Abstract
Cancer cells reprogram metabolism to maintain rapid proliferation under often stressful conditions. Glycolysis and glutaminolysis are two central pathways that fuel cancer metabolism. Allosteric regulation and metabolite driven post-translational modifications of key metabolic enzymes allow cancer cells glycolysis and glutaminolysis to respond to changes in nutrient availability and the tumor microenvironment. While increased aerobic glycolysis (the Warburg effect) has been a noted part of cancer metabolism for over 80 years, recent work has shown that the elevated levels of glycolytic intermediates are critical to cancer growth and metabolism due to their ability to feed into the anabolic pathways branching off glycolysis such as the pentose phosphate pathway and serine biosynthesis pathway. The key glycolytic enzymes phosphofructokinase-1 (PFK1), pyruvate kinase (PKM2) and phosphoglycerate mutase 1 (PGAM1) are regulated by upstream and downstream metabolites to balance glycolytic flux with flux through anabolic pathways. Glutamine regulation is tightly controlled by metabolic intermediates that allosterically inhibit and activate glutamate dehydrogenase, which fuels the tricarboxylic acid cycle by converting glutamine derived glutamate to α-ketoglutarate. The elucidation of these key allosteric regulatory hubs in cancer metabolism will be essential for understanding and predicting how cancer cells will respond to drugs that target metabolism. Additionally, identification of the structures involved in allosteric regulation will inform the design of anti-metabolism drugs which bypass the off-target effects of substrate mimics. Hence, this review aims to provide an overview of allosteric control of glycolysis and glutaminolysis.
Keywords: Allosteric, glutamate dehydrogenase, glutaminolysis, glycolysis, PFK1, PGAM1, PKM2
Anabolic carbons derived from glycolytic intermediates feedback to control glycolytic flux
Cancer metabolism is rewired to meet the demands induced by rapid proliferation in often adverse conditions. The most noted change in cancer metabolism is increased aerobic glycolysis. First documented by Otto Warburg in the early 20th century, glucose is predominantly converted to lactate by many cancers even in the presence of oxygen (Koppenol et al., 2011). In aerobic, non-cancerous cells, glucose is primarily converted to pyruvate in the cytosol. Pyruvate then enters the mitochondrial tricarboxylic acid (TCA) cycle after pyruvate dehydrogenase converts it to acetyl-CoA. While glucose oxidation generates much more adenosine triphosphate (ATP) per molecule of glucose, aerobic glycolysis in cancer cells permits shunting of glycolytic intermediates toward anabolic synthesis of cellular building blocks at the expense of ATP synthesis by fermentation. The pentose phosphate pathway (PPP), which begins with glucose 6-phosphate, produces the five carbon sugar ribose for nucleotide synthesis and NADPH (nicotinamide adenine dinucleotide phosphate). The NADPH generated by the PPP plays several key roles in cancer metabolism. NADPH provides the reducing power necessary for lipid synthesis (Wamelink et al., 2008). NADPH is also required to generate reduced glutathione (GSH) from oxidized glutathione (GSSG) for reactive oxygen species (ROS) homeostasis. The glycolytic intermediate 3-phosphoglycerate can be shunted into the serine and glycine biosynthesis pathway. Serine participates in single-carbon metabolism through its conversion to glycine. Levels of the these anabolic substrates allosterically modulate the activity of key glycolytic enzymes, ensuring that adequate carbon backbones and reducing equivalents to generate NADPH are siphoned from glycolysis for macromolecular synthesis and redox balance in the growing cancer cell. Also, metabolically driven post-translational modifications provide an additional level of the control of these key metabolic enzymes
Phosphofructokinase 1 (PFK1) allosteric regulation
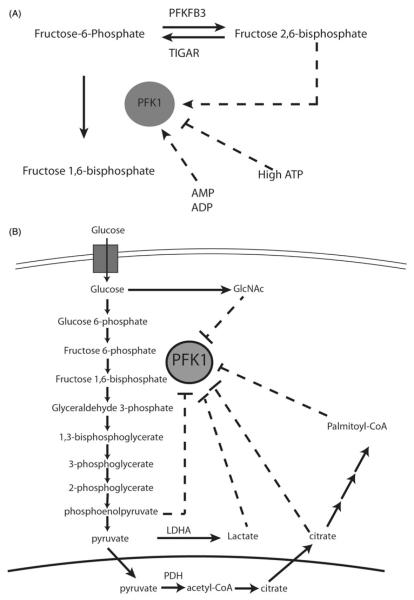
Phosphofructokinase 1 (PFK1) catalyzes the ATP-dependent first committed step of glycolysis and is a key point of regulation of the rate of glycolysis. PFK1 forms an active tetramer, but can be dissociated into inactive dimer and monomer forms (Uyeda, 1979). Multiple metabolic intermediates regulate PFK1 activity in feedback loops. PFK1 can be inhibited by high concentrations of ATP, which can result from high levels of oxidative phosphorylation in a well fed state (Hers & Van Schaftingen, 1982). Although ATP is a substrate of PFK1, a second ATP allosteric binding site, dependent upon mouse Arg 429 and Arg 433, negatively regulates PFK1 activity (Kemp & Gunasekera, 2002). By contrast, PFK1 can be activated by metabolites which are elevated with energy deprivation. AMP (adenosine monophosphate) and ADP (adenosine diphosphate) activate PFK1 via a binding site that is dependent upon the amino acids Ser 377, Lys 678, Asn 341 (Bruser et al., 2012).
During glycolysis, PFK1 phosphorylates the first carbon of fructose 6-phosphate to form fructose 1,6-bisphosphate (Fru 1,6-BP). Alternatively, fructose 6-phosphate can be phosphorylated on the second carbon to make fructose 2,6-bisphosphate (Fru 2,6-BP), a potent allosteric activator of PFK1 which promotes PFK1 activity by enhancing active tetramer formation (Reinhart, 1983) (Figure 1A). To stimulate PFK1 activity, Fru 2,6-BP binds to a highly conserved site which, in mammals, contains Arg 566, Arg 655 and His 661. These amino acids play a key role in the binding of Fru 2,6-BP to PFK1 (Ferreras et al., 2009). Fru 2,6-BP is produced by the group of enzymes termed 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatases (PFKFB), which are encoded by the genes PFKFB1-4. The four PFKFB enzymes can catalyze both the forward (kinase) and reverse (phosphatase) reactions and have differing ratios of kinase to phosphatase activity and expression patterns. Accumulation of fructose 6-phosphate can lead to an increase in Fru 2,6-BP synthesis, which, in turn, stimulates PFK1 activity to relieve the buildup of fructose 6-phosphate.
Figure 1.
Allosteric regulation of phosphofructokinase 1 (PFK1). (A) PFK1 is allosterically activated by fructose 2,6-bisphosphate, which is created by PFKFB3 and converted back to fructose-6-phosphate by TIGAR. PFK1 is activated by high AMP and ADP and inhibited by high ATP levels. (B) PFK1 is inhibited by glycolytic products GlcNAc, lactate, citrate and palmitoyl-CoA. AMP, ADP, ATP, adenosine mono-, di and tri-phosphate, respectively; PFKFB3, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3; TIGAR, TP53-induced glycolysis and apoptosis regulator; LDHA, lactate dehydrogenase; GlcNAc, N-acetylglucosamine; PDH, pyruvate dehydrogenase. (see colour version of this figure at www.informahealthcare.com/bmg).
The role of Fru 2,6-BP in cancer metabolism remains poorly understood. While cancer cells exhibit high glycolytic flux, cancer cells also benefit from the enhanced flux through anabolic pathways branching-off of glycolysis. PFKFB3, which favors the formation of Fru 2,6-BP and thus promotes PFK1 activity, has been shown to be overexpressed in cancer and induced by the hypoxia inducible factor-1 (HIF-1) (Atsumi et al., 2002; Kessler et al., 2008). Being developed for phase 1 clinical trials, a PFKFB3 inhibitor induces apoptosis in transformed cells and slows the development of xenografts in vivo (Clem et al., 2013).
Some controversy and confusion surrounds the role of PFKFB4 in cancer metabolism. PFKFB4 expression is associated with prostate cancer metastasis and with shorter survival of patients with gliomas (Goidts et al., 2012; Ros et al., 2012). PFKFB4 is also a prognostic marker for bladder cancer progression (Yun et al., 2012). While experimental data show that PFKFB4 favors the formation of Fru 2,6-BP with a kinase to phosphatase activity ratio of 4.1 (Sakakibara et al., 1997), confusion has been added by a review paper that states PFKFB4 may slightly favor the conversion of Fru 2,6-BP back to fructose 6-phosphate with a kinase/bisphosphatase ratio of 0.9 (Okar et al., 2001). However, knockdown of PFKFB4 in prostate cancer cells increases the levels of Fru 2,6-BP (Ros et al., 2012), although it is not clear if this is due directly to PFKFB4 kinase/phosphatase activity or from cellular adaption. Knockdown of the hypoxia induced PFKFB4 increased ROS and cell death in prostate cancer cells, consistent with increased Fru 2,6-BP causing reduced PPP flux through PFK1 activation (Minchenko et al., 2004; Ros et al., 2012).
Intriguingly, p53 has been documented to upregulate the protein TP53-induced glycolysis and apoptosis regulator (TIGAR) (Bensaad et al., 2006), which has high Fru 2,6-bisphosphatase activity. P53 induction of TIGAR is presumably also for the redox homeostasis upon p53 activation (Wanka et al., 2012). However, p53 status in human cancer cells does not correlate with TIGAR expression, which does not appear to have tumor suppressive effects in vivo (Cheung et al., 2013). In aggregate, many recent studies document a complex regulatory role of Fru 2,6-BP in tumorigenesis as well as tumor maintenance.
When it is not converted to lactate, pyruvate derived from glycolysis can be converted to acetyl-CoA, which is combined with oxaloacetate in the TCA cycle to form citrate in the mitochondria. Citrate can be exported from the mitochondria to the cytosol and then re-converted back to acetyl-CoA for fatty acid synthesis by the enzyme ATP citrate lyase (ACLY). Randle et al. (1963) demonstrated in the 1960s that fatty acids can slow glucose utilization in muscle, a phenomenon termed the “Randle Cycle”. Both citrate and activated fatty acids (acyl-CoA) can regulate glycolysis in feedback loops. Citrate is capable of inhibiting PFK1, slowing glycolysis and linking TCA cycle activity with rate of glycolysis when there is sufficient contribution of glucose to citrate synthesis (Poorman et al., 1984) (Figure 1B). Inhibition of PFK1 by citrate is dependent on the key amino acids Lys 577 and Lys 617 in human PFK1 (Usenik & Legisa, 2010).
Many cancer cells depend on de novo lipogenesis to provide lipids for energy storage and membrane formation, increasing levels of acyl-CoA and other lipogenesis intermediates. A recent study (Jenkins et al., 2011) shows that long chain acyl-CoAs, including the lipogenesis intermediate palmitoyl-CoA, can feedback to inhibit PFK1 activity. Four cysteines (Cys 114, Cys 170, Cys 351, Cys 577) in PFK1 can be acylated to mediate inhibition by acyl-CoA. However, acyl-CoA can also non-covalently interact with PFK1, causing a conformational change in PFK1 that promotes an interaction with its inhibitor calmodulin (Jenkins et al., 2011). Since lipogenesis requires reducing equivalents from NADPH for fatty acid chain elongation, palmitoyl-CoA synthesized during lipogenesis could stimulate NADPH production by shunting glucose away from glycolysis via PFK1 inhibition and toward PPP. This feedback inhibition of PFK1 by citrate and acyl-CoAs may explain many of the observations described by the Randle cycle.
The activity of PFK1 is regulated by glycosylation in response to changes in nutrient availability or cellular stress, allowing the dynamic balancing of glycolysis and the PPP as cellular conditions change (Yi et al., 2012). N-acetylglucosamine (GlcNAc) can modify proteins at serine and threonine residues via O-linked conjugation in a process termed O-linked GlcNAcylation. Since its synthesis requires glucose, glutamine, ATP, acetyl-CoA and uridine, UDP-GlcNAc synthesis is sensitive to the availability of different nutrients. Both GlcNAc and the enzyme responsible for O-linked GlcNAcylation (O-linked N-acetylglucosaminyltransferase; OGT), are acutely upregulated by glucose deprivation and other cellular stresses (Taylor et al., 2008; Zachara et al., 2004). A recent study found that UDP-GlcNAc is used to glycosylate Ser 529 on PFK1, resulting in reduced PFK1 activity in a variety of human cancer cells (Yi et al., 2012). Ser 529 is a key residue at the site where Fructose 2,6-BP binds PFK1, forming a hydrogen bond with the 2-phosphate group on Fructose 2,6-BP (Ferreras et al., 2009; Yi et al., 2012). Glucose deprivation induces OGT dependent PFK1 Ser 529 glycosylation, slowing glycolytic flux to allow glucose to be channeled elsewhere in low glucose conditions. Additionally, hypoxia induces rapid PFK1 Ser 529 glycosylation and presumably PPP flux, allowing cancer cells to curb hypoxia-induced ROS by increasing NADPH necessary to reduce glutathione. Hypoxia induced glycosylation may be due to increased uptake of glucose, which can lead to increased GlcNAc (Mattaini & Vander Heiden, 2012). However, how different cellular stresses and changes in nutrient availability affect OGT, GlcNAc levels and PFK1 glycosylation remain poorly understood.
Lactate generated through aerobic glycolysis can modulate glycolytic rate by directly regulating PFK1 activity in muscle cells. The allosteric modulation of PFK1 by lactate appears to be independent of pH (Costa Leite et al., 2007). Lactate seems to exert its effect on PFK1 activity through inhibition of PFK1 active tetramer formation. Lactate-dependent allosteric regulation of PFK1 is, however, inhibited by the presence of the PFK1 activator Fru 2,6-BP (Costa Leite et al., 2007). Lactate can additionally inhibit the activating phosphorylation of PFK1 (Leite et al., 2011). The role of lactate-dependent PFK1 inhibition in cancer cells remains to be explored, as lactate transporter extrusion of this waste product into the tumor microenvironment has been shown to be critical for some cancers (Pinheiro et al., 2012). Intriguingly, hypoxic and aerobic cancer cells sometimes show a commensal relationship, where lactate extruded by hypoxic cells appears to be taken up by aerobic cells and converted back to pyruvate for oxidation in mitochondria (Sonveaux et al., 2008). In this regard, it seems possible that lactate mediated inhibition of PFK1 in aerobic cells could permit them to oxidize imported lactate derived pyruvate rather than competing with hypoxic cells for glucose. It is also possible that high lactate in hypoxic cancer cells slows glycolysis to promote flux through the PPP for redox homeostasis, consistent with the increase in ROS observed when the production of lactate is decreased by the depletion of lactate dehydrogenase A (LDHA) activity (Le et al., 2010).
Pyruvate kinase M2 (PKM2) allosteric regulation balances glycolysis with anabolic metabolism
Pyruvate kinase catalyzes the final step of glycolysis, converting phosphoenolpyruvate to pyruvate. The two isozymes of muscle pyruvate kinase are PKM1 and PKM2. PKM1 and PKM2 are encoded by the same gene, and differ only in the alternative splicing of two mutually exclusive exons (Noguchi et al., 1986). PKM2 has been intensely studied for its role in cancer, but confusion exists as to whether PKM2 is overexpressed in cancer. PKM2 is expressed in proliferating cells and most adult tissues, while PKM1 is expressed in many non-cancerous cells requiring high energy (Wong et al., 2013). While PKM2 is the primary pyruvate kinase in many cancers, proteomic analysis shows that PKM2 may not be upregulated in tumors compared to normal tissues (Bluemlein et al., 2011). Oncogenes, such as Myc, can promote the alternative splicing pyruvate kinase mRNA to form PKM2 (David et al., 2010). The single exon difference has a large functional impact, rendering PKM2 sensitive to allosteric regulation. PKM2 has both metabolic and non-metabolic functions not performed by PKM1 (Chaneton et al., 2012; Christofk et al., 2008; Keller et al., 2012; Luo et al., 2011; Yang et al., 2011, 2012). Seemingly paradoxically, PKM2 has slower catalytic activity than PKM1 but is required for aerobic glycolysis. PKM2 appears to be another major glycolytic nexus susceptible to complex regulatory feedback loops. Indeed, recent work reveals that PKM2 is regulated by the products of both glycolysis and the anabolic pathways that branch off of glycolysis (Figure 2).
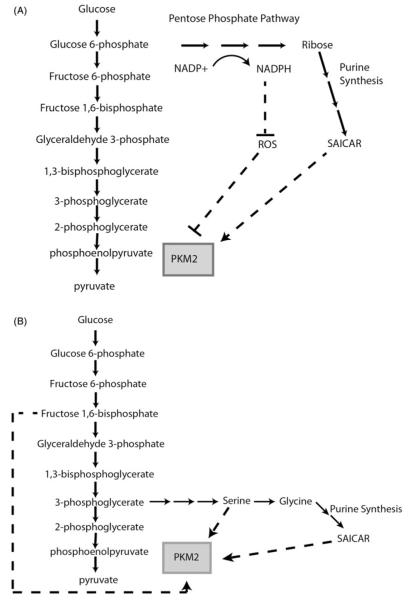
Figure 2.
PKM2 is allosterically regulated by products of glucose derived glycolytic intermediates. (A) Reactive oxygen species (ROS) inhibit the activity of PKM2 via oxidation of Cys 358. Pentose phosphate pathway (PPP) derived NAPDH represses reactive oxygen species (ROS) through the reduction of glutathione. Ribose derived from the PPP contributes to the formation of the PKM2 activator SAICAR (succinylaminoimidazolecarboxamide ribose-5′-phosphate), a purine synthesis intermediate. (B) The glycolytic intermediate fructose 1,6-bisphosphate is a strong activator of PKM2. Serine can feedback to activate PKM2. Serine can also be converted to glycine, which contributes to the formation of PKM2 activator SAICAR.
PKM2 plays a key role in balancing glycolytic flux with flux through the PPP and the serine/glycine biosynthesis pathway, which both play a critical role in nucleotide synthesis. The PPP contributes ribose to nucleotide synthesis, while the serine and glycine synthesis pathway provide tetrahydrofolate and glycine to nucleotide synthesis. One of the intermediates of the purine synthesis pathway, succinylaminoimidazolecarboxamide ribose-5′-phosphate (SAICAR) has been shown to co-purify with PKM2 in glucose limited conditions (Keller et al., 2012). SAICAR quickly accumulates in glucose-deprived cancer cells but not in normal cells, stimulating PKM2 activity and promoting cancer cell survival in low glucose conditions (Keller et al., 2012). The amino acid residue Gln 393 of PKM2 is required for SAICAR stimulation. SAICAR, however, does not appear to bind PKM1. This isoform specific feedback loop likely can control PKM2 activity so that when there is sufficient ribose to generate nucleotides, PKM2 is activated, thereby increasing glycolytic flux in nutrient replete states. Decreased SAICAR levels lead to decreased PKM2 activity, resulting in increased PPP flux and, presumably, increased SAICAR levels.
PKM2, but not PKM1, is also under allosteric control from the product of PFK1, fructose 1,6-bisphosphate (Fru 1,6-BP) (Ashizawa et al., 1991a,b). Fru 1,6-BP binds and allosterically activates PKM2 by stimulating the formation of active tetramers from less active monomers. Hence, increased PFK1 activity can induce PKM2 activity and enhance glycolytic flux through a feed-forward loop that diminishes downstream glycolytic intermediates used for macromolecular synthesis. The stimulation of PKM2 by Fru 1,6-BP is potentially disadvantageous for biomass synthesis in rapidly proliferating cancer cells. This disadvantage, however, may be overcome by oncogenic tyrosine kinase signaling through, for example, fibroblast growth factor receptor 1 (FGFR1) activation, which can dampen Fru 1,6-BP dependent PKM2 activation. FGFR1 dependent phosphorylation of PKM2 on Tyr 105 blocks its ability to bind Fru 1,6-BP, rendering PKM2 less active and resulting in the accumulation of glycolytic intermediates (Hitosugi et al., 2009). Mutation of Tyr 105 increases PKM2 activity and leads to decreased cell proliferation in hypoxia (Hitosugi et al., 2009). In addition to regulation of PKM2 by direct phosphorylation, phosphotyrosine peptides can bind and displace Fru 1,6BP from PKM2 (Christofk et al., 2008). Interestingly, these phosphotyrosine peptides cannot bind to PKM1 (Christofk et al., 2008). The inhibition of PKM2 by phosphotyrosine peptides should also shunt carbons for biomass synthesis. A recently developed PKM2 activator can mitigate PKM2 inhibition by phosphotyrosine peptides, resulting in decreased hypoxic cell proliferation (Anastasiou et al., 2012).
The interplay between Fru 1,6-BP and phosphotyrosine in the regulation of PKM2 under various conditions is complex and our understanding of this interplay is further complicated by studies that use cancer cells rather than normal cells. For example, most normal cells tend to cease proliferation under hypoxic conditions, triggering high flux of glucose to lactate via hypoxia inducible factor 1 (HIF-1) mediated anaerobic glycolysis. In normal cells, the high levels of Fru 1,6-BP under hypoxia would stimulate PKM2 without the opposing effects of tyrosine kinase signaling. However, in cancer cells that have evolved to proliferate under hypoxia, oncogenic tyrosine kinase activation could trigger PKM2 inhibition and diminished glycolytic flux, favoring cell proliferation over high direct flux of glucose to lactate. Fully clarifying these issues will require comparative metabolomics studies of normal cells and their oncogenic transformed counterparts.
PKM2 activity is also regulated by acetylation in response to glucose replete conditions (Lv et al., 2011). In cells cultured at high glucose concentrations, PKM2 displays increased acetylation in a p300/CBP-associated factor (PCAF) dependent manner (Lv et al., 2011). The acetylation of PKM2 at Lys 305 leads to decreased enzymatic activity and stimulates PKM2 chaperone mediated degradation. The decrease in both PKM2 levels and enzymatic activity in the presence of high glucose can lead to the accumulation of glycolytic intermediates, thereby promoting cell growth and proliferation. A recently developed PKM2 activator can reverse the inhibitory effects of Lys 305 acetylation on PKM2 activity (Anastasiou et al., 2012), offering the potential to inhibit high glucose induced anabolic metabolism in cancer cells.
The metabolic regulatory nexus at PKM2 allows for the shunting of carbons out of glycolysis via the 3-phosphoglycerate junction with the serine and glycine synthesis pathway, a key anabolic pathway in cancer (Locasale et al., 2011; Possemato et al., 2011). In cells where PKM1, which lacks serine allosteric regulation, is substituted with PKM2, serine synthesis level increase (Ye et al., 2012). In contrast, cancer cells lacking PKM2 expression show increased sensitivity to serine and glycine deprivation (Chaneton et al., 2012). Intriguingly, serine produced from glycolytic intermediates plays a direct role in regulating its own synthesis (Chaneton et al., 2012). Serine binds and allosterically activates human PKM2, while glycine has no effect on PKM2 activity (Chaneton et al., 2012). Conversely, serine deprivation decreases PKM2 activity, allowing PKM2 to act as a rheostat controlling serine synthesis. Serine, however, has little effect on PKM1 or PKMLR (Pyruvate Kinase Liver and RBC – red blood cell) enzymatic activity (Chaneton et al., 2012). The serine activation of PKM2 is dependent on His 464 (Chaneton et al., 2012), and a small molecule activator of PKM2 renders cancer cells auxotrophic for serine, supporting the central role of PKM2 in balancing glycolysis and serine synthesis (Kung et al., 2012).
As a critical metabolic regulatory enzyme of cancer cell growth and survival, PKM2 can also function as a sensor of oxidative stress. PKM2 activity is inhibited by ROS via oxidation of a sulfhydryl group on Cys 358, leading to an accumulation of glycolytic intermediates and increased PPP flux and NADPH production (Anastasiou et al., 2011). ROS species play a complicated role in cancer, capable of both promoting and inhibiting cancer growth. Cancer cells expressing oxidation resistant PKM2 with a Cys 358 to Ser 358 substitution show impaired tumor formation through increased sensitivity to oxidative stress (Anastasiou et al., 2011). By fine-tuning PPP flux, NADPH production can be altered to regulate ROS levels through the production of reduced glutathione. Thus, inhibition of PKM2 by ROS can elevate NADPH-dependent increases of reduced glutathione, which can neutralize and titrate ROS.
In summary, many studies support a key role for PKM2 in fine tuning cancer metabolism in response to stress. However, a recent study suggests that PKM2 is dispensable for tumor development (Cortes-Cros et al., 2013) and a pyruvate kinase independent glycolysis pathway has been reported (Vander Heiden et al., 2010). Although additional studies are necessary to sort out the complex role of PKM2 in cancer, metabolic heterogeneity of cancers suggests that some cancers may require PKM2 while it may be dispensable in other cancers.
PGAM1 controls glycolysis and anabolic synthesis by controlling levels of allosteric regulators
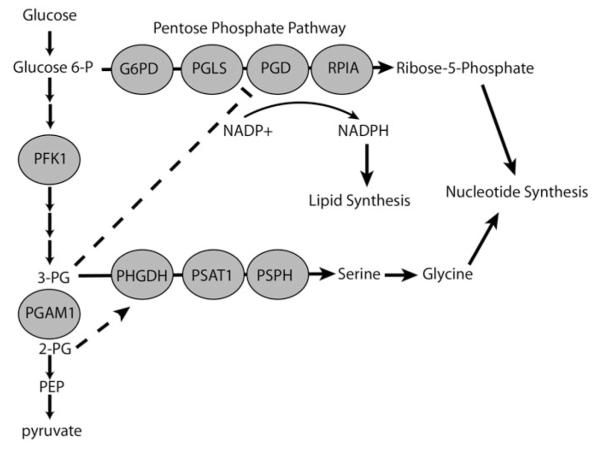
Phosphoglycerate Mutase 1 (PGAM1) is a glycolytic enzyme that converts 3-phosphoglycerate (3-PG), the glycolytic intermediate that can branch off into serine and glycine synthesis, to 2-phosphoglycerate (2-PG). PGAM1 is upregulated in cancers, potentially due to the loss of the PGAM1 negative regulator p53 (Corcoran et al., 2006; Ren et al., 2010; Ruiz-Lozano et al., 1999). A recent study has elegantly shown that PGAM1 activity controls the levels of its substrate (3-PG) and product (2-PG), and that these molecules allosterically regulate key enzymes to control the flux through the PPP and serine and glycine synthesis pathway (Hitosugi et al., 2012) (Figures 3 and 4). When PGAM1 activity is low, 3-PG builds up and then inhibits the PPP enzyme 6-phosphogluconate dehydrogenase (PGD) (Hitosugi et al., 2012). Co-crystallization of PGD with 3-PG shows that 3-PG interacts with several key residues. When PGAM1 activity is high, there is an increase in the product, 2-PG. 2-PG activates phosphoglycerate dehydrogenase (PHGDH), the first enzyme of the serine and glycine synthesis pathway, and thus increases serine/glycine synthesis (Hitosugi et al., 2012). In addition to reducing downstream products derived from glycolysis, stable knockdown of PGAM1 reduced both PPP flux and serine/glycine synthesis by increasing 3-PG levels (inhibiting PGD) and decreasing 2-PG levels (decreasing PHGDH activation) (Hitosugi et al., 2012). A small molecule inhibitor of PGAM1 showed similar effects on metabolism that could be rescued by addition of exogenous 2-PG, which activates PHGDH (phosphoglycerate dehydrogenase) to relieve the buildup of 3-PG and thus increase the flux through the PPP. Further, inhibition of PGAM1 leads to a reduction in tumor size in mouse xenograft models (Hitosugi et al., 2012).
Figure 3.
Phosphoglycerate mutase 1 (PGAM1) controls the levels of allosteric regulators 3-phosphoglycerate and 2-phosphoglycerate. Low PGAM1 activity causes a buildup 3-phosphoglcyerate (3-PG) which slows pentose phosphate pathway (PPP) flux inhibits 6-phosphogluconate dehydrogenase (PGD). High PGAM1 activity causes an increase in the levels of 2-phosphoglycerate (2-PG), which allosterically activates PHGDH, the first enzyme in serine/glycine synthesis. Inhibiting PGAM1 increases 3-PG and decreases 2-PG, decreasing flux through both the serine/glycine pathway and PPP.
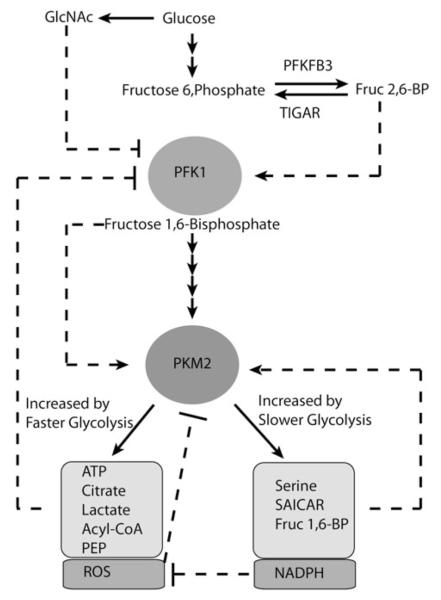
Figure 4.
Summary of allosteric regulation of glycolysis enzymes PFK1 and PKM2.
PGAM1 appears to play another critical role in PKM2 expressing cancer cells. It is seemingly paradoxical that the decreased enzymatic activity of PKM2 leads to increased lactate production. However, the identification of an alternative PGAM1 dependent pathway for converting phosphoenolpyruvate to pyruvate in PKM2 expressing cells provides a possible explanation (Vander Heiden et al., 2010). The phosphate group from PEP is transferred onto His 11 of PGAM1, creating pyruvate from PEP independent of pyruvate kinase activity. Phosphorylation of PGAM1 His 11 is activating, allowing PEP levels to feed back to regulate PGAM1 activity and glycolytic flux independent of pyruvate kinase (Vander Heiden et al., 2010). The dispensability of PKM2 in some tumors suggests that the role of PGAM1 in cancer metabolism is only beginning to be understood (Cortes-Cros et al., 2013).
Fine-tuning of glutamine metabolism
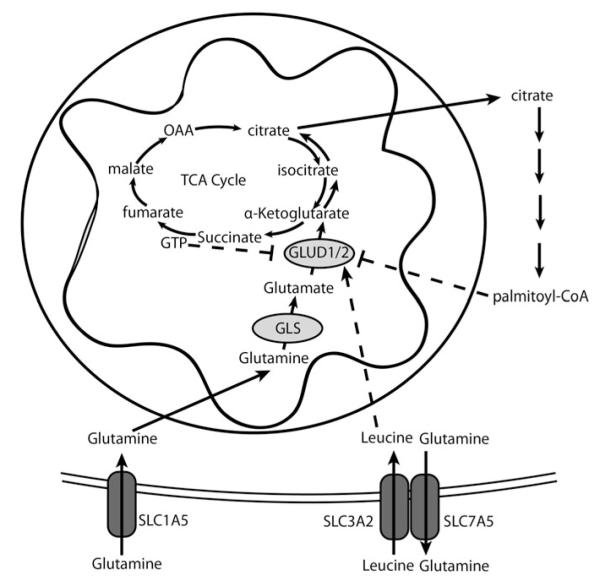
Like glucose metabolism, glutamine metabolism can be fine tuned in cancer cells to respond to conditions. In cancer, glutamine metabolism plays a key role in anabolic synthesis, and its inhibition can slow tumor growth (Wang et al., 2010; Ward & Thompson, 2012). Glutamine can be converted to glutamate by the enzyme glutaminase (Friday et al., 2012). Glutamate can then be converted to α-ketoglutarate by the enzyme Glutamate Dehydrogenase (GLUD) or transaminases (glutamate–pyruvate transaminase, GPT; glutamic-oxaloacetic transaminase, GOT; phosphoserine aminotransferase, PSAT1). Alpha-ketoglutarate can enter the TCA cycle where it is catabolized in a series of reactions to malate and ultimately to oxaloacetate. Malate contributes to NADPH production when malic enzyme uses NADP+ and oxidizes it to pyruvate. Oxaloacetate can be converted to aspartate, which directly contributes to purine synthesis. Otherwise, oxaloacetate continues through the TCA cycle, where it conjugates with acetyl-CoA to produce citrate, which can be extruded into the cytosol for lipid synthesis. Glutamine also contributes to citrate synthesis via reductive carboxylation, particularly in hypoxic conditions. Reductive carboxylation involves the addition of a carbon from carbon dioxide to α-ketoglutarate by the reverse isocitrate dehydrogenase reaction, which had been previously thought to be essentially irreversible upon release of carbon dioxide through the canonical forward reaction (Metallo et al., 2012; Mullen et al., 2012; Wise et al., 2011; Yoo et al., 2008). Not only is glutamine an important contributor of anabolic carbons, it also plays a key role in the activation of mTORC1, a master regulator of cancer metabolism, through enhancing the import of branched-chain amino acids (Nicklin et al., 2009). Through allosteric regulation of GLUD and glutaminase, glutamine metabolism can be fine tuned (Figure 5; Table 1).
Figure 5.
Glutamate dehydrogenase (GLUD1/2) is allosterically regulated to regulate glutamine metabolism. Glutamine enters the cell through glutamine transporters such as SLC1A5. Glutaminase (GLS) converts glutamine to glutamate, and GLUD1/2 converts glutamate to α-ketoglutarate which fuels the TCA cycle, or undergoes reductive carboxylation (counter clockwise arrows in TCA cycle) to form citrate. Citrate can be used to form palmitoyl-CoA, which can inhibit GLUD1/2. Canonical TCA cycle converts α-ketoglutarate to downstream product, yielding GTP which can feedback to inhibit GLUD1/2. Glutamine can be exchanged through SLC3A2/SLC7A5 transporter for leucine, which in turn allosterically activates GLUD1/2.
Table 1.
Summary of allosteric regulators and metabolism controlled post-translational modifications that modify glycolysis and glutaminolysis.
| Enzyme | Allosteric Regulator/PTM | Effect |
|---|---|---|
| PFK1 | AMP | Activating |
| ADP | Activating | |
| Fructose 2,6-bisphosphate | Activating | |
| ATP | Inactivating | |
| Citrate | Inactivating | |
| Palmitoyl-CoA | Inactivating | |
| GlcNAc | Inactivating | |
| Lactate | Inactivating | |
| PKM2 | Fructose 1,6-bisphosphate | Activating |
| SAICAR | Activating | |
| Serine | Activating | |
| Phosphotyrosines | Inactivating | |
| Acetylation | Inactivating | |
| Reactive Oxygen Species | Inactivating | |
| PHGDH | 2-phosphoglycerate | Activating |
| PGD | 3-phosphoglycerate | Inactivating |
| GLS | Phosphate | Activating |
| GLUD | ADP | Activating |
| Leucine | Activating | |
| ADP-Ribosylation | Inactivating | |
| GTP | Inactivating | |
| Palmitoyl-CoA | Inactivating |
PTM = Post-translational Modification. PFK1 = phosphofructokinase 1. PKM2 = pyruvate kinase muscle 2. PHGDH = phosphoglycerate dehydrogenase. PGD = 6-phosphogluconate dehydrogenase. GLUD = glutamate dehydrogenase. GLS = kidney type glutaminase. SAICAR = Succinylaminoimidazolecarboxamide ribose-5′-phosphate
Phosphate-activated glutaminase
Glutaminase (GLS) plays a pivotal role in the use of glutamine by the mitochondrion for anabolic metabolism. Two key forms, kidney type glutaminase (GLS) and liver type glutaminase (GLS2), are encoded by two separate genes in mammals. GLS is broadly expressed and is found at high levels in the normal brain and kidney, while GLS2 is primarily expressed in the liver. Less is known about the role of GLS2 in cancer. While induced by p53 and p63, contradictory roles for GLS2 in cancer have been proposed as it has been shown to act both as a tumor suppressor and a radiation resistance protein (Giacobbe et al., 2013; Hu et al., 2010; Suzuki et al., 2010; Xiang et al., 2013). However, a role for GLS in cancer metabolism has more clearly documented. GLS gives rise to two spliced variants with alternative polyadenylation sites, which produce GAC and KGA (Porter et al., 2002). GAC has been shown to be mitochondrial, whereas KGA is thought to exist in the cytosol – although the localization of KGA is currently controversial (Cassago et al., 2012). Intriguingly, GAC activity is inducible by high phosphate concentrations, while KGA is relatively less sensitive to phosphate levels (Cassago et al., 2012). It has been surmised, based on these observations, that GAC is designed to function in the mitochondrion, which is reported to have a high (mM) phosphate concentration (Akerboom et al., 1978). Although the in vitro activity of GAC is quite dependent on phosphate, it is currently unclear whether phosphate sensitivity provides a regulatory strategy for glutaminase activity in vivo. Very little is currently known about the intra-mitochondrial levels of free phosphate, ADP and ATP under different conditions of cell growth and nutrient availability. GLS activity, however, appears to be regulated by phosphorylation downstream of mitogenic signaling (Thangavelu et al., 2012), but the details of the pathways involved have not been documented.
Glutamate dehydrogenase
Glutamate dehydrogenase (GLUD) is an enzyme that plays a key role in cancer metabolism by converting glutamate to α-ketoglutarate, which is further catabolized through the TCA cycle. GLUD forms a homohexamer, using NAD+ or NADP+ as a co-factor. GLUD plays an important physiological role in insulin release from pancreatic beta cells. Decreased glucose levels can increase NAD+ levels in beta cells, activating Sirtuin 4 (SIRT4) (Haigis et al., 2006). SIRT4 was shown to ADP-ribosylate GLUD and inhibit insulin release by blunting glutamine catabolism, tightly controlling insulin release in a poorly fed state (Haigis et al., 2006). Activating GLUD mutations are associated with hyperinsulinemia in humans (Stanley et al., 1998). The role of GLUD in cancer is suggested by the fact that inhibitors of GLUD, such as Epigallocatechin gallate (EGCG), can curb tumorigenesis in animal models (Liao et al., 1995). The loss of SIRT4, a tumor suppressor, increases GLUD activity in cancer cells (Jeong et al., 2013). In addition to post-translational modifications in mammals and GLUD’s transcriptional regulation in mammals and other kingdoms, glutamate dehydrogenase is highly regulated by allosteric modifiers in animals. In fact, GLUD has multiple allosteric regulators, including ADP, GTP, leucine and palmitoyl-CoA (Figure 5).
GLUD is activated by ADP and inhibited by GTP. Since it fuels the TCA cycle, glutamine metabolism contributes to ATP production. Hence low energetic states could induce GLUD through ADP, increasing glutamine flux into the TCA cycle (Li et al., 2012). GLUD can form abortive complexes when the product is replaced by substrate before the altered co-factor is removed (i.e. if glutamate binds the complex before the complex removes NADH). The abortive complex leads to decreased enzymatic activity by tight cofactor binding (Li et al., 2012). ADP is believed to bind the NAD+ allosteric binding domain and activate GLUD at least partially through the reduction of abortive complexes (Li et al., 2012). The GLUD inhibitor EGCG inhibits enzyme activity by blocking the ADP binding site (Li et al., 2011). GTP is produced in the TCA cycle when succinyl-CoA, which is derived from α-ketoglutarate, is converted to succinate by Succinyl-CoA synthetase. Thus, it is possible that increased glutamate flux through the TCA cycle can elevate GTP, which would be expected to curb GLUD activity in a putative feedback loop. GTP inhibits GLUD allosterically by binding to the enzyme complex in the closed formation, slowing product release and ultimately enzymatic activity. Through this mechanism, glucose flux through the TCA cycle could diminish the entry of glutamine into the TCA cycle. Activating GLUD mutations that cause hyperinsulinemia have been shown to disrupt GTP regulation of GLUD activity (Stanley et al., 1998). Other means by which GTP levels are changed, such as GTPase enzymes, could also affect GLUD, and hence additional studies will be required to delineate whether other pathways could be linked to glutaminolysis via GLUD allosteric regulation.
mTOR activation is essential for cancer cell growth, and the amino acid leucine is capable of activating mTORC1 (mTOR complex 1). It has been shown that glutamine is essential for the import of leucine through the SLC7A5/SLC3A2 (LAT1) exchanger, which extrudes glutamine in exchange for the import of leucine (Nicklin et al., 2009). Leucine has been shown to directly stimulate mTORC1 via its association with lysosomes through a protein complex (Bar-Peled et al., 2012; Zoncu et al., 2011). However, a recent study suggests an additional role for leucine, which may contribute to mTORC1 activation through stimulation of glutamine metabolism (Duran et al., 2012). Leucine can stimulate GLUD enzymatic activity, with leucine likely playing a role in subunit interface environment through interaction with Arg 151 and Asp 185 in human GLUD2 (Erecinska & Nelson, 1990; Tomita et al., 2011). Thus, leucine increases GLUD activity and enhances glutaminolysis, activating mTORC1 (Duran et al., 2012). Inhibition of glutaminolysis reduced mTORC1 activation (Duran et al., 2012). Thus glutamine and leucine metabolism appear highly connected, and one could surmise that a critical nutrient for cell growth (glutamine), would be tightly linked to another mediator of cell growth (leucine) which acts through mTORC1. Given that other mechanisms of leucine dependent mTORC1 activation have also been proposed, including the role of leucyl-tRNA synthetase and leucyl-tRNA in mTORC1 activation (Han et al., 2012), this area of research is still in flux. Hence some of the proposed mechanisms will need verification by additional independent studies.
GLUD is also inhibited by palmitoyl-CoA (Fahien & Kmiotek, 1981), a long chain fatty acid that is a product of fatty acid synthesis and a substrate for beta-oxidation. Shorter chain fatty acids show less efficient inhibition of GLUD (Lai et al., 1993). Palmitoyl-CoA inhibits GLUD by causing the formation of inactive dimers (Kawaguchi & Bloch, 1976). Little is known about where palmitoyl-CoA binds GLUD to inhibit its function, but recent studies show that four GLUD cysteine residues may be required for palmitoyl-CoA GLUD inhibition (Son et al., 2012). Palmitoyl-CoA inhibition of GLUD represents a key point of crosstalk between amino acid and fatty acid metabolism. As Palmitoyl-CoA is a substrate for beta-oxidation, this regulation could allow cells to downregulate glutaminolysis when the TCA cycle is being used for fatty acid oxidation versus its use for anaplerotic flux and glutaminolysis. Additionally, glutamine can be a key fuel for lipogenesis either via the forward TCA cycling or through reductive carboxylation (Metallo et al., 2012; Mullen et al., 2012; Wise et al., 2011; Yoo et al., 2008). This suggests that production of palmitoyl-CoA from glutamine might have a negative feedback loop that inhibits glutaminolysis when palmitoyl-CoA is produced in excess. It should be noted that these speculations need additional studies to verify or refute them, and that cellular localization of palmitoyl-CoA and GLUD would alter this regulation.
Targeted therapy to disrupt allosteric regulation
The field of cancer metabolism research has blossomed over the last decade, providing hope that new classes of drugs will be developed for cancer therapy. Indeed, this rapidly expanding field has created many opportunities at the leading edge of research, which, by its nature, is turbulent. As the wake behind the leading edge settles with time and studies are replicated, we hope that additional drugs will result from targeting the allosteric regulation of key enzymes that are reviewed here.
We speculate that this class of drug will have higher target specificity, since active site inhibitors could have off-target effects by inhibiting enzymes that utilize the same substrates. For example, active site inhibitors of glutaminase such as 6-diazo-5-oxo-l-norleucine (DON) have been generated previously. DON is a glutamine analog that inhibits glutaminase as well as other enzymes that use glutamine as a substrate, leading to toxicity (Wise & Thompson, 2010). Its biological activity is diverse and could not be attributed to its inhibition of glutaminase alone. By contrast, phosphate activation of GLS can be potently inhibited by the allosteric inhibitor, BPTES (bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide 3). Co-crystal structures of GLS and BPTES reveal that BPTES binds the tetramerization interface of GLS that is involved in phosphate activation (DeLaBarre et al., 2011; Thangavelu et al., 2012). Indeed, derivatives of BPTES have also been shown to have significant biological activity and companies are beginning to generate lead compounds that have developed into clinically usable drugs (Shukla et al., 2012). Although an allosteric inhibitor of GLUD, EGCG, has been co-crystallized with GLUD and has considerable pre-clinical effect on tumor growth (Li et al., 2011, 2012; Liao et al., 1995), whether EGCG decreases tumor growth through specifically inhibiting GLUD has not been conclusively established. Hence, development of highly specific allosteric inhibitors of GLUD could well produce new drugs that target cancer metabolism. The discovery of Fru 1,6-BP as an activator of PKM2 led to an extensive effort to identify pharmacological activators and inhibitors of PKM2 (Anastasiou et al., 2012; Kung et al., 2012). Indeed, one such activator demonstrates pre-clinical in vivo activity against tumor xenografts, presumably by diminishing the ability of treated cells to accumulate glycolytic intermediates to build biomass (Anastasiou et al., 2012). Whether these PKM2 activators will prove to be clinically useful remains to be determined. Additionally, since PFKFB3 produces a potent activator of PFK1, a PFKFB3 inhibitor that indirectly alters allosteric regulation of PFK1 has been developed and destined for clinical trials (Clem et al., 2013). In conclusion, we hope that this overview of allosteric regulation of cancer metabolism will give pause for this field of research to explore new opportunities for cancer therapy.
Acknowledgements
We would like to thank the Dang lab members and R. Stine for helpful feedback.
This work was supported by NCI/NIH grant R01 CA057341.
Footnotes
Declaration of interest The authors report no declarations of interest.
References
- Akerboom TP, Bookelman H, Zuurendonk PF, et al. Intramitochondrial and extramitochondrial concentrations of adenine nucleotides and inorganic phosphate in isolated hepatocytes from fasted rats. Eur J Biochem. 1978;84:413–20. doi: 10.1111/j.1432-1033.1978.tb12182.x. [DOI] [PubMed] [Google Scholar]
- Anastasiou D, Poulogiannis G, Asara JM, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–83. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou D, Yu Y, Israelsen WJ, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8:839–47. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashizawa K, McPhie P, Lin KH, Cheng SY. An in vitro novel mechanism of regulating the activity of pyruvate kinase M2 by thyroid hormone and fructose 1, 6-bisphosphate. Biochemistry. 1991a;30:7105–11. doi: 10.1021/bi00243a010. [DOI] [PubMed] [Google Scholar]
- Ashizawa K, Willingham MC, Liang CM, Cheng SY. In vivo regulation of monomer-tetramer conversion of pyruvate kinase subtype M2 by glucose is mediated via fructose 1,6-bisphosphate. J Biol Chem. 1991b;266:16842–6. [PubMed] [Google Scholar]
- Atsumi T, Chesney J, Metz C, et al. High expression of inducible 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (iPFK-2; PFKFB3) in human cancers. Cancer Res. 2002;62:5881–7. [PubMed] [Google Scholar]
- Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–20. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Bluemlein K, Gruning NM, Feichtinger RG, et al. No evidence for a shift in pyruvate kinase PKM1 to PKM2 expression during tumorigenesis. Oncotarget. 2011;2:393–400. doi: 10.18632/oncotarget.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruser A, Kirchberger J, Kloos M, et al. Functional linkage of adenine nucleotide binding sites in mammalian muscle 6-phosphofructokinase. J Biol Chem. 2012;287:17546–53. doi: 10.1074/jbc.M112.347153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassago A, Ferreira AP, Ferreira IM, et al. Mitochondrial localization and structure-based phosphate activation mechanism of Glutaminase C with implications for cancer metabolism. Proc Natl Acad Sci U S A. 2012;109:1092–7. doi: 10.1073/pnas.1112495109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaneton B, Hillmann P, Zheng L, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491:458–62. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung EC, Athineos D, Lee P, et al. TIGAR Is Required for Efficient Intestinal Regeneration and Tumorigenesis. Dev Cell. 2013;25:463–77. doi: 10.1016/j.devcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Wu N, et al. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–6. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- Clem BF, O’Neal J, Tapolsky G, et al. Targeting 6-phosphofructo-2-kinase (PFKFB3) as a therapeutic strategy against cancer. Mol Cancer Ther. 2013;12:1461–70. doi: 10.1158/1535-7163.MCT-13-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran CA, Huang Y, Sheikh MS. The regulation of energy generating metabolic pathways by p53. Cancer Biol Ther. 2006;5:1610–3. doi: 10.4161/cbt.5.12.3617. [DOI] [PubMed] [Google Scholar]
- Cortes-Cros M, Hemmerlin C, Ferretti S, et al. M2 isoform of pyruvate kinase is dispensable for tumor maintenance and growth. Proc Natl Acad Sci U S A. 2013;110:489–94. doi: 10.1073/pnas.1212780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Leite T, Da Silva D, Guimaraes Coelho R, et al. Lactate favours the dissociation of skeletal muscle 6-phosphofructo-1-kinase tetramers down-regulating the enzyme and muscle glycolysis. Biochem J. 2007;408:123–30. doi: 10.1042/BJ20070687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Chen M, Assanah M, et al. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–8. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delabarre B, Gross S, Fang C, et al. Full-length human glutaminase in complex with an allosteric inhibitor. Biochemistry. 2011;50:10764–70. doi: 10.1021/bi201613d. [DOI] [PubMed] [Google Scholar]
- Duran RV, Oppliger W, Robitaille AM, et al. Glutaminolysis activates Rag-mTORC1 signaling. Mol Cell. 2012;47:349–58. doi: 10.1016/j.molcel.2012.05.043. [DOI] [PubMed] [Google Scholar]
- Erecinska M, Nelson D. Activation of glutamate dehydrogenase by leucine and its nonmetabolizable analogue in rat brain synaptosomes. J Neurochem. 1990;54:1335–43. doi: 10.1111/j.1471-4159.1990.tb01967.x. [DOI] [PubMed] [Google Scholar]
- Fahien LA, Kmiotek E. Regulation of glutamate dehydrogenase by palmitoyl-coenzyme A. Arch Biochem Biophys. 1981;212:247–53. doi: 10.1016/0003-9861(81)90364-7. [DOI] [PubMed] [Google Scholar]
- Ferreras C, Hernandez ED, Martinez-Costa OH, Aragon JJ. Subunit interactions and composition of the fructose 6-phosphate catalytic site and the fructose 2,6-bisphosphate allosteric site of mammalian phosphofructokinase. J Biol Chem. 2009;284:9124–31. doi: 10.1074/jbc.M807737200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friday E, Oliver R, Turturro F, Welbourne T. Role of glutamate dehydrogenase in cancer growth and homeostasis. In: Canuto RA, ed. Dehydrogenases. Rijeka, Croatia: InTech. 2012:181–90. [Google Scholar]
- Giacobbe A, Bongiorno-Borbone L, Bernassola F, et al. p63 regulates glutaminase 2 expression. Cell Cycle. 2013;12:1395–405. doi: 10.4161/cc.24478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goidts V, Bageritz J, Puccio L, et al. RNAi screening in glioma stem-like cells identifies PFKFB4 as a key molecule important for cancer cell survival. Oncogene. 2012;31:3235–43. doi: 10.1038/onc.2011.490. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–54. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Han JM, Jeong SJ, Park MC, et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149:410–24. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- Hers HG, Van Schaftingen E. Fructose 2,6-bisphosphate 2 years after its discovery. Biochem J. 1982;206:1–12. doi: 10.1042/bj2060001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T, Kang S, Vander Heiden MG, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T, Zhou L, Elf S, et al. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell. 2012;22:585–600. doi: 10.1016/j.ccr.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Zhang C, Wu R, et al. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci U S A. 2010;107:7455–60. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins CM, Yang J, Sims HF, Gross RW. Reversible high affinity inhibition of phosphofructokinase-1 by acyl-CoA: a mechanism integrating glycolytic flux with lipid metabolism. J Biol Chem. 2011;286:11937–50. doi: 10.1074/jbc.M110.203661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SM, Xiao C, Finley LW, et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell. 2013;23:450–63. doi: 10.1016/j.ccr.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi A, Bloch K. Inhibition of glutamate dehydrogenase and malate dehydrogenases by palmitoyl coenzyme A. J Biol Chem. 1976;251:1406–12. [PubMed] [Google Scholar]
- Keller KE, Tan IS, Lee YS. SAICAR stimulates pyruvate kinase isoform M2 and promotes cancer cell survival in glucose-limited conditions. Science. 2012;338:1069–72. doi: 10.1126/science.1224409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp RG, Gunasekera D. Evolution of the allosteric ligand sites of mammalian phosphofructo-1-kinase. Biochemistry. 2002;41:9426–30. doi: 10.1021/bi020110d. [DOI] [PubMed] [Google Scholar]
- Kessler R, Bleichert F, Warnke JP, Eschrich K. 6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB3) is up-regulated in high-grade astrocytomas. J Neurooncol. 2008;86:257–64. doi: 10.1007/s11060-007-9471-7. [DOI] [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–37. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- Kung C, Hixon J, Choe S, et al. Small molecule activation of PKM2 in cancer cells induces serine auxotrophy. Chem Biol. 2012;19:1187–98. doi: 10.1016/j.chembiol.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JC, Liang BB, Jarvi EJ, et al. Differential effects of fatty acyl coenzyme A derivatives on citrate synthase and glutamate dehydrogenase. Res Commun Chem Pathol Pharmacol. 1993;82:331–8. [PubMed] [Google Scholar]
- Le A, Cooper CR, Gouw AM, et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A. 2010;107:2037–42. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite TC, Coelho RG, Da Silva D, et al. Lactate downregulates the glycolytic enzymes hexokinase and phosphofructokinase in diverse tissues from mice. FEBS Lett. 2011;585:92–8. doi: 10.1016/j.febslet.2010.11.009. [DOI] [PubMed] [Google Scholar]
- Li C, Li M, Chen P, et al. Green tea polyphenols control dysregulated glutamate dehydrogenase in transgenic mice by hijacking the ADP activation site. J Biol Chem. 2011;286:34164–74. doi: 10.1074/jbc.M111.268599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Li C, Allen A, et al. The structure and allosteric regulation of mammalian glutamate dehydrogenase. Arch Biochem Biophys. 2012;519:69–80. doi: 10.1016/j.abb.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Umekita Y, Guo J, et al. Growth inhibition and regression of human prostate and breast tumors in athymic mice by tea epigallocatechin gallate. Cancer Lett. 1995;96:239–43. doi: 10.1016/0304-3835(95)03948-v. [DOI] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–74. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Hu H, Chang R, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–44. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L, Li D, Zhao D, et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42:719–30. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaini KR, Vander Heiden MG. Cancer. Glycosylation to adapt to stress. Science. 2012;337:925–6. doi: 10.1126/science.1227513. [DOI] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–4. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchenko O, Opentanova I, Minchenko D, et al. Hypoxia induces transcription of 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase-4 gene via hypoxia-inducible factor-1alpha activation. FEBS Lett. 2004;576:14–20. doi: 10.1016/j.febslet.2004.08.053. [DOI] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–8. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–34. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Inoue H, Tanaka T. The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J Biol Chem. 1986;261:13807–12. [PubMed] [Google Scholar]
- Okar DA, Manzano A, Navarro-Sabate A, et al. PFK-2/FBPase-2: maker and breaker of the essential biofactor fructose-2,6-bisphosphate. Trends Biochem Sci. 2001;26:30–5. doi: 10.1016/s0968-0004(00)01699-6. [DOI] [PubMed] [Google Scholar]
- Pinheiro C, Longatto-Filho A, Azevedo-Silva J, et al. Role of monocarboxylate transporters in human cancers: state of the art. J Bioenerg Biomembr. 2012;44:127–39. doi: 10.1007/s10863-012-9428-1. [DOI] [PubMed] [Google Scholar]
- Poorman RA, Randolph A, Kemp RG, Heinrikson RL. Evolution of phosphofructokinase – gene duplication and creation of new effector sites. Nature. 1984;309:467–9. doi: 10.1038/309467a0. [DOI] [PubMed] [Google Scholar]
- Porter LD, Ibrahim H, Taylor L, Curthoys NP. Complexity and species variation of the kidney-type glutaminase gene. Physiol Genomics. 2002;9:157–66. doi: 10.1152/physiolgenomics.00017.2002. [DOI] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–50. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randle PJ, Garland PB, Hales CN, Newsholme EA. The glucose fatty-acid cycle. Its role in insulin sensitivity and the metabolic disturbances of diabetes mellitus. Lancet. 1963;1:785–9. doi: 10.1016/s0140-6736(63)91500-9. [DOI] [PubMed] [Google Scholar]
- Reinhart GD. Influence of fructose 2,6-bisphosphate on the aggregation properties of rat liver phosphofructokinase. J Biol Chem. 1983;258:10827–30. [PubMed] [Google Scholar]
- Ren F, Wu H, Lei Y, et al. Quantitative proteomics identification of phosphoglycerate mutase 1 as a novel therapeutic target in hepatocellular carcinoma. Mol Cancer. 2010;9:81. doi: 10.1186/1476-4598-9-81. doi:10.1186/1476-4598-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros S, Santos CR, Moco S, et al. Functional metabolic screen identifies 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 as an important regulator of prostate cancer cell survival. Cancer Discov. 2012;2:328–43. doi: 10.1158/2159-8290.CD-11-0234. [DOI] [PubMed] [Google Scholar]
- Ruiz-Lozano P, Hixon ML, Wagner MW, et al. p53 is a transcriptional activator of the muscle-specific phosphoglycerate mutase gene and contributes in vivo to the control of its cardiac expression. Cell Growth Differ. 1999;10:295–306. [PubMed] [Google Scholar]
- Sakakibara R, Kato M, Okamura N, et al. Characterization of a human placental fructose-6-phosphate, 2-kinase/fructose-2,6-bisphosphatase. J Biochem. 1997;122:122–8. doi: 10.1093/oxfordjournals.jbchem.a021719. [DOI] [PubMed] [Google Scholar]
- Shukla K, Ferraris DV, Thomas AG, et al. Design, synthesis, and pharmacological evaluation of bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide 3 (BPTES) analogs as glutaminase inhibitors. J Med Chem. 2012;55:10551–63. doi: 10.1021/jm301191p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son HJ, Ha SC, Hwang EY, et al. Roles of cysteine residues in the inhibition of human glutamate dehydrogenase by palmitoyl-CoA. BMB Rep. 2012;45:707–12. doi: 10.5483/BMBRep.2012.45.12.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonveaux P, Vegran F, Schroeder T, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest. 2008;118:3930–42. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley CA, Lieu YK, Hsu BY, et al. Hyperinsulinism and hyperammonemia in infants with regulatory mutations of the glutamate dehydrogenase gene. N Engl J Med. 1998;338:1352–7. doi: 10.1056/NEJM199805073381904. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Tanaka T, Poyurovsky MV, et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci U S A. 2010;107:7461–6. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RP, Parker GJ, Hazel MW, et al. Glucose deprivation stimulates O-GlcNAc modification of proteins through up-regulation of O-linked N-acetylglucosaminyltransferase. J Biol Chem. 2008;283:6050–7. doi: 10.1074/jbc.M707328200. [DOI] [PubMed] [Google Scholar]
- Thangavelu K, Pan CQ, Karlberg T, et al. Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism. Proc Natl Acad Sci U S A. 2012;109:7705–10. doi: 10.1073/pnas.1116573109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita T, Kuzuyama T, Nishiyama M. Structural basis for leucine-induced allosteric activation of glutamate dehydrogenase. J Biol Chem. 2011;286:37406–13. doi: 10.1074/jbc.M111.260265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usenik A, Legisa M. Evolution of allosteric citrate binding sites on 6-phosphofructo-1-kinase. PLoS One. 2010;5:e15447. doi: 10.1371/journal.pone.0015447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda K. Phosphofructokinase. Adv Enzymol Relat Areas Mol Biol. 1979;48:193–244. doi: 10.1002/9780470122938.ch4. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Locasale JW, Swanson KD, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329:1492–9. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamelink MM, Struys EA, Jakobs C. The biochemistry, metabolism and inherited defects of the pentose phosphate pathway: a review. J Inherit Metab Dis. 2008;31:703–17. doi: 10.1007/s10545-008-1015-6. [DOI] [PubMed] [Google Scholar]
- Wang JB, Erickson JW, Fuji R, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18:207–19. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanka C, Steinbach JP, Rieger J. Tp53-induced glycolysis and apoptosis regulator (TIGAR) protects glioma cells from starvation-induced cell death by up-regulating respiration and improving cellular redox homeostasis. J Biol Chem. 2012;287:33436–46. doi: 10.1074/jbc.M112.384578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–33. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Ward PS, Shay JE, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108:19611–16. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong N, De Melo J, Tang D. PKM2, a central point of regulation in cancer metabolism. Int J Cell Biol. 2013;2013:242513. doi: 10.1155/2013/242513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang L, Xie G, Liu C, et al. Knock-down of glutaminase 2 expression decreases glutathione, NADH, and sensitizes cervical cancer to ionizing radiation. Biochim Biophys Acta. 2013;1833:2996–3005. doi: 10.1016/j.bbamcr.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Yang W, Xia Y, Hawke D, et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150:685–96. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Xia Y, Ji H, et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–22. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Mancuso A, Tong X, et al. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc Natl Acad Sci U S A. 2012;109:6904–9. doi: 10.1073/pnas.1204176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W, Clark PM, Mason DE, et al. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science. 2012;337:975–80. doi: 10.1126/science.1222278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H, Antoniewicz MR, Stephanopoulos G, Kelleher JK. Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. J Biol Chem. 2008;283:20621–7. doi: 10.1074/jbc.M706494200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun SJ, Jo SW, Ha YS, et al. PFKFB4 as a prognostic marker in non-muscle-invasive bladder cancer. Urol Oncol. 2012;30:893–9. doi: 10.1016/j.urolonc.2010.08.018. [DOI] [PubMed] [Google Scholar]
- Zachara NE, O’Donnell N, Cheung WD, et al. Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells. J Biol Chem. 2004;279:30133–42. doi: 10.1074/jbc.M403773200. [DOI] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–83. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]