SUMMARY
Mutations in KRAS are prevalent in human cancers and universally predictive of resistance to anti-cancer therapeutics. Although it is widely accepted that acquisition of an activating mutation endows RAS genes with functional autonomy, recent studies suggest that the wild-type forms of Ras may contribute to mutant Ras-driven tumorigenesis. Here we show that downregulation of wild-type H-Ras or N-Ras in mutant K-Ras cancer cells leads to hyperactivation of the Erk/p90RSK and PI3K/Akt pathways, and consequently, the phosphorylation of Chk1 at an inhibitory site, Ser 280. The resulting inhibition of ATR/Chk1 signaling abrogates the activation of the G2 DNA damage checkpoint and confers specific sensitization of mutant K-Ras cancer cells to DNA damage chemotherapeutic agents in vitro and in vivo.
INTRODUCTION
In mammals three closely related RAS oncogenes, HRAS, NRAS, and KRAS, have been identified. These genes encode small GTPases that function as molecular switches governing the activation of a vast network of signaling pathways. Growth factor signaling activates Ras by recruiting guanine nucleotide exchange factors (GEFs) that catalyze the exchange of GDP for GTP (Bos et al., 2007). In turn, Ras activity is terminated through GTP hydrolysis which is greatly enhanced by GTPase accelerating proteins (GAPs). Hyperactivation of Ras, which largely occurs through the acquisition of mutations that hinder GTP hydrolysis, has been implicated in the etiology of a wide number of human cancers. Overall, mutations in the RAS genes have been associated with ~30% of all human tumors. Such mutations are generally limited to one of the RAS genes, with KRAS being the most frequently mutated and with the highest incidence in adenocarcinomas of the pancreas (57%), colon (33%), and lung (17%) (Pylayeva-Gupta et al., 2011).
The critical role of oncogenic K-Ras as a driving mutation in the pathogenesis of cancer is supported by several genetically engineered mouse models. Accordingly, expression of mutant K-Ras alone is sufficient to drive malignant progression, whereas its elimination from established tumors leads to tumor regression (Chin et al., 1999; Fisher et al., 2001; Haigis et al., 2008; Jackson et al., 2001; Li et al., 2011; Ying et al., 2012). Because of its capacity to constitutively engage downstream effector pathways, oncogenic K-Ras was initially thought to drive the tumorigenic process independently of the wild-type forms. However, it is becoming increasingly evident that the biological outputs of oncogenic K-Ras are subject to a complex and context-dependent modulation by wild-type Ras proteins. Studies in chemically-induced models of lung or skin tumorigenesis have demonstrated that the acquisition of an activating mutation in a KRas or HRas allele is associated with allelic loss of the KRas wild-type or HRas wild-type allele, respectively (Bremner and Balmain, 1990; Hegi et al., 1994; Zhang et al., 2001). Zhang et al. further demonstrated that loss of the wild-type KRas allele enhanced mutant K-Ras driven tumorigenesis (Zhang et al., 2001). Together these results suggest a tumor suppressive effect of the wild-type Ras allele. Conversely, a recent study reported that in mutant K-Ras-driven colorectal cancer, wild-type K-Ras plays a tumor promoting role through counteracting mutant K-Ras-induced apoptosis by mediating signaling from mutant K-Ras-dependent autocrine-activated EGFR (Matallanas et al., 2011).
Mutant K-Ras-driven cancers also retain the wild-type products of the remaining RAS genes, H- and NRAS, which appear to synergize with mutant K-Ras in tumors of various tissues. For example, enhanced levels of GTP-bound H-Ras and N-Ras, due to mutant K-Ras dependent nitrosylation of wild-type H- and N-Ras, were shown to be required for the proliferation of mutant K-Ras cancer cells (Lim et al., 2008). A role for wild-type H-Ras and N-Ras proteins in mediating RTK signaling and proliferation of cancer cells that harbor mutant K-Ras has also been demonstrated (Young et al., 2012). Moreover, SON OF SEVENLESS (SOS), a guanine nucleotide exchange factor (GEF) for Ras and Rho GTPases has been implicated in mutant Ras-driven tumorigenesis (Jeng et al., 2012).
In the current study we sought to determine the mechanisms by which wild-type H-Ras and N-Ras proteins promote the mutant K-Ras-driven tumorigenic phenotype.
RESULTS
Mutant K-Ras cancer cells require wild-type H-Ras for proliferation and progression through mitosis
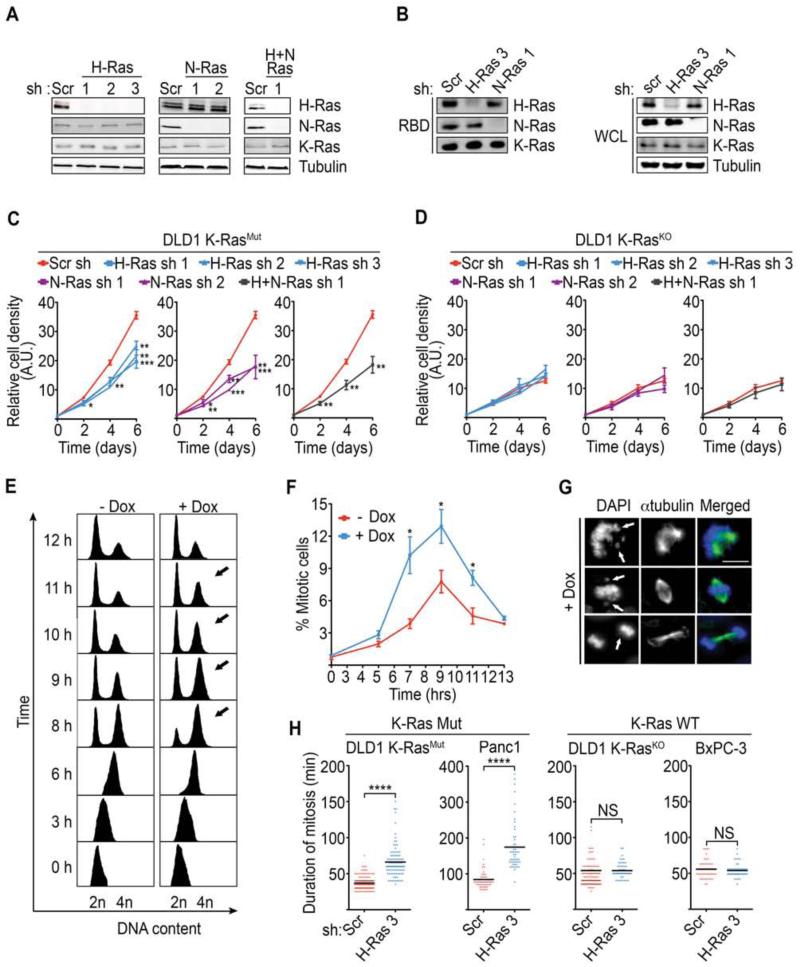
To investigate the functional relationship between mutant K-Ras and wild-type (WT) H/N-Ras, we took the approach of specifically suppressing the expression of WT-H-Ras and/or WT-N-Ras, in cancer cells positive or negative for mutant K-Ras. To this end, we initially employed the isogenic colon cancer cells DLD-1 K-RasWT/G12D (DLD1 K-RasMut) and DLD1 K-RasWT/- (DLD1 K-RasKO) where the K-RASG12D allele has been knocked out by homologous recombination (Luo et al., 2009a; Shirasawa et al., 1993). These cell lines were engineered to harbor doxycycline (Dox)-inducible shRNAs directed at H-Ras, N-Ras, or both H- and N-Ras. Accordingly, doxycycline treatment specifically suppressed expression and activity of the targeted isoforms, with no effect on the remaining isoforms (Figure 1A-1B and Figure S1A). As shown in Figure 1C individual knockdown of WT-H-Ras or WT-N-Ras in DLD1 K-RasMut cells led to slower growth. Of note, no synergy was observed upon knockdown of both WT-H-Ras and WT-N-Ras suggesting that the two WT-isoforms converge on the same signaling module that regulates growth of DLD1 K-RasMut cells (Figure 1C). In contrast, knockdown of either WT-H-Ras or WT-N-Ras, or the two combined, in DLD1 K-RasKO cells, had no effect on cell growth indicating that the dependence on WT-H- and/or N-Ras for cell growth is a unique property of mutant K-Ras cancer cells (Figure 1D and Figure S1A).
Figure 1. WT-H-Ras knockdown perturbs the mitotic progression of K-Ras mutant cancer cells.
(A) Isoform specific knockdown of WT-H-Ras, WT-N-Ras, or WT-H- and N-Ras combined. DLD1 K-RasMut cells that harbor doxycycline inducible shRNAs directed at H-Ras (H-Ras sh 1, 2, 3), N-Ras (N-Ras sh 1, 2), H-Ras and N-Ras combined (H-Ras sh 1 and N-Ras sh 1), or scramble shRNA (Scr sh), were treated with doxycycline for 96 hr. Whole cell lysates (WCL) were immunoblotted for H-Ras, N-Ras, K-Ras, and tubulin (loading control).
(B) Effect of WT-H-Ras or WT-N-Ras suppression on the GTP-bound status of the remaining Ras isoforms. WCLs were subjected to GST-bound CRAF Ras-binding domain (RBD) pulldowns and immunoblotted with the indicated antibodies.
(C-D) Effect of suppression of WT-H-Ras, WT-N-Ras, or WT-H- and N-Ras combined on the proliferation of DLD1 K-RasMut cells (C) and DLD1 K-RasKO cells (D). The relative cell density was measured using a Syto60 stain and is expressed in arbitrary units (A.U.).
(E) Representative FACS histograms showing cell cycle progression of synchronized control (-Dox) and H-Ras-suppressed (+Dox) DLD1 K-RasMut cells that harbor inducible H-Ras sh 3. Cells were released from a double thymidine block, fixed at the indicated time points, and FACS sorted for DNA content. Arrows indicate the G2/M fraction at relevant time points. Data are representative of at least 3 independent experiments.
(F) Detection of mitotic fraction by FACS analysis of phosphorylated histone H3 (P-HH3) positive cells in double-thymidine-release experiments as in (E).
(G) Representative images of mitotic aberrations (arrows) seen in WT-H-Ras-depleted DLD1 K-RasMut cells following a double-thymidine-release experiment as in (E). Immunofluorescence images of cells stained for α-tubulin and DNA. Scale bar, 10μm.
(H) Scatter plots show the duration of mitosis, as determined by phase-contrast time-lapse microscopy, in asynchronous K-Ras Mut (DLD1 K-RasMut cells and Panc-1) and K-Ras WT (DLD1 K-RasKO and BxPC-3) cancer cells expressing scramble or H-Ras shRNAs. ****p<0.0001 by student’s t-test; NS, no significant difference.
All experiments: error bars, mean ± SEM, n=3, *p<0.05, **p<0.005, ***p<0.0005. See also Movie S1, Movie S2, and Figure S1.
We next investigated whether the attenuated cell growth observed upon WT-H-Ras and/or N-Ras knockdown in DLD1 K-RasMut cells could be the result of a slower progression through the cell cycle. Initially, we examined the cell cycle progression of WT-H-Ras-suppressed DLD1 K-RasMut cells that were synchronized at the G1/S border by double thymidine treatment. Six hours after release, both WT-H-Ras-suppressed (+Dox) and WT-H-Ras-intact (-Dox) DLD1 K-RasMut cells had completed replication and were predominantly in G2 as determined by the accumulation of cells with 4N DNA content (Figure 1E-1F and Figure S1B). However, whereas the majority of WT-H-Ras-intact cells completed mitosis and cell division and reached G1 over the next 4 hours, WT-H-Ras-suppressed cells showed a delayed transition through G2/M, as evident by the persistence of cells with 4N DNA content (Figure 1E, arrows). FACS analysis of phospho-histone H3 positive cells revealed an increased mitotic index of WT-H-Ras-suppressed cells relative to control (7 hr - 11 hr), suggesting that the elevated fraction of 4N DNA content cells associated with WT-H-Ras knockdown was due to a mitotic delay (Figure 1E-1F). Consistent with this interpretation, WT-H-Ras-suppressed DLD1 K-RasMut cells showed mitotic defects that would preclude the timely satifaction of the spindle checkpoint including misaligned and damaged chromosomes (Figure 1G). To quantitatively measure the mitotic delay within individual cells and rule out the possibility that the observed mitotic aberrations were due to the synchronization technique, we analyzed the duration of mitosis in asynchronous cells by time-lapse phase-contrast microscopy. We observed that WT-H-Ras knockdown delayed mitotic progression in DLD1 K-RasMut cells (duration of mitosis was ~2 times longer compared to control) but not in DLD1 K-RasKO cells (Figure 1H and Figure S1C-S1D). Similar results were obtained when this analysis was extended to the pancreatic cell line pair Panc-1 (K-Ras mutant) and BxPC-3 (K-Ras WT) (Figure 1H, Movie S1, Movie S2, and Figure S1C-S1D). Taken together these data indicate that K-Ras mutant cells specifically require WT-H-Ras for the timely progression of mitosis.
Wild-type H/N-Ras knockdown enhance DNA damage in mutant K-Ras cancer cells
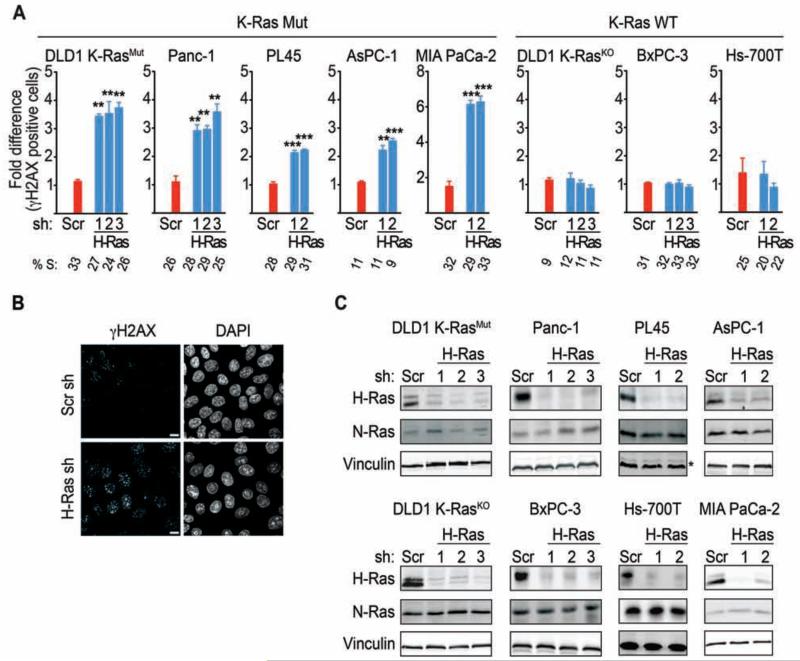
In principle, the delay in mitosis and damaged chromosomes, induced by WT-H-Ras knockdown, can be explained by misregulation of the DNA damage response (DDR) (Rieder and Maiato, 2004; Brown and Baltimore, 2000; Lam et al., 2004; Loffler et al., 2006; Zachos and Gillespie, 2007). A defective DDR would compromise the ability of the cell to repair DNA damage thereby leading to an enhancement in the levels of DNA strand breaks (Syljuasen et al., 2005; Toledo et al., 2011b). Evaluation of DNA strand breaks by monitoring γH2AX staining revealed a significant accumulation of γH2AX-positive cells upon knockdown of WT-H-Ras in the DLD1-K-RasMut cells and a panel of K-Ras mutant (Mut) pancreatic cancer cells (Panc-1, AsPC-1, PL45, MIA PaCa-2) (Figure 2A-2C). In contrast, γH2AX levels remain unaltered when WT-H-Ras was knocked down in the DLD1-K-RasKO and K-Ras WT pancreatic cancer cell lines (BxPC-3 and Hs-700T), consistent with K-Ras Mut cancer cells being uniquely dependent on WT-H-Ras for modulating DNA damage and cell cycle progression (Figure 2A-2C). A similar dependency was observed upon knockdown of WT-N-Ras, and no synergistic effect was observed when both WT-H-Ras and WT-N-Ras were knocked down (Figure S2A-S2C). The elevated γH2AX levels were not due to an accumulation of cells in S-phase because there was no significant difference in the S-phase profiles between control and WT-H-Ras, WT-N-Ras, or WT-H- and N-Ras-suppressed cells (Figure 2A and Figure S2B). Of note, we have observed no correlation between enhanced γH2AX levels due to WT-H- and/or N-Ras knockdown and the proliferative rate or basal γH2AX levels of the cancer cells analyzed (Figure S1D and data not shown). This suggests that the elevated DNA damage induced by WT-H- and/or N-Ras knockdown is not due to a faster proliferation rate or higher basal DNA damage of K-Ras mutant cells, but instead is a consequence of a perturbed DDR. Moreover, knockdown of WT-H-Ras in melanoma cells that harbor an activating N-Ras mutation (NRas Q61L) also enhanced γH2AX levels, indicating that WT-Ras could be required for the regulation of the basal levels of DNA damage in cancer cells with activating mutations in any of the Ras isoforms (Figure S2E).
Figure 2. WT-H-Ras knockdown enhances DNA damage in K-Ras Mut but not in K-Ras WT cancer cells.
(A) Quantification of γH2AX positive cells in K-Ras Mut and K-Ras WT cancer cells upon WT-H-Ras knockdown. K-Ras Mut and K-Ras WT cells from cancers of the colon (DLD1) and pancreas (Panc-1, AsPC-1, PL45, MIA PaCa-2, BxPC-3, Hs-700T), depleted of WT-H-Ras, were co-stained for γH2AX and DAPI and the percentage of cells positive for γH2AX foci (>10 per cell) was determined. At least 500 cells were scored per condition. Data are presented relative to the values obtained for scramble shRNA cells in each cell line, respectively. Error bars, mean ± SEM, n=3, **p<0.005, ***p<0.0005. S-phase percentages as determined by flow cytometry analysis of the percentage of BrdU incorporating cells are indicated below the bar graphs.
(B) Representative images showing γH2AX levels in WT-H-Ras-suppressed DLD1 K-RasMut cells. Cells were co-stained for γH2AX and DAPI. Scale bars, 5 μm.
(C) Isoform specific knockdown of H-Ras in the cancer cells lines shown in (A). * indicates Erk2 as a loading control instead of vinculin. See also Figure S2.
Dependence of mutant K-Ras cancer cells on wild-type H/N-Ras for the activation of the G2 DNA damage checkpoint
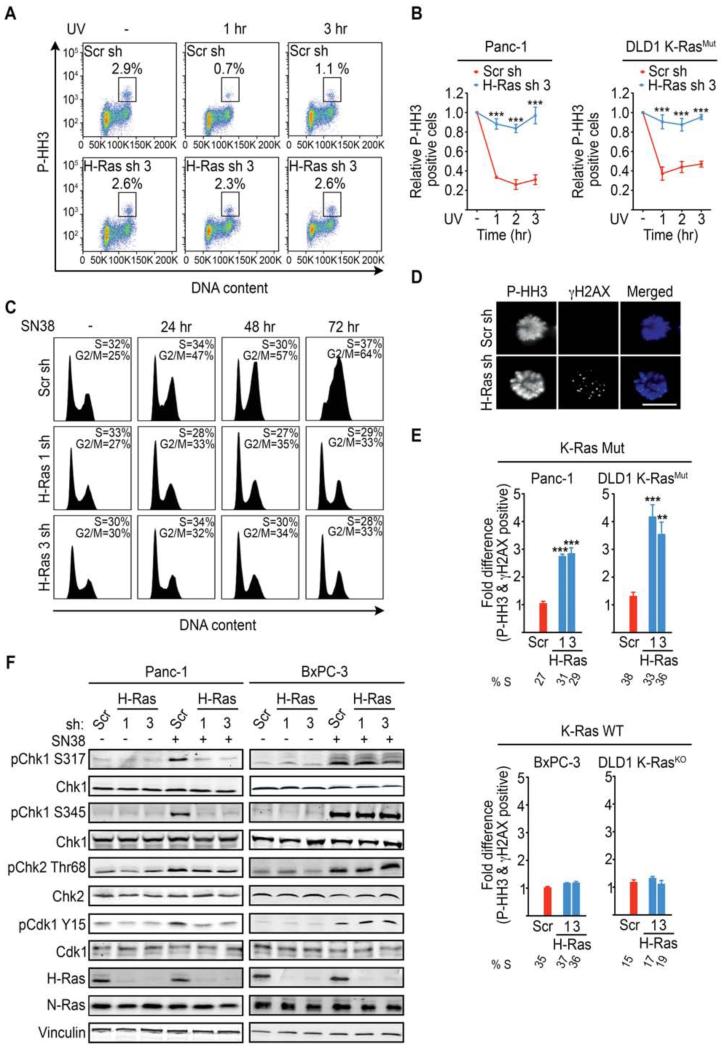
To directly evaluate whether WT-H-Ras knockdown impacted the activation of the DNA damage checkpoint, we subjected K-Ras Mut cells to UV-C irradiation and monitored mitotic entry at 1, 2, and 3 hr intervals after UV-C induced damage. Panc-1 and DLD1-K-RasMut cells expressing scramble shRNA displayed a block in mitotic entry in response to UV-C-induced DNA damage indicating a functional G2 DNA damage checkpoint (Figure 3A-3B and Figure S3A). In contrast, progression into mitosis following UV-C treatment was unperturbed in Panc-1 and DLD1-K-RasMut cells depleted of WT-H-Ras, indicating a defective G2 DNA damage checkpoint (Figure 3A-3B). This defect was not specific to UV-C-induced damage as Panc-1 (Figure 3C) and DLD1-K-RasMut cells (Figure S3B) depleted of WT-H-Ras failed to initiate and maintain cell cycle arrest in response to the topoisomerase I inhibitor SN38. A similar defect was observed in Panc-1 cells depleted of WT-N-Ras (Figure S3C).
Figure 3. K-Ras mutant cancer cells selectively depend on WT-H-Ras for the activation of the ATR-Chk1 mediated G2 DNA damage checkpoint.
(A) Representative FACS plots showing the P-HH3-positive population in untreated (−) and (+) UV-C treated (at 1 hr and 3 hr post treatment) Panc-1 cells expressing scramble (Scr) or WT-H-Ras shRNA (n=3). The boxed area represents the % P-HH3-positive cells.
(B) Quantifications of the experiments described in (A) for K-Ras Mut cell lines. Data are presented as relative to the values obtained for the respective (scramble or WT-H-Ras shRNA) untreated cells.
(C) FACS histograms showing the cell cycle profile of WT-H-Ras-suppressed Panc-1 cells following SN38 (4 nM) treatment. The S and G2/M fractions are indicated. Data are representative of 3 independent experiments.
(D-E) K-Ras Mut and K-Ras WT cancer cells expressing the indicated shRNAs were treated with UV-C, placed in nocodazole containing media for 4 hr to trap mitotic cells, and co-stained for P-HH3 (Blue) and γH2AX (Green). Representative immunofluorescence images of mitotic DLD1 K-RasMut cells treated as indicated are shown in (D). Scale bar, 10μm. Quantifications of the fraction of mitotic cells (P-HH3) expressing the indicated shRNAs that are positive for γH2AX foci (>10 per cell) are shown in (E). Data are presented as relative to the values obtained for the scramble shRNA (Scr sh) cells. At least 50 mitotic cells were analyzed per experiment. S-phase percentages are indicated below the bar graphs.
(F) Representative immunoblots for the indicated proteins in mock or SN38 treated (4nM SN38 for 2h) Panc-1 (K-Ras Mut) and BxPC-3 (K-Ras WT) cancer cells are shown.
All experiments: error bars, mean ± SEM, n=3, **p<0.005, ***p<0.0005. See also Figure S3.
A predictable outcome of a defective G2 DNA damage checkpoint is entry into mitosis with unresolved DNA breaks following damage (Jiang et al., 2009). In agreement with this prediction, a significant fraction of WT-H- and/or N-Ras depleted Panc-1 and DLD1-K-RasMut cancer cells that entered mitosis following UV-C exposure, also stained positive for γH2AX (Figure 3D-3E and Figure S3D). Notably, no such differences were observed in K-Ras WT cancer cell lines (Figure 3E and Figure S3D). These data demonstrate that WT-H- and/or N-Ras are critical for the establishment of a functional G2 DNA damage checkpoint selectively in K-Ras Mut cancer cells.
Wild-type H/N-Ras knockdown impairs Chk1 activation in K-Ras mutant cancer cells
To gain insight into the molecular basis for the perturbation of the G2 DNA damage checkpoint by WT-H/N-Ras knockdown, we examined the DNA-damage specific activation of ATR/Chk1 and ATM/Chk2 in response to SN38 or UV-C treatment (Bartek and Lukas, 2003; Zhou and Elledge, 2000). Notably, WT-H-Ras knockdown was accompanied by a defective Chk1 activation in response to either SN38 or UV-C treatment in K-Ras Mut cell lines Panc-1 and DLD1-K-RasMut (Figure 3F and Figure S3E-S3F), and MIA PaCa-2 (Figure S4A) as evidenced by impaired phosphorylation of Chk1 at Ser 317 and Ser 345. The requirement for WT H-Ras for ATR/Chk1 activation was a specific property of K-Ras mutant cells as WT-H-Ras knockdown had no effect on Chk1 activation in the K-Ras WT cell lines BxPC-3 and DLD1-K-RasKO (Figure 3F and Figure S3E-S3F). The defect in Chk1 activation upon WT H-Ras knockdown in K-Ras mutant cells was also reflected in the impaired inhibitory phosphorylation of Cdk1 at Tyr 15 (Figure 3F and Figure S3E). By comparison, Chk2 activation, as monitored by phosphorylation of Chk2 at threonine 68 (Thr68), was not affected by WT-H-Ras knockdown in either K-Ras Mut (Panc-1) or K-Ras WT (BxPC-3) cancer cell lines (Figure 3F and Figure S3E); Of note, Chk2 protein levels in DLD1 cells were too low to reliably measure its activation status. WT-N-Ras knockdown also led to a selective and similar impairment of ATR/Chk1 activation in K-Ras mutant cells (Figure 4). Taken together, these results indicate that the defective G2 DNA damage checkpoint caused by WT-H/N-Ras knockdown in K-Ras mutant cells is due to the failure to properly activate Chk1.
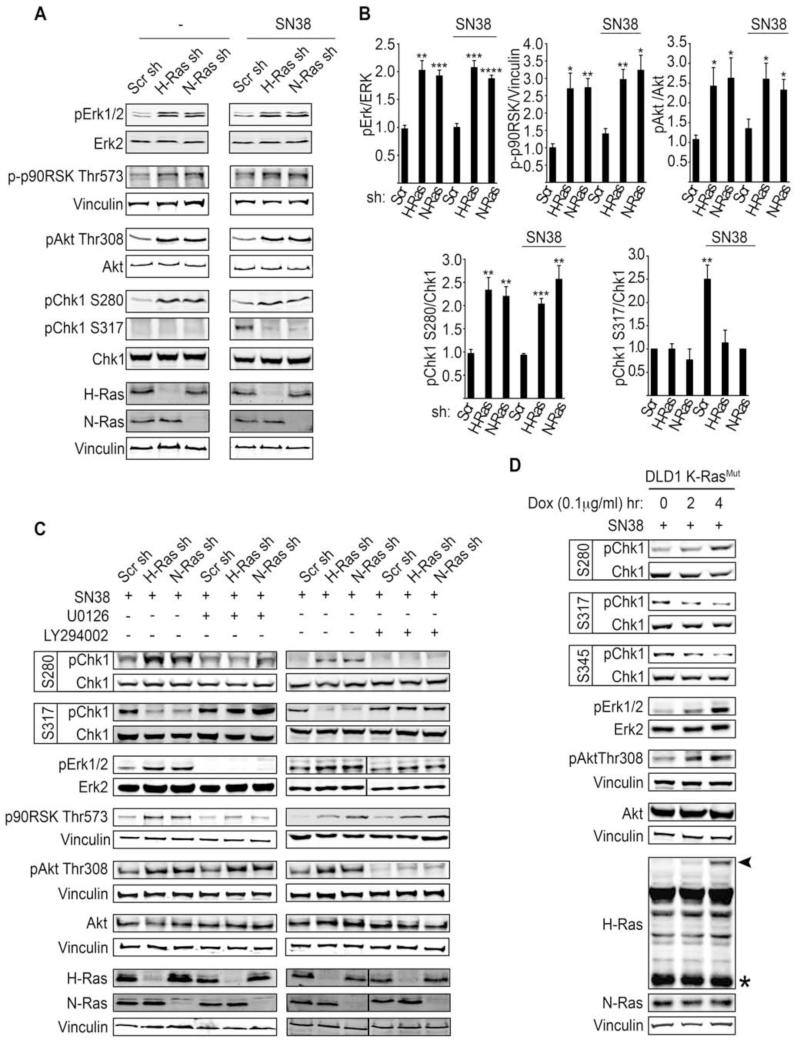
Figure 4. WT-H/N-Ras knockdown enhances Erk/p90RSK and PI3K/Akt activitation to promote phosphorylation of Chk1 at Ser 280 and inhibit its activity.
(A) Activation status of Erk, p90RSK, Akt, and Chk1 in WT-H/N-Ras-suppressed Panc-1 cells in the basal state or upon SN38-induced damage (8 nM SN38 for 2 hr).
(B) The fold change in the amount of pErk1/2, p-p90RSK Thr573, pAkt Thr308, pChk1 S280, and pChk1 S317 in WT-H/N-Ras shRNA as compared to scramble shRNA expressing cells is indicated. Quantification of pErk1/2, pAkt Thr308, pChk1 S280, pChk1 S317 and p-p90RSK Thr573 was carried out by densitometry scanning and normalized to the total levels of Erk, Akt, and Chk1, respectively, and vinculin for p-p90RSK Thr573. Error bars, mean ± SD, n=3, *p<0.05, **p<0.005, ***p<0.0005, ****p<0.00005
(C) Panc-1 cells expressing the indicated shRNAs were treated with 8 nM SN38 for 2 hr and in the presence or absence of the MAPK inhibitor (U0126, 10 μM) or the PI3K inhibitor (LY294002, 5 μM). WCLs were analyzed to determine ERK/p90RSK or Akt inhibition and their respective effect on Chk1 phosphorylation and activation status.
(D) Isogenic derivatives of DLD1 K-RasMut cells engineered to inducibly express GFP-H-RasG12V (DLD1 K-RasMut Flp-IN TREX GFP-H-RasV12) upon addition of doxycycline were induced for 0, 2, 4 hr and subjected to SN38 (8 nM) for 2 hr (for the 2 hr Dox induction, Dox and SN38 were added simultaneously; for the 4 hr Dox induction, SN38 was added 2 hr post induction and the cells were incubated for an additional 2 hr). Representative immunoblots for the indicated proteins are shown. Arrowhead indicates GFP-H-RasG12V and * indicates endogenous H-Ras. See also Figure S4.
Wild-type H/N-Ras negatively regulate MAPK and Akt signaling to control Chk1 phosphorylation
The Ras effector signaling pathways, Raf/Erk and PI3K/Akt, have been shown to play a key role in Chk1 activation and the G2/M phase of the cell cycle. PI3K/Akt was reported to override DNA-damage-induced G2 arrest through repression of Chk1 activation via Akt-mediated inhibitory phosphorylation of Chk1 at Ser 280 (King et al., 2004; Puc and Parsons, 2005; Shtivelman et al., 2002). More recently, the Raf/MAPK pathway has been shown to impair Chk1 activity through Chk1 Ser 280 phosphorylation by MAPK-activated protein kinase RSK (p90 ribosomal S6 kinase) (Li et al., 2012; Ray-David et al., 2012). As wild-type Ras proteins have been reported to antagonize Ras effector signaling output in cancer cells that harbor mutant Ras (Young et al., 2012), we next investigated whether impaired Chk1 activation upon WT-H- or N-Ras knockdown was due to enhanced activation of Ras effector pathways. Knockdown of WT-H- or N-Ras in mutant K-Ras cancer cells was associated with elevated Erk/p90RSK and Akt activation, which correlated with enhanced inhibitory phosphorylation of Chk1 at Ser 280 both in the basal state and following SN38-induced DNA damage (Figure 4A-4B). Conversely, suppression of Erk or Akt signaling via treatment with MAPK or Akt inhibitors overturned the hyperphosphorylation of Chk1 at Ser 280 in WT-H- or N-Ras-depleted mutant K-Ras cancer cells. Importantly, under these conditions, the activation of Chk1 in response to SN38-induced DNA damage was restored as shown by Chk1 Ser 317 phosphorylation (Figure 4C). Altogether these results support a model whereby in K-Ras mutant cells, the downregulation WT-H/N-Ras leads to the enhancement of Erk/p90RSK and Akt signaling, which in turn represses Chk1 activation through Chk1 Ser 280 phosphorylation.
To rule out the possibility that WT-H/N-Ras may also specifically prevent Chk1 Ser 280 phosphorylation, we generated an isogenic derivative of the DLD1-K-RasMut cell line that can inducibly express GFP-H-RasG12V upon doxycycline administration (DLD1-K-RasMut Flip-IN TREX GFP-H-RasV12) (Girdler et al., 2006). As illustrated in Figure 4D, expression of GFP-H-RasG12V at sub-endogenous levels led to a concomitant increase in Erk and AKT signaling despite the presence of WT-H/N-Ras in these cells. Moreover, this was accompanied by the phosphorylation of Chk1 at the inhibitory site Ser 280, and an impairment of Chk1 phosphorylation at the activation sites Ser 317 and Ser 345 (Figure 4D). Altogether, our data are consistent with a model in which the enhanced phosphorylation of Chk1 at Ser 280 and inhibition of Chk1 activity observed under conditions of WT-H-Ras deficiency in mutant K-Ras cells is a consequence of an increase in Erk/Akt signaling.
If the activation of Chk1 is directly linked to the presence of mutant K-Ras, then the acute expression of mutant K-Ras in an otherwise K-Ras wild-type cancer cell line should render Chk1 activation in these cells dependent on WT-H/N-Ras. To test this idea, we silenced WT-H-Ras in BxPC-3 cells (K-Ras WT) that had been engineered to inducibly express K-RasG12V (BxPC-3 K-RasV12) (Figure S4C). Whereas, knockdown of WT-H-Ras in the parental BxPC-3 cell line had no effect on Chk1 Ser280 phosphorylation and Chk1 activation (Figure S4B and Figure 3F), knockdown of WT-H-Ras in BxPC-3 cells induced to express K-RasG12V led to elevated Erk activation, induction of Chk1 Ser 280 phosphorylation and impairment of Chk1 phosphorylation at Ser 317 (Figure S4D). These results suggest that the hyperactivation of Erk/Akt pathways, the enhancement of Chk1 Ser 280 phosphorylation and the impairment of Chk1 activation induced by WT-H-Ras knockdown represent a set of responses that are specifically dictated by the mutational status of K-Ras.
Mutant K-Ras cancer cells depleted of wild-type H/N-Ras are highly sensitive to DNA damage-inducing agents
The underlying premise for the therapeutic use of DNA damaging agents is that susceptibility of cancer cells is linked to the lack of G1/S and G2 checkpoints (Ma et al., 2011; Zhou and Bartek, 2004). Therefore, we reasoned that the abrogation of the ATR/Chk1-induced DNA damage checkpoint in K-Ras mutant cells by WT-H- and/or N-Ras knockdown could enhance the therapeutic efficacy of DNA damaging agents. Assessment of cell viability following treatment with SN38 indicated that the K-Ras mutant cells DLD1 K-RasMut and Panc-1 expressing shRNAs targeting H-Ras, were on average ~90-fold and ~50-fold more sensitive to SN38 respectively, as compared to DLD1 K-RasMut and Panc-1 cells expressing scramble shRNA (Figure 5A). Similarly, WT-H-Ras knockdown sensitized these cells (~13-fold for DLD1 K-RasMut and ~25 fold for Panc-1) to the DNA intrastrand crosslinker oxaliplatin (Figure 5B). Notably, knockdown of WT-H-Ras or N-Ras in K-Ras WT cancer cells did not lead to a sensitization to SN38 or oxaliplatin treatment (Figure 5A-5B). Analysis of the apoptotic index, as measured by FACS detection of cleaved caspase 3-positive cells, demonstrates an exacerbation of cell death by WT-H-Ras knockdown in response to SN38 treatment (Figure 5C-5D). Knockdown of WT-N-Ras also sensitized these cells to SN38 and oxaliplatin (Figure S5). Together, these results show that WT-H-Ras or N-Ras knockdown specifically sensitizes K-Ras Mut cancer cells to DNA damaging agents. A similar sensitization pattern was observed upon the treatment of cells with the Chk1/Chk2 inhibitor AZD7726 (Figure 6A-6B and Figure S6), supporting the hypothesis that WT-H/N-Ras downregulation selectively sensitizes K-Ras mutant cells to DNA damage-inducing agents by abrogating Chk1 activity.
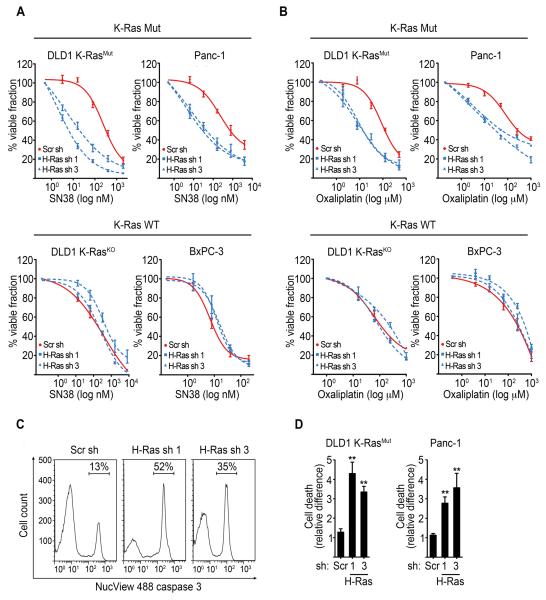
Figure 5. Knockdown of WT-H-Ras selectively sensitizes mutant K-Ras cancer cells to DNA damage.
(A-B) MTT cell viability assays of K-Ras Mut cancer cells (DLD1 K-RasMut and Panc-1) and K-Ras WT cancer cells (DLD1 K-RasKO and BxPC-3) induced to express scramble or WT-H-Ras shRNAs, and following a 72 hr treatment with SN38 (A) or oxaliplatin (B). Viable fraction is expressed as a percentage of the viability values obtained for the respective untreated conditions. p<0.005 for H-Ras sh 1 or H-Ras sh 3 plus SN38 or oxaliplatin versus Scr sh plus SN38 or oxaliplatin in K-Ras mutant cells, respectively.
(C) Detection of cell death by FACS analysis of the Nucview Alexa-488 caspase 3-positive population in SN38 treated K-Ras Mut cells. Representative profiles of Nucview Alexa-488 caspase 3 fractions in DLD1 K-RasMut cells inducibly expressing scramble or WT-H-Ras shRNAs and treated with SN38 (16 nM) for 72 hr.
(D) Quantification of cell death as determined by the Nucview Alexa-488 caspase 3-positive fraction in WT-H-Ras-depleted K-Ras mutant cells treated with SN38 as in (C). Data are presented relative to the values obtained for the respective scramble shRNA expressing cells. Error bars, mean ± SEM, n=3, **p<0.005. See also Figure S5.
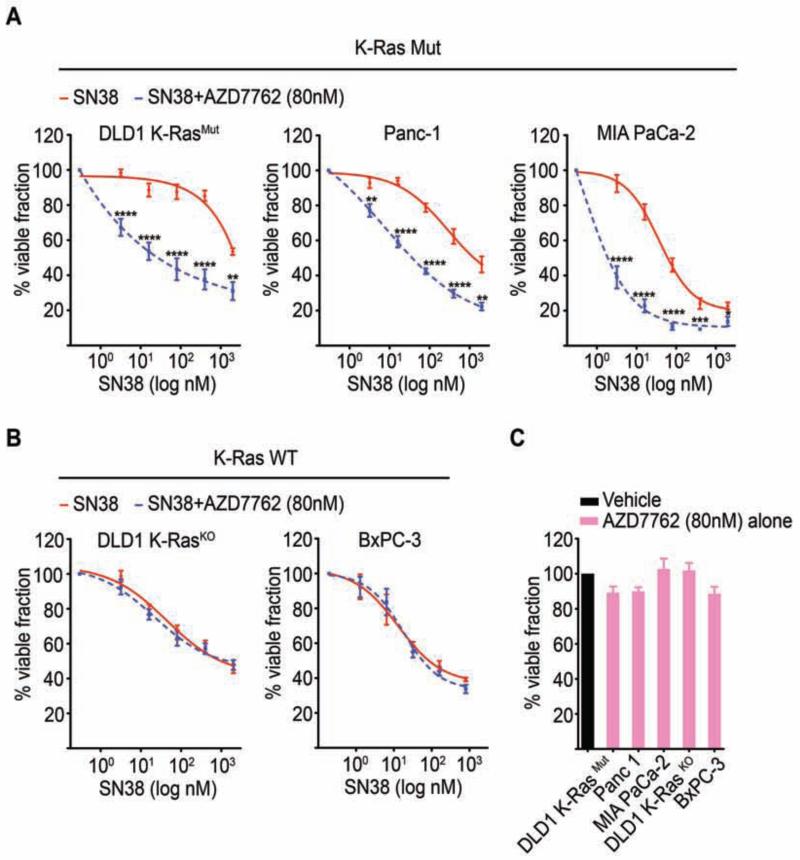
Figure 6. The Chk1/Chk2 inhibitor AZD7762 potentiates SN38 cytotoxicity selectively in K-Ras mutant cancer cells.
(A-B) K-Ras mutant cancer cells (DLD1 K-RasMut, Panc-1, and MIA PaCa-2) (A), or K-Ras WT cancer cells (DLD1 K-RasKO and BxPC-3) (B), were treated with SN38 alone or a combination of SN38 and AZD7762 (SN38 + AZD7762) for 48 hr at the indicated concentrations and analyzed for cell viability by the MTT assay. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 for SN38 + AZD7762-treated versus SN38-treated alone.
(C) K-Ras mutant and K-Ras WT cells were treated with AZD7762 alone for 48 hr and assessed for cell viability by the MTT assay.
(A-C) Viable fraction is expressed as a percentage mean ± SEM of the viability values obtained for respective vehicle only treated conditions from 3 independent experiments each performed in triplicate. See also Figure S6.
Knockdown of wild-type H-Ras sensitizes K-Ras mutant tumors to DNA damage-inducing chemotherapy and leads to tumor regression
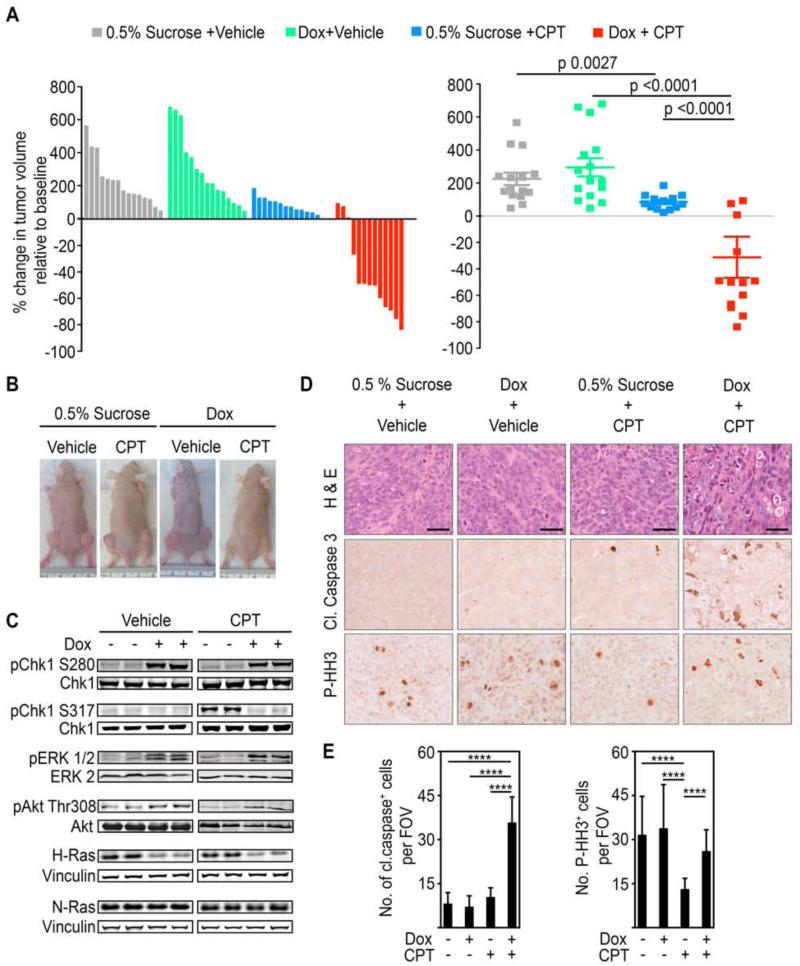
To test the in vivo effects of WT-H-Ras knockdown on the sensitivity of K-Ras mutant tumors to DNA damaging agents, we established xenografts of DLD1-K-Ras mutant cells that inducibly express WT-H-Ras shRNA upon exposure to doxycycline in athymic nu/nu mice. Administration of doxycycline or vehicle was initiated after tumors had reached ~100mm3. Seven days post-induction (tumor size ~ 250 mm3), efficient knockdown of WT-H-Ras was confirmed in the established tumors and treatment with irinotecan (CPT), a topoisomerase I inhibitor that is FDA-approved for the treatment of colorectal cancer, was initiated (Figure S7A). In the absence of treatment with irinotecan, tumors arising from the WT-H-Ras-suppressed cells grew similarly to those arising from uninduced control cells (Figure 7A-7B). Thus, distinct from our in vitro studies, WT-H-Ras knockdown in vivo was not associated with any delay in growth or mitotic progression under these conditions (Figure 7A and Figure 7D-7E). However, similar to our observations in the synchronization studies, we did note an elevation in aberrant mitotic figures (chromosome misalignment and lagging chromosomes), which is an indication of perturbed mitosis (Figure 1G and data not shown). Hence, the lack of a higher mitotic index despite aberrant mitosis, may reflect clearance of the aberrant mitotic cells in the in vivo setting. In agreement with previously reported studies, treatment with irinotecan alone resulted in a reduction of tumor growth (Figure 7A-7B) (Harris et al., 2005; Zabludoff et al., 2008). Notably, five days following the termination of irinotecan administration, WT-H-Ras-suppressed tumors had undergone regression, which was maintained for the duration of the study, up to 18 days post treatment (Figure 7A-7B). In contrast, the control irinotecan-treated tumors showed in large part a modest growth over the same time period (Figure S7B). Consistent with our cell-based studies, abrogation of WT-H-Ras in mutant K-Ras tumors led to Erk and Akt hyperactivation and the inhibition of Chk1 activation, as reflected by an elevated Chk1 Ser 280 phosphorylation and an impaired Chk1 Ser 317 phosphorylation in both mock and irinotecan-treated tumors (Figure 7C). Assessment of the extent of apoptosis revealed that the combination of WT-H-Ras knockdown and irinotecan treatment induced a significant increase in tumor cell apoptosis compared to WT-H-Ras knockdown or irinotecan treatment alone (Figure 7D-7E). Importantly, irinotecan treatment failed to induce cell cycle arrest of WT-H-Ras-suppressed tumors as evident by the significant increase in the number of cells staining positive for phosphorylated histone H3 compared to WT-H-Ras-intact tumors (Figure 7D-7E). These results indicate that WT-H-Ras knockdown in K-Ras mutant cells compromises the DNA damage checkpoint-mediated cell cycle arrest in vivo.
Figure 7. WT-H-Ras knockdown in established mutant K-Ras tumors impairs cell cycle arrest induced by DNA damaging chemotherapeutic agents and leads to tumor regression.
(A) Waterfall plot and scatter plot showing percentage change in the volume of subcutaneous DLD1 K-RasMut tumors in nude mice five days after the last dose of irinotecan (CPT) administration. Percentage change was determined relative to the tumor volume at the start of irinotecan (CPT) treatment for each individual tumor. Mice engrafted with 2×106 DLD1 K-RasMut cells stable for the doxycycline-inducible H-Ras 1 sh were given either doxycycline or vehicle-only (0.5% sucrose) as a control via their drinking water once tumors attained ~100 mm3. Irinotecan (CPT) administration (50 mg/kg every other day for 6 days (q2dx3)) was initiated when tumors reached ~250 mm3. Error bars, mean ± SEM.
(B) Representative xenograft tumors are shown.
(C) WCLs of tumor tissue obtained from the indicated animals 24 hr after the initiation of irinotecan (CPT) administration were immunoblotted with antibodies for the indicated proteins.
(D-E) Tumor sections from mice in (A) treated as indicated were stained for hematoxylin and eosin (H&E), anti-cleaved caspase 3, or anti-P-HH3 antibody. Representative images are shown in (D) and quantifications are shown in (E). Scale bars, 40 μm. Cleaved caspase 3 or P-HH3 positive cells were counted per field of view (FOV) at 20x magnification. Error bars, mean ± SD, n=3 mice per group, 4 FOV per mouse. ****p<0.0001. See also Figure S7.
To further substantiate the in vivo analyses of the consequences of WT-H-Ras knockdown, we administered irinotecan or vehicle to xenograft tumors derived from either MIA PaCa-2 (K-Ras Mutant) or BxPC-3 (K-Ras WT) cells that were engineered to inducibly (+Dox) express WT-H-Ras shRNA (Figure S7C-S7D). Similar to DLD1-K-RasMut xenografts, combination of WT-H-Ras knockdown and irinotecan treatment led to MIA PaCa-2 tumor regression, whereas irinotecan alone led to a growth delay and WT-H-Ras knockdown alone had no effect (Figure S7C). Importantly, we found no synergy between WT-H-Ras knockdown and irinotecan treatment in BxPC-3 xenograft tumors (Figure S7D). Collectively, these observations establish a role for WT-H-Ras in maintaining a functional Chk1-dependent DNA damage checkpoint in established K-Ras mutant tumors.
DISCUSSION
Effective targeting of oncogenic K-Ras-driven tumors has remained a major challenge in cancer therapy. Considerable evidence indicates that cancer cells develop dependencies on normal functions of certain genes that can potentially be exploited to improve therapeutic strategies (De Raedt et al., 2011; Kumar et al., 2012; Luo et al., 2009a; Luo et al., 2009b). In the present study we demonstrate that K-Ras mutant cancers display a dependency on WT-H/N-Ras for the activation of the ATR/Chk1 mediated DNA damage response (DDR) and therefore can be sensitized to DNA damaging chemotherapeutics through the suppression of WT-H/N-Ras. The activation of DDR has been shown to play distinct context-dependent roles in the course of malignant transformation (Toledo et al., 2011a). In premalignant lesions DDR activation is triggered by oncogenic stress and commonly leads to cell death or senescence thereby functioning as an intrinsic barrier to malignancy (Bartkova et al., 2005; Gilad et al., 2010; Schoppy et al., 2012). Full malignant transformation, however, is accompanied by a weakening of the DDR barrier through the selective acquisition of mutations within critical DDR signaling modules (for example p53 mutations and abrogation of ATM/Chk2/p53 signaling). As such, advanced tumors become highly reliant on the remaining functional DDR pathway (ATR/Chk1) for coping with the high levels of oncogene-induced genotoxic stress. In this context, the role of WT-H/N-Ras in coordinating the activation of ATR/Chk1 is critical for supporting the tumorigenic phenotype of K-Ras mutant tumors by preventing catastrophic DNA damage and enhancing tumorigenic fitness and survival. Consistent with this model, wild-type Ras has been shown to play a tumor-promoting role in cell lines from established tumors (Young et al., 2012).
Our results identify a role for WT-H/N-Ras in facilitating Chk1 activation by suppressing the Erk/p90RSK and PI3K/Akt pathways that inhibit Chk1 via Ser 280 phosphorylation. The capacity of WT-Ras to negatively regulate effector pathway signaling in mutant Ras cancer cells is consistent with earlier reports showing that the levels of WT-Ras proteins in mutant Ras cancer cell lines are inversely correlated to the activation status of Ras-effector molecules (Young et al., 2012; Zhang et al., 2001). It is noteworthy that the knockdown of either wild-type isoform alone is sufficient to hyperactivate Erk and Akt and inhibit Chk1 activity and checkpoint function. This suggests a tightly controlled threshold for the WT-H/N-Ras-mediated attenuation of Ras-effector signaling in mutant K-Ras cancers. The mechanisms underlying the WT-H/N-Ras-mediated antagonism of Ras effector signaling in mutant K-Ras cancers remain to be delineated. Of potential relevance to this question are the seemingly contradictory observations that while the knockdown of WT-H/N-Ras in mutant K-Ras cancer cells induces Erk and Akt hyperactivation, the knockdown of Sos1, a guanine nucleotide exchange factor for Ras GTPases, instead impairs Erk and Akt activity (Jeng et al., 2012). A fundamental difference between these two scenarios is that whereas Sos knockdown would affect the levels of GTP-bound Ras, the knockdown of WT-H/N-Ras would inevitably lessen both GDP and GTP-bound Ras levels. Since the WT-isoforms exist predominantly in the GDP-bound form, knockdown of WT-H/N-Ras is likely to significantly alter the stoichiometry of GDP- to GTP-Ras molecules. This may provide a plausible explanation for the observed hyperactivation of Erk and Akt, as GDP-bound Ras molecules have been suggested to play an inhibitory role in Ras signaling (Singh et al., 2005). Furthermore, in the case of Raf, activation depends on Ras-mediated homo- and/or heterodimerization of Raf proteins, which likely require at least two Ras-GTP molecules (Heidorn et al., 2010; Inouye et al., 2000; Poulikakos et al., 2011; Rushworth et al., 2006; Weber et al., 2001). Since Ras dimerization appears to be constitutive and non-selective for GDP or GTP-bound Ras, depletion of GDP-bound Ras, as in the case of knockdown of WT-H-Ras or N-Ras, would stoichiometrically favor Ras-GTP dimer formation and consequently lead to Raf hyperactivation (Heidorn et al., 2010; Inouye et al., 2000).
There is now a large body of pre-clinical evidence showing that inhibition of the ATR/Chk1 pathway enhances the efficacy of standard chemotherapy. Indeed, several Chk1 inhibitors are being tested in clinical trials (Ma, 2012). As such, the ability of WT-H/N-Ras to determine Chk1 activation in mutant K-Ras tumors may warrant further exploration into the development of a therapeutic approach that utilizes inhibition of wild-type Ras-ATR/Chk1 signaling in combination with DNA damaging agents for the selective targeting of K-Ras driven cancers.
EXPERIMENTAL PROCEDURES
Cell culture and lentiviral transduction
Human pancreatic cancer cell lines Panc-1, PL45, AsPC-1, CFPAC-1, MIA PaCa-2, BxPC-3, and Hs700T were obtained from American Type Culture Collection. The isogenic colon cancer cells DLD1-K-RasMut and DLD1-K-RasKO were a kind gift from Dr. Mark Philips. Lentiviral particles were generated according to standard protocols. For knockdown experiments cells were transduced with lentiviral particles (multiplicity of infections (MOI) for Hs700T, 15; all other cell lines, 7) containing pTripz scramble shRNA, H-Ras shRNA, or N-Ras shRNA, and selected with puromycin (Calbiochem; for AsPC-1, 4 μg/ml; for all others, 2 μg/ml) for 3 days. Unless otherwise indicated all experiments were performed on day 4 of doxycycline (1 μg/ml) induction. DLD1 K-RasMut Flip-IN TREX GFP-H-RasV12 cells were generated through Flp recombinase mediated homologous recombination between the FRT sites in the DLD1 K-RasMut cell line and the pcDNA3/FRT/TO/GFP-HRasG12V expression vector. To generate BxPC-3-K-RasV12 cells, BxPC-3 cells were transduced with a TET inducible lentiviral vector to express K-RasG12V, pLenti-TO-K-RasG12V (MOI=1). Following puromycin selection, cells that had efficiently integrated the K-RasG12V lentiviral construct (BxPC-3-K-RasV12) were expanded and subsequently transduced with a pTRIPZ-H-Ras sh construct. As both vectors are Dox inducible, induction of the expression of K-RasV12 also induces the knockdown of WT-H-Ras. Following a 48 hr induction, BxPC-3-K-RasV12 cells that also expressed the H-Ras sh were obtained by fluorescent sorting (RFP: pTRIPZ-H-Ras sh also expresses RFP upon Dox induction). The obtained cells were then cultured for an additional 48h in the presence of Dox and assayed for Chk1 activation upon treatment with SN38.
Cell viability assays
For viability assays, cells were treated with doxycycline to induce shRNA expression for 4 days and then seeded at a density of 4000 cells/well in a 96-well plate in doxycycline containing media. 24 hours post plating, either SN-38 (Tocris Biosciences), or oxaliplatin (Tocris Biosciences), or vehicle was added. Following a 72h treatment, cell viability was assessed by the MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide; Sigma-Aldrich) assay according to the manufacturer’s protocol. Viable fraction is expressed as the percentage of vehicle treated control cells. EC50 was calculated using Graphpad Prism v4.0 software.
Cell synchronization and flow cytometry
Cells were synchronized at the G1/S transition by a double thymidine block. Cells were treated with 2 mM thymidine for 22 hours, and released from thymidine block by washing twice with PBS followed by incubation in fresh medium. 14 hours after release, thymidine was added for another 20 hours. Induction of H-Ras shRNA expression was initiated with the first thymidine block. For the G2/M checkpoint analysis in response to UV-C, cells were irradiated with UV-C (25 J/m2) during the exponential growth phase. Cells were harvested 1 hr, 2 hr, and 3 hr later, fixed with ice-cold 80% ethanol/PBS, and incubated overnight at −20°C. Fixed cells were washed with PBS and permeabilized with 0.25% Triton X-100/PBS on ice for 10 min. Cells were stained with anti-phospho histone H3 to detect mitotic cells and TO-PRO 3 for DNA content. For G2/M checkpoint activation in response to SN-38, cells were treated with 4 nM SN-38 and fixed at the indicated time points. Staining for cleaved caspase 3 positive cells was performed using the Nucview-488 Caspase 3 Kit (Biotium). Flow cytometry was performed on an LSRII (BD Biosciences) at NYU School of Medicine Flow Cytometry Core Facility, and data were analyzed using FlowJo software.
Animal Studies
For xenograft studies we subcutaneously implanted 2 × 106 DLD1 K-RasMut, MIA PaCa-2, or BxPC-3 cells stable for pTripz-H-Ras shRNA (1:1 in Matrigel, BD Biosciences) in both flanks of 8-week-old female athymic nude (NCRNU, Taconic) mice. When tumor size reached ~100 mm3 mice were given drinking water containing either doxycycline (0.2 mg/ml) / 0.5% sucrose or 0.5% sucrose alone. Water was replaced every 3 days. Tumor volume was determined using electronic calipers to measure length (l), width (w), and the formula (w2 × l)/2. Tumor volume was measured twice a week. We treated mice bearing H-Ras depleted (dox/sucrose) or H-Ras intact (sucrose) DLD1 K-RasMut tumors with either irinotecan or vehicle, once tumors reached 250 mm3. Irinotecan hydrochloride (CPT) powder was dissolved into solution as previously described (Harris et al., 2005; Zabludoff et al., 2008). This solution was diluted with 5% dextrose for intraperitoneal (i.p.) injection at a dose of 50 mg/kg every other day for 3 rounds of treatment (q2dx3). Irinotecan, or the combinatorial H-Ras knockdown and irinotecan treatments, were well tolerated as no weight loss above 10% body mass was observed. Body mass was measured using an electronic scale every 2 days. The percent change in tumor volume from day 0 of irinotecan treatment to tumor volume five days after the last dose of irinotecan administration was measured. Mice were euthanized by carbon dioxide-induced narcosis. To prepare lysates tumor tissue was homogenized in RIPA buffer and sonicated to shear genomic DNA. All animal work was approved by New York University Langone Medical Center Institutional Animal Care and Use Committee.
Statistical Analyses
Data were analyzed by the Graphpad Prism built-in test (unpaired, two-tailed), and results were considered significant at P<0.05.
Supplementary Material
HIGHLIGHTS.
-
■
Mutant K-Ras cancer cells require WT-H/N-Ras for G2 DNA damage checkpoint activation
-
■
The activation of ATR/Chk1 in mutant K-Ras cancer cells is dependent on WT-H/N-Ras
-
■
WT-H-Ras suppression leads to regression of K-Ras tumors in response to DNA damage
SIGNIFICANCE.
This study defines a functional dependence of K-Ras driven tumors on wild-type H- and N-Ras for the DNA damage response and reveals a promising therapeutic strategy for the treatment of mutant K-Ras tumors. We demonstrate that mutant K-Ras cancer cells require wild-type H-Ras and N-Ras for the activation of the ATR-Chk1 mediated DNA damage checkpoint, and that this dependence can be exploited to specifically sensitize K-Ras-driven cancers to DNA damage-inducing agents.
Acknowledgments
We thank Dr. Cosimo Commisso for critical reading of the manuscript and helpful discussions. We are grateful to Dr. Stephen Taylor for providing the DLD1 Flip-IN TRex cell line.
Grant Support
This work was supported by research grants from National Institutes of Health to D.B.-S. (GM078266 and CA 055360) and E.G. (1F32CA13922). The NYU Cancer Institute Cytometry and Cell Sorting and Immunohistochemistry Core Facilities were supported in part by grant 5P30CA016087-32 from the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing financial interests
The authors declare no competing financial interests.
REFERENCES
- Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer cell. 2003;3:421–429. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Bremner R, Balmain A. Genetic changes in skin tumor progression: correlation between presence of a mutant ras gene and loss of heterozygosity on mouse chromosome 7. Cell. 1990;61:407–417. doi: 10.1016/0092-8674(90)90523-h. [DOI] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes & development. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, Shen Q, O’Hagan R, Pantginis J, Zhou H, et al. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400:468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- De Raedt T, Walton Z, Yecies JL, Li D, Chen Y, Malone CF, Maertens O, Jeong SM, Bronson RT, Lebleu V, et al. Exploiting cancer cell vulnerabilities to develop a combination therapy for ras-driven tumors. Cancer cell. 2011;20:400–413. doi: 10.1016/j.ccr.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher GH, Wellen SL, Klimstra D, Lenczowski JM, Tichelaar JW, Lizak MJ, Whitsett JA, Koretsky A, Varmus HE. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes & development. 2001;15:3249–3262. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad O, Nabet BY, Ragland RL, Schoppy DW, Smith KD, Durham AC, Brown EJ. Combining ATR suppression with oncogenic Ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner. Cancer research. 2010;70:9693–9702. doi: 10.1158/0008-5472.CAN-10-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girdler F, Gascoigne KE, Eyers PA, Hartmuth S, Crafter C, Foote KM, Keen NJ, Taylor SS. Validating Aurora B as an anti-cancer drug target. Journal of cell science. 2006;119:3664–3675. doi: 10.1242/jcs.03145. [DOI] [PubMed] [Google Scholar]
- Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, Niwa-Kawakita M, Sweet-Cordero A, Sebolt-Leopold J, Shannon KM, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nature genetics. 2008;40:600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SM, Mistry P, Freathy C, Brown JL, Charlton PA. Antitumour activity of XR5944 in vitro and in vivo in combination with 5-fluorouracil and irinotecan in colon cancer cell lines. British journal of cancer. 2005;92:722–728. doi: 10.1038/sj.bjc.6602403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegi ME, Devereux TR, Dietrich WF, Cochran CJ, Lander ES, Foley JF, Maronpot RR, Anderson MW, Wiseman RW. Allelotype analysis of mouse lung carcinomas reveals frequent allelic losses on chromosome 4 and an association between allelic imbalances on chromosome 6 and K-ras activation. Cancer research. 1994;54:6257–6264. [PubMed] [Google Scholar]
- Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer CJ, Pritchard C, Marais R. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye K, Mizutani S, Koide H, Kaziro Y. Formation of the Ras dimer is essential for Raf-1 activation. The Journal of biological chemistry. 2000;275:3737–3740. doi: 10.1074/jbc.275.6.3737. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & development. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng HH, Taylor LJ, Bar-Sagi D. Sos-mediated cross-activation of wild-type Ras by oncogenic Ras is essential for tumorigenesis. Nature Communications. 2012;3:1168. doi: 10.1038/ncomms2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Reinhardt HC, Bartkova J, Tommiska J, Blomqvist C, Nevanlinna H, Bartek J, Yaffe MB, Hemann MT. The combined status of ATM and p53 link tumor development with therapeutic response. Genes & development. 2009;23:1895–1909. doi: 10.1101/gad.1815309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King FW, Skeen J, Hay N, Shtivelman E. Inhibition of Chk1 by activated PKB/Akt. Cell cycle. 2004;3:634–637. [PubMed] [Google Scholar]
- Kumar MS, Hancock DC, Molina-Arcas M, Steckel M, East P, Diefenbacher M, Armenteros-Monterroso E, Lassailly F, Matthews N, Nye E, et al. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell. 2012;149:642–655. doi: 10.1016/j.cell.2012.02.059. [DOI] [PubMed] [Google Scholar]
- Lam MH, Liu Q, Elledge SJ, Rosen JM. Chk1 is haploinsufficient for multiple functions critical to tumor suppression. Cancer cell. 2004;6:45–59. doi: 10.1016/j.ccr.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Li P, Goto H, Kasahara K, Matsuyama M, Wang Z, Yatabe Y, Kiyono T, Inagaki M. P90 RSK arranges Chk1 in the nucleus for monitoring of genomic integrity during cell proliferation. Molecular biology of the cell. 2012;23:1582–1592. doi: 10.1091/mbc.E11-10-0883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Haigis KM, McDaniel A, Harding-Theobald E, Kogan SC, Akagi K, Wong JC, Braun BS, Wolff L, Jacks T, Shannon K. Hematopoiesis and leukemogenesis in mice expressing oncogenic NrasG12D from the endogenous locus. Blood. 2011;117:2022–2032. doi: 10.1182/blood-2010-04-280750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KH, Ancrile BB, Kashatus DF, Counter CM. Tumour maintenance is mediated by eNOS. Nature. 2008;452:646–649. doi: 10.1038/nature06778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffler H, Rebacz B, Ho AD, Lukas J, Bartek J, Kramer A. Chk1-dependent regulation of Cdc25B functions to coordinate mitotic events. Cell cycle. 2006;5:2543–2547. doi: 10.4161/cc.5.21.3435. [DOI] [PubMed] [Google Scholar]
- Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong KK, Elledge SJ. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009a;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009b;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CX, et al. Targeting Chk1 in p53-deficient triple-negative breast cancer is therapeutically beneficial in human-in-mouse tumor models. Journal of Clinical Investigation. 2012;122:1541–1552. doi: 10.1172/JCI58765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CX, Janetka JW, Piwnica-Worms H. Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends in molecular medicine. 2011;17:88–96. doi: 10.1016/j.molmed.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matallanas D, Romano D, Al-Mulla F, O’Neill E, Al-Ali W, Crespo P, Doyle B, Nixon C, Sansom O, Drosten M, et al. Mutant K-Ras activation of the proapoptotic MST2 pathway is antagonized by wild-type K-Ras. Molecular cell. 2011;44:893–906. doi: 10.1016/j.molcel.2011.10.016. [DOI] [PubMed] [Google Scholar]
- Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puc J, Parsons R. PTEN loss inhibits CHK1 to cause double stranded-DNA breaks in cells. Cell cycle. 2005;4:927–929. doi: 10.4161/cc.4.7.1795. [DOI] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nature reviews Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-David H, Romeo Y, Lavoie G, Deleris P, Tcherkezian J, Galan JA, Roux PP. RSK promotes G2 DNA damage checkpoint silencing and participates in melanoma chemoresistance. Oncogene. 2012 doi: 10.1038/onc.2012.472. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Maiato H. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev cell. 2004;7(5):637–651. doi: 10.1016/j.devcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Rushworth LK, Hindley AD, O’Neill E, Kolch W. Regulation and role of Raf-1/B-Raf heterodimerization. Molecular and cellular biology. 2006;26:2262–2272. doi: 10.1128/MCB.26.6.2262-2272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoppy DW, Ragland RL, Gilad O, Shastri N, Peters AA, Murga M, Fernandez-Capetillo O, Diehl JA, Brown EJ. Oncogenic stress sensitizes murine cancers to hypomorphic suppression of ATR. The Journal of clinical investigation. 2012;122:241–252. doi: 10.1172/JCI58928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260:85–88. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- Shtivelman E, Sussman J, Stokoe D. A role for PI 3-kinase and PKB activity in the G2/M phase of the cell cycle. Current biology: CB. 2002;12:919–924. doi: 10.1016/s0960-9822(02)00843-6. [DOI] [PubMed] [Google Scholar]
- Singh A, Sowjanya AP, Ramakrishna G. The wild-type Ras: road ahead. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2005;19:161–169. doi: 10.1096/fj.04-2584hyp. [DOI] [PubMed] [Google Scholar]
- Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, Helleday T, Sehested M, Lukas J, Bartek J. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Molecular and cellular biology. 2005;25:3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo LI, Murga M, Fernandez-Capetillo O. Targeting ATR and Chk1 kinases for cancer treatment: a new model for new (and old) drugs. Molecular oncology. 2011a;5:368–373. doi: 10.1016/j.molonc.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, Oyarzabal J, Pastor J, Bischoff JR, Fernandez-Capetillo O. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nature structural & molecular biology. 2011b;18:721–727. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CK, Slupsky JR, Kalmes HA, Rapp UR. Active Ras induces heterodimerization of cRaf and BRaf. Cancer research. 2001;61:3595–3598. [PubMed] [Google Scholar]
- Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A, Lou D, McCormick F. Oncogenic and wild-type Ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer discovery. 2012 doi: 10.1158/2159-8290.CD-12-0231. [DOI] [PubMed] [Google Scholar]
- Zabludoff SD, Deng C, Grondine MR, Sheehy AM, Ashwell S, Caleb BL, Green S, Haye HR, Horn CL, Janetka JW, et al. AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Molecular cancer therapeutics. 2008;7:2955–2966. doi: 10.1158/1535-7163.MCT-08-0492. [DOI] [PubMed] [Google Scholar]
- Zachos G, Gillespie DA. Exercising restraints: role of Chk1 in regulating the onset and progression of unperturbed mitosis in vertebrate cells. Cell cycle. 2007;6:810–813. doi: 10.4161/cc.6.7.4048. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wang Y, Vikis HG, Johnson L, Liu G, Li J, Anderson MW, Sills RC, Hong HL, Devereux TR, et al. Wildtype Kras2 can inhibit lung carcinogenesis in mice. Nature genetics. 2001;29:25–33. doi: 10.1038/ng721. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Bartek J. Targeting the checkpoint kinases: chemosensitization versus chemoprotection. Nature reviews Cancer. 2004;4:216–225. doi: 10.1038/nrc1296. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.