Abstract
lndole-3-acetic acid (IAA), the most important natural auxin in plants, is mainly synthesized from the amino acid tryptophan (Trp). Recent genetic and biochemical studies in Arabidopsis have unambiguously established the first complete Trp-dependent auxin biosynthesis pathway. The first chemical step of auxin biosynthesis is the removal of the amino group from Trp by the TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS (TAA) family of transaminases to generate indole-3-pyruvate (IPA). IPA then undergoes oxidative decarboxylation catalyzed by the YUCCA (YUC) family of flavin monooxygenases to produce IAA. This two-step auxin biosynthesis pathway is highly conserved throughout the plant kingdom and is essential for almost all of the major developmental processes. The successful elucidation of a complete auxin biosynthesis pathway provides the necessary tools for effectively modulating auxin concentrations in plants with temporal and spatial precision. The progress in auxin biosynthesis also lays a foundation for understanding polar auxin transport and for dissecting auxin signaling mechanisms during plant development.
INTRODUCTION
Auxin has long been recognized as a hormone essential for almost every aspect of plant growth and development (Zhao, 2010). However, an understanding of its biosynthetic mechanisms in plants had remained elusive until very recently. For a long time, the physiological roles of auxin were mainly inferred from studies on how plants responded to exogenous auxin treatments. These studies were also the foundation for elucidating the auxin signaling and polar transport mechanisms. However, to precisely define the physiological roles of auxin, we need to characterize auxin deficient mutants, a goal that becomes feasible only when we understand how auxin is synthesized in plants. Understanding of auxin biosynthesis will also reveal the sites of auxin production in plants, thereby allowing us to define auxin sources/sinks and to better understand polar auxin transport. Knowledge in auxin biosynthesis will greatly facilitate our understanding of the molecular mechanisms by which auxin controls various developmental processes. Progress in auxin biosynthesis research lays the foundation for improving agriculturally important traits such as branching and flower development by allowing us to regulate auxin levels in specific tissues/cells. Therefore, a clear understanding of auxin biosynthesis will ultimately have many significant impacts on agriculture and will also greatly extend our knowledge of fundamental plant biology.
Auxin biosynthesis can be divided into two general categories: de novo auxin biosynthesis and the release from auxin conjugates [see recent reviews (Normanly, 2010; Ludwig-Muller, 2011; Mano and Nemoto, 2012; Brumos et al., 2013; Ljung, 2013; Zhao, 2013; Tivendale et al., 2014)]. Indole-3-acetic acid (IAA), the main natural auxin in plants, exists in both free and conjugated forms. Free IAA is the active form of auxin and the conjugated auxins are considered storage forms or intermediates destined for degradation (Woodward and Bartel, 2005; Korasick et al., 2013). Free IAA can be released from IAA conjugates such as IAA esters, IAA-sugar, and IAA-amino acid conjugates by hydrolysis (Davies et al., 1999; Rampey et al., 2004; Ludwig-Muller, 2011; Korasick et al., 2013). Free IAA can also be produced from indole-3-butyric acid by a process similar to fatty acid β-oxidation in the peroxisomes (Zolman et al., 2000; Zolman et al., 2008). In this chapter, I focus on the recent progresses in de novo auxin biosynthesis. Mechanisms regarding the release of free auxin from conjugates and IBA have been reviewed elsewhere (Woodward and Bartel, 2005; Ludwig-Muller, 2011; Korasick et al., 2013).
Trp is a known precursor for auxin biosynthesis and it has been demonstrated that feeding plants with labeled Trp leads to the production of labeled IAA (Wright et al., 1991; Normanly et al., 1993). Two decades ago, isotope-labeling experiments in combination with using Trp biosynthetic mutants led to the proposal that IAA is also synthesized in a Trp-independent fashion (Wright et al., 1991; Normanly et al., 1993). So far, however, the molecular components of the Trp-independent pathway have not been identified. In this chapter, I will not discuss the Trp-independent auxin biosynthesis pathway. Instead, I will concentrate on the discovery of the first complete plant auxin biosynthetic pathway in which Trp is converted into IAA in two steps using indole-3-pyruvate (IPA) as the intermediate (Figure 1). This two-step auxin biosynthesis pathway plays an essential role in almost all of the major developmental processes including embryogenesis, seedling growth, root elongation, vascular patterning, gravitropism, and flower development. The pathway is highly conserved throughout the plant kingdom and has been functionally characterized in several plant species in both monocots and dicots.
Figure 1.
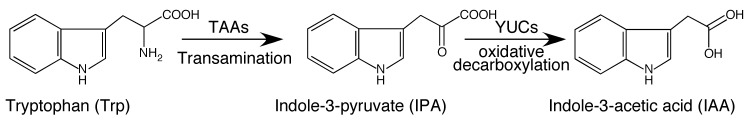
A complete tryptophan-dependent auxin biosynthesis pathway in plants.
Auxin is synthesized from the amino acid Trp in two chemical steps. The first step is the removal of the amino group by the TAA family of aminotransferases to produce IPA. The second step is the oxidative decarboxylation of IPA catalyzed by the YUC family of flavin-containing monooxygenases to generate IAA, CO2 and water.
Besides IPA, several other compounds including Indole-3-acetonitrile and Indole-3-acetamide have also been proposed as intermediates in auxin biosynthesis. Because the other pathways are less well defined and they have been reviewed extensively elsewhere (Woodward and Bartel, 2005; Zhao, 2010; Brumos et al., 2013; Korasick et al., 2013; Tivendale et al., 2014), I will not elaborate further on those alternative pathways. In this chapter, I discuss the genetic and biochemical data that led to the establishment of the first complete Trp-dependent auxin biosynthetic pathway. I also discuss the implications of the breakthrough in auxin biosynthesis in the context of studying polar auxin transport and auxin signaling. Knowledge of auxin biosynthesis allows us to manipulate auxin concentrations in plant cells, thereby enabling us to investigate the molecular mechanisms of auxin-regulated plant developmental processes from a different perspective.
GENETIC DISSECTION OF AUXIN BIOSYNTHESIS
Early auxin overproduction mutants and Trp biosynthesis mutants
Genetic approaches have been widely used to dissect the biosynthetic and signaling pathways of plant hormones. But genetic dissection of auxin biosynthesis has been a very long and difficult journey. Early attempts were centered on the characterization of Trp biosynthesis mutants, which display pleiotropic developmental phenotypes. Interestingly, free IAA concentrations in some of the Trp biosynthetic mutants do not differ significantly from those in wild type plants (Normanly et al., 1993). Some trp mutants actually contain elevated levels of total IAA (Normanly et al., 1993). The studies on Trp biosynthesis mutants led to the discovery of a Trp-independent auxin biosynthesis pathway, but revealed very little about how Trp is converted into IAA in plants.
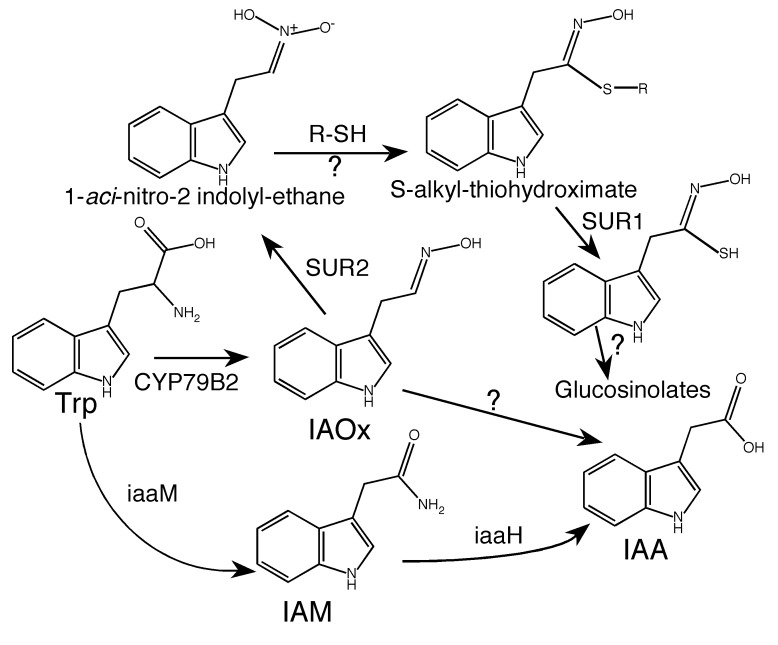
The first major advance in the genetic dissection of auxin biosynthesis in plants was the isolation of the auxin overproduction mutants superroot1 (sur1: At2g20610) and superroot2 (sur2: At4g31500) in Arabidopsis (Boerjan et al., 1995; Delarue et al., 1998). When grown in light, both sur1 and sur2 have much longer hypocotyls than do wild type plants. Light-grown sur1 and sur2 plants also display epinastic cotyledons and true leaves. In total darkness, sur1 and sur2 seedlings lack an apical hook and have short hypocotyls (Boerjan et al., 1995; Delarue et al., 1998). The most striking phenotype of sur1 and sur2 is the development of adventitious roots from their hypocotyls. SUR1 encodes a C-S lyase and SUR2 encodes the Cytochrome P450 monooxygenase CYP83B1 (Earlier et al., 2000; Mikkelsen et al., 2004). Both SUR1 and SUR2 are now known key components of glucosinolate biosynthesis (Figure. 2). Disruption of either SUR1 or SUR2 blocks the biosynthesis of glucosinolates and leads to the accumulation of the metabolic intermediate indole-3-acetaloxime (IAOx), which can be converted into IAA through an undefined mechanism (Figure 2). IAOx is produced from Trp in Arabidopsis by the Cytochrome P450 monooxygenases CYP79B2 (AT4G39950) and B3 (AT2G22330) (Figure 2) (Hull et al., 2000; Zhao et al., 2002). Overexpression of CYP79B2 leads to the accumulation of IAOx and consequently auxin overproduction phenotypes (Hull et al., 2000; Zhao et al., 2002). Characterization of CYP79B2/B3 overexpression lines and the sur1 and sur2 mutants firmly establishes that IAOx can function as an auxin biosynthesis intermediate in Arabidopsis. However, it is now clear that IAOx is probably not a key auxin biosynthesis intermediate used throughout the plant kingdom (Zhao et al., 2002; Sugawara et al., 2009).
Figure 2.
Two proposed metabolic pathways for the amino acid Tryptophan (Trp).
Trp is oxidized into indole-3-acetaloxime (IAOx) by the Cytochrome P450 CYP79B2/B3. IAOx is a substrate for another P450, CYP83B1, which is also called SUPERROOT2 (SUR2). SUR2 converts IAOx into 1-aci-nitro-2 indolyl-ethane, which is further converted to Indol-3-ylmethyl S-alkyl-thiohydroximate by an unknown mechanism. SUPERROOT1 (SUR1) is a C-S lyase that converts S-alkyl-thiohydroximate to thiohydroximate. Both SUR1 and SUR2 are important for glucosinolate biosynthesis. Inactivation either SUR1 or SUR2 leads to the accumulation of IAOx, which is converted into IAA by an unknown mechanism. Trp can also be oxidized to indole-3-acetamide (IAM) by the bacterial auxin biosynthesis enzyme iaaM Trp-2-monooxygenase. IAM is hydrolyzed into IAA by the bacterial enzyme iaaH. Overexpression of iaaM or CYP79B2 leads to auxin overproduction phenotypes. Question mark indicates that enzymes for the steps are not known.
Although the analysis of the sur1 and sur2 auxin overproduction mutants and the CYP79B2 overexpression lines did not uncover the key auxin biosynthesis mechanisms used by the majority of plants, these early studies established the phenotypic characteristics of auxin overproduction in Arabidopsis. Light grown auxin overproduction mutants have long hypocotyls and epinastic cotyledons. When grown in the dark, auxin overproduction mutants have short hypocotyls and lack apical hooks. These characteristic auxin overproduction phenotypes are also observed in transgenic plants in which the bacterial auxin biosynthesis gene iaaM is overexpressed under the control of the Cauliflower Mosaic Virus (CaMV) 35S promoter (Romano et al., 1995). The iaaM gene encodes the Trp-2-monooxygenase that catalyzes the conversion of Trp into indole-3-acetamide (IAM), which is subsequently hydrolyzed into IAA by the bacterial hydrolase iaaH (Figure 2) (Yamada et al., 1985). Interestingly, the auxin overproduction phenotypes observed in sur1, sur2, CYP79B2 overexpression lines, and the iaaM overexpression lines are quite different from those displayed in plants treated with exogenous IAA, the synthetic auxin 1-naphthaleneacetic acid (NAA), and 2,4-dichlorophenoxyacetic acid (2,4-D). The prominent phenotype of Arabidopsis seedlings grown on IAA-containing media is that the plants develop short roots (Lincoln et al., 1990; Hobbie and Estelle, 1994). The characteristic phenotype of the auxin overproduction mutants is that they all have long hypocotyls and epinastic cotyledons.
Identification of YUCCA genes by activation tagging
The development of activation tagging technology in Arabidopsis in the late 1990s made it possible to identify important genes by isolating gain-of-function mutants (Weigel et al., 2000). When copies of the CaMV 35S enhancers are inserted near a gene, the enhancers cause elevated expression of the gene. Joanne Chory's group isolated long hypocotyl mutants by activation tagging in an attempt to identify genes involved in light signaling. One of the mutants named yucca (late renamed as yuc1-D, At4g32540) displays long hypocotyl and epinastic cotyledons when grown in light (Figure 3). In total darkness, yuc1-D has short hypocotyl and lacks an apical hook (Figure 3) (Zhao et al., 2001). The phenotypes of yuc1-D are almost identical to those observed in sur1, sur2, CYP79B2 overexpression lines, and the iaaM overexpression lines, suggesting that yuc1-D phenotypes are probably caused by auxin overproduction. The yuc1-D mutants also have short primary roots, and develop longer and more root hairs than wild type plants (Zhao et al., 2001). Moreover, yuc1-D plants display increased apical dominance, which is often associated with elevated auxin levels. All of the growth and developmental phenotypes of yuc1-D plants indicate auxin overproduction (Zhao et al., 2001).
Figure 3.
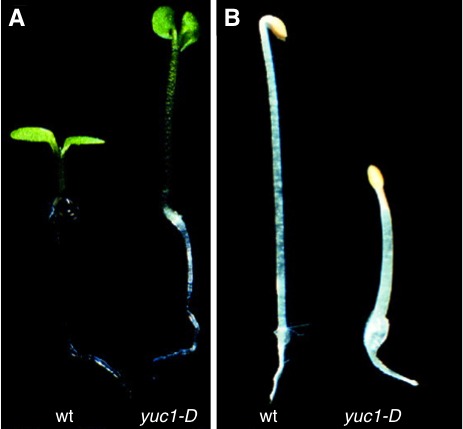
The characteristic phenotypes of auxin overproduction in Arabidopsis.
Overexpression of YUC1 (yuc1-D) leads to long hypocotyl and epinastic cotyledons in light grown seedlings (A). In total darkness, yuc1-D has short hypocotyl and lacks an apical hook (B). This figure is reprinted with permission from Science. The original figure was published in Zhao, Y., Christensen, S.K., Fankhauser, C., Cashman, J.R., Cohen, J.D., Weigel, D., and Chory, J. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291,306–309.
Physiological and genetic evidence demonstrates that yuc1-D indeed is an auxin overproduction mutant (Zhao et al., 2001). Expiants of yuc1-D develop a large number of roots on plates containing Murashige and Skoog (MS) media. The yuc1-D explants develop into calluses on auxin-free media, suggesting that yuc1-D produces sufficient auxin to support the regeneration of an Arabidopsis plant. In yuc1-D, the known auxin inducible genes such as GH3, Aux/IAA, and the SAUR genes are up regulated. The auxin reporter DR5-GUS is also expressed at elevated levels (Zhao et al., 2001). Furthermore, the phenotypes of yuc1-D can be partially reversed by overexpressing the bacterial gene iaaL, which encodes an enzyme that conjugates free IAA with the amino acid lysine (Glass and Kosuge, 1986, 1988). Direct auxin measurements show that yuc1-D contains 50% more free IAA than wild type plants. All of the physiological, genetic, and analytic biochemical data provide evidence that yuc1-D phenotypes are caused by auxin overproduction (Zhao et al., 2001).
The yuc1-D phenotypes are caused by the insertion of copies of CaMV 35S enhancers into Chromosome IV. The 35S enhancers lead to overexpression of the YUCCA (YUC1: AT4G32540) gene, which encodes a putative flavin-containing monooxygenase (Zhao et al., 2001). YUC belongs to a large gene family with 11 members in the Arabidopsis genome. YUC genes have been identified in all of the sequenced plant genomes, suggesting that YUCs are involved in an evolutionary conserved auxin biosynthesis pathway. It has been shown that overexpression of YUC genes in Arabidopsis (Zhao et al., 2001; Woodward et al., 2005; Cheng et al., 2006; Kim et al., 2007; Hentrich et al., 2013;), tomato (Exposito-Rodriguez et al., 2011), tobacco (Zhao et al., 2001), petunia (Tobena-Santamaria et al., 2002), potato (Kim et al., 2012), strawberry (Liu et al., 2012), and rice (Yamamoto et al., 2007) leads to auxin overproduction.
The YUC genes are key auxin biosynthesis genes and essential for plant development
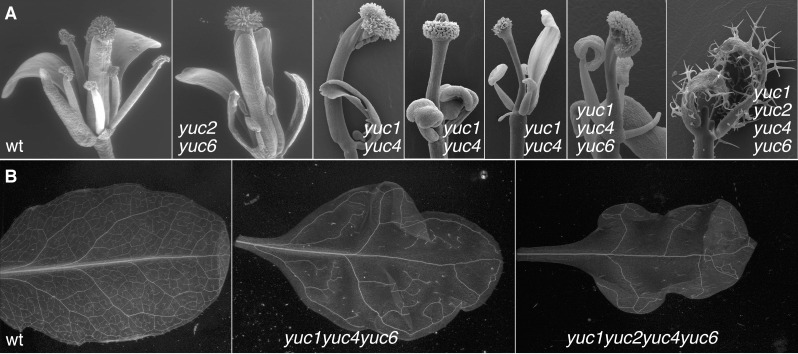
The YUC genes were not identified from previous genetic screens for loss-of-function mutants in Arabidopsis because of genetic redundancy among the YUC genes. Inactivation of a single YUC gene in Arabidopsis does not result in any obvious developmental defects. Characterization of various combinations of yuc mutants demonstrates that YUC genes are essential for embryogenesis, seedling growth, vascular pattern formation, and flower development (Figure 4) (Cheng et al., 2006, 2007a). The yuc1 yuc4 (yuc1: At4g32540; yuc4: At5g11320) double mutants fail to make tertiary veins in leaves and produce discontinuous veins in flowers (Figure 4) (Cheng et al., 2006). The yuc1 yuc4 phenotypes are further enhanced by disrupting YUC2 (AT4G13260) and YUC6 (AT5G25620) (Cheng et al., 2006). The yuc1 yuc4 yuc10 yuc11 (yuc10: At1g48910; yuc11: At4g32540) quadruple mutants do not have the basal part of the embryo (Cheng et al., 2007a), a phenotype that is also observed in auxin signaling mutants such as monopteros (mp: At1g19850) (Przemeck et al., 1996; Hardtke and Berleth, 1998), bodenlos (bdl: At1g04550) (Hamann et al., 1999), and tir1 afb1 (tir1:At3g62980; afb1: At4g03190) mutants (Dharmasiri et al., 2005). When YUC3 (AT1G04610), YUC5 (AT5G43890), YUC7 (AT2G33230), YUC8 (AT4G28720), and YUC9 (AT1G04810), which form a clade in a phylogenetic tree, are simultaneously inactivated, the resulting quintuple mutants (yucQ) develop very short and agravitropic roots (Chen et al., 2014).
Figure 4.
The YUC genes play essential roles in flower (A) and vascular development (B) in Arabidopsis.
Arabidopsis WT flowers usually have 4 sepals, 4 petals, 6 stamens, and two fused carpels (Note that 2 sepals and 2 petals are removed in WT to reveal the inner organs). Disruption of YUC2 and YUC6 leads to very short stamens. The yuc1 yuc4 double mutants produce sterile flowers with dramatically reduced number of floral organs. In addition, yuc1 yuc4 flowers display variations in terms of the type and number of floral organs. Note that the three yuc1 yuc4 flowers shown are from the same plant and that they have different number and type of floral organs. The yuc1 yuc2 yuc4 yuc6 quadruple mutants sometimes fail to produce flowers. Instead, pin-like structures are produced in the quadruple mutants. B) Inactivation of YUC genes prevents plants from forming tertiary and higher order veins in leaves. This figure is reprinted with permission from Genes & Development. The original figure was published in Cheng, Y., Dai, X., and Zhao, Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20, 1790–1799.
Although overexpression of YUC genes causes auxin overproduction and disruption of YUC genes leads to dramatic developmental phenotypes, it was challenging to demonstrate that the developmental defects of the yuc mutants were caused by auxin deficiency. The yuc1 yuc4 double mutants that displayed severe vascular and floral defects (Figure 4) could not be rescued by adding auxin to the growth media or by spraying auxin directly onto the plants (Cheng et al., 2006). It was hypothesized that auxin needs to be synthesized in specific cells at the right time in order for proper plant growth and development to proceed. The essential role of YUC genes in auxin biosynthesis was eventually demonstrated by using a genetic approach. Auxin biosynthesis by YUCs can be mimicked by expressing the bacterial auxin biosynthetic gene iaaM under the control of a specific YUC promoter. Expression of the iaaM gene under the control of the YUC1 promoter rescued the yuc1 yuc4 double mutants (Cheng et al., 2006). YUC6 promoterdriven iaaM expression was able to completely rescue the sterile phenotypes of yuc2 yuc6 double mutants (Cheng et al., 2006). Recently, the yucQ quintuple mutants were discovered to have dramatic defects in root development (Chen et al., 2014). Interestingly, the yucQ phenotypes are rescued by adding a very low concentration (5nM) of IAA into the growth media (Chen et al., 2014). The difference in response to exogenous auxin between yucQ and yuc1 yuc4 double mutants may be caused by different auxin transport capacity among different tissues.
In summary, the essential roles of the YUC genes in auxin biosynthesis and in Arabidopsis development have been unambiguously demonstrated. Overexpression of YUCs leads to auxin overproduction and disruption of YUC genes causes dramatic developmental defects, which can be rescued by adding auxin in growth media (Chen et al., 2014) or by expressing the iaaM gene under the control of a YUC promoter (Cheng et al., 2006).
YUC genes in petunia and maize have also been shown to play essential roles in development (Tobena-Santamaria et al., 2002; Gallavotti et al., 2008; LeCLere et al., 2010; Bernardi et al., 2012). Interestingly, the genetic redundancy of YUC genes appears to be less pronounced in petunia and in maize than in Arabidopsis. The single yuc mutant in petunia called floozy has defects in the formation of floral organ primordia and bracts at the base of the flower. Additionally, floozy mutants fail to produce secondary veins in leaves and bracts and display a decreased apical dominance in the inflorescence (Tobena-Santamaria et al., 2002). The floozy phenotypes are similar to those observed in Arabidopsis yuc1 yuc4 double mutants. The maize yuc mutant sparse inflorescencel (spi1) has defects in the initiation of axillary meristems and lateral organs during both vegetative and reproductive development (Gallavotti et al., 2008). The maize mutant defective endosperm18 (del8) is another yuc mutant (Zmyuc1) that has dramatically reduced free IAA levels and has approximately 40% less dry mass than the wild type (LeCLere et al., 2010; Bernardi et al., 2012).
Identification of TAA aminotransferases as key auxin biosynthetic enzymes
Three groups independently identified the TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 (TAA1: AT1G70560) gene in Arabidopsis from three forward genetic screens (Stepanova et al., 2008; Tao et al., 2008; Yamada et al., 2009). In a genetic screen for mutants with altered responses to shade growth conditions, Joanne Chory's group isolated a mutant called sav3 that had short hypocotyls in shade compared to wild type plants, which had elongated hypocotyls under the same conditions (Tao et al., 2008). Jose Alonso's group isolated a weak ethylene insensitive 8 (wei8) mutant, which has elongated primary roots in the presence of the ethylene biosynthetic precursor ACC (Stepanova et al., 2008). It was later found out that wei8 is allelic to the sav3 mutants. Mark Estelle's group identified additional sav3/wei8 alleles from a screen for mutants resistant to the auxin transport inhibitor NPA (tir2 mutants) (Yamada et al., 2009). SAV3/WEI8/TIR2 were renamed as TAA1, which encodes an aminotransferase that catalyzes the conversion of Trp to IPA.
TAA1 is the founding member of a large family of aminotransferases. Like the YUC genes, TAA genes also have overlapping functions. Although taa1 mutants display altered responses to shade and are resistant to both ethylene and NPA, they do not show dramatic developmental defects under normal growth conditions. However, simultaneous inactivation of TAA1 and its two close homologs TAA RELATED1 (TAR1: AT1G23320) and TAA RELATED 2 (TAR2: AT4G24670) leads to embryogenesis defects. The wei8 tar1 tar2 triple mutants display mp-like phenotypes: the basal part of the embryo fails to develop (Stepanova et al., 2008). The wei8 tar2 double mutants have dramatic defects in vascular and floral development and are completely sterile (Stepanova et al., 2008).
That the TAA genes are key auxin biosynthesis genes is supported by several observations. First, the tea mutants have lower auxin concentrations than do wild type plants (Stepanova et al., 2008; Tao et al., 2008). Second, the expression levels of the auxin reporter DR5-GUS/GFP are decreased in the taa mutants (Stepanova et al., 2008; Tao et al., 2008). Third, exogenous IAA can rescue the root phenotypes of wei8 (Stepanova et al., 2008). The shade avoidance defects of sav3 are also rescued by overexpressing the bacterial auxin biosynthetic gene iaaM or by using the synthetic auxin picloram (Tao et al., 2008).
TAA genes are highly conserved throughout the plant kingdom. The orthologs of the Arabidopsis TAA1 in maize, in rice, and in Brachypodium have been shown to play important roles in auxin biosynthesis and in development (Phillips et al., 2011; Pacheco-Villalobos et al., 2012; Yoshikawa et al., 2014). The genetic redundancy of the TAA genes in monocots appears to be less than that of the TAA genes in Arabidopsis. Inactivation of a single TAA1-like gene in maize (vt2 mutants) leads to dramatic defects in both vegetative and reproductive development (Phillips et al., 2011). Disruption of the rice TAA1 gene (fish bone mutant) results in pleiotropic phenotypes including small panicles and abnormal vascular development (Yoshikawa et al., 2014). Mutations in the Brachypodium TAR2-like gene counter-intuitively cause an increase in auxin levels. Consequently the Bdtar2 mutants have dramatically elongated seminal roots because of enhanced cell elongation (Pacheco-Villalobos et al., 2012). In Bdtar2 mutants, several YUC genes are up regulated, which may account for the observed increase of auxin levels (Pacheco-Villalobos et al., 2012). Unlike overexpression of YUCs, overexpression of TAAs does not lead to any obvious developmental phenotypes, suggesting that TAAs-catalyzed reaction may not be a rate-limiting step in auxin biosynthesis (Stepanova et al., 2008; Tao et al., 2008).
Establishment of a complete two-step auxin biosynthesis pathway
TAA genes and YUCs were previously placed in two independent auxin biosynthesis pathways (Zhao et al., 2001; Stepanova et al., 2008; Tao et al., 2008). However, some yuc mutant combinations display phenotypes similar to those observed in the taa mutant combinations. The phenotypic similarities between yuc and taa mutants suggest that TA As and YUCs may participate in the same auxin biosynthesis pathway (Strader and Bartel, 2008; Mashiguchi et al., 2011; Stepanova et al., 2011; Won et al., 2011). Both wei8 tar1 tar2 triple mutants and yuc1 yuc4 yuc10 yuc11 quadruple mutants develop embryos without the basal part (Cheng et al., 2006, 2007a; Stepanova et al., 2008). Similar vascular and floral defects have also been observed in wei8 tar2 and yuc1 yuc4 double mutants (Figure 4) (Cheng et al., 2006, 2007a; Stepanova et al., 2008). In fact, yuc mutants display all of the characteristic phenotypes of the taa mutants (Won et al., 2011). For example, yucQ mutants are resistant to ACC and NPA in root elongation, a characteristic phenotype of taa1 mutants (Won et al., 2011). The yuc1 yuc4 double mutants have altered shade avoidance responses, a phenotype that is also observed in taa1 mutants (Won et al., 2011). Interestingly, Arabidopsis plants appear to use different sets of YUC genes for auxin biosynthesis in the shoot and in the root despite using the same set of TAA genes in both shoots and roots (Won et al., 2011).
On the other hand, all of the characteristic phenotypes of yuc mutants are also displayed in taa mutants (Won et al., 2011). For example, inactivation of the protein kinase gene PINOID (PID: AT2G34650) in yuc1 yuc 4 backgrounds completely abolishes the development of cotyledons (Cheng et al., 2007b). The wei8 tar2 pid triple mutants also fail to develop cotyledons (Won et al., 2011). The NAKED PINS IN YUC MUTANTS (NPY) (NPY1: AT4G31820; NPY2: AT2G14820; NPY3: AT5G67440; NPY4: AT2G23050; NPY5: AT4G37590) genes were identified from a genetic screen for yuc1 yuc4 enhancers and the yuc1 yuc4 npy1 triple mutants developed pin-like inflorescences (Cheng et al., 2007b, 2008). Disruption of NPY1 in wei8 tar2 background also leads to the formation of pin-like inflorescences (Won et al., 2011).
Several key pieces of evidence have unambiguously placed TAAs and YUCs in the same Trp-dependent auxin biosynthesis pathway. First, the auxin overproduction phenotypes displayed in YUC overexpression lines are dependent on the TAA genes (Mashiguchi et al., 2011; Stepanova et al., 2011; Won et al., 2011). Overexpression of YUC genes in wei8 tar2 plants does not lead to the typical auxin overproduction phenotypes (Stepanova et al., 2011; Won et al., 2011). In contrast, overexpression of the iaaM gene in wei8 tar2 still leads to long hypocotyls and epinastic cotyledons, two characteristic auxin overproduction phenotypes, indicating that wei8 tar2 specifically block the auxin overproduction caused by overexpression of YUCs (Won et al., 2011). Second, the taa mutants contain a decreased concentration of IPA while yuc mutants have elevated concentrations of IPA, indicating that TAAs participate in IPA production while YUCs are involved in IPA consumption (Mashiguchi et al., 2011; Won et al., 2011). The hypothesis that YUCs function downstream of TAAs is also consistent with the findings that overexpression of YUC1 can partially rescue the shade avoidance phenotypes of taa1/sav3 (Won et al., 2011). Moreover overexpression of YUC1 also partially rescues the fertility defects in the weak we 18 tar2-2 mutants (Won et al., 2011). The conversion of the residual IPA in the weak taa mutants is probably accelerated in YUC overexpression lines, providing sufficient IAA for partially rescuing the weak taa mutant phenotypes. Genetic studies in maize also suggest that TAAs and YUCs participate in the same auxin biosynthesis pathway. The taa1/vt2 mutants and the yuc/spi1 mutants have similar phenotypes. The spi1 vt2 double mutants are similar to vt2 in terms of auxin levels and developmental phenotypes (Phillips et al., 2011).
BIOCHEMICAL MECHANISMS OF AUXIN BIOSYNTHESIS
Biochemical mechanisms of the TAA-catalyzed reaction.
Recombinant TAA1 protein fused with either an N-terminal GST-tag or C-terminal His-tag is able to catalyze a PLP-dependent transaminase reaction in vitro (Stepanova et al., 2008; Tao et al., 2008). TAA1 catalyzes the transfer of the amino group from Trp to an α-ketoacid such as α-ketoglutarate to produce IPA and the amino acid Glutamate (Stepanova et al., 2008; Tao et al., 2008). TAA1 has an apparent KM of 290 µM for Trp and a kcat of 1.85 s-1 (Tao et al., 2008).
All 20 amino acids except Glu have been tested as amino donors in the TAA1-catalyzed reaction with α-ketoglutarate as the acceptor of the amino group (Tao et al., 2008). Besides Trp, TAA1/SAV3 can also use Phe, Tyr, Leu, Ala, Met, and Gln as an amino group donor (Tao et al., 2008). However, Trp appears to be the best amino donor because the KM values for Tyr (4.7 mM) and Phe (9.35 mM) are much higher than that for Trp (0.29 mM). The kcat values are similar when Trp, Phe, and Tyr are used as substrates (Tao et al., 2008). Docking experiments using the TAA1/SAV3 crystal structures reveal that IPA has the best score in docking followed by Trp and then by Phe, Tyr, and His (Tao et al., 2008). Therefore, Trp appears to be the best amino donor among the 20 amino acids, a finding that is consistent with the physiological roles of TAA1 in Trp-dependent auxin biosynthesis.
The preference for the amino acceptor in the TAA1-catalyzed reaction has not yet been systematically analyzed. So far, only pyruvate and α-ketoglutarate have been tested as the in vitro amino-acceptor (Stepanova et al., 2008; Tao et al., 2008). A systematic analysis of the α-ketoacids corresponding to the 20 amino acids is still needed to determine the best amino acceptor for TAAs. Interestingly, IPA performed better than Trp in the docking experiments, suggesting that the release of the product IPA could be the rate-limiting step of the TAA1-catalyzed reaction (Tao et al., 2008). It is also likely that the TAA-catalyzed reaction may be subject to product inhibition (Tao et al., 2008). It will also be interesting to test whether TAAs can catalyze the transfer of the amino group from other amino acids to IPA to produce Trp and the other α-ketoacids. Although genetic studies have demonstrated that TAA1 participates in Trp-dependent auxin biosynthesis by producing IPA, it should be recognized that TAA1-catalyzed reaction is coupled with homeostasis of another amino acid/α-ketoacid. Disruption of TAA genes not only abolishes IPA production, but also affects the metabolism of other α-ketoacids and amino acids. Therefore, phenotypes of taa mutants may be caused by a combination of IAA deficiency and defects in homeostasis of other amino acids.
So far, the quantitative biochemical studies on TAA family proteins have been limited to the Arabidopsis TAA1 (Stepanova et al., 2008; Tao et al., 2008, He et al., 2011). The TAAs (PsTAR1 and PsTAR2) from the pea plants (Pisum sativum) have been shown qualitatively to convert Trp to IPA and 4-chlora-Trp to 4-chlora-IPA (Tivendale et al. 2012). It will be interesting to determine whether TAA1 and TARs differ in substrate specificity and kinetic properties.
The biochemical mechanisms of YUC flavin monooxygenases
YUC family proteins showed significant sequence homologies to the family of flavin-containing monooxygenases, which is one of the largest groups of monooxygenases that have been identified. Monooxygenases take one oxygen atom from a molecular oxygen and insert it into an organic compound (Cashman, 1995; Cashman et al., 1995). Flavin-containing monooxygenases play key roles in the synthesis of many physiologically important molecules in various organisms, and are responsible for the degradation of a large variety of aromatic and heteroatom-containing compounds. Flavin-containing monooxygenases usually use NADPH and FAD as cofactors and molecular oxygen as a co-substrate. YUC proteins contain two GXGXXG motifs that function as binding sites for FAD and NADPH (Hou et al., 2011). Mutations in the predicted FAD or NADPH binding sites completely abolish YUC functions in Arabidopsis, suggesting that YUCs probably also use FAD and NADPH as cofactors (Hou et al., 2011).
Flavin-containing monooxygenases are notorious for being very difficult enzymes for in vitro studies. Our current understanding of flavin-containing monooxygenases is mainly based on studies of the mammalian enzymes, which are purified from animal tissues (Cashman, 1995; Cashman et al., 1995). The fungal enzyme N5-L-ornithine monooxygenase (OMO), a flavin-containing monooxygenase, was discovered over 25 years ago, but it was not until recently that the biochemical details of OMO were resolved, largely due to the recent success in expressing OMO in E. coli and purifying OMO to near homogeneity (Mayfield et al., 2010). In general, flavin-containing monooxygenases use a relatively stable C4a-(hydro)peroxyflavin intermediate to conduct oxidative reactions (Ziegler, 1988, 2002). Therefore, flavin-containing monooxygenases are usually promiscuous and the mammalian microsome flavin-containing monooxygenases are known to catalyze the hydroxylation of a large class of nitrogen and sulfur-containing compounds (Ziegler, 1988, 2002). In addition, mammalian flavin-containing monooxygenases have also been shown to catalyze Baeyer-Villiger type of reactions, in which the C4a intermediate makes a nucleophilic attack on a carbonyl group to form the Criegee intermediate, which then undergoes rearrangement to generate the final products (Lai et al., 2010). The difference between Baeyer-Villiger type of reactions and N-hydroxylation is that the C4a intermediate serves as a nucleophile in Baeyer-Villiger type of reactions while the C4a intermediate is an electrophile in N-hydroxylation reactions. Because of the catalytic mechanisms of flavin-containing monooxygenases, it is often very difficult to pin down the exact physiological substrates. Interestingly, some of the flavin-containing monooxygenases display remarkable substrate specificity. For example, OMO catalyzes hydroxylation of ornithine, but does not hydroxylate lysine, which has one more methylene than ornithine (Mayfield et al., 2010).
Solving the biochemical mechanisms of the YUC enzymes has been a challenging journey. Initially, it was proposed based on in vitro assays that YUCs catalyze the N-hydroxylation of tryptamine (Zhao et al., 2001; Expósito-Rodríguez et al., 2007, Kim et al., 2007; LeCLere et al., 2010). However, the early biochemical work had several caveats. In the early studies, YUC proteins with an N-terminal or C-terminal tag expressed in and purified from E. coli were not biochemically characterized. It was not clear what cofactors were required for the YUC-catalyzed reactions. The recombinant YUC proteins lacked any yellow color, an indication that the recombinant YUC proteins did not contain the flavin cofactor. The YUC proteins purified from E. coli were probably not folded correctly. Early enzyme assays of YUC proteins were all qualitative and only tryptamine was tested as a substrate (Zhao et al., 2001; Expósito-Rodríguez et al., 2007, Kim et al., 2007; LeCLere et al., 2010). In addition, the early enzymatic assays required the addition of free FAD to the reaction mixture, probably causing off-enzyme reactions. Recent studies no longer support the hypothesis that YUCs use tryptamine as a physiological substrate for auxin biosynthesis (Tivendale et al., 2010; Mashiguchi et al., 2011; Stepanova et al., 2011; Won et al., 2011; Kriechbaumer et al., 2012).
Recent genetic studies have clearly indicated that YUCs and TAAs participate in the same Trp-dependent auxin biosynthesis pathway (Mashiguchi et al., 2011; Stepanova et al., 2011; Won et al., 2011). The findings that yuc mutants accumulate IRA and taa mutants are IPA deficient suggested that IPA was a substrate for YUC flavin-containing monooxygenases (Mashiguchi et al., 2011; Won et al., 2011). YUC enzymatic assays were then conducted to determine whether YUCs could use IPA as a substrate in vitro (Mashiguchi et al., 2011; Kriechbaumer et al., 2012). Recombinant GST-tagged YUC2, expressed in and purified from E. coli, was able to convert IPA to IAA in an NADPH-dependent manner (Mashiguchi et al., 2011). Interestingly, GST-YUC2 did not convert IPA into Indole-3-acetaldehyde, which had long been hypothesized as an auxin biosynthesis intermediate (Mashiguchi et al., 2011). Two isoforms of Arabidopsis YUC4, produced from in vitro transcription and translation using the PUREexpressTM kit, were also shown to use IPA as a substrate (Kriechbaumer et al., 2012).
In order to elucidate the catalytic mechanisms and the biochemical properties of YUC flavin monooxygenases, large quantities of YUC proteins are required. This was achieved for the Arabidopsis YUC6, which was expressed in and purified to near homogeneity from E. coli (Dai et al., 2013). The recombinant YUC6 displayed bright yellow color (Figure 5) and an UV-Visible spectrum resembled those of FAD and FMN (Dai et al., 2013). This was the first time that large quantities of a plant flavin-containing monooxygenase had been purified to near homogeneity with the flavin co-factor bound. Several factors contributed to the success: 1) The YUC6-expressing E. coli cells were grown at low temperature for two days before induction and two days after induction; 2) high concentrations of glycerol (30%) were added to the purification buffer to stabilize the YUC protein (Dai et al., 2013).
Figure 5.
Purification of recombinant YUC6 with the flavin-cofactor bound.
YUC6 tagged with His6 at the N-terminus was expressed in E. coli and purified with Nickel NTA agrose. Note that the flavin cofactor in YUC6 gave the bright yellow color.
The availability of large quantities of the purified YUC6 enables the detailed characterization of the biochemical properties and the catalytic mechanisms of YUC6. YUC6 is an FAD-containing protein, as demonstrated by several pieces of experimental data: 1) The UV-Visible spectrum of YUC6 is very similar to those of FAD and FMN; 2) denatured YUC6 protein releases a small molecule that has the same retention time in HPLC as the standard FAD; 3) The flavin cofactor released from YUC6 has very little fluorescence, but upon treatment with a phosphodiesterase that converts FAD to FMN, the fluorescence increased more than 10 times (Dai et al., 2013). The experimental data also are consistent with sequence-based prediction that YUCs are FAD-containing enzymes (Dai et al., 2013).
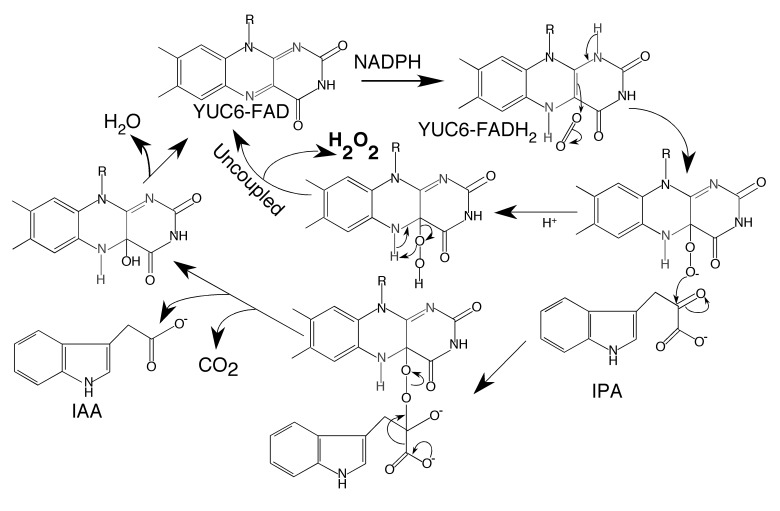
Kinetic and spectroscopic characterization of the YUC6 protein has established the basic framework for YUC-mediated catalysis (Figure 6) (Dai et al., 2013). YUC6 as purified contains an oxidized FAD, which has peaks at 448 nm and 376 nm in the UV-visible spectrum (Dai et al., 2013). The FAD cofactor in YUC6 readily takes two electrons from NADPH to become FADH2, which no longer has the peak at 448 nm in the UV-visible spectrum (Dai et al., 2013). Reduction of FAD to FADH2 by NADPH is the first step of the YUC-catalyzed reaction (Figure 6). The second step is the reaction of FADH2 with molecular oxygen to form the C4a-(hydro)peroxyflavin intermediate (Dai et al., 2013). Mechanistically, flavin-containing monooxygenases use either a C4a-hydroperoxyflavin or a C4a-peroxyflavin as the species that carries out the oxygenation reaction. Because it is generally difficult to distinguish these two forms spectroscopically, the intermediate observed in YUC6-mediated catalysis is designated C4a-(hydro) peroxyflavin to reflect the uncertainty about the actual form of the intermediate. The YUC6 C4a-(hydro)peroxyflavin intermediate displays a distinct UV-Visible spectrum with a peak at 381 nm (Dai et al., 2013). The last step of the YUC6-catalyzed reaction is the oxidative decarboxylation of IPA by the C4a-(hydro)peroxyflavin to produce IAA (Figure 6). The proposed catalytic mechanism is well supported by the kinetic and spectroscopic results (Dai et al., 2013). The oxidized, reduced, and the C4a intermediate of YUC6 display characteristic UV-Visible spectra and have been observed experimentally. Overall, the YUC6-catalyzed reaction employs a catalytic strategy that is similar to those used by other well-characterized flavin-containing monooxygenases.
Figure 6.
The mechanism of YUC6-catalyzed reaction.
YUC6 undergoes reduction and then binds to oxygen to form the C4a-(hydro)peroxyflavin intermediate. The C4a intermediate reacts with IPA to generate IAA, CO2 and water. The C4a intermediate can also decompose into H2O2. H2O2 produced from the uncoupled reaction may kill the enzymatic activities of YUC6. The reaction center of the flavin cofactor is marked red.
If the flavin-containing monooxygenases turn over NADPH without oxidizing the substrates, NADPH is wasted. There are several general strategies to prevent flavin-containing monooxygenases from becoming an NADPH oxidase: 1) Reduction of FAD by NADPH only takes place in the presence of the substrate; 2) Reaction of the reduced flavin with oxygen is regulated by substrate binding; 3) The C4a intermediate is stabilized in the absence of the substrate. However, the last strategy often leads to promiscuous enzymatic reactions. YUC6 is readily reduced by NADPH regardless of the presence of the substrate IPA. IPA also does not affect the rate of YUC6 reduction by NADPH (Dai et al., 2013). Furthermore, IPA does not affect the kinetic pattern and rate of formation of the C4a intermediate (Dai et al., 2013). In the presence of IPA, the decomposition of the C4a intermediate is greatly accelerated, suggesting that YUC6 uses the third strategy to minimize the NADPH oxidase activity. Interestingly, the C4a-intermediate in YUC6 appears much less stable than the counterpart intermediates in mammalian flavin-containing monooxygenases, which catalyze both N-hydroxylation and Baeyer-Villiger type reactions (Dai et al., 2013). The half-life values of the C4a intermediate in mammalian flavin-containing monooxygenases are usually more than 30 minutes whereas the halflife of the C4a intermediate of YUC6 is less than 20 seconds (Dai et al., 2013). Although it is not clear what significance of the short half-life of the YUC6 C4a intermediate, it probably increases the substrate specificity of YUC proteins.
YUC6 is a good NADPH oxidase with a turnover number of 2.4 per minute (Dai et al., 2013). The NADPH oxidase activity uses the electrons from NADPH to convert oxygen into hydrogen peroxide, which is a component of the reactive oxygen species (ROS). ROS has been implicated in many diverse physiological and pathological processes. The fact that YUC6 has NADPH oxidase activity indicates that YUC activities have to be tightly regulated.
Genetic studies have clearly demonstrated that YUCs use IPA as their physiological substrate. However, YUC6 can also use Phenylpyruvate (PPA) as a substrate in vitro (Dai et al., 2013). It still remains to be investigated whether YUCs can use all of the α-ketoacids as substrates. Because YUC6 forms the active C4a intermediate, it is reasonable to hypothesize that any α-ketoacids that can enter the active sites of YUCs probably will be able to react with the C4a intermediate. This raises the question of how substrate specificity of YUCs is achieved in plants. So far, only YUC6 has been analyzed both kinetically and spectroscopically. It is still a challenge to determine the biochemical properties of the other YUCs from Arabidopsis and other species.
REGULATION OF AUXIN BIOSYNTHESIS
Auxin is synthesized in two chemical steps using IPA as the intermediate (Figure 1). De Novo auxin biosynthesis can be effectively regulated by using three strategies: 1) control of the IPA production; 2) Regulation of the conversion of IPA into IAA; 3) Diversion of IPA to other metabolic pathways
Metabolic regulation of auxin biosynthesis by VAS1
Recently, Zheng et al. isolated an aminotransferase gene named VAS1 (AT1G80360) from a genetic screen for mutations that can suppress the defects of sav3/taa1 in shade avoidance responses (Zheng et al., 2013). Under shade conditions, wild type plants have elongated hypocotyls and sav3 fails to elongate its hypocotyl. The vas1 sav3 double mutants display long hypocotyl phenotypes under shade conditions. VAS1 is a predicted aminotransferase based on its sequence homology to known PLP-dependent aminotransferase. VAS1 has been systematically analyzed to identify its amino donors and acceptors (Zheng et al., 2013). It turns out that IPA is the most suitable amino acceptor for the VAS 1-catalyzed transamination reaction. PPA or 4-hydroxyphenylpyruvate (two ketoacids from the amino acids Phe and Tyr, respectively) is only 3% as active as IPA in a VAS1-catalyzed reaction (Zheng et al., 2013). Other α-ketoacids including glyoxylate, pyruvate, 2-keto butyrate, 2-keto-4-methyl-thiobutyric acid (KMBA), 2-oxoglutarate, and oxaloacetate, fail to function in vitro as amino acceptors in the VAS1 catalyzed transamination reaction (Zheng et al., 2013). For amino donors, VAS1 prefers Met to all other amino acids. The second most active amino donor is Phe. Flowever, the relative activity with Phe as a substrate is only 21 % that of Met. Other amino acids including Val, Ile, Tyr and Leu are less than 1 % as active as Met in vitro (Zheng et al., 2013).
The in vitro biochemical analysis reveals that VAS1 has unusually high substrate specificity. It essentially only catalyzes the transfer of the amino group from Met to IPA (Zheng et al., 2013). Met and IPA are intermediates for ethylene biosynthesis and auxin biosynthesis, respectively. Therefore VAS 1-catalyzed reaction metabolically links the biosynthesis of the two important plant hormones. Disruption of VAS1 leads to the accumulation of IPA and IAA in Arabidopsis. Light grown vas1 mutants have longer hypocotyls than wild type plants, a phenotype that is observed in auxin overproduction mutants. The other substrate of VAS1 is Met, which is converted into KMBA in the VAS1 catalyzed reaction. Both Met and KMBA are precursors for ethylene biosynthesis in the Yang Cycle. The vas1 mutants have elevated levels of ACC, the immediate precursor of ethylene biosynthesis (Zheng et al., 2013). It is reported that the elevated ethylene levels account for the exaggerated petiole elongation of the vas1 mutants (Zheng et al., 2013). Trp is the other product of the VAS1-catalyzed reaction. It is not clear whether VAS1 also plays a key role in maintaining Trp homeostasis. The identification of VAS1 as a key component for maintaining the homeostasis of IPA, ethylene, and auxin demonstrated the complexity of regulating auxin biosynthesis.
Transcriptional regulation of auxin biosynthesis
One of the surprising findings in auxin biosynthesis is that the expression of auxin biosynthetic genes is often restricted to discrete groups of cells (Cheng et al., 2006, 2007a; Stepanova et al., 2008; Tao et al., 2008; Won et al., 2011). For a long time, it was believed that the location of auxin biosynthesis was not very important because auxin could reach any regions via the polar auxin transport system (Grieneisen et al., 2007). Computer models demonstrated that polar auxin transport is necessary and sufficient for generating auxin gradients for plant development (Grieneisen et al., 2007). However, recent progress in auxin biosynthesis has unequivocally demonstrated that localized auxin biosynthesis also plays an essential role in virtually every major developmental processes including embryogenesis, seedling growth, vascular patterning, root development, phyllotaxis, and flower development (Cheng et al., 2006, 2007a; Stepanova et al., 2008; Pinon et al., 2013).
Auxin biosynthesis is regulated at the transcriptional level by developmental signals and environmental cues. The specific expression patterns of TA A genes and YUC genes are determined by transcription factors that bind to the regulatory regions of the genes. Several groups of transcription factors have been identified by their ability to bind directly to the regulatory regions of TA A genes and/or YUC genes. The SHORT INTENOTES/STYLISH (SHI/STY) (SHI: AT5G66350; STY1: AT3G51060) family of transcription factors plays an important role in leaf and flower development by regulating the expression of YUC genes (Sohlberg et al., 2006; Eklund et al., 2010). STY1, one of the SHI/STY proteins, binds directly to the short motif of ACTCTAC in the YUC4 promoter activating the expression of YUC4 (Eklund et al., 2010). STY1 also binds to the promoter region of YUC8 and activates YUC8 expression (Eklund et al., 2010). The NGATHA (NGA1: AT2G46870) family of transcription factors, which redundantly controls style development in a dosage-dependent manner, positively regulates YUC2 and YUC4 in the apical domain of Arabidopsis gynoecium (Alvarez et al., 2009; Trigueros et al., 2009). But it is not clear whether the regulation by NGA is direct or not. LEAFY COTYLEDON2 (LEC2: AT1G28300), a central regulator of embryogenesis, binds directly to the promoters of YUC2, YUC4, and YUC10 to activate the expression of the YUC genes (Wojcikowska et al., 2013; Stone et al., 2008). The Arabidopsis IDD14 (AT1G68130), IDD15 (AT2G01940), and IDD16 (AT1G25250) are important regulators for lateral organ morphogenesis and gravitropism. It has been shown that the IDDs directly target YUC5 and TAA1 (Cui et al., 2013). Interestingly, the class III HD-Zip transcription factor REVOLUTA (REV: AT5G60690) also directly binds to the promoters of TAA1 and YUC5, suggesting that part of REV's roles in shoot development and organ polarity is probably mediated by altering auxin biosynthesis (Brandt et al. 2012). PLETHORA transcription factors (PLT3: AT5G10510; PLT5: AT5G57390; PLT7: AT5G65510) control phyllotaxis in Arabidopsis by regulating the expression of YUC genes (Pinon et al., 2013).
Environmental signals have a profound effect on auxin biosynthesis. TAA1 was identified from a screen for mutants that were defective in shade avoidance responses, a light-mediated signaling process (Tao et al., 2008). Shade avoidance responses require the phytochrome photoreceptors and the Phytochrome-Interacting Factors (PIFs), which are bHLH transcription factors (Ballare, 1999). Recent studies have shown that part of the light signaling response is to modulate auxin homeostasis. Some of the auxin biosynthetic genes are direct targets of the PIFs (Li et al., 2012). Under shade conditions, auxin levels in Arabidopsis are elevated more than 50% compared to the levels in plants grown under normal white light conditions (Li et al., 2012). The increased auxin concentrations are caused by the up regulation of the expression of several YUC genes (Li et al., 2012). PIF7 (AT5G61270) is one of the key transcription factors downstream of the photoreceptor phytochrome B. Disruption of PIF7 leads to short hypocotyls and expanded cotyledons under shade, suggesting that PIF7 plays a positive role in shade avoidance response. It is reported that PIF7 binds to the promoters of YUC2, YUC5, YUC8, and YUC9 and activates the expression of the YUC genes under shade conditions (Li et al., 2012).
In addition to participating in light signaling, PIFs have recently been shown to contribute to temperature-mediated and sugar-induced auxin biosynthesis (Franklin et al., 2011; Sun et al., 2012). It has been well documented that high temperatures stimulate hypocotyl elongation and auxin over-accumulation (Gray et al., 1998). The high temperature-induced developmental changes are mediated through PIF4 (AT2G43010). PIF4 binds directly to the promoters of YUC8, TAA1, and CYP79B2, thereby controlling auxin levels (Franklin et al., 2011; Sun et al., 2012). Sugar (glucose and sucrose) treatments cause elevated auxin biosynthesis, probably through PIF-dependent activation of YUC expression (Lilley et al., 2012; Sairanen et al., 2012). Circadian rhythm also regulates plant development through modulating auxin levels. For example, RVE1 (AT5G17300), a MYB-like transcription factor and clock-regulated transcription factor appears to positively regulate YUC8 (Rawat et al., 2009).
The majority of transcription factors including STY1, PIFs, and REV1 that bind to promoters of auxin biosynthesis genes are positive regulators. In contrast, SPOROCYTELESS (SPL: AT4G27330) is a known negative regulator of auxin biosynthesis. Overexpression of SPL represses the expression of YUC2 and YUC6 (Li et al., 2008). LEAFY (LFY: AT5G61850) is another negative regulator of auxin biosynthesis. LFY binds directly to the YUC4 promoter and inhibits YUC4 expression (Li et al., 2013).
Control enzymatic activities of TAAs and YUCs
YUCs catalyze the rate-limiting step of auxin biosynthesis and the enzymatic activities of YUCs have to be tightly regulated. However, factors that regulate YUC activities are not understood. It is also not clear whether YUC protein stability is regulated. Interestingly, about 4% of the YUC6-catalyzed reaction is un-coupled (Figure 6) (Dai et al., 2013). The uncoupled YUC6-catalyzed reaction uses the electrons from NADPH to partially reduce oxygen to H2O2. Production of hydrogen peroxide by the uncoupled reaction potentially provides a built-in deactivation mechanism for YUCs. After several rounds of catalysis, H2O2 produced from the uncoupled reaction may deactivate the YUC enzymatic activities. Currently, very little is known regarding how TAA enzymatic activities are regulated.
MANIPULATION OF AUXIN CONCENTRATIONS WITH SPATIAL AND TEMPORAL CONTROL
The successful elucidation of the TAA/YUC auxin biosynthesis pathway provides tools for us to modulate auxin concentrations in plants with temporal and spatial control. Auxin concentrations in cells or tissues can now be increased or decreased by regulating auxin biosynthesis in plants. Auxin conjugation and degradation can also be manipulated to alter auxin concentrations in plants (Glass and Kosuge, 1986; Peer et al., 2013; Pencik et al., 2013; Zhao et al., 2013). In this chapter, I focus on discussing two general strategies to control auxin biosynthesis in plants: 1) use auxin biosynthesis inhibition by chemicals; 2) genetic activation or deactivation of auxin biosynthetic genes.
Inhibitors for auxin biosynthesis
Two inhibitors have been developed to block the activities of TAA aminotransferases. L-amino-oxyphenylpropionic acid (AOPP) was identified by a genomics-based approach as an effective inhibitor for Trp aminotransferases (Soeno et al., 2010). Plants grown on 50 µM L-AOPP displayed defects in root elongation, gravitropism, and root hair formation (Soeno et al., 2010). Adding exogenous auxin in the growth media largely reversed the phenotypes caused by L-AOPP. L-AOPP treatments lowered free IAA levels in plants (Soeno et al., 2010). Although L-AOPP is an effective inhibitor for the TAA family of aminotransferases, further characterization of the compound is still needed. L-AOPP targets PLP-dependent enzymes and it probably also inhibits other nonauxin biosynthesis enzymes.
The other characterized TAA/TAR inhibitor is L-Kynurenine, which is a degradation product of Trp in animal systems. L-Kynurenine is a competitive inhibitor of TAA with a Ki of 11.52 µM (He et al., 2011). L-Kynurenine treatments can phenocopy the seedling phenotypes of wei8 tar2. Results from molecular modeling and computational docking experiments suggest that L-Kynurenine is a specific and highly selective auxin biosynthesis inhibitor (He et al., 2011).
Recently, an inhibitor for YUC flavin-containing monooxygenases has been reported. The inhibitor named yucasin, 5-(4-chlorophenyl)-4H-1,2,4-triazole-3-thiol, was initially identified as a potent inhibitor of auxin biosynthesis in maize coleoptile tips (Nishimura et al., 2013). Late yucasin was shown to suppress the auxin overproduction phenotypes displayed in YUC1 overexpression Arabidopsis plants (Nishimura et al., 2013). It was shown that yucasin competitively inhibits the oxidative decarboxylation of IPA in vitro catalyzed by the YUC flavin monooxygenases (Nishimura et al., 2013). L-Kynurenine and yucasin synergistically inhibit auxin biosynthesis and plant growth (Nishimura et al., 2013). The inhibitors can be used to modulate auxin biosynthesis in plants with temporal control.
Modulate auxin biosynthesis by genetic approaches
IAA is synthesized from Trp in two chemical steps and the YUC-catalyzed reaction is the rate-liming step (Figure 1). Expression of one of the YUC genes under the control of an appropriate promoter can increase auxin levels in specific cells/tissues. If we are concerned that the YUC substrate IPA may not be available in certain cells, we can co-express both TAA and YUC genes under the control of the same promoter. The YUC and TAA genes can also be placed downstream of an inducible promoter so that auxin production can be temporally controlled. A two-component system can also be used to control the expression of auxin biosynthetic genes, thus providing precise temporal and spatial control of auxin biosynthesis.
RNAi, artificial microRNAs, and the recently developed CRISPR genome editing technology make it possible to disrupt auxin biosynthesis in specific cells/tissues. By choosing the appropriate promoters, YUC genes and TAA genes can be disrupted in the targeted cells/tissues such that auxin biosynthesis can be blocked both spatially and temporally. Previous studies analyzed the gene expression profiles in response to auxin treatments that led to elevated high auxin concentrations in cells. Disruption of auxin biosynthesis using genetic or chemical approaches will allow us to conduct gene expression profiling in response to auxin deficiency.
In summary, much progress has been made in the field of auxin biosynthesis. The two-step pathway from Trp to IAA catalyzed by the TAA aminotransferases and the YUC flavin-containing monooxygenases is the first identified complete auxin biosynthetic pathway, which is essential for almost all of the major developmental events. The identification of the TAA/YUC pathway offers novel tools to modulate auxin concentrations in plants and thus facilitates the elucidation of the molecular mechanisms by which auxin controls various developmental processes. The various auxin biosynthetic mutants are also very useful for further dissecting the roles of auxin in plant growth and development. The advancements in auxin biosynthesis reveal that localized auxin biosynthesis provides an essential and effective means to control auxin concentrations in plants.
ACKNOWLEDGEMENT
I would like to thank Dr. Hiroyuki Kasahara, Dr. Kiyoshi Mashiguchi, Dr. Yongxia Guo, Dr. Zuyu Zheng, and members of the Zhao lab for comments on the manuscript. Work in my lab is supported by the NIH (R01GM068631 to Y.Z.) and the NSF (Plant Genome DBI-0820729 to Y. Z.).
Footnotes
Citation: Yunde Zhao. (2014) Auxin Biosynthesis. The Arabidopsis Book 11:e0173. doi:10.1199/tab.0173
elocation-id: e0173
First published on June 13, 2014: e0173. doi: 10.1199/tab.0173
REFERENCES
- Alvarez J.P., Goldshmidt A., Efroni I., Bowman J.L., Eshed Y. The NGATHA distal organ development genes are essential for style specification in Arabidopsis. Plant Cell. 2009;21:1373–1393. doi: 10.1105/tpc.109.065482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballare C.L. Keeping up with the neighbours: phytochrome sensing and other signalling mechanisms. Trends Plant Sci. 1999;4:97–102. doi: 10.1016/s1360-1385(99)01383-7. [DOI] [PubMed] [Google Scholar]
- Barlier I., Kowalczyk M., Marchant A., Ljung K., Bhalerao R., Bennett M., Sandberg G., Bellini C. The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc Natl Acad Sci U S A. 2000;97:14819–14824. doi: 10.1073/pnas.260502697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi J., Lanubile A., Li Q.B., Kumar D., Kladnik A., Cook S.D., Ross J.J., Marocco A., Chourey P.S. Impaired auxin biosynthesis in the defective endosperm18 mutant is due to mutational loss of expression in the ZmYuc1 gene encoding endosperm-specific YUCCA1 protein in maize. Plant Physiol. 2012;160:1318–1328. doi: 10.1104/pp.112.204743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W., Cervera M.T., Delarue M., Beeckman T., Dewitte W., Bellini C., Caboche M., Van Onckelen H., Van Montagu M., Inze D. Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell. 1995;7:1405–1419. doi: 10.1105/tpc.7.9.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt R., Salla-Martret M., Bou-Torrent J., Musielak T., Stahl M., Lanz C., Ott F., Schmid M., Greb T., Schwarz M., Choi S.B., Barton M.K., Reinhart B.J., Liu T., Quint M., Palauqui J.C., Martinez-Garcia J.F., Wenkel S. Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J. 2012;72:31–42. doi: 10.1111/j.1365-313X.2012.05049.x. [DOI] [PubMed] [Google Scholar]
- Brumos J., Alonso J.M., Stepanova A.N. Genetic aspects of auxin biosynthesis and its regulation. Physiol Plant. 2013. doi: 10.1111/ppl.12098. [Epub ahead of print] [DOI] [PubMed]
- Cashman J.R. Structural and catalytic properties of the mammalian flavin-containing monooxygenase. Chem Res Toxicol. 1995;8:166–181. doi: 10.1021/tx00044a001. [DOI] [PubMed] [Google Scholar]
- Cashman J.R., Park S.B., Berkman C.E., Cashman L.E. Role of hepatic flavin-containing monooxygenase 3 in drug and chemical metabolism in adult humans. Chem Biol Interact. 1995;96:33–46. doi: 10.1016/0009-2797(94)03581-r. [DOI] [PubMed] [Google Scholar]
- Chen Q., Dai X., De-Paoli H., Cheng Y., Takebayashi Y., Kasahara H., Kamiya Y., Zhao Y. Auxin Overproduction in Shoots Cannot Rescue Auxin Deficiencies in Arabidopsis Roots. Plant Cell Physiol. 2014. doi: 10.1093/pcp/pcu039. [DOI] [PMC free article] [PubMed]
- Cheng Y., Dai X., Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006;20:1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell. 2007a;19:2430–2439. doi: 10.1105/tpc.107.053009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Qin G., Dai X., Zhao Y. NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc Natl Acad Sci U S A. 2007b;104:18825–18829. doi: 10.1073/pnas.0708506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Qin G., Dai X., Zhao Y. NPY genes and AGC kinases define two key steps in auxin-mediated organogenesis in Arabidopsis. Proc Natl Acad Sci U S A. 2008;105:21017–21022. doi: 10.1073/pnas.0809761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui D., Zhao J., Jing Y., Fan M., Liu J., Wang Z., Xin W., Hu Y. The arabidopsis IDD14, IDD15, and IDD16 cooperatively regulate lateral organ morphogenesis and gravitropism by promoting auxin biosynthesis and transport. PLoS Genet. 2013;9:e1003759. doi: 10.1371/journal.pgen.1003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Mashiguchi K., Chen Q., Kasahara H., Kamiya Y., Ojha S., DuBois J., Ballou D., Zhao Y. The biochemical mechanism of auxin biosynthesis by an arabidopsis YUCCA flavin-containing monooxygenase. J Biol Chem. 2013;288:1448–1457. doi: 10.1074/jbc.M112.424077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies R.T., Goetz D.H., Lasswell J., Anderson M.N., Bartel B. IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell. 1999;11:365–376. doi: 10.1105/tpc.11.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M., Prinsen E., Onckelen H.V., Caboche M., Bellini C. Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 1998;14:603–611. doi: 10.1046/j.1365-313x.1998.00163.x. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Weijers D., Lechner E., Yamada M., Hobbie L., Ehrismann J.S., Jurgens G., Estelle M. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Eklund D.M., Staldal V., Valsecchi I., Cierlik I., Eriksson C., Hiratsu K., Ohme-Takagi M., Sundstrom J.F., Thelander M., Ezcurra I., Sundberg E. The Arabidopsis thaliana STYLISH1 protein acts as a transcriptional activator regulating auxin biosynthesis. Plant Cell. 2010;22:349–363. doi: 10.1105/tpc.108.064816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expósito-Rodríguez M., Borges A.A., Borges-Pérez A., Hernández M., Pérez J.A. Cloning and biochemical characterisation of ToFZY, a tomato gene encoding a flavin monooxygenase involved in a tryptophan -dependent auxin biosynthesis pathway. Journal of plant growth regulation. 2007;26:329–340. [Google Scholar]
- Exposito-Rodriguez M., Borges A.A., Borges-Perez A., Perez J.A. Gene structure and spatiotemporal expression profile of tomato genes encoding YUCCA-like flavin monooxygenases: the ToFZY gene family. Plant Physiol Biochem. 2011;49:782–791. doi: 10.1016/j.plaphy.2011.02.022. [DOI] [PubMed] [Google Scholar]
- Franklin K.A., Lee S.H., Patel D., Kumar S.V., Spartz A.K., Gu C., Ye S., Yu P., Breen G., Cohen J.D., Wigge P.A., Gray W.M. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci U S A. 2011;108:20231–20235. doi: 10.1073/pnas.1110682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A., Barazesh S., Malcomber S., Hall D., Jackson D., Schmidt R.J., McSteen P. sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc Natl Acad Sci U S A. 2008;105:15196–15201. doi: 10.1073/pnas.0805596105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass N.L., Kosuge T. Cloning of the gene for indoleacetic acid-lysine synthetase from Pseudomonas syringae subsp. savastanoi. J Bacteriol. 1986;166:598–603. doi: 10.1128/jb.166.2.598-603.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass N.L., Kosuge T. Role of indoleacetic acid-lysine synthetase in regulation of indoleacetic acid pool size and virulence of Pseudomonas syringae subsp. savastanoi. J Bacteriol. 1988;170:2367–2373. doi: 10.1128/jb.170.5.2367-2373.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W.M., Ostin A., Sandberg G., Romano C.P., Estelle M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci U S A. 1998;95:7197–7202. doi: 10.1073/pnas.95.12.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieneisen V.A., Xu J., Maree A.F., Hogeweg P., Scheres B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature. 2007;449:1008–1013. doi: 10.1038/nature06215. [DOI] [PubMed] [Google Scholar]
- Hamann T., Mayer U., Jurgens G. The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development. 1999;126:1387–1395. doi: 10.1242/dev.126.7.1387. [DOI] [PubMed] [Google Scholar]
- Hardtke C.S., Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Brumos J., Li H., Ji Y., Ke M., Gong X., Zeng Q., Li W., Zhang X., An F., Wen X., Li P., Chu J., Sun X., Yan C., Yan N., Xie D.Y., Raikhel N., Yang Z., Stepanova A.N., Alonso J.M., Guo H. A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell. 2011;23:3944–3960. doi: 10.1105/tpc.111.089029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentrich M., Bottcher C., Duchting P., Cheng Y., Zhao Y., Berkowitz O., Masle J., Medina J., Pollmann S. The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J. 2013;74:626–37. doi: 10.1111/tpj.12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie L., Estelle M. Genetic approaches to auxin action. Plant Cell Environ. 1994;17:525–540. doi: 10.1111/j.1365-3040.1994.tb00147.x. [DOI] [PubMed] [Google Scholar]
- Hou X., Liu S., Pierri F., Dai X., Qu L.J., Zhao Y. Allelic analyses of the Arabidopsis YUC1 locus reveal residues and domains essential for the functions of YUC family of flavin monooxygenases. J Integr Plant Biol. 2011;53:54–62. doi: 10.1111/j.1744-7909.2010.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull A.K., Vij R., Celenza J.L. Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci U S A. 2000;97:2379–2384. doi: 10.1073/pnas.040569997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.I., Sharkhuu A., Jin J.B., Li P., Jeong J.C., Baek D., Lee S.Y., Blakeslee J.J., Murphy A.S., Bohnert H.J., Hasegawa P.M., Yun D.J., Bressan R.A. yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol. 2007;145:722–735. doi: 10.1104/pp.107.104935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.I., Baek D., Park H.C., Chun H.J., Oh D.H., Lee M.K., Cha J.Y., Kim W.Y., Kim M.C., Chung W.S., Bohnert H.J., Lee S.Y., Bressan R.A., Lee S.W., Yun D.J. Overexpression of Arabidopsis YUCCA6 in potato results in high-auxin developmental phenotypes and enhanced resistance to water deficit. Mol Plant. 2012;6:337–349. doi: 10.1093/mp/sss100. [DOI] [PubMed] [Google Scholar]
- Korasick D.A., Enders T.A., Strader L.C. Auxin biosynthesis and storage forms. J Exp Bot. 2013;64:2541–2555. doi: 10.1093/jxb/ert080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriechbaumer V., Wang P., Hawes C., Abell B.M. Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartimentation. Plant J. 2012;70:292–302. doi: 10.1111/j.1365-313X.2011.04866.x. [DOI] [PubMed] [Google Scholar]
- Lai W.G., Farah N., Moniz G.A., Wong Y.N. A Baeyer-Villiger oxidation specifically catalyzed by human flavin-containing monooxygenase 5. Drug Metab Dispos. 2010;39:61–70. doi: 10.1124/dmd.110.035360. [DOI] [PubMed] [Google Scholar]
- LeCLere S., Schmelz E.A., Chourey P.S. Sugar levels regulate tryptophan-dependent auxin biosynthesis in developing maize kernels. Plant Physiol. 2010;153:306–318. doi: 10.1104/pp.110.155226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Ljung K., Breton G., Schmitz R.J., Pruneda-Paz J., Cowing-Zitron C., Cole B.J., Ivans L.J., Pedmale U.V., Jung H.S., Ecker J.R., Kay S.A., Chory J. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012;26:785–790. doi: 10.1101/gad.187849.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.C., Qin G.J., Tsuge T., Hou X.H., Ding M.Y., Aoyama T., Oka A., Chen Z., Gu H., Zhao Y., Qu L.J. SPOROCYTELESS modulates YUCCA expression to regulate the development of lateral organs in Arabidopsis. New Phyto I. 2008;179:751–764. doi: 10.1111/j.1469-8137.2008.02514.x. [DOI] [PubMed] [Google Scholar]
- Li W., Zhou Y., Liu X., Yu P., Cohen J.D., Meyerowitz E.M. LEAFY controls auxin response pathways in floral primordium formation. Sci Signal. 2013;6:ra23. doi: 10.1126/scisignal.2003937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley J.L., Gee C.W., Sairanen I., Ljung K., Nemhauser J.L. An endogenous carbon-sensing pathway triggers increased auxin flux and hypocotyl elongation. Plant Physiol. 2012;160:2261–2270. doi: 10.1104/pp.112.205575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C., Britton J.H., Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell. 1990;2:1071–1080. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Ying Y.Y., Zhang L., Gao Q.H., Li J., Zhang Z., Fang J.G., Duan K. Isolation and characterization of two YUCCA flavin monooxygenase genes from cultivated strawberry (Fragaria x ananassa Duch.). Plant Cell Rep. 2012;31:1425–1435. doi: 10.1007/s00299-012-1258-4. [DOI] [PubMed] [Google Scholar]
- Ljung K. Auxin metabolism and homeostasis during plant development. Development. 2013;140:943–950. doi: 10.1242/dev.086363. [DOI] [PubMed] [Google Scholar]
- Ludwig-Muller J. Auxin conjugates: their role for plant development and in the evolution of land plants. J Exp Bot. 2011;62:1757–1773. doi: 10.1093/jxb/erq412. [DOI] [PubMed] [Google Scholar]
- Mano Y., Nemoto K. The pathway of auxin biosynthesis in plants. J Exp Bot. 2012;63:2853–2872. doi: 10.1093/jxb/ers091. [DOI] [PubMed] [Google Scholar]
- Mashiguchi K., Tanaka K., Sakai T., Sugawara S., Kawaide H., Natsume M., Manada A., Yaeno T., Shirasu K., Yao H., McSteen P., Zhao Y., Hayashi K., Kamiya Y., Kasahara H. The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108:18512–18517. doi: 10.1073/pnas.1108434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield J.A., Frederick R.E., Streit B.R., Wencewicz T.A., Ballou D.P., DuBois J.L. Comprehensive spectroscopic, steady state, and transient kinetic studies of a representative siderophore-associated flavin monooxygenase. J Biol Chem. 2010;285:30375–30388. doi: 10.1074/jbc.M110.157578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen M.D., Naur P., Halkier B.A. Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 2004;37:770–777. doi: 10.1111/j.1365-313x.2004.02002.x. [DOI] [PubMed] [Google Scholar]
- Nishimura T., Hayashi K., Suzuki H., Gyohda A., Takaoka C., Sakaguchi Y., Matsumoto S., Kasahara H., Sakai T., Kato J., Kamiya Y., Koshiba T. Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis. Plant J. 2013;77:352–366. doi: 10.1111/tpj.12399. [DOI] [PubMed] [Google Scholar]
- Normanly J. Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb Perspect Biol. 2010;2:a001594. doi: 10.1101/cshperspect.a001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J., Cohen J.D., Fink G.R. Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc Natl Acad Sci U S A. 1993;90:10355–10359. doi: 10.1073/pnas.90.21.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco-Villalobos D., Sankar M., Ljung K., Hardtke C.S. Disturbed local auxin homeostasis enhances cellular anisotropy and reveals alternative wiring of auxin-ethylene crosstalk in Brachypodium distachyon seminal roots. PLoS Genet. 2012;9:e1003564. doi: 10.1371/journal.pgen.1003564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer W.A., Cheng Y., Murphy A.S. Evidence of oxidative attenuation of auxin signalling. J Exp Bot. 2013;64:2629–2639. doi: 10.1093/jxb/ert152. [DOI] [PubMed] [Google Scholar]
- Pencik A., Simonovik B., Petersson S.V., Henykova E., Simon S., Greenham K., Zhang Y., Kowalczyk M., Estelle M., Zazimalova E., Novak O., Sandberg G., Ljung K. Regulation of auxin homeostasis and gradients in Arabidopsis roots through the formation of the indole-3-acetic acid catabolite 2-oxindole-3-acetic acid. Plant Cell. 2013;25:3858–3870. doi: 10.1105/tpc.113.114421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips K.A., Skirpan A.L., Liu X., Christensen A., Slewinski T.L., Hudson C., Barazesh S., Cohen J.D., Malcomber S., McSteen P. vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell. 2011;23:550–566. doi: 10.1105/tpc.110.075267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon V., Prasad K., Grigg S.P., Sanchez-Perez G.F., Scheres B. Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis. Proc Natl Acad Sci U S A. 2013;110:1107–1112. doi: 10.1073/pnas.1213497110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przemeck G.K., Mattsson J., Hardtke C.S., Sung Z.R., Berleth T. Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta. 1996;200:229–237. doi: 10.1007/BF00208313. [DOI] [PubMed] [Google Scholar]
- Rampey R.A., LeClere S., Kowalczyk M., Ljung K., Sandberg G., Bartel B. A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 2004;135:978–988. doi: 10.1104/pp.104.039677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat R., Schwartz J., Jones M.A., Sairanen I., Cheng Y., Andersson C.R., Zhao Y., Ljung K., Harmer S.L. REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc Natl Acad Sci U S A. 2009;106:16883–16888. doi: 10.1073/pnas.0813035106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano C.P., Robson P.R., Smith H., Estelle M., Klee H. Transgene-mediated auxin overproduction in Arabidopsis: hypocotyl elongation phenotype and interactions with the hy6-1 hypocotyl elongation and axr1 auxin-resistant mutants. Plant Mol Biol. 1995;27:1071–1083. doi: 10.1007/BF00020881. [DOI] [PubMed] [Google Scholar]
- Sairanen I., Novak O., Pencik A., Ikeda Y., Jones B., Sandberg G., Ljung K. Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis. Plant Cell. 2012;24:4907–4916. doi: 10.1105/tpc.112.104794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeno K., Goda H., Ishii T., Ogura T., Tachikawa T., Sasaki E., Yoshida S., Fujioka S., Asami T., Shimada Y. Auxin biosynthesis inhibitors, identified by a genomics-based approach, provide insights into auxin biosynthesis. Plant Cell Physiol. 2010;51:524–536. doi: 10.1093/pcp/pcq032. [DOI] [PubMed] [Google Scholar]
- Sohlberg J.J., Myrenas M., Kuusk S., Lagercrantz U., Kowalczyk M., Sandberg G., Sundberg E. STY1 regulates auxin homeostasis and affects apical-basal patterning of the Arabidopsis gynoecium. Plant J. 2006;47:112–123. doi: 10.1111/j.1365-313X.2006.02775.x. [DOI] [PubMed] [Google Scholar]
- Stepanova A.N., Yun J., Robles L.M., Novak O., He W., Guo H., Ljung K., Alonso J.M. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell. 2011;23:3961–3973. doi: 10.1105/tpc.111.088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A.N., Robertson-Hoyt J., Yun J., Benavente L.M., Xie D.Y., Dolezal K., Schlereth A., Jurgens G., Alonso J.M. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- Stone S.L., Braybrook S.A., Paula S.L., Kwong L.W., Meuser J., Pelletier J., Hsieh T.F., Fischer R.L., Goldberg R.B., Harada J.J. Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: Implications for somatic embryogenesis. Proc Natl Acad Sci U S A. 2008;105:3151–3156. doi: 10.1073/pnas.0712364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L.C., Bartel B. A new path to auxin. Nat Chem Biol. 2008;4:337–339. doi: 10.1038/nchembio0608-337. [DOI] [PubMed] [Google Scholar]
- Sugawara S., Hishiyama S., Jikumaru Y., Hanada A., Nishimura T., Koshiba T., Zhao Y., Kamiya Y., Kasahara H. Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc Natl Acad Sci U S A. 2009;106:5430–5435. doi: 10.1073/pnas.0811226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Qi L., Li Y., Chu J., Li C. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet. 2012;8:e1002594. doi: 10.1371/journal.pgen.1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Ferrer J.L., Ljung K., Pojer F., Hong F., Long J.A., Li L., Moreno J.E., Bowman M.E., Ivans L.J., Cheng Y., Lim J., Zhao Y., Ballare C.L., Sandberg G., Noel J.P., Chory J. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133:164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivendale N.D., Ross J.J., Cohen J.D. The shifting paradigms of auxin biosynthesis. Trends Plant Sci. 2014;19:44–51. doi: 10.1016/j.tplants.2013.09.012. [DOI] [PubMed] [Google Scholar]
- Tivendale N.D., Davies N.W., Molesworth P.P., Davidson S.E., Smith J.A., Lowe E.K., Reid J.B., Ross J.J. Reassessing the role of N-hydroxytryptamine in auxin biosynthesis. Plant Physiol. 2010;154:1957–1965. doi: 10.1104/pp.110.165803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivendale N.D., Davidson S.E., Davies N.W., Smith J.A., Dalmais M., Bendahmane A.I., Quittenden L.J., Sutton L., Bala R.K., Le Signor C., Thompson R., Horne J., Reid J.B., Ross J.J. Biosynthesis of the halogenated auxin, 4-chloroindole-3-acetic acid. Plant Physiol. 2012;159:1055–1063. doi: 10.1104/pp.112.198457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobena-Santamaria R., Bliek M., Ljung K., Sandberg G., Mol J.N., Souer E., Koes R. FLOOZY of petunia is a flavin monooxygenase-like protein required for the specification of leaf and flower architecture. Genes Dev. 2002;16:753–763. doi: 10.1101/gad.219502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigueros M., Navarrete-Gomez M., Sato S., Christensen S.K., Pelaz S., Weigel D., Yanofsky M.F., Ferrandiz C. The NGATHA genes direct style development in the Arabidopsis gynoecium. Plant Cell. 2009;21:1394–1409. doi: 10.1105/tpc.109.065508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., Ahn J.H., Blazquez M.A., Borevitz J.O., Christensen S.K., Fankhauser C., Ferrandiz C., Kardailsky I., Malancharuvil E.J., Neff M.M., Nguyen J.T., Sato S., Wang Z.Y., Xia Y., Dixon R.A., Harrison M.J., Lamb C.J., Yanofsky M.F., Chory J. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcikowska B., Jaskola K., Gasiorek P., Meus M., Nowak K., Gaj M.D. LEAFY COTYLEDON2 (LEC2) promotes embryogenic induction in somatic tissues of Arabidopsis, via YUCCA-mediated auxin biosynthesis. Planta. 2013;238:425–440. doi: 10.1007/s00425-013-1892-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C., Shen X., Mashiguchi K., Zheng Z., Dai X., Cheng Y., Kasahara H., Kamiya Y., Chory J., Zhao Y. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANS-FERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108:18518–18523. doi: 10.1073/pnas.1108436108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward A.W., Bartel B. Auxin: regulation, action, and interaction. Ann Bot. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward C., Bemis S.M., Hill E.J., Sawa S., Koshiba T., Torii K.U. Interaction of auxin and ERECTA in elaborating Arabidopsis inflorescence architecture revealed by the activation tagging of a new member of the YUCCA family putative flavin monooxygenases. Plant physiology. 2005;139:192–203. doi: 10.1104/pp.105.063495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A.D., Sampson M.B., Neuffer M.G., Michalczuk L., Slovin J.P., Cohen J.D. lndole-3-Acetic Acid Biosynthesis in the Mutant Maize orange pericarp, a Tryptophan Auxotroph. Science. 1991;254:998–1000. doi: 10.1126/science.254.5034.998. [DOI] [PubMed] [Google Scholar]
- Yamada M., Greenham K., Prigge M.J., Jensen P.J., Estelle M. The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol. 2009;151:168–179. doi: 10.1104/pp.109.138859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Palm C.J., Brooks B., Kosuge T. Nucleotide sequences of the Pseudomonas savastanoi indoleacetic acid genes show homology with Agrobacterium tumefaciens T-DNA. Proc Natl Acad Sci U S A. 1985;82:6522–6526. doi: 10.1073/pnas.82.19.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Kamiya N., Morinaka Y., Matsuoka M., Sazuka T. Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol. 2007;143:1362–1371. doi: 10.1104/pp.106.091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T., Ito M., Sumikura T., Nakayama A., Nishimura T., Kitano H., Yamaguchi I., Koshiba T., Hibara K.I., Nagato Y., Itoh J.I. The rice FISH BONE gene encodes a tryptophan aminotransferase, which affects pleiotropic auxin-related processes. Plant J. 2014. doi: 10.1111/tpj.12517. [Epub ahead of print] [DOI] [PubMed]
- Zhao Y. Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol. 2010;61:49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol Plant. 2013;5:334–338. doi: 10.1093/mp/ssr104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Christensen S.K., Fankhauser C., Cashman J.R., Cohen J.D., Weigel D., Chory J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Hull A.K., Gupta N.R., Goss K.A., Alonso J., Ecker J.R., Normanly J., Chory J., Celenza J.L. Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 2002;16:3100–3112. doi: 10.1101/gad.1035402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Zhang Y., Liu X., Zhang X., Liu S., Yu X., Ren Y., Zheng X., Zhou K., Jiang L., Guo X., Gai Y., Wu C., Zhai H., Wang H., Wan J. A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev Cell. 2013;27:113–122. doi: 10.1016/j.devcel.2013.09.005. [DOI] [PubMed] [Google Scholar]
- Zheng Z., Guo Y., Novak O., Dai X., Zhao Y., Ljung K., Noel J.P., Chory J. Coordination of auxin and ethylene biosynthesis by the aminotransferase VAS1. Nat Chem Biol. 2013;9:244–246. doi: 10.1038/nchembio.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler D.M. Flavin-containing monooxygenases: catalytic mechanism and substrate specificities. Drug Metab Rev. 1988;19:1–32. doi: 10.3109/03602538809049617. [DOI] [PubMed] [Google Scholar]
- Ziegler D.M. An overview of the mechanism, substrate specificities, and structure of FMOs. Drug Metab Rev. 2002;34:503–511. doi: 10.1081/dmr-120005650. [DOI] [PubMed] [Google Scholar]
- Zolman B.K., Yoder A., Bartel B. Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics. 2000;156:1323–1337. doi: 10.1093/genetics/156.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B.K., Martinez N., Millius A., Adham A.R., Bartel B. Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics. 2008;180:237–251. doi: 10.1534/genetics.108.090399. [DOI] [PMC free article] [PubMed] [Google Scholar]