Abstract
A majority of patients with bladder cancer present with superficial disease and subsequently, some patients show progression to muscle invasive or metastatic disease. Bladder cancer has a complex genetic process and identification of the genetic alterations which occur during progression may lead to the understanding of the nature of the disease and provide the possibility of early treatment. The aim of the present study was to compare the structural and numerical chromosomal differences and changes in the p16 and p53 genes between low-grade (LG) and high-grade (HG) bladder cancer (BC) using cytogenetic and molecular cytogenetic methods. Between March 2009 and March 2010, cytogenetic analyses were carried out on tumor and blood samples in 34 patients with transitional cell type BC, and on blood samples of 34 healthy patients as a control group. Fluorescence in situ hybridization probes for the p16 and p53 genes were also used to screen the alterations in these genes in 32 patients with BC. The patients were divided into two groups (LG and HG) and the findings were compared. A total of 11 (32.3%) patients exhibited LGBC, 22 (64.7%) exhibited HGBC and one (3%) patient exhibited carcinoma in situ. There were no differences between the LGBC and HGBC groups according to the number of chromosomal aberrations (P=0.714); however, differences between alterations of the p16 and p53 genes were significant (P=0.002 and P=0.039). Almost all structural abnormalities were found to be located to the 1q21, 1q32, 3p21 and 5q31 regions in patients with HG tumors. In conclusion, the p16 and p53 genes were altered more prominently in patients with HG tumors compared with LG tumors. The structural abnormalities in the 1q21, 1q32, 3p21 and 5q31 regions were observed more frequently in patients with HG tumors. These regions may play significant roles in the progression of BC, but further studies are required to find candidate genes for a panel of BC.
Keywords: low grade, high grade, bladder carcinoma, chromosomal aberrations, p16, p53, fluorescence in situ hybridization
Introduction
An estimated 386,300 new cases and 150,200 fatalities from bladder cancer (BC) occurred worldwide in 2008 (1). BC has a number of known risk factors, including age, cigarette smoking, exposure to chemicals, chronic infections or irritations and exposure to pelvic radiation. However, numerous patients with BC have no history of exposure to carcinogens (2). The identification of genetic events during tumorigenesis may lead to an understanding of the genetic mechanism underlying BC.
In total, ~75% of patients present with superficial disease (Ta and T1) and 20% with T2 or higher disease. Overall, 70% of treated tumors recur, with 30% of recurrent tumors progressing to metastatic disease of the non-muscle-invasive lesions. Approximately 10% of low-grade (LG) papillary tumors subsequently develop muscle-invasive or metastatic cancer, whereas roughly a third of high-grade (HG) tumors progress, if not already, to muscle-invasive at the time of diagnosis (3). Therefore, the determination of the ideal biomarkers for predicting progression to invasion or metastatic disease is important.
The molecular and genetic changes in urothelial carcinoma (UC) of the bladder are grouped into three processes: i) Chromosomal alteration, which activates the initial carcinogenic event; ii) tumor proliferation, due to a loss of cell-cycle regulation and derangements in normal apoptotic turnover; and iii) metastasis, which involves the initial tumor migration and other processes, including angiogenesis and loss of cell adhesions (4). Since studies have revealed the association between genetic changes and BC, numerous genes have been studied for their connection to BC (5–7). It is known that p53 plays a key role in the regulation of the cell cycle, and mutations in p53 result in chromosomal instability. Alterations in the p53 gene are more frequently observed in invasive HG tumors compared with LG tumors (6). The cyclin-dependent kinase inhibitors p21 and p16 are correlated with an increased disease recurrence and progression. Additionally, the genesis and/or progression of BC has been shown to be a consequence of genetic instability, and chromosomes 3, 7, 9 and 17 are frequently involved in uroepithelial oncogenesis (8,9).
In the present study, cytogenetic methods and fluorescence in situ hybridization (FISH) were used to investigate the frequencies of chromosomal aberrations (CAs) and alterations (amplifications and deletions) of the p53 and p16 genes, alone or in combination, in Turkish patients with BC. The results were compared between cases of HGBC and LGBC.
Materials and methods
Patients
Between March 2009 and March 2010, following approval of the study by the ethics committee of the Medical Faculty of Çukurova University (Adana, Turkey), blood and tissue samples were collected from 34 patients with BC. Written informed consent was obtained from all patients. Tissue samples were removed by transurethral resection or from radical cystectomy specimens, and blood samples were drawn simultaneously during these surgical procedures. A small piece of the tumor sample was obtained for genetic study. The remainders of the tissue samples were evaluated in the Department of Pathology, Çukurova University (Adana, Turkey) by the same pathologist. Structural and numerical abnormalities of chromosomes were detected in the blood and tissue samples from patients with BC by cytogenetic methods. The blood samples from 34 healthy patients were collected and analyzed as the control group. The p16 and p53 genes were also identified in the bladder tumor samples using FISH. The numbers of CAs, including deletion, amplification, fragility, chromosome break, chromatin break and translocation, were compared among the patient and control groups. The patients with BC were divided into two groups: LG and HG. This was performed according to the histopathological type of tumors present, based on World Health Organisation histological criteria (10). Subsequently, the two groups were compared according to age, body mass index (BMI), smoking history, number of chromosomal abnormalities and differences in p16 and p53 genes. Finally, the values were assessed using statistical methods.
Cytogenetic examination
The peripheral blood from 34 patients was obtained for culture and FISH studies. The expression of folate-sensitive fragile sites (FSs) and cytogenetic abnormalities (CAs) in each sample was examined in the genetic laboratory of the Department of Medical Biology and Genetics, Faculty of Medicine, Çukurova University. A 0.3-ml blood sample was incubated at 37°C for 72 h in two types of media; RPMI-1640 (Sigma-Aldrich, St. Louis, MO, USA) and M199 without folic acid (Biological Industries Israel Beit-Haemek, Ltd., Kibbutz Beit-Haemek, Israel). Standard cytogenetic techniques were used for harvesting and slide preparation. The slides were first stained only with Giemsa prior to the examination to avoid missing any gaps. For a detailed analysis of the FSs, a few slides were prepared by GTG-banding, and 50 metaphases were scored for each assay. A CA was defined when it was present in 1% of the cells analyzed and in ≥50% of the individuals studied (11). All gaps and breaks were recorded and localized according to the International System for Human Cytogenetic Nomenclature (1995) (12). The classification of CAs was carried out according to the nomenclature established in the 11th International Workshop on Human Gene Mapping (13).
Tumoral tissues
Bladder tumor samples were obtained from 32 patients by transurethral resection or from radical cystectomy specimens. All samples were mechanically minced and enzymatically disaggregated by digestion with trypsin-EDTA (Biological Industries Israel Beit-Haemek Ltd.) for 1 h. Following the digestion, BioAMF1 medium (Biological Industries Israel Beit-Haemek Ltd.) supplemented with supplement, penicillin-streptomycin and gentamycin (all Biological Industries Israel Beit-Haemek Ltd.) was used for culture. A long-term cell culturing method was performed for proliferation of tumor and normal cells. Once enough proliferation (average, 10 days) had occurred, standard cytogenetic techniques were used for harvesting and slide preparation. GTG-banding was achieved by trypsin-Giemsa treatment. The karyotype was determined by analyzing ≥25 metaphases from the normal and tumor bladder epithelium cells for each individual patient. If there were not enough metaphases observed, the slides were evaluated. For eliminating inherited CAs, lymphocyte cultures were also performed and 25 metaphases were counted for each patient.
Slide preparation and FISH analysis
Cytogenetic analysis of BC cells has remained difficult as these cells have a risk of infection and limited proliferative capacity in vitro, which precludes analysis by metaphase cytogenetics. Therefore, interphase FISH was used to study p53 and p16 genes in non-dividing cells. Standard cytogenetic techniques were used for harvesting and preparation of slides for FISH (14). To observe the p53 and p16 genes, bladder tissues from 32 patients were examined by interphase FISH. Poseidon Repeat-Free FISH Probe p16 (on chromosome 9p21/9q21) and Poseidon Repeat-Free FISH Probe p53 (on chromosome 17p13/SE 17) probes purchased from Kreatech Diagnostics (Amsterdam, The Netherlands) were used.
Statistical analysis
Comparisons between groups were applied using Student’s t-test and one-way analysis of variance for normally distributed data. The Mann-Whitney U-test and Kruskal-Wallis test were used to compare data that were not normally distributed. The categorical variables between groups were analyzed using the χ2 test. Results are presented as the mean ± standard deviation and the median (range). P<0.05 was considered to indicate a statistically significant difference. Statistical analyses were performed using SPSS, version 18.0 (SPSS, Inc., Chicago, IL, USA).
Results
Demographic data of the patients
A total of 30 (88.2%) male and four (11.2%) female patients with BC were recruited for the present study, with a mean age of 60.6±14.2 years (range, 26–81 years). Histopathological examinations revealed that 11 (32.3%) patients had LGUC, 22 (64.7%) patients had HGUC and one (3%) patient had carcinoma in situ (CIS). The patient with CIS was added to the HG-tumor group. The mean values of age, BMI and smoking time for the LG-cancer group were 58.9±18.51 years (range, 26–81 years), 25.5±3.51 kg/m2 (range, 22.2–32.5 kg/m2) and 20.6±15.8 packs/year (range, 0–40 packs/year), respectively. These same parameters were calculated for the HG-cancer group as 61.5±12.03 years (range, 43–81 years), 28.1±4.73 kg/m2 (range, 20.5–37.8 kg/m2) and 25.5±16.94 packs/year (range, 0–60 packs/year) (Table I). There were no statistically significant differences between the patients with LG-UC or HG-cancer with regard to age, BMI and smoking time (P=0.971, P=0.106 and P=0.561).
Table I.
Demographic data, p16 and p53 statuses, and blood and tissue culture results of the patients.
| P no. | Agea/gender | G | S | Tobacco useb | p16 | p53 | RPMI | M199 | Tissue |
|---|---|---|---|---|---|---|---|---|---|
| P1 | 76/M | H | T3b | 20 | 22 del | 5 amp 20 del |
gap(3p21) | 9qh+ ×3;fra(5q31); chbr(14q?);hsr(2q?)x2;45? | 47,XY,+2×2; 45,XY,−1;92,XXYY |
| P2 | 57/M | L | T1 | 40 | 2 amp 4 del |
6 amp | 92,XXYYx3;fra(1q32);fra(6p21); 47,XY,+21;9qh+ ×2 | 92,XXYYx4;fra(7q?);45,XY,izo(Xq/p),−21 | 47,XY,+21;42,XY,−2,−11,−12,−22 |
| P3 | 61/M | H | T2a | 45 | 10 del | 15 del | 47,XX,+3p,+ace;45,XX,−14;44, X,−Y, −8; 45,XY,−16; 44,X,−Y,−18,−13,+12 | 45,XY,−16;45,XY,−8;chtb(1q11); del(10)(q24-qter); 45,X–Y;chrb(2q31); del(17)(q11-qter);45,XY,−22 | 43,Y,−11,−17,−X;45,XY,−19;45,X,−Y; 44,X,t(14;18)(p11;p11); 45,XY,−13 |
| P4 | 53/F | H | T1 | 0 | 16 del | 18 del | 9qh+ ×54 48,XX,+4,+21;45,XX−10; 46,XX,?15p+; inv(13)(p13;q14) | 9qh+ ×2;44,XX,−12,−21;chbr(4q?); fra(Xq26); 15p+x52; 45,X,−X; 9qh+x2; 15p+,fra(3p25)x2;15p +,chtbr(3p21)x2; fra(12p13); fra(3p21) | 46,XX,+17,−20,9qh+;gap(6q15); gap(5q31) ×2; del(3)(p23-pter) |
| P5 | 77/M | L | T1 | 0 | 3 del | 3 del | no cultered | no cultered | no cultered |
| P6 | 81/M | L | T1 | 2 | 10 del | 13 del | 9qh+ ×2;fra(4q33);43,X,−1,−10,−Y;45, XY,−17; 44,XY,−21,−22;43,Y,9qh+, −22[2],−X | 92,XXYYx2;9qh+ ×2;45,X,−Yx2;45,XY,−20; 44,XY,−3,−18;del(5)(p14-pter),del(X) (p21-pter); chtbr(2q23);chtbr(3p21.3), chtbr(5p13) | no cultered |
| P7 | 42/M | L | T1 | 25 | 5 del | 5 del | 45,X-Y ×2;45,XY, −10; del(9)(q12-qter) | del(7)(q11–q12);del(3)(p25-pter);del(5)(q13–q15); 45,XY,−22;chtb(3q26.2);chtb(12q13) | 45,X,−Y |
| P8 | 73/M | H | T3b | 30 | 16 del | 10 del 3 amp |
fra(1p36);del(1)(q41-qter);45,XY,−17; hsr(3q11–q13); 45,XY,−3 | far(Xp22.1); t(14;22)(q32;q11.2);45,XY, del(17) (q21-qter),−19 | 47,XYY; 47,XX,+ace |
| P9 | 73/M | H | T1 | 60 | 9 del | 12 del | 92,XXYY | 45,XY,−22×2;45,XY,gap(1q21)x2,−18;del(13) (q32-qter);9qh+ | 45,XY,−17;44,XY,−12,−20; 45,X,−Y;chtb(2p15) |
| P10 | 53/M | H | T4a | 40 | 18 del | 10 del | 45,XY,−21; 44,XY,−5,−17 | 45,XY, chtb(5q15), −18 | 92, XXYY |
| P11 | 38/M | L | T1 | 10 | 9 del | 12 del | 46,XYY,−22; 45,XY,−D 47XY, (?); 45,XY,−14 | 46,XY, fra(1p36.1); 45,XY, −22; 47,XXY; chbr(9q32); del(3)(p13–p14) | chtb(3q26.2);chtb(12q13) |
| P12 | 79/M | L | Ta | 0 | 2 del | 5 del | 46,XY,−10, +3 | 46,XY,fra(3p25); 45,XY,−3 | 45,XX,−21 |
| P13 | 74/F | H | T1 | 32 | 8 del | 5 del | 45,XX,−12 | 45,XX,−11×2;48,XX,+15,+16[2],−12;45,XX,−8; 45,XX,−4,−10;43,XX,+15,−8,−10,−19,−21 | 92,XXXX;92,XXYY;47,XY,+14 |
| P14 | 50/M | CIS | Tis | 30 | 23 del | 18 del | 44,XY,−17,−20; 45,XY,−22 | 45,XY,−8;47,XY,+mar;chtb(3p21.2); chtb(11q13.4) | no cultered |
| P15 | 26/M | L | Ta | 20 | 15 del | 22 del | 45,XY,−22; 45,XY,−8;45,XY,−10; chtb(5q33) | 45X,−Y;47,XY,+mar;44,XY,−21,−22;45,XY,−19 | 45,X,−Yx6;45,X,+4q+,−Y; 43,X,−6, −8,−Y;43,X,−18,−20,−Y; 92,XXYY |
| P16 | 81/M | H | T1 | 0 | 8 del | 10 del | 45,XY,−22 ×2; 47,XY, +mar | 45,XY,gap(6q23),−14;fra(5q31);fra(2q35); 45,XY, gap(4q27),−Y; 47,XY,+4;45,XY;fra(Xq13), −22; fra(5q31); fra(12q22); fra(Xq26); 45,XY,−15; 45,XY,−22;43,X,−Y,−7,−18; fra(5q31) | del(X)(q21-qter);fra(12q24); del(9) (q22-qter) |
| P17 | 51/M | L | T1 | 20 | 9 del | 6 del | Yq+; chrb(4q31) | 47,XY,+21;del(14)(q11.2-pter);fra(11q23);45,XY,−19 | no cultered |
| P18 | 56/M | L | T1 | 35 | 4 del | 3 del | 44,XY,−18,−21;43,XY,−15,−18,−19;45, XY,−21; chrb(8q22);inv(9)(p11;q12)x3; del(2)(p23-pter) | del(5)(q31-qter);45,X,−8,−9,−Y,+21;47,XY,?+(p); 45,XY,del(1)(p32-pter),−19;del(1)(p24-pter); chrb(8q23); 45,XY,−19 | no cultered |
| P19 | 44/M | H | T3b | 0 | 9 del 3 amp |
6 del 8 amp |
t(2;5)(p25;q14); gap(2q23); gap(2q35); gap(4q31.3), gap(5q31); gap(1q36.1); gap(5q31) | 45,XY,t(13;22)(p11;p11);45,XY,del(9)(q22-qter),−21; del(2)(p24-pter);t(12;17)(p13;q12);45,XY,−8; 45,XX, −21×2 | no cultered |
| P20 | 60/F | H | T2a | 0 | 3 del | 2 del | 47,XX,+mar;del(1)(q32-qter);chtb (2q32.2); 45,XX,−22; chtb(4q31) | t(3;12)(p26;q15);del(10)(q23-qter); 47,XXX; del(3)(q11-qter); 47,XX,+ace | 15p+x10;47,XXX,15p+;mar(10<); chtb(12q14) |
| P21 | 55/M | H | T1 | 25 | 16 del | 10 del | 45,XY,−22;44,XY,−13,−22;44,XY,−7, −22;45,XY,−13; 45,XY,−16 | t(10;14)(q26;q13);47,XY,+15;del(2) (p13-pter);44, XY,+21,−13 | 92,XXYYx13;92,XXYY,t(19;19) (q13.4;q13.4), t(21;22)(p13;p13); 45, XY,−14; 92,XXYY, t(21;22)XY, −(p13;p13),t(7;15)(p22;q26);45,22; 43,XYY,−2,−4,−8,−15;45,XY,−12; 44,XY,−4,−14 |
| P22 | 65/M | H | T4 | 15 | 12 del | 16 del | fra(2q21);chtb(19q13);44,XY,chbr (2q31),−2×2 | 47,XY,+ace;48,XY,+1,+19,+22;chtb(1q21)(q32) | 69,XXYx2;92,XXYYx2;45,XY, −20;chtb(2p15); 45,X,−Y |
| P23 | 65/M | L | T1 | 40 | no tissue | no tissue | 46,XY | 45,XY,−14;45,XY,−8;chtb(5q33) | no cultered |
| P24 | 49/M | H | T1 | 30 | no tissue | no tissue | 16qh+ ×14 | 47,XY,+ace;fra(9q32) | no cultered |
| P25 | 46/M | H | T2a | 30 | 12 del | 7 del | no cultered | 46,XY | 46,XY |
| P26 | 76/M | L | T1 | 35 | 0 | 2 del | 46,XY | 45,XY,−17×7 | no cultered |
| P27 | 72/M | H | T4a | 45 | 17 del | 14 del | 45.X,−Y;chbt (5q33) | 45,XY,−8,11p+;47,XY,+21;fra(1q32)x2; fra(1q42); fra(1q22) | no cultered |
| P28 | 72/M | H | T3b | 20 | 21 del | 18 del | 47,XY,+ace;fra(2q33);fra(2q31);fra (3p21);fra(6p21)x2; fra (1q21)x4;22p+x4 | 45,X,−Y;46,XY,−11,+mar,del(11)(q?); fra(6q23), fra(12q22); fra(2p23) | no cultered |
| P29 | 49/M | H | T1 | 35 | 14 del | 8 del | 45,XY,−22; 42,? | 45,XY,−9;45,XY,−10;chtb(5q33) | 44,XY,−3,−21 |
| P30 | 79/F | H | T1 | 0 | 9 del 3 amp |
5 del 4 amp |
inv(9)(p11;q13) ×50;45,X ×40; t(7;14)(p15;q24) | inv(9)(p11;q13) ×50;45,X ×40; chtb(16q22) | no cultered |
| P31 | 43/M | H | T1 | 20 | 0 | 0 | 45,XY,−10;43.X,−21,−Y;45,XY,−21; 45.X,−Y | 45,XY,−20;45,X,−Yx2;45,XY,−21 | no cultered |
| P32 | 69/M | H | T2a | 30 | 13 del | 12 del | 46,XY | 46,XY | no cultered |
| P33 | 63/M | H | T2a | 45 | 16 del | 21 del | chtb(17p11); hsr (2q22) ×3 | no cultered | no cultered |
| P34 | 54/M | H | T3a | 35 | 12 del | 15 del | 46,XY | no cultered | no cultered |
package/year.
P, patient; M, male; F, female; G, grade; L, low grade; H, high grade; S, stage; del, deletion; amp, amplification; fra, fragility; chbr, chromosome break; chtb, chromatin break; hsr, homogen staining region; t, translocation; inv, inversion; mar, marker; ace, acentric; ter, terminal; izo, izochromosome.
According to the tumor-node-metastasis classification, there were three (8.8%) patients in the Ta stage, 17 (50%) in T1, five (14.7%) in T2a, one (2.9%) in T3a, four (11.8%) in T3b, three (8.8%) in T4a and one (2.9%) in Tis stage.
Cytogenetic findings
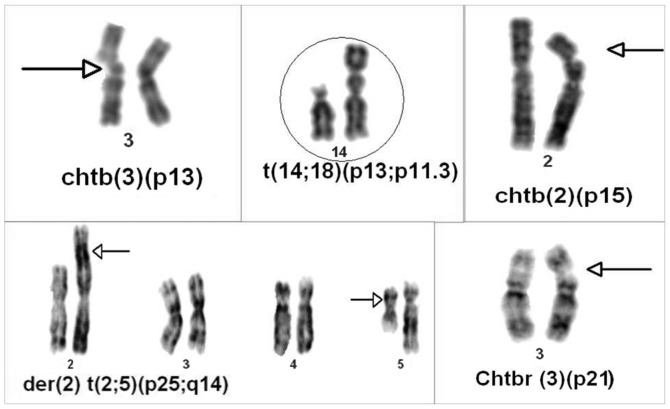
CAs were identified in 576 (24.6%) of the 2,344 cells analyzed in peripheral blood [363 (15.5%) and 213 (9.1%) of the cells had structural and numerical aberrations, respectively], and 62 (19.5%) of the 318 cells analyzed in tumoral tissues [24 (7.5%) and 38 (11.9%) of the cells had structural and numerical aberrations, respectively]. Structural aberrations predominated and usually consisted of deletions, translocations, breaks and fragilities in various chromosomes. In particular, deletions in 1p24-pter, 1p32-pter, 1q41-qter, 1q32-qter, 2p13-pter, 2p23-pter, 2p24-pter, 3p13-p14, 3p23-pter, 3p25-pter, 3q11-qter, 5p14-pter, 5q13-q15, 5q31-qter, 7q11-q12, 9q12-qter, 9q22-qterx2, 10q23-qter, 10q24-qter, 11q?, 13q32-qter, 14q11.2-pter, 17q11-qter, 17q21-qter and Xp21-pterx2; translocations between t(2;5)(p25;q14), t(3;12)(p26;q15), t(7;15)(p22;q26), t(7;14)(p15;q24), t(10;14)(q26;q13), t(12;17)(p13;q12), t(13;22)(p11;p11), t(14;22)(q32;q11.2), t(19;19)(q13.4;q13.4) and t(21;22)(p13;p13) ×2; and inversions in izo(Xq/p) and inv(13)(p13;q14) were more frequently observed. In patient 30, inversion of chromosome 9 [inv(9)(p11;q13)] was found in the blood (Table I, Fig. 1). Autosomal monosomies were observed as common findings (chromosomes X, Y, 22, 21, 17 and 8; and trisomies 21, Y, 4 and 15). In the control group, chromosomal aberrations were only found in 33 (2.6%) of 1,250 analyzed cells. The mean number of chromosomal abnormalities in patients with BC compared with the healthy control group was 20±36.2 (range, 0–182) and 1.3±1.6 (range, 0–5), respectively, and the difference between these values was significant (P=0.0001). Also, chromosomal abnormalities were overviewed and compared between the two groups (HG and LG) and almost all of the structural abnormalities found at 1q21, 1q32, 3p21 and 5q31 were detected in patients with HG tumors. Other structural abnormalities were found not only in patients with HG tumors but also in patients with LG tumors.
Figure 1.
Partial metaphase figures showing chromosomal abnormalities.
FISH findings
A total of 32 patients with multiple copies of the p53 and p16 signals were identified by an interphase FISH screening program using the Poseidon probe. A genetic alteration (amplification and mostly deletion) of p16 was observed in 6.30±4.47 cells (range, 0–60 cells) in the LG group and in 13.8±5.65 cells (range, 0–23 cells) in the HG group, and the difference was significant (P=0.002). Similarly, an alteration (amplification and mostly deletion) of p53 was detected in 7.7±6.21 cells (range, 0–23 cells) in the LG group and 12.4±5.99 cells (range, 0–25 cells) in the HG group, and these differences were also significant (P=0.039). When the cut-off value of 10 altered cells was considered, 19 patients had a positive result for p16 and 17 of these 19 patients had a HG tumor [odds ratio, 13.6; 95% confidence interval (CI), 2.2–85.8]. In addition, 19 patients had a positive result with the same cut-off value for p53, and 16 of these 19 patients had a HG tumor (odds ratio, 6.22; 95% CI, 1.2–32.2) (Table I, Fig. 2).
Figure 2.
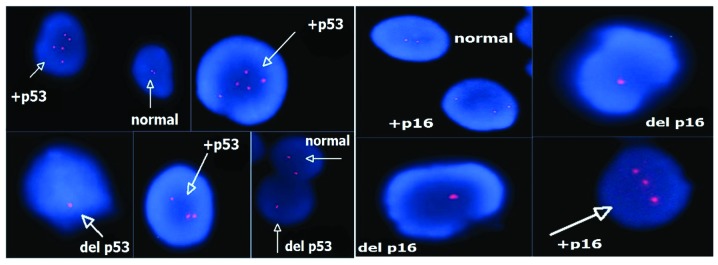
Partial fluorescence in situ hybridization images showing amplification of p53 and p16. del, deletion.
Although the number of chromosomal abnormalities was higher in the HG group compared with the LG group [23.26±43.19 (range, 0–182) vs. 12.5±4.89 (range, 4–18)], this difference was not significant (P=0.714). However, when the changes of the p16 and p53 genes specifically are considered, these differences were significant (P=0.002 and P=0.039) (Table I).
When all patients were considered, the majority of structural abnormalities were observed on chromosomes 1, 2, 3, 5 and 9, and the majority of numerical abnormalities were observed on chromosomes 8, 17, 21, 22, X and Y. The regions of 1p24–36, 1q21, 1q32, 2q31, 3p21, 3p25–26, 4q31, 5q31, 5q33, 6p21 and 9p-q were detected as being the most affected areas (Table I).
Discussion
In the present study, the risk factors of age, BMI and smoking time were compared between LGBC and HGBC patient groups. There were no statistically significant differences between the two groups in terms of these factors. Associations between HGBC and older age and longer smoking time were predicted, but no significant differences were found. This may be due to the small study population.
BC is a multistep and complex genetic process and mainly presents as one of two distinct tumor entities: Genetically stable LG tumors and genetically unstable HG tumors (15). While LG tumors are less aggressive, HG tumors can be highly aggressive (6). In the present study, the mean number of chromosomal abnormalities in patients with BC was significantly higher compared with the control group (P=0.0001). In addition, chromosomal abnormalities were detected more frequently in HG tumors compared with LG tumors, but the difference was not significant (P=0.714). Chromosomal abnormalities are more frequently detected in higher-stage than lower-stage BC (16). This means that genetic changes are necessary for the development of cancer and that there is a linear correlation between the aggressiveness of the tumor and the genetic aberrations present.
The 1p24, 1p36, 1q21 and 1q32 regions on chromosome 1 were identified as being the most affected areas in all patients with BC. However, the 1q21 and 1q32 regions were found to be affected more prominently in patients with HGBC compared with LGBC (Table II). A study by Tommasi et al (17) isolated the NORE1 gene at 1q32.1 that is homologous to the tumor suppressor gene RASSF1A, and advocated that this gene may be involved with the signal transmission of Ras or Ras-like proteins. Caramazza et al (18) reported that specific genes located at 1q21 were associated with myeloproliferative neoplasms, and that this region may contain oncogenes or tumor suppressor genes. According to the results of the present study, the 1q21 and 1q32 regions may contain certain oncogenes or tumor suppressor genes that play a significant role in the development of invasive BC.
Table II.
Comparison of structural and numerical abnormalities for each chromosome in low and high-grade bladder cancers.
| Chrom. no. | Structural abnormalities | Numerical abnormalities | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Location (ratio) | Low grade | High grade | ||||
|
|
|
|
||||
| Low grade | High grade | n | Ratio | n | Ratio | |
| 1 | 1p24(1/50), 1p32(1/50), 1p36.1(1/50), 1q32 (1/27) | 1p36(1/50), 1q11(1/50), 1q21(7/150), 1q22(1/100), 1q32(4/174), 1q36.1(1/50), 1q41(1/50), 1q42(1/100) | 1 | (1/32) | 2 | (2/69) |
| 2 | 2p23 (1/50), 2q23(1/50) | 2p13(1/50), 2p15(2/80), 2p23(1/50), 2p24(1/50), 2p25(1/50), 2q21(1/50), 2q22(3/100), 2q23(1/50), 2q31(3/150), 2q32.2(1/24), 2q33(1/50), 2q35(2/105), 2q?(2/50) | 1 | (1/12) | 4 | (4/105) |
| 3 | 3p13(1/50), 3p21.3(1/50), 3p25(2/100), 3q26.2(2/60) | 3p21(6/261), 3p23(1/50), 3p25(2/52), 3p26(1/30), 3q11(1/50),3q11(1/30), +3p(1/50) | 3 | (3/103) | 2 | (2/60) |
| 4 | 4q31(1/6), 4q33(1/32), +4q(1/14) | 4q27(1/55), 4q31.3(2/74), 4q(1/52) | 0 | - | 5 | (5/234) |
| 5 | 5p13(1/50), 5p14(1/50), 5q13(1/50), 5q31(1/50), 5q33(2/63) | 5q15(1/10), 5q31(8/365), 5q33(2/35) | 0 | - | 1 | (1/10) |
| 6 | 6p21(1/27) | 6p21(2/50), 6q15(1/50), 6q23(2/105) | 1 | (1/14) | 0 | (0/0) |
| 7 | 7q11(1/50), 7q?(1/32) | 7p15(1/50), 7p22(1/36) | 0 | - | 2 | (2/105) |
| 8 | 8q23(1/50), 8q22(1/50) | 4 | (4/127) | 8 | (8/449) | |
| 9 | 9p11(3/50), 9q12(1/50), 9q32(1/50), 9qh+(6/109) | 9p11(100/100), 9q22(2/60), 9q32(1/60), 9qh+(64/358) | 1 | (1/50) | 1 | (1/10) |
| 10 | 10q23(1/30), 10q24(1/50), 10q26(1/50) | 4 | (4/138) | 4 | (4/170) | |
| 11 | 11q23(1/22) | 11q?(1/50), 11q13.4(1/57) | 1 | (1/12) | 4 | (4/123) |
| 12 | 12q13(2/60) | 12p13(2/102), 12q14.1(1/50), 12q22(2/105), 12q24(1/10) | 1 | (1/12) | 6 | (6/257) |
| 13 | 13p13(1/54), 13q32(1/52) | 0 | - | 5 | (5/220) | |
| 14 | 14q11.2(1/22) | 14q32(1/50), 14q?(1/50) | 2 | (2/40) | 5 | (5/194) |
| 15 | 15p+(68/218) | 1 | (1/50) | 5 | (5/247) | |
| 16 | 16q22(1/50), 16qh+(14/14) | 0 | - | 4 | (4/203) | |
| 17 | 17p11(2/150), 17q21(1/25) | 8 | (8/39) | 6 | (6/208) | |
| 18 | 4 | (4/164) | 4 | (4/167) | ||
| 19 | 19q13(1/50) | 5 | (5/205) | 4 | (4/148) | |
| 20 | 2 | (2/64) | 4 | (4/158) | ||
| 21 | 9 | (9/268) | 10 | (10/429) | ||
| 22 | 22p+(4/50) | 8 | (8/292) | 15 | (15/526) | |
| X | Xp21(1/50), Xq/p(1/32) | Xp22.1(1/50), Xq13(1/55), Xq21(1/10), Xq26(2/107) | 2 | (2/82) | 84 | (84/202) |
| Y | Yq+(1/16) | 17 | (17/250) | 8 | (8/382) | |
Chrom. no., chromosome number; n, number of cells with aberration; -, choromosomal abnormality was not detected.
Specific alterations were found at 3p21, 3p25 and 3p26 in the patients of the present study. When these findings were compared between patients with HG and LG tumors, the 3p21 loci was dominantly altered in the HG group (Table II). There are a number of reported genes at 3q21 that are associated with either genitourinary or other tumors in the literature. The GPX1 gene was reported as a selenium-dependent detoxifying enzyme gene located at chromosome 3p21, and a study by Ichimura et al (19) showed that the GPX1 Pro/Leu genotype was associated with an increased risk of BC and may also be associated with the development of high-stage tumors. The TU3A gene, located on 3p21.2, was reported as a candidate tumor suppressor gene in renal cell carcinoma (RCC). Additionally, Awakura et al (20) advocated that this gene is involved in primary cancers of the bladder and testis. The histone methyltransferase gene SETD2/HYPB, located at 3p21.31, was identified as a novel tumor suppressor gene in RCC (21). The RASSF1 gene, located at 3p21.3, is silenced in a variety of human cancers, including lung, bladder, prostate and kidney cancers (22). Jarmalaite et al (23) studied promoter hypermethylation of the p16, RARβ, RASSF1A, DAPK and MGMT genes in patients with BC, and hypermethylation of the RASSF1A gene was more frequently detected in muscle-invasive tumors compared with non-invasive tumors. A high frequency of RASSF1A methylation, or the inactivation of RASSF1A, was correlated with an advanced tumor stage and poor prognosis in cases of BC, and hypermethylation of the RASSF1A gene was detected in urine samples with high specificity (24). In conclusion, the 3p21 gene location contains numerous cancer-related genes, and certain genes may be candidates for a panel of markers for BC.
The regions of 5q31 and 5q33 on chromosome 5 were also detected as highly affected areas in the present study, and 5q31 was more frequently altered in patients with a HG tumor rather than LG tumor (Table II). Specific studies have previously reported that in a variety of cancers, certain tumor suppressor genes were located to region 5q31. Dallasso et al (25) reported that protocadherin genes that are located to region 5q31 could be tumor suppressor genes in Wilms’ tumor. An association between the sprouty homolog 4 gene at 5q31 and testicular cancer was shown in a study by Kanetsky et al (26). These results indicate that the 5q31 gene location requires further study to elucidate its role in BC.
It is known that the p16 gene, located at 9p21, regulates the cell cycle and prevents abnormal cell proliferation. Statistically significant alterations in p16 were detected in HGBC in the present study. The alteration of p16 is concluded to be strongly correlated with the advanced tumor grade. In a previous study, the validity of p16 expression was evaluated in urine cytological and histological samples, and the study reported that a high incidence of p16 overexpression in HGUC was noted in cytological samples and that immunocytological analysis of p16 is a useful method for detecting UC and the tumor infiltrating potential (27). In another study, investigators researched the genetic alterations of the p16 and p14 genes in BC, and they did not find any association between tumor grade/stage and p16 alterations. However, the deletion of the p14 gene was more frequently observed in poorly differentiated tumors. This study also noted that p16 plays a role in early tumorigenesis (28). Conversely, in the present study it was found that the p16 gene was more frequently altered in patients with HGBC. Krüger et al (29) assessed the prognostic effect of p16 alterations in patients with T1 stage BC and concluded that there is a significant correlation between the status of p16 and progression-free survival. However, they did not find any significant correlations between p16 status and the tumor grade. The latter finding does not agree with the data of the present study. Currently, there is no consensus regarding p16 status associating to tumor grade, stage and prognosis.
Alterations in the p53 tumor suppressor gene are correlated with a number of varied malignancies. The association between p53 changes and a higher cancer grade, stage, recurrence, progression and mortality has been shown in a number of studies (30,31). Despite these studies, there is conflicting data regarding the p53 status. Malats et al (32) overviewed 168 publications from 117 studies and reported that changes in p53 are weakly predictive of recurrence, progression and mortality in BC. In the present study, alterations of p53 were more frequently observed in HGBC rather than LGBC. This difference was statistically significant. Furthermore, this result was similar to that of the p16 gene. Depending on the frequency of p53 alterations in HGBC, the expression of p53 in combination with other markers has also been researched. Shariat et al (33) studied four cell cycle regulators (p53, pRb, p21 and p27) in patients with locally advanced BC and advocated that the combination of multiple molecular markers was more informative than examining a single molecular marker. These results indicate that the study of the p53 and p16 genes has had predictive value in the clinic.
Currently used prognostic markers may be inadequate for effective treatment decisions. In the literature, there are numerous studies focused on determining prognostic markers. Although there is currently no consensus about molecular markers for BC, certain genes have been frequently detected in research. In the present study, alterations of p16 and p53 were more frequently detected in HG-cancer patients, and these genes may have predictive values for BC. Aside from these genes, novel chromosomal locations were searched for that may be responsible for the progression of BC. Chromosomal abnormalities of two patient groups were overviewed and compared. Almost all structural abnormalities were detected in the 1q21, 1q32, 3p21 and 5q31 regions in patients with HG tumors. Other structural abnormalities were found not only in patients with HG tumors, but also in patients with LG tumors. Based on this result, it was predicted that these regions may have a significant role in the progression of BC. Aberrations in these areas may be observed as a late event in BC pathogenesis and certain tumor suppressor genes or oncogenes may be located in these regions.
Numerous studies have advocated that the decision of BC management should not be made according to only one prognostic marker. In the present study, the p16 and p53 genes were assessed in patients with BC and it was revealed that these genes were altered more prominently in patients with HG tumors compared with patients with LG tumors, and this difference was statistically significant. In addition to these genes, the structural and numerical abnormalities of chromosomes were also assessed in blood and tissue samples. Certain structural abnormalities were mostly detected in the chromosomal regions of 1q21, 1q32, 3p21 and 5q31 in patients with HG tumors rather than LG tumors. These areas must be further studied to find candidate genes for a panel of BC markers.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Jacobs BL, Lee CT, Montie JE. Bladder cancer in 2010: how far have we come? CA Cancer J Clin. 2010;60:244–272. doi: 10.3322/caac.20077. [DOI] [PubMed] [Google Scholar]
- 3.Sullivan PS, Chan JB, Levin MR, Rao J. Urine cytology and adjunct markers for detection and surveillance of bladder cancer. Am J Trans Res. 2010;2:412–440. [PMC free article] [PubMed] [Google Scholar]
- 4.Vrooman OP, Witjes JA. Molecular markers for detection, surveillance and prognostication of bladder cancer. Int J Urol. 2009;16:234–243. doi: 10.1111/j.1442-2042.2008.02225.x. [DOI] [PubMed] [Google Scholar]
- 5.Habuchi T, Marberger M, Droller MJ, et al. Prognostic markers for bladder cancer: International Consensus Panel on bladder tumor markers. Urology. 2005;66(Suppl 1):64–74. doi: 10.1016/j.urology.2005.08.065. [DOI] [PubMed] [Google Scholar]
- 6.Proctor I, Stoeber K, Williams GH. Biomarkers in bladder cancer. Histopathology. 2010;57:1–13. doi: 10.1111/j.1365-2559.2010.03592.x. [DOI] [PubMed] [Google Scholar]
- 7.Lin HH, Ke HL, Huang SP, Wu WJ, Chen YK, Chang LL. Increase sensitivity in detecting superficial, low grade bladder cancer by combination analysis of hypermethylation of E-cadherin, p16, p14, RASSF1A genes in urine. Urol Oncol. 2010;28:597–602. doi: 10.1016/j.urolonc.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Knowles MA. The genetics of transitional cell carcinoma: progress and potential clinical application. BJU Int. 1999;84:412–427. doi: 10.1046/j.1464-410x.1999.00217.x. [DOI] [PubMed] [Google Scholar]
- 9.Cianciulli AM, Leonardo C, Guadagni F, et al. Genetic instability in superficial bladder cancer and adjacent mucosa: an interphase cytogenetic study. Hum Pathol. 2003;34:214–221. doi: 10.1053/hupa.2003.30. [DOI] [PubMed] [Google Scholar]
- 10.Eble JN, Sauter G, Epstein JI, Sesterhenn I, editors. WHO Classification of Tumours Pathology and Genetics of Tumours of the Urinary System and Male Genital Organs. IARC Press; Lyon: 2004. pp. 90–91. [Google Scholar]
- 11.Fundia AF, Larripa IB. Coincidence in fragile site expression with fluorodeoxyuridine and bromodeoxyuridine. Cancer Genet Cytogenet. 1989;41:41–48. doi: 10.1016/0165-4608(89)90106-4. [DOI] [PubMed] [Google Scholar]
- 12.Mitelman F, editor. ISCN: An International System for Human Cytogenetics Nomenclature. S Karger; Basel: 1995. [Google Scholar]
- 13.McAlpine PJ, Shows TB, Boucheli C, Huebner M, Anderson WA. The 1991 catalog of mapped genes and report of the nomenclature committee, Human Gene Mapping 11. Cytogenet Cell Genet. 1991;58:5–102. [Google Scholar]
- 14.Trask B, Pinkel D. Fluorescence in situ hybridization with DNA probes. Methods Cell Biol. 1990;33:383–400. doi: 10.1016/s0091-679x(08)60542-7. [DOI] [PubMed] [Google Scholar]
- 15.Junker K, van Oers JM, Zwarthoff EC, Kania I, Schubert J, Hartmann A. Fibroblast growth factor receptor 3 mutations in bladder tumors correlate with low frequency of chromosome alterations. Neoplasia. 2008;10:1–7. doi: 10.1593/neo.07178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolff DJ. The genetics of bladder cancer: a cytogeneticist’s perspective. Cytogenet Genome Res. 2007;118:177–181. doi: 10.1159/000108298. [DOI] [PubMed] [Google Scholar]
- 17.Tommasi S, Dammann R, Jin SG, Zhang XF, Avruch J, Pfeifer GP. RASSF3 and NORE1: identification and cloning of two human homologues of the putative tumor suppressor gene RASSF1. Oncogene. 2002;18: 21:2713–2720. doi: 10.1038/sj.onc.1205365. [DOI] [PubMed] [Google Scholar]
- 18.Caramazza D, Hussein K, Siragusa S, et al. Chromosome 1 abnormalities in myeloid malignancies: a literature survey and karyotype-phenotype associations. Eur J Haematol. 2010;84:191–200. doi: 10.1111/j.1600-0609.2009.01392.x. [DOI] [PubMed] [Google Scholar]
- 19.Ichimura Y, Habuchi T, Tsuchiya N, et al. Increased risk of bladder cancer associated with a glutathione peroxidase 1 codon 198 variant. J Urol. 2004;172:728–732. doi: 10.1097/01.ju.0000130942.40597.9d. [DOI] [PubMed] [Google Scholar]
- 20.Awakura Y, Nakamura E, Ito N, Kamoto T, Ogawa O. Methylation-associated silencing of TU3A in human cancers. Int J Oncol. 2008;33:893–899. [PubMed] [Google Scholar]
- 21.Duns G, van den Berg E, van Duivenbode I, et al. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res. 2010;1: 70:4287–4291. doi: 10.1158/0008-5472.CAN-10-0120. [DOI] [PubMed] [Google Scholar]
- 22.Angeloni D. Molecular analysis of deletions in human chromosome 3p21 and the role of resident cancer genes in disease. Brief Funct Genomic Proteomic. 2007;6:19–39. doi: 10.1093/bfgp/elm007. [DOI] [PubMed] [Google Scholar]
- 23.Jarmalaite S, Jankevicius F, Kurgonaite K, Suziedelis K, Mutanen P, Husgafvel-Pursiainen K. Promoter hypermethylation in tumour suppressor genes shows association with stage, grade and invasiveness of bladder cancer. Oncology. 2008;75:145–151. doi: 10.1159/000158665. [DOI] [PubMed] [Google Scholar]
- 24.Dammann R, Schagdarsurengin U, Seidel C, et al. The tumor suppressor RASSF1A in human carcinogenesis: an update. Histol Histopathol. 2005;20:645–663. doi: 10.14670/HH-20.645. [DOI] [PubMed] [Google Scholar]
- 25.Dallosso AR, Hancock AL, Szemes M, et al. Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms’ tumor. PLoS Genet. 2009;5:1000745. doi: 10.1371/journal.pgen.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanetsky PA, Mitra N, Vardhanabhuti S, et al. Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat Genet. 2009;41:811–815. doi: 10.1038/ng.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakazawa K, Murata S, Yuminamochi T, et al. p16(INK4a) expression analysis as an ancillary tool for cytologic diagnosis of urothelial carcinoma. Am J Clin Pathol. 2009;132:776–784. doi: 10.1309/AJCP61KNVHJVHAFN. [DOI] [PubMed] [Google Scholar]
- 28.Chang LL, Yeh WT, Yang SY, Wu WJ, Huang CH. Genetic alterations of p16INK4a and p14ARF genes in human bladder cancer. J Urol. 2003;170:595–600. doi: 10.1097/01.ju.0000067626.37837.3e. [DOI] [PubMed] [Google Scholar]
- 29.Krüger S, Mahnken A, Kausch I, Feller AC. P16 immunoreactivity is an independent predictor of tumor progression in minimally invasive urothelial bladder carcinoma. Eur Urol. 2005;47:463–467. doi: 10.1016/j.eururo.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 30.Moonen PM, van Balken-Ory B, Kiemeney LA, Schalken JA, Witjes JA. Prognostic value of p53 for high risk superficial bladder cancer with long-term followup. J Urol. 2007;177:80–83. doi: 10.1016/j.juro.2006.08.110. [DOI] [PubMed] [Google Scholar]
- 31.Smith ND, Rubenstein JN, Eggener SE, Kozlowski JM. The p53 tumor suppressor gene and nuclear protein: basic science review and relevance in the management of bladder cancer. J Urol. 2003;169:1219–1228. doi: 10.1097/01.ju.0000056085.58221.80. [DOI] [PubMed] [Google Scholar]
- 32.Malats N, Bustos A, Nascimento CM, et al. P53 as a prognostic marker for bladder cancer: a meta-analysis and review. Lancet Oncol. 2005;6:678–686. doi: 10.1016/S1470-2045(05)70315-6. [DOI] [PubMed] [Google Scholar]
- 33.Shariat SF, Chade DC, Karakiewicz PI, et al. Combination of multiple molecular markers can improve prognostication in patients with locally advanced and lymph node positive bladder cancer. J Urol. 2010;183:68–75. doi: 10.1016/j.juro.2009.08.115. [DOI] [PubMed] [Google Scholar]