Abstract
Chromosomal rearrangements account for all erythroblast transformation specific (ETS) family member gene fusions that have been reported in prostate cancer and have clinical, diagnostic and prognostic implications. Androgen-regulated genes account for the majority of the 5’ genomic regulatory promoter elements fused with ETS genes. TMPRSS2-ERG, TMPRSS2-ETV1 and SLC45A3-ERG rearrangements account for roughly 90% of ETS fusion prostate cancer. ELK4, another ETS family member, is androgen-regulated, involved in promoting cell growth, and highly expressed in a subset of prostate cancer, yet the mechanism of ELK4 over-expression is unknown. In this study, we identified a novel ETS family fusion transcript, SLC45A3-ELK4, and found it to be expressed in both benign prostate tissue and prostate cancer. We found high levels of SLC45A3-ELK4 mRNA restricted to a subset of prostate cancer samples. SLC45A3-ELK4 transcript can be detected at high levels in urine samples from men at risk for prostate cancer. Characterization of the fusion mRNA revealed a major variant in which SLC45A3 exon 1 is fused to ELK4 exon 2. Based on quantitative PCR analyses of DNA, unlike other ETS fusions described in prostate cancer, the expression of SLC45A3-ELK4 mRNA is not exclusive to cases harbouring a chromosomal rearrangement. Treatment of LNCaP cancer cells with a synthetic androgen (R1881) revealed that SLC45A3-ELK4, and not endogenous ELK4, mRNA expression is androgen-regulated. Altogether, our findings show that SLC45A3-ELK4 mRNA expression is heterogeneous, highly induced in a subset of prostate cancers, androgen-regulated, and most commonly occurs through a mechanism other than chromosomal rearrangement (e.g., trans-splicing).
Keywords: prostate cancer, ETS genes, splicing, SLC45A3, ELK4
INTRODUCTION
Emerging data suggests that ETS rearranged prostate cancer, similar to other translocation tumors, represents a distinct subclass of prostate cancer based on studies demonstrating varying morphologic features (1), survival (2, 3), and a specific expression profile (4, 5). Androgen-regulated genes account for the majority of the 5’ genomic regulatory promoters elements fused with ETS genes in prostate cancer (6). The promoter of the androgen-regulated transmembrane protease, serine 2 (TMPRSS2) gene is fused to the coding region of members of the ETS family of transcription factors, most commonly v-ets erythroblastosis virus E26 oncogene homolog (avian) (ERG) (7). SLC45A3 (solute carrier family 45, member 3), also referred to as prostein, is a prostate-specific, androgen-regulated gene that has been shown to be a 5’ partner with ETV1 and ETV5 (6, 8) and more recently with the coding sequence of ERG (9).
Interestingly, ELK4 (ETS-domain protein (SRF accessory protein 1), a member of the ETS family of transcription factors, has recently been described as a novel androgen receptor target in LNCaP cells promoting cell growth and is highly expressed in a subset of prostate cancer samples in comparison to benign prostate tissues. (10). Herein we report the expression of novel SLC45A3-ELK4 transcripts in prostate cancer. We provide data characterizing different SLC45A3-ELK4 mRNA variants and evidence that this transcript does not primarily arise from a chromosomal rearrangement as seen for other ETS fusion events in prostate cancer.
MATERIALS AND METHODS
Sample preparation
Tissue Samples were collected as part of an Institutional Review Board approved protocol and RNA was extracted (See supplemental methods).
Conventional RT-PCR Sequencing
RT-PCR was performed using primers to SLC45A3 exon 1 (5-CCGCGGAGTAACCTGGAGATTT-3) and ELK4 exon 2 (5-TGCCCATCATTAGAGGTCCAACAG-3, see Supplemental methods for details).
Quantitative RT-PCR using Taqman technology
We used TaqMan Gene Expression Assays (See Supplemental methods for details).
Chromosome 1q32, SLC45A3 to ELK4 region assessment was performed using qPCR and primers specific 13 regions on chrom.1 (See Supplemental methods for details).
Functional studies were performed using R1881 on LNCaP cells (See Supplemental Methods for details).
RESULTS
SLC45A3-ELK4 mRNA is expressed in prostate cancer, benign prostate tissue and the LNCaP cancer line
We developed a Taqman assay targeting SLC45A3 exon 1 and ELK4 exon 2 (Figure 1). We initially screened RNA from 31 prostate cancer samples, 6 benign prostate tissue samples and 11 cell lines including malignant prostate (LNCaP, PC-3, 22Rv1, VCaP, NCI-H660, DU-145), non-prostate (ACHN, Caki-1, A-498, HK-2) and non-malignant prostate (RWPE-1) epithelial cell lines. All samples yielded detectable, albeit low SLC45A3-ELK4 mRNA transcript expression levels (Figure 2a). Three prostate cancer samples demonstrated high SCL45A3-ELK4 expression with levels greater than 10-fold over the median level calculated from benign prostate tissue. Levels of endogenous ELK4 mRNA varied widely in all prostate samples tested. While we found a good overall correlation between endogenous ELK4 mRNA and SLC45A3-ELK4 mRNA levels (r = 0.86), several samples yielded significantly different expression values between the 2 transcripts. We also found relative increased levels of SLC45A3-ELK4 in PC-3 and LNCaP cells and the human epithelial-like kidney adenocarcinoma cell line ACHN.
Figure 1.
Schematic of chromosome 1q32.1 (Chr.1q32.1) demonstrating the orientation and relative distance of SLC45A3 and ELK4. Red arrows and bar indicate the SLC45A3-ELK4 Taqman assay primers and probe, respectively.
Figure 2.
Taqman expression data of SLC45A3-ELK4 and ELK4 mRNA levels in 31 prostate cancer samples (red bar) relative median of the values obtained from the 6 benign samples (green bar) in which cases yielding higher than 10 fold relative SLC45A3-ELK4 mRNA levels are indicated in dark red; and 10 cell lines (9 cancer and 1 benign, HK-2) relative to RWPE-1 (purple) (a). Schematic of the sequencing results obtained from PCR (primers are indicated in red) and 5’RACE (primer indicate in blue) that correspond to the different SLC45A3-ELK4 mRNA variants (v, see Supplemental Information for junction sequence, b). Taqman assay results from RNA extracted from 5 samples (C08, C03, C33 and C30 corresponding to cancer positive biopsies and C13 corresponded to cancer negative biopsy (c)). Contingency table of the 14 samples that yielded adequate TCFL1 values (inset).
SLC45A3-ELK4 mRNA variants in prostate cancer
We performed conventional RTPCR followed by cDNA sequencing to characterize the composition of the SLC45A3-ELK4 transcripts of amplified products obtained from 35 prostate cancer samples, 6 benign samples, 6 prostate cancer cell lines and 1 benign cell line (Supplemental Figure 1). Given the lower sensitivity of this approach only the majority of the samples yielded a major product that consisted of SLC45A3 exon 1 fused to ELK4 exon 2 (Figure 2b, see Supplemental Information for junction sequence). Three less common products were detected consisting of a portion of SLC45A3 exon 2 fused to ELK4 exon 2. Interestingly, we found one amplified product that consisted of 84 base pairs of intergenic sequence separating SLC45A3 exons 1, 2, to ELK4 exon 2. Using the unbiased approach 5’ RNA ligase-mediated rapid amplification of cDNA ends (RACE), we confirmed another SLC45A3-ELK4 mRNA variant consisting of SLC45A3 exons 1–3 fused to the same 84-bp sequence described above followed by ELK4 exon 2 in sample 1701_A.
SLC45A3-ELK4 mRNA can be detected using a non-invasive assay
We screened 14 pre-biopsy, post-digital exam urine specimens from men who were at risk for having prostate cancer using our SLC45A3-ELK4 Taqman assay. According to pathology reports of the biopsied prostate tissue, 8 out of the 14 were diagnosed with prostate cancer (Figure 2c). Detectable levels of SLC45A3-ELK4 transcript were measured in 6 out of 8 corresponding urine specimens and 2 out of the 6 specimens from men whose biopsies did not reveal prostate cancer (sensitivity of 75% and a specificity of 67%). Interestingly, as seen in the prostate tissue, high levels (> 10-fold) were detected in only a few of the prostate cancer-associated samples.
Chromosome rearrangement does not account for SLC45A3-ELK4 expression
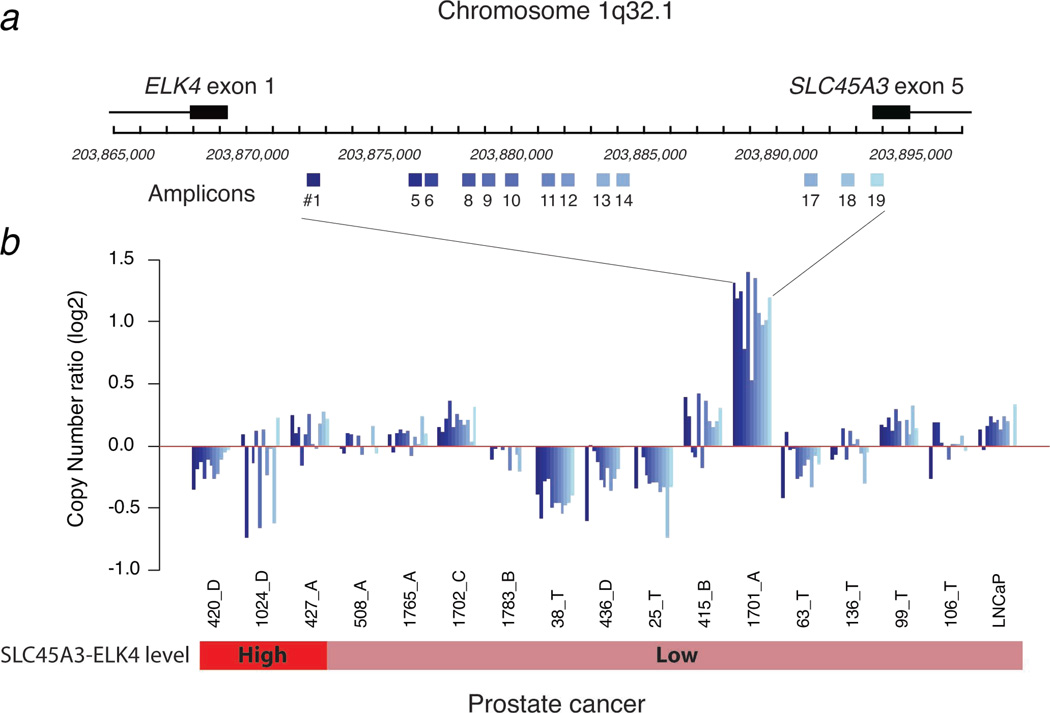
The development of a standard FISH break-apart assay requires using BACs which usually span 100–150 kb. The distance from SLC45A3 to ELK4 is 25 kb and thus was not suitable for detecting a possible deletion between these genes (Supplemental Figure 2). To explore for genomic loss within the region separating SLC45A3 and ELK4, we analyzed 13 loci on chromosome covering this region (Figure 3). The resulting amplicon raw data was normalized to a region on chromosome 1 (within ARHGEF) that is not altered from HapMap SNP data (11). Deletion or partial deletion of this region was observed in several samples with both high (420_D and 1024_D) and low (38_T, 436_D and 25_T) SLC45A3-ELK4 transcript levels. The majority of samples were assessed as copy number neutral or demonstrated genomic gain in this region. This included 1 sample (427_A) with high levels of SLC45A3-ELK4 mRNA but copy number neutral and 1 sample (1701_A) that had low SLC45A3-ELK4 mRNA and high DNA amplification in this region. Taken together, we did not observe a consistent loss of genomic DNA in cases with SLC45A3-ELK expression.
Figure 3.
Schematic of the region Chr.1q32 demonstrating the position of the primer pairs (blue boxes, a). SLC45A3 exon 5 and ELK4 exon 1 positions are indicated. qPCR results obtained for 16 prostate cancer samples (ordered from left to right as a function of the level of SLC45A3-ELK4 mRNA levels) and from LNCaP cells (b). Colored bars indicates samples with over 10-fold higher (red) or benign-like (light red) SLC45A3-ELK4 mRNA levels. All qPCR experiments were run in triplicate. Bars indicate the average calibrated values.
SLC45A3-ELK4 is androgen-regulated
To address the confounding results from Makkonnen et al (10), who recently reported that ELK4 is a novel androgen receptor target in LNCaP cells, we repeated their experiment using our assay for the SLC45A3-ELK4 transcript in addition to an assay for ELK4 that does not target the fusion transcript. As anticipated, twelve hours following treatment with a synthetic androgen (R1881, 1nM), we observed a 25-fold induction of SLC45A3-ELK4 but no change in ELK4 (Figure 4). This induction was abrogated in the presence of the androgen antagonist Flutamide. As a control, we also measured the levels of KLK3 (PSA) mRNA and observed a similar profile.
Figure 4.
Median fold induction of SLC45A3-ELK4 (a), ELK4 (b) and KLK3 (PSA, c) mRNA in LNCaP cells treated with 1nM R1881 in the absence or presence of 10 µM flutamide at the indicated time points. All experiments were run in triplicate (SEM indicated by the error bars).
DISCUSSION
This is the first description of novel multiple ETS family fusion (SLC45A3-ELK4) transcripts, high levels of which are restricted to a subset of prostate cancer samples. Characterization of the fusion mRNA revealed a major variant in which SLC45A3 exon 1 is fused to ELK4 exon 2. Other minor variants include other downstream exons of both genes and more interestingly, an 84-bp chromosome sequence separating the two genes. Chromosome 1q32.1 has been cited as a region that is involved in chromosome loss in prostate cancer (12). Interestingly, exon 1 of SLC45A3 is located roughly 50 Kb telomeric on 1q32.1 from ELK4 exon 2 and is transcribed in the same direction. We observed a chromosome deletion of the interstitial region separating TMPRSS2 and ERG in 60% of TMPRSS2-ERG fusion prostate cancers (13). However, our data suggests that this is a less common event and that the expression of SLC45A3-ELK4 fusion transcript may more commonly occur through another mechanism. This is consistent with a recent report analyzing ETS genes and known 5’ fusion partners using FISH (9).
Chimeric mRNA resulting from trans-splicing has been observed between premRNAs from the same gene (homotypic trans-splicing) (14) and pre-mRNAs from different genes (intergenic trans-splicing) (15) as well as reported from computational analyses (16). We have also noted numerous trans-splicing events in RNA-sequencing data generated thus far on prostate cancer samples (MA Rubin, unpublished observations).
In prostate cancer cases with known TMPRSS2-ERG or SLC45A3-ERG fusions, we do not see a mutually exclusive expression of SLC45A3-ELK4 as observed with the other prostate cancer fusions (7). Interestingly, the 3 samples that yielded high SLC45A3-ELK4 transcript levels were negative for ERG rearrangement by FISH.
Makkonen et al. reported an induction of ELK4 mRNA variants upon androgen stimulation which was most pronounced in metastatic, hormone-refractory prostate cancer. Our current study confirms that ELK4 is over-expressed in prostate cancer but only in a subset of tumors and correlated with high SLC45A3-ELK4 mRNA. Only SLC45A3-ELK4 mRNA, and not endogenous ELK4 mRNA, is up-regulated upon treatment of LNCaP cells with R1881. This finding suggests that Makkonen et al. were measuring in SLC45A3-ELK4 expression and not the wild type ELK4. This has direct implications to the putative oncogenic properties of ELK4. Recent work from our group identified an estrogen-mediated activation of TMPRSS2-ERG transcription (4). Non-androgen regulated mechanisms for SLC45A3 will also need to be explored in future work. Based on our data, we found increased levels of SLC45A3-ELK4 in the human epithelial-like kidney adenocarcinoma cell line ACHN. This suggests that this transcript is not prostate specific. Large-scale screening of multiple cancer and non-cancer tissue types is needed to determine the specificity and the extent of SLC45A3-ELK4 expression.
Finally, the SLC45A3-ELK4 fusion transcript may also have potential clinical applications. Similar to TMPRSS2-ERG and PCA3 (17, 18), SLC45A3-ELK4 is detected at higher levels in prostate cancer than in benign prostate tissue. We demonstrated a proof-of-principle that SLC45A3-ELK4 transcript can be detected at high levels urine samples from patients with prostate cancer and therefore might be a useful biomarker. Clearly, larger cohorts of urine samples will need to analyzed to be able to determine if detection of this transcript will provide added diagnostic utility.
In summary, we describe for the first time novel fusion events between the androgen regulated 5’ prime promoter, SLC45A3, and the ETS gene, ELK4. We were unable to confirm if some of these fusions are due to genomic rearrangement or if trans-splicing explains this phenomenon. A combination of the two mechanisms is also possible. The SLC45A3-ELK4 transcripts can be detected in a range of prostate cancers and benign prostate tissues but due to higher levels in cancer, we also propose SLC45A3-ELK4 transcripts as a putative prostate cancer biomarker. The biologic implications of this fusion transcript are yet to be determined.
Supplementary Material
ACKNOWLEMENTS
National Institutes of Health (NIH)/National Cancer Institute grant R01 CA125612-01 (M. A. Rubin and F Demichelis), and the Heinrich Warner Stiftung (D. Pflueger).
Footnotes
While this manuscript was in review, a paper reported the presence of the SLC45A3-ELK4 transcripts in prostate cancer tumor cell lines and metastatic samples (Maher CA, et al. Nature 2009).
REFERENCES
- 1.Mosquera JM, Perner S, Demichelis F, et al. Morphological features of TMPRSS2-ERG gene fusion prostate cancer. J Pathol. 2007;212:91–101. doi: 10.1002/path.2154. [DOI] [PubMed] [Google Scholar]
- 2.Attard G, Clark J, Ambroisine L, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27:253–263. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demichelis F, Fall K, Perner S, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–4599. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 4.Setlur SR, Mertz KD, Hoshida Y, et al. Estrogen-Dependent Signaling in a Molecularly Distinct Subclass of Aggressive Prostate Cancer. J Natl Cancer Inst. 2008;100:815–825. doi: 10.1093/jnci/djn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomlins SA, Rhodes DR, Yu J, et al. The role of SPINK1 in ETS rearrangement-negative prostate cancers. Cancer Cell. 2008;13:519–528. doi: 10.1016/j.ccr.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomlins SA, Laxman B, Dhanasekaran SM, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 7.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 8.Helgeson BE, Tomlins SA, Shah N, et al. Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer Res. 2008;68:73–80. doi: 10.1158/0008-5472.CAN-07-5352. [DOI] [PubMed] [Google Scholar]
- 9.Han B, Mehra R, Dhanasekaran SM, et al. A fluorescence in situ hybridization screen for E26 transformation-specific aberrations: identification of DDX5-ETV4 fusion protein in prostate cancer. Cancer Res. 2008;68:7629–7637. doi: 10.1158/0008-5472.CAN-08-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makkonen H, Jaaskelainen T, Pitkanen-Arsiola T, et al. Identification of ETS-like transcription factor 4 as a novel androgen receptor target in prostate cancer cells. Oncogene. 2008 doi: 10.1038/onc.2008.125. [DOI] [PubMed] [Google Scholar]
- 11.McCarroll SA, Kuruvilla FG, Korn JM, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 12.Wolf M, Mousses S, Hautaniemi S, et al. High-resolution analysis of gene copy number alterations in human prostate cancer using CGH on cDNA microarrays: impact of copy number on gene expression. Neoplasia. 2004;6:240–247. doi: 10.1593/neo.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perner S, Demichelis F, Beroukhim R, et al. TMPRSS2:ERG Fusion-Associated Deletions Provide Insight into the Heterogeneity of Prostate Cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 14.Takahara T, Tasic B, Maniatis T, Akanuma H, Yanagisawa S. Delay in synthesis of the 3' splice site promotes trans-splicing of the preceding 5' splice site. Mol Cell. 2005;18:245–251. doi: 10.1016/j.molcel.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 15.Pradet-Balade B, Medema JP, Lopez-Fraga M, et al. An endogenous hybrid mRNA encodes TWE-PRIL, a functional cell surface TWEAK-APRIL fusion protein. EMBO J. 2002;21:5711–5720. doi: 10.1093/emboj/cdf565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akiva P, Toporik A, Edelheit S, et al. Transcription-mediated gene fusion in the human genome. Genome Res. 2006;16:30–36. doi: 10.1101/gr.4137606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hessels D, Smit FP, Verhaegh GW, Witjes JA, Cornel EB, Schalken JA. Detection of TMPRSS2-ERG fusion transcripts and prostate cancer antigen 3 in urinary sediments may improve diagnosis of prostate cancer. Clin Cancer Res. 2007;13:5103–5108. doi: 10.1158/1078-0432.CCR-07-0700. [DOI] [PubMed] [Google Scholar]
- 18.Laxman B, Morris DS, Yu J, et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008;68:645–649. doi: 10.1158/0008-5472.CAN-07-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.