to Fas-mediated apoptosis and is a poor prognostic factor of patients with colorectal carcinoma. Carcinogenesis. 2006;27:1113–1120. doi: 10.1093/carcin/bgi351. [DOI] [PubMed] [Google Scholar]
- 28.Bannikov GA, Karelina TV, Collier IE, Marmer BL, Goldberg GI. Substrate binding of gelatinase B induces its enzymatic activity in the presence of intact propeptide. J. Biol. Chem. 2002;277:16022–16027. doi: 10.1074/jbc.M110931200. [DOI] [PubMed] [Google Scholar]
- 29.Rosenblum G, Meroueh S, Toth M, Fisher JF, Fridman R, Mobashery S, Sagi I. Molecular structures and dynamics of the stepwise activation mechanism of a matrix metalloproteinase zymogen: challenging the cysteine switch dogma. J. Am. Chem. Soc. 2007;129:13566–13574. doi: 10.1021/ja073941l. [DOI] [PubMed] [Google Scholar]
- 30.Yu WH, Woessner JF., Jr Heparin-enhanced zymographic detection of matrilysin and collagenases. Anal. Biochem. 2001;293:38–42. doi: 10.1006/abio.2001.5099. [DOI] [PubMed] [Google Scholar]
- 31.Griffith MJ. Heparin-catalyzed inhibitor/protease reactions: kinetic evidence for a common mechanism of action of heparin. Proc. Natl. Acad. SciUSA. 1983;80:5460–5464. doi: 10.1073/pnas.80.18.5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W, Johnson DJ, Esmon CT, Huntington JA. Structure of the antithrombin-thrombin-heparin ternary complex reveals the antithrombotic mechanism of heparin. Nat Struct Mol Biol. 2004;11:857–862. doi: 10.1038/nsmb811. [DOI] [PubMed] [Google Scholar]
- 33.Conrad HE. Heparin-Binding Proteins. San Diego: Academic Press; 1998. [Google Scholar]
- 34.Khan S, Gor J, Mulloy B, Perkins SJ. Semi-Rigid Solution Structures of Heparin by Constrained X-ray Scattering Modelling: New Insight into Heparin–Protein Complexes. J. Mol. Biol. 2010;395:504–521. doi: 10.1016/j.jmb.2009.10.064. [DOI] [PubMed] [Google Scholar]
- 35.Mulloy B, Forster MJ, Jones C, Davies DB. N.m.r and molecular-modeling studies of the solution conformation of heparin. Biochem. J. 1993;293:849–858. doi: 10.1042/bj2930849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gunning AP, Kirby AR, Fuell C, Pin C, Tailford LE, Juge N. Mining the “glycocode”—exploring the spatial distribution of glycans in gastrointestinal mucin using force spectroscopy. FASEB J. 2013;27:2342–2354. doi: 10.1096/fj.12-221416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milz F, Harder A, Neuhaus P, Breitkreuz-Korff O, Walhorn V, Lübke T, Anselmetti D, Dierks T. Cooperation of binding sites at the hydrophilic domain of cell-surface sulfatase Sulf1 allows for dynamic interaction of the enzyme with its substrate heparan sulfate. Biochim. Biophys. Acta. 2013;1830:5287–5298. doi: 10.1016/j.bbagen.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 38.Cha J, Auld DS. Site-directed mutagenesis of the active site glutamate in human matrilysin: investigation of its role in catalysis. Biochemistry. 1997;36:16019–16024. doi: 10.1021/bi972223g. [DOI] [PubMed] [Google Scholar]
- 39.Olson ST, Björk I. Predominant contribution of surface approximation to the mechanism of heparin acceleration of the antithrombin-thrombin reaction. Elucidation from salt concentration effects. J. Biol. Chem. 1991;266:6353–6364. [PubMed] [Google Scholar]
- 40.Neumann U, Kubota H, Frei K, Ganu V, Leppert D. Characterization of Mca-Lys-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2, a fluorogenic substrate with increased specificity constants for collagenases and tumor necrosis factor converting enzyme. Anal. Biochem. 2004;328:166–173. doi: 10.1016/j.ab.2003.12.035. [DOI] [PubMed] [Google Scholar]
- 41.Palmier MO, Van Doren SR. Rapid determination of enzyme kinetics from fluorescence: overcoming the inner filter effect. Anal. Biochem. 2007;371:43–51. doi: 10.1016/j.ab.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z, Kienetz M, Cherney MM, James MN, Bromme D. The crystal and molecular structures of a cathepsin K:chondroitin sulfate complex. J. Mol. Biol. 2008;383:78–91. doi: 10.1016/j.jmb.2008.07.038. [DOI] [PubMed] [Google Scholar]
- 43.Van Doren SR. Structural Basis of Extracellular Matrix Interactions with Matrix Metalloproteinases, in. In: Parks WC, Mecham RP, editors. Extracellular Matrix Degradation. Berlin: Springer-Verlag; 2011. pp. 123–144. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.