SUMMARY
Actin assembly beneath enterohemorrhagic E. coli (EHEC) attached to its host cell is triggered by the intracellular interaction of its translocated effector proteins Tir and EspFU with human IRSp53 family proteins and N-WASP. Here, we report the structure of the N-terminal I-BAR domain of IRSp53 in complex with a Tir-derived peptide, in which the homodimeric I-BAR domain binds two Tir molecules aligned in parallel. This arrangement provides a protein scaffold linking the bacterium to the host cell's actin polymerization machinery. The structure uncovers a specific peptide-binding site on the I-BAR surface, conserved between IRSp53 and IRTKS. The Tir Asn-Pro-Tyr (NPY) motif, essential for pedestal formation, is specifically recognized by this binding site. The site was confirmed by mutagenesis and in vivo-binding assays. It is possible that IRSp53 utilizes the NPY-binding site for additional interactions with as yet unknown partners within the host cell.
INTRODUCTION
Enterohemorrhagic E. coli (EHEC) are food-born human pathogenic bacteria that cause severe diarrhea and the hemolytic uremic syndrome (HUS), defined by the triad of acute kidney failure, low platelet count, and destruction of red blood cells. EHEC strains of the O157:H7 serotype are the best studied, and in Europe, North America, and Japan clinically most relevant pathogenic E. coli (Nataro and Kaper, 1998). In the intestinal mucosa, attachment of EHEC O157:H7 bacteria to epithelial cells results in loss of microvilli and localized actin polymerization, leading to the formation of pseudopod-like “actin pedestals” beneath the attached bacteria. Pedestal formation is critical for the pathogenicity of EHEC and the closely related enteropathogenic E. coli (EPEC). Upon attachment, they utilize a cascade of specific protein-protein interactions to connect with the host cell's actin polymerization machinery. Although closely related, distinct pathways are exploited for inducing actin assembly by the EHEC O157:H7 and EPEC O127:H6 or by other, more recently identified EHEC and EPEC strains (Frankel and Phillips, 2008).
The initial step during infection for both, EHEC and EPEC, is tight attachment to the intestinal epithelium. This is mediated by translocation of the effector protein Tir (translocated intimin receptor) into the host cell via the type III secretion system (TTSS) (Hayward et al., 2006). Tir is integrated into the plasma membrane, and its extracellular domain serves as a receptor for the bacterial outer membrane protein intimin, thereby anchoring the bacterium to the host cell (Campellone and Leong, 2003). In this arrangement both termini of Tir project into the host cell cytoplasm providing the recruitment sites for the host's actin polymerization machinery. The N-terminal cytoplasmic domain is not absolutely required for pedestal formation but modulates pedestal length (Campellone et al., 2006). Its exact link to the actin cytoskeleton is still largely unclear but may involve α-actinin (Goosney et al., 2000) and cortactin (Cantarelli et al., 2006). In contrast the C-terminal cytoplasmic domain (TirC) is essential for pedestal formation (Campellone et al., 2006).
The Tir molecule is well conserved between EPEC, EHEC, and also other related bacteria such as Citrobacter rodentium. Only a short stretch of TirC around Tyrosine 474 is specific for EPEC-Tir. Phosphorylation of EPEC Y474 by host cell kinases (Phillips et al., 2004; Swimm et al., 2004) triggers binding of the SH2 domain of Nck proteins (Frese et al., 2006; Gruenheid et al., 2001), resulting in recruitment and activation of N-WASP and stimulation of Arp2/3-dependent actin polymerization (reviewed in Hayward et al., 2006). In contrast the signaling leading to EHEC-induced actin-pedestal formation is different due to the lack of this phosphorylation site in EHEC-Tir. Instead, a second virulence factor, EspFU, is delivered into the host cell via the TTSS. EspFU is able to recruit and activate N-WASP leading to Arp2/3-dependent actin polymerization. Finally, the EspFU:N-WASP complex is bridged to EHEC-Tir by another host cell factor, either IRSp53 (insulin receptor substrate p53) or its paralogue IRTKS (insulin receptor tyrosine kinase substrate). IRSp53 and IRTKS comprise an N-terminal IRSp53-MIM homology domain (IMD), and a central Src homology 3 (SH3) domain (Figure 1A). In addition, IRSp53 but not IRTKS harbors a central Cdc42/Rac interactive-binding (CRIB) motif. Although IRSp53 is subject to excessive C-terminal splicing and can encode C termini as divergent as PDZ domain-binding motifs or WH2 domains, IRTKS terminates with an actin-binding WH2 domain (Millard et al., 2007). EspFU consists of variable numbers of almost identical unspaced 47 amino acid repeats, each carrying a hydrophobic helix that binds to and triggers N-WASP (Cheng et al., 2008; Campellone et al., 2004) and a proline-rich domain that binds to the SH3 domain of IRSp53 (Weiss et al., 2009; Vingadassalom et al., 2009).
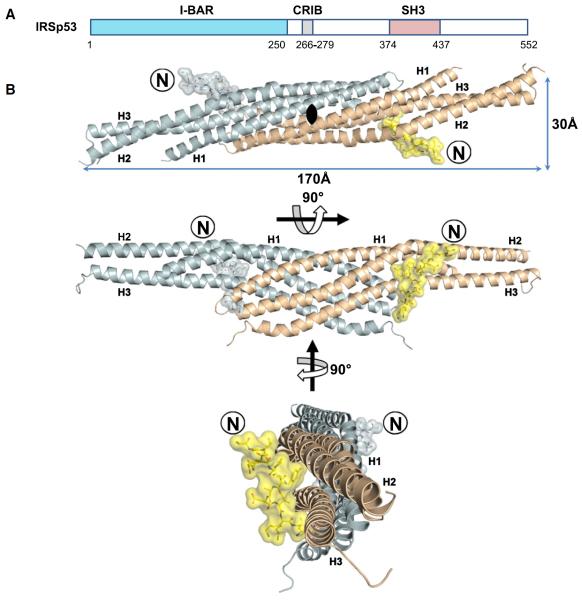
Figure 1. Complex of the I-BAR Domain of IRSp53 with the Tir-Derived Peptide.
(A) Location of the IRSp53 I-BAR and SH3 domains and CRIB motif.
(B) I-BARIRSp53 binds a 12 residue peptide derived from the EHEC virulence factor Tir incorporating the NPY motif. Ribbon diagrams of the two subunits of the I-BAR homodimer are drawn in cyan and wheat. Two identical peptide molecules are colored blue and yellow, respectively. Peptide N termini are labeled with an encircled N.
See also Figure S2.
The interaction surface of IRSp53/IRTKS with EHEC-Tir was identified to lie within the IMD (Weiss et al., 2009; Vingadassalom et al., 2009). A sequence of EHEC TirC that links to actin assembly has been mapped down to a stretch of nine amino acids with a central NPY motif (Campellone et al., 2006). Substitutions with alanine demonstrated that each position of the motif, but none of the neighboring positions, is critical for actin-pedestal formation. Interestingly, a conservative substitution of the tyrosine by phenylalanine attenuated pedestal formation but could not entirely abrogate it (Brady et al., 2007).
Ectopic expression of IRSp53 mediates the formation of filopodia (Lim et al., 2008), whereas cytoskeletal changes induced by IRTKS are more subtle (Millard et al., 2007). In any case these proteins link changes of plasma membrane morphology to actin cytoskeleton dynamics (Millard et al., 2005; reviewed by Scita et al., 2008; Suetsugu et al., 2006). IMDs are distant members of the Bin-Amphiphysin-Rvs (BAR) domain super-family. Whereas canonical banana-shaped BAR domains bind membranes with their concave surface and induce invagination, IMDs are cigar-shaped and promote outward protrusions like filopodia and are, therefore, referred to as inverse BAR or I-BAR domains (Frost et al., 2009; Mattila et al., 2007). Interestingly, I-BAR domains also directly bind to and bundle F-actin (Disanza et al., 2006; Millard et al., 2005; Suetsugu et al., 2006). However, two recent studies suggested that the isolated I-BAR domain itself does not crosslink actin filaments under physiological conditions (Mattila et al., 2007; Lee et al., 2007). The actin-binding sites overlap, at least in part, with those that target lipid membranes (Millard et al., 2005; Suetsugu et al., 2006). It should be noted, though, that the basic cluster of amino acids that confers lipid and potentially actin binding to I-BAR domains (Millard et al., 2005; Suetsugu et al., 2006) is not required for the formation of pedestals by EHEC bacteria (Weiss et al., 2009). Thus, in the context of EHEC infection, the function of IRSp53/IRTKS is to physically link Tir and EspFU/N-WASP.
In the current study we describe the structure of an EHEC Tir-derived peptide containing the NPY motif in complex with the I-BAR domain of IRSp53. The structure allows a detailed description of the I-BAR:Tir interaction and explains the critical role of the NPY motif in providing a central molecular scaffold, linking the bacterium and the host cell actin polymerization machinery. Furthermore, the novel binding site for NPY motifs described here has implications for IRSp53/IRTKS interactions and candidate proteins that may utilize this interaction surface are identified by data mining. Finally, the crystal structure was used to design IBAR mutants that are abrogated in binding, in vitro and in vivo.
RESULTS
Both Tir Molecules Bound by I-BAR Point into the Same Direction
The crystal structure of the complex of the I-BAR domain of IRSp53 with a dodecapeptide derived from EHEC Tir (GTVQNPYADVKT, residues 452–463) was solved by molecular replacement and refined at a resolution of 2.1Å (Table 1). In the resulting structure the I-BAR domain forms a 170Å long, cigar-shaped homodimer formed by a bundle of parallel coiled coils of kinked a helices (Figure 1). The two protomers of the dimer measure about 120Å in length and 30Å in diameter and bury a surface area of 2600Å2. The structure is in accord with previously published I-BAR structures (Millard et al., 2005; Suetsugu et al., 2006; Lee et al., 2007), with a calculated root-mean-square deviation (rmsd) of 0.95Å to Protein Data Bank (PDB) entry 1Y2O (218 common Cα positions aligned).
Table 1.
Summary of Crystallographic Analysis
| Data Collection | |
|---|---|
| Space group | C2221 |
| Cell dimensions (Å) | |
| a | 88.35 |
| b | 187.36 |
| c | 36.81 |
| X-ray source | Beamline14.2, BESSY II, Berlin |
| Wavelength (Å) | 0.9184 |
| Resolution range (Å) | 9.44–2.11 |
| Last shell (Å) | 2.16–2.11 |
| Rmerge (%) | 7.7 (53.2) |
| Observations | 231,684 (9,729) |
| Unique reflections | 18,190 (1,343) |
| Mean (I)/SD (I) | 20.13 (3.13) |
| Completeness | 99.8 (100) |
| Multiplicity | 12.7 (7.2) |
|
| |
| Structure Refinement | |
|
| |
| Resolution range (Å) | 9.44–2.11 (2.16–2.11) |
| Rwork (%) | 23.3 |
| Rfree (%) | 27.6 |
| Total Number of | |
| Nonhydrogen atoms | 1,991 |
| Protein atoms | 1,837 |
| Peptide atoms | 103 |
| Water molecules | 46 |
| Sulfate molecules | 1 |
| Rmsd | |
| Bond length (Å) | 0.022 |
| Bond angle (°) | 1.90 |
| Main-chain B factors (Å2) | 39.4 |
| Side-chain B factors (Å2) | 43.5 |
| Wilson B factor (Å2) | 41.8 |
| Average B factor protein atoms (Å2) | 41.5 |
| Average B factor peptide atoms (Å2) | 27.0 |
| Average B factor solvent atoms (Å2) | 40.8 |
| Ramachandran statistics | |
| Most favored regions (%) | 99.1 |
| Allowed regions (%) | 0.9 |
| Disallowed regions (%) | 0 |
Values in parentheses are for the highest resolution shell.
The electron density map clearly reveals the position of the bound Tir-derived peptide (Figure 2A). The I-BAR homodimer binds two Tir-derived peptides, one at the center of each protomer (Figure 1). The peptides point into the same direction, almost parallel to the 2-fold rotation axis of the dimer and perpendicular to the I-BAR cigar shape. C termini of the peptides point to the side of the domain that is assumed to face the cytoplasm in the membrane-bound state. Peptides are located far apart with 91 and 61Å distances between their C and N termini, respectively. Each peptide buries 564Å2 of the I-BAR surface, which is close to the average of buried surface areas in peptide-protein interactions (London et al., 2010). The whole length of the Tir-derived peptide is in close proximity to the domain, with the two Tir valine side chains (V454, V461) oriented inward, thus maximizing solvent entropy upon binding. All amino acid residues of the peptide and the domain are well defined in the electron density map, except for the loop between helices H2 and H3 and residues 234–250 encoding helix H4.
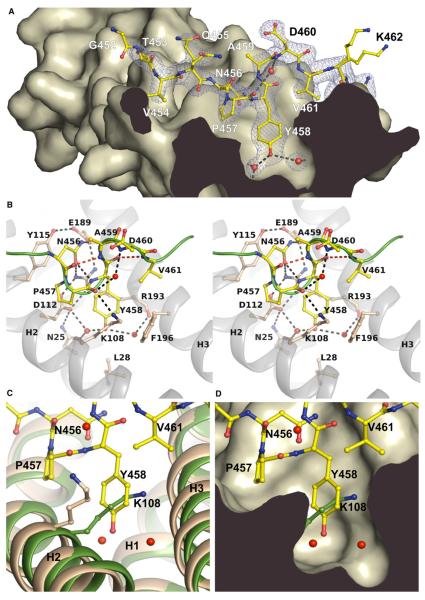
Figure 2. Atomic Details of the I-BARIRSp53:Tir Complex.
(A) Profile of the binding pocket. The Tir-derived peptide 452GTVQNPYADVKT463 is drawn in yellow with its electron density map in blue. Dashed lines indicate hydrogen bonds. Tyr458 of the NPY motif is centered by two water molecules (red) in the interior of the binding pocket.
(B) Stereo view of the interactions of the Tir-derived peptide comprising the NPY motif (456–458). Residues involved in the complex interface and in β-turn formation are labeled. The green line represents the backbone of the peptide. Hydrogen bonds involved in β-turn formation are shown as red dashed lines (see also Movie S1). (C and D) Superposition of the free I-BAR structure of Millard et al. (2005) (green, PDB 1Y2O) and the peptide-bound domain (wheat). The side chain of Lys108 blocks the binding pocket in the free structure. The conformation of the side chain of Lys108 in the free I-BAR structure of Suetsugu et al. (2006) is similar to the complex (1WDZ, not shown).
See also Movie S1.
A Novel Peptide-Binding Site of IRSp53 Recognizes the NPY Motif
In the structure of the I-BAR:Tir complex, the NPY motif of Tir interacts with a large hydrophobic-binding pocket in the central region of the I-BARIRSp53 protomer (Figure 2A). This pocket is also present in the free structure of the I-BAR domain, as reported by Suetsugu et al. (2006) (PDB entry 1WDZ). On the contrary, in the structure reported by Millard et al. (2005) (PDB entry 1Y2O), the binding pocket of one of the two protomers is partly occluded by the side chain of Lys108 (Figures 2C and 2D). The reason for this difference remains unclear.
All three side chains of the NPY motif (456–458) are specifically bound by the I-BAR domain of IRSp53. The conformation of the peptide backbone is defined by two overlapping type I β-turns (456–459, 458–461), which are stabilized by three intramolecular hydrogen bonds and an ordered water molecule (Figures 2B and 3; see Movie S1 available online). The pyrrolidine ring of Pro457 of the NPY motif further rigidifies the backbone and is accommodated in a shallow hydrophobic groove on the I-BAR surface (Figures 2A and 2D).
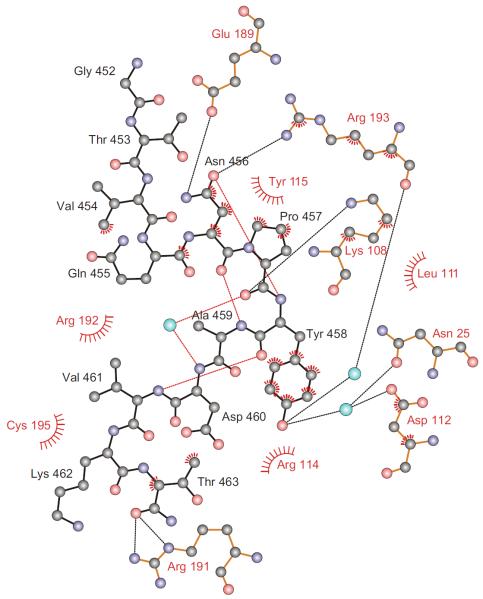
Figure 3. Schematic Diagram of Atomic Interactions.
EHEC Tir and the I-BAR's binding pocket are drawn with black and red labels and bonds, respectively. Cyan spheres represent water and dashed lines hydrogen bonds. Hydrophobic interactions are indicated by spoked arcs and atoms with spokes. An ordered water molecule and three intramolecular hydrogen bonds (red) stabilize the peptide's two overlapping β-turns. Oxygen atoms are colored red, nitrogen atoms blue, and carbon atoms black. See also Figure S1.
The side chain of Tyr458 of the NPY motif is accommodated in a deep hydrophobic pocket and is sandwiched between the hydrophobic portions of the I-BAR Arg193 and Lys108 side chains (Figures 2A and 2B; Movie S1). Two ordered water molecules are bound in the interior of the pocket and link the Tyr458 hydroxyl group to helices H1, H2, and H3 of the I-BAR domain (Figures 2B and 3; Movie S1). Substitution of Tyr458 by phenylalanine would compromise these water-mediated interactions, which rationalizes the reduced pedestal formation of the EHEC-Tir Y458F mutant (Brady et al., 2007).
Recognition of Asn456 of the NPY motif is based on the unique hydrogen bonding potential of its side chain (Figure 3). The side chain's carbonyl oxygen is hydrogen bonded to a nitrogen of the guanidinium group of I-BAR Arg193 and accepts an additional, intrapeptide hydrogen bond from the amide nitrogen of Tir Tyr458, an i to i+2 interaction that often stabilizes Asn-Pro-containing β-turns (Wilson and Finlay, 1997). The side-chain amide nitrogen of Asn456 is hydrogen bonded to I-BAR Glu189. Hydrophobic contacts of the Asn456 side chain to Tyr115 further strengthen the interaction. Clearly, asparagine is essential at this position because no other residue could fully satisfy all of the observed bonding constraints.
Arg193 of the I-BAR domain of IRSp53 is part of the signature sequence 189EERRR193 of the IMD family (Yamagishi et al., 2004) and appears to be the most critical residue for the recognition of the NPY motif. In the free domain its side chain is already oriented in the correct conformation for binding to Tir Asn456 and Tyr458 by a salt bridge between its guanidinium group and Asp112. This is in contrast to Lys108, which has a variable side-chain conformation in the absence of the peptide.
In the structure of the free I-BAR homodimer of Millard et al. (2005) (PDB 1Y2O), Arg191 binds to Asp232 of the loop linking the opposing protomer's helices H3 and H4 (Figure S1). In complex to Tir the Arg191 guanidinium group contacts the oxygens of the peptide's terminal carboxylic amide group and of Ala229 of the opposing protomer. This induced an altered conformation of the loop linking helix H3 and the last helix, H4, which was in turn not observed in the structure. These differences may be causative of the entirely different crystal packing of the complex (Figure S2).
The Tir-Binding Site Is Conserved
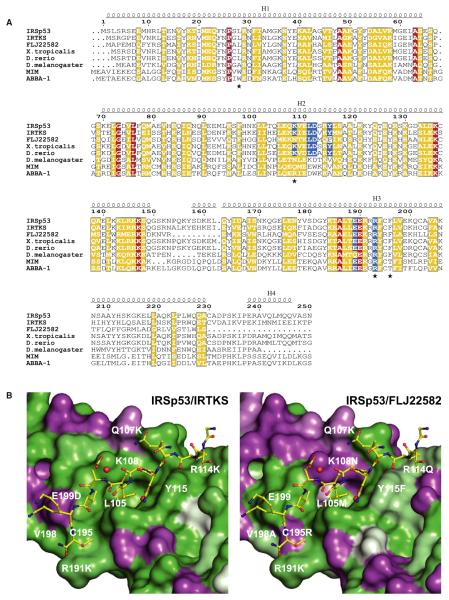
The paralogues IRSp53 and IRTKS interact with both EHEC effectors Tir and EspFU via their I-BAR and SH3 domains, respectively. In contrast the third family member in mammals, FLJ22582, only binds EspFU (Weiss et al., 2009). The sequence identity for the I-BARs of IRSp53 and IRTKS is 47%, and all residues contacting the NPY motif are conserved (Figure 4A). However, the I-BAR domain of FLJ22582 has only a sequence identity of 28% to IRSp53, and the residues forming the binding pocket differ markedly. The most prominent difference is FLJ22582 Asn108, which replaces Lys108 of IRSp53 (Figure 4B). Lys108 is one of most important residues in the binding pocket, stabilizing Tyr458 of the NPY motif (Figure 2B). Thus, the lack of Tir binding in FLJ22582 may be explained by its absence. The Tir-binding site is highly conserved within the group of IRSp53 and IRTKS orthologs of evolutionary distant vertebrates including claw frog and zebrafish, but not in invertebrate orthologs. The site is also not conserved in the human I-BAR proteins MIM and ABBA-1 (Figure 4A).
Figure 4. I-BAR Sequence Conservation.
(A) Sequence alignment of the known human I-BAR domains of IRSp53, IRTKS, FLJ22582, MIM, and ABBA-1 and IRSp53 orthologs from Xenopus tropicalis (claw frog), Danio rerio (zebrafish), and Drosophila melanogaster (fruit fly) (GenBank accession numbers NP_059344, NP_061330, NP_079321, NP_055566, NP_612392, NP_001121436, NP_001035335, NP_729679). Blue boxes indicate IRSp53 residues in contact to the Tir NPY motif. Red and yellow boxes highlight identical and similar residues, respectively. Positions that were mutated in this study are marked by asterisks.
(B) Sequence conservation between the IRSp53 residues constituting the hydrophobic-binding pocket and IRTKS (left) or FLJ22582 (right). Deep green indicates identity, whereas purple represents differing residues.
Effect of Mutations on Complex Formation
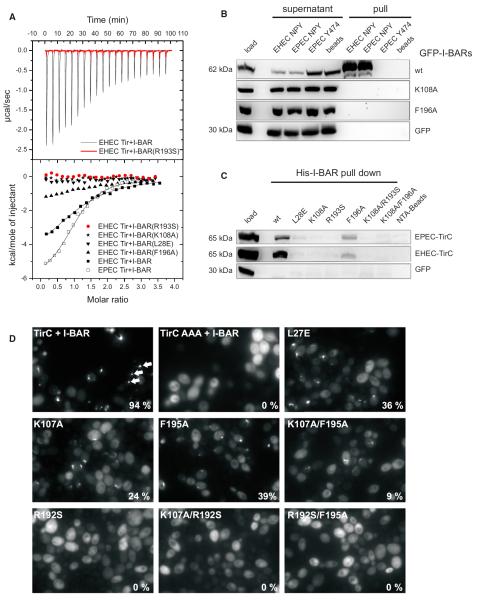
Complex formation between the I-BAR domain of IRSp53 and the Tir-derived peptide was studied by isothermal titration calorimetry (ITC). The stoichiometry of two peptide molecules per I-BAR homodimer was confirmed, and binding was apparently noncooperative. A dissociation constant of 59 ± 10 μM was determined (Figure 5A), which indicates a weak interaction, comparable to SH3 domain interactions with proline-rich motifs (Mayer and Saksela, 2004; Ball et al., 2005; Kuriyan and Cowburn, 1997). A corresponding 12-mer peptide was derived from EPEC-Tir (SEVVNPYAEVGG), and a similar dissociation constant of 37 ± 14 μM was obtained. Binding of the I-BAR domain of IRSp53 to additional peptides with motifs related to NPY was studied by ITC, including the 456NPY458→NPF mutant described above and a peptide derived from the insulin receptor comprising the related NPXY motif. No binding was detectable by ITC, demonstrating preference of the I-BAR for the NPY motif. To interpret these results, it should be noted that the protein concentration used for ITC (100 μM) was close to the dissociation constant of the wild-type domain and the Tir-derived peptide (60 μM), and it would not have been possible to detect binding at lower affinities.
Figure 5. Interaction of Tir with Wild-Type and Mutant I-BAR Domains.
(A) ITC of the binding reaction of I-BARIRSp53 and four I-BAR point mutants with peptides derived from EHEC and EPEC Tir, comprising the NPY motif. The
upper panel shows considerable heat release upon titration of the wild-type I-BARIRSp53 (black) with the EHEC Tir-derived peptide, in contrast to the R193S mutant (red). The heat data, corrected for dilution, are plotted against the molar ratio of peptide molecules and I-BAR protomers in the lower panel. Sigmoidal curves typical of exothermic-binding reactions were obtained for the wild-type I-BARIRSp53 domain and the mutant F196A, but not for the R193S, K108A, and L28E mutants.
(B) Pull-down assays of Tir-derived peptides from lysates of mammalian cells ectopically expressing GFP-tagged I-BAR variants of human IRSp53 as indicated on the right. Shown are cell lysates before (load) and after (supernatant) pull-downs and precipitates (pull) of three different peptides as indicated above. EHEC Tir-derived peptides that bind to Nck instead of IRSp53 (Y474) as well as streptavidin beads were employed as bait-control, and GFP alone in the cell lysate was the prey-control.
(C) Pull-down assays of recombinant His-tagged I-BAR domains from lysates of COS7 cells ectopically expressing the cytoplasmic C termini of EPEC or EHEC Tir as indicated on the right. Shown is the cell lysate before the pull-down (load) and the precipitates (His-I-BAR pull-down) of wild-type I-BAR (wt) and six variants as indicated above. Note the weak binding to mutant F196A in accordance with the ITC measurements.
(D) Interaction of Tir with wild-type and mutant I-BAR domain of IRTKS in vivo. Yeast cells coexpressing μNS-TirC and wild-type or mutant GFP-tagged I-BAR domains of IRTKS were visualized by fluorescence microscopy 4–5 hr post-induction of fusion protein expression. Exemplary GFP inclusions are marked by arrows. The percentage of cells containing GFP inclusions was determined visually and is indicated at the bottom right of each image.
Close inspection of the binding site led us to design a set of IRSp53 point mutants that were predicted to be impaired in Tir binding without disturbing the overall I-BAR structure (Figure 4A). Substitution of Leu28 by glutamic acid (L28E) was predicted to fill the hydrophobic pocket. The residue Lys108 forms a hydrogen bond and hydrophobic contacts with the peptide. It is changed to an asparagine in FLJ22582, the IRSp53 paralogue that does not recognize Tir. The conformation of the Lys108 side chain is stabilized by Phe196. Arg193 may be critically important for recognizing Tir because it is the residue with most interactions with the peptide (Figure 3; Movie S1), and because the salt bridge to Asp112 appears to stabilize the binding pocket. No binding of the Tir-derived peptide was detected by ITC when Leu28, Lys108, or Arg193 was substituted in the I-BAR domain of IRSp53 (L28E, K108A, R193S in Figure 5A). Binding of the Phe196 point mutant F196A to the peptide was weaker than for the wild-type domain but still clearly detectable.
Next, we probed with pull-down assays the ability of the above-described I-BAR mutants to interact with Tir-derived peptides or the entire cytoplasmic C-terminal tail of EPEC- and EHEC-Tir (TirC). We performed pull-down assays from lysates of COS7 cells ectopically expressing the I-BAR variants as N-terminal GFP fusions using biotinylated synthetic Tir-derived peptides immobilized on streptavidin-agarose beads as bait. Under these conditions, wild-type EPEC and EHEC-derived peptides showed significant binding to the I-BAR domain of IRSp53, and could effectively deplete GFP-tagged I-BAR domains from the lysate and enrich in the precipitate (Figure 5B). In contrast, neither I-BAR mutants K108A, F196A (Figure 5B), nor L28E, R193S, or the I-BAR double mutants K108A/R193S and K108A/F196A (not shown) could be detected in the precipitates. Vice versa experiments were performed by using recombinant His-tagged wild-type and mutant I-BARs as baits in pull-down assays from lysates of COS7 cells ectopically expressing the entire TirC. The recombinant wild-type I-BAR domain effectively bound to TirC from both EPEC and EHEC. In accordance with the data obtained by ITC, weak but reproducible binding of TirC was also observed for the F196A mutant of I-BAR domain of IRSp53, whereas all other I-BAR mutants were abrogated for binding (Figure 5C).
Finally, Protein Interaction Platform (PIP) assays (Schmitz et al., 2009) were used to investigate whether the mutations analyzed above within the I-BAR of IRSp53 would also affect the interaction of Tir with IRTKS. Given the strong conservation of the Tir-binding site, IRSp53 and IRTKS most likely bind to the Tir in the same fashion. EHEC TirC and a 456NPY458→AAA substitution mutant (TirC AAA) (Vingadassalom et al., 2009) were fused to the scaffolding protein μNS that forms large focal inclusions (“platforms”) in yeast cells. Coexpression of GFP-tagged I-BAR of IRTKS with the TirC fusion protein resulted in complex assembly and formation of fluorescent foci in almost all cells (Figure 5D). The TirC AAA mutant was not able to alter the diffuse intracellular localization of I-BARIRTKS, as expected. Substitution of IRTKS Lys107 or Phe195 (corresponding to IRSp53 Lys108 and Phe196) partially inhibited the interaction with TirC (K107A and F195A, Figure 5D). This was exacerbated by combining both mutations (K107A/F195A). Strong inhibition was also observed for the substitution of Leu27 by glutamic acid (L27E). Finally, substitution of Arg192 in IRTKS completely abolished the Tir:I-BARIRTKS interaction (R192S), as the double mutations R192S/F195A and K107A/R192S did (Figure 5D).
DISCUSSION
The crystal structure presented here reveals the molecular details of the interaction between EHEC Tir and the I-BAR of human IRSp53. It elucidates why the highly conserved NPY motif of EHEC Tir is critical for interaction with the I-BAR domains of IRSp53 and IRTKS, whereas neighboring residues can be substituted with alanine without any effect (Brady et al., 2007; Weiss et al., 2009). Specificity of recognition of Tir by the I-BAR is determined by the characteristic features of the NPY motif's three residues, 456–458. The Asn456 side chain is bound by specific hydrogen bonds. The Tyr458 phenyl ring projects into a hydrophobic pocket, forming extensive hydrophobic interactions to the I-BAR domain, whereas the phenolic hydroxyl group is bound by two water molecules at the bottom of the pocket. Correspondingly, substitution of Tyr458 with alanine virtually abolished pedestal formation by EHEC bacteria, whereas substitution with phenylalanine led to a milder defect (Brady et al., 2007). The pyrrolidine ring of Pro457 of the NPY motif fits into a shallow hydrophobic cavity on the I-BAR surface. It rigidifies the backbone and supports the formation of two overlapping type I β-turns in the Tir polypeptide chain. The crucial role of certain residues within the I-BAR NPY-binding site revealed by the structure was confirmed by site-directed mutagenesis combined with binding studies. The interaction of the I-BAR domains of IRSp53 and IRTKS with Tir in vitro and in living cells was impaired by substitution of residues constituting the peptide-binding site (Figure 5). The results were conclusive, also concerning residual binding that was always found to be strongest for the F196A or F195A mutants, respectively. The tight interaction of Arg193 with Tir in the crystal structure was reflected by the complete loss of binding upon substitution of the corresponding residues in IRSp53 and IRTKS. Substitution of Lys107/108 in IRTKS or IRSp53 considerably reduced or abolished binding. The homologous FLJ22582 has an asparagine at this position, which is likely causative of its lack of binding to Tir (Weiss et al., 2009).
Full-length IRSp53 was shown to equally well precipitate with both EPEC and EHEC Tir-derived peptides, containing this NPY motif. Moreover, both GFP-tagged IRSp53 and endogenous IRSp53 can be readily detected in both EPEC and EHEC pedestals. However, in contrast to EHEC, that essentially require IRSp53, it is dispensable for EPEC pedestal formation as exemplified by using IRSp53 knockout cells (Weiss et al., 2009). Notwithstanding this, introduction of EspFU into EPEC drastically increases Nck-independent pedestal formation (Brady et al., 2007). Thus, IRSp53 effectively binds to EPEC Tir, but no information is available to date on the state of EPEC Tir Y454 phosphorylation, when bound to IRSp53. Together, our current data confirm a virtually complete conservation of the binding characteristics between IRSp53 and IRTKS to the C termini of EPEC and EHEC Tir.
The mode of binding of the NPY motif is strikingly similar to the recognition of NPXY and NPF motifs by phosphotyrosine binding (PTB) and Eps15 homology (EH) domains, respectively. The NPXY and NPF motifs also form stabilized Asn-Pro type I β-turns, their asparagine side chains are specifically hydrogen bonded, and their aromatic side chains enter hydrophobic pockets in the respective domains (de Beer et al., 2000; Eck et al., 1996). Binding of peptides containing an NPXY or NPF motif by I-BAR domains was not detected by ITC, indicating preference for NPY over related motifs.
The striking specificity of the recognition of the NPY motif by the I-BAR domain suggests that the domain has evolved to recognize this motif, a feature that just becomes usurped by pathogenic E. coli. Thus, it is tempting to speculate that this binding site may have cellular ligands that harbor NPY motifs, which occur in up to 3% of human proteins. Among these proteins, numerous candidates can be found that are related to cellular processes that may involve IRSp53 (Table S3). It will be exciting to probe these interactions and analyze the recently described IRSp53 knockout mouse (Sawallisch et al., 2009) for potential phenotypes that may relate to defective I-BAR:NPY interactions. However, up until now, no binding partners for the I-BAR domains of IRSp53 and IRTKS other than Tir have been pinned down unambiguously because the in vivo relevance of its binding to both Rac1 (Disanza et al., 2006; Krugmann et al., 2001; Miki et al., 2000) and actin (Mattila et al., 2007; Millard et al., 2005) is still the subject of debate. Insights derived from the structure described here may pave the way to identify new interactions potentially involving the NPY motif.
The structure described here adds the I-BAR to the list of known peptide-binding domains. The binding energy of protein-protein and peptide-protein interactions generally depends on only a small number of “hot spot” residues (London et al., 2010), obviously represented here by the NPY motif, including the tightly bound Tyr458. Interestingly, tyrosine ranks in third position of the most frequent amino acid residues identified in peptide-protein complexes (London et al., 2010). Specific recognition of phosphorylated Tyr474 by Nck proteins is essential for pedestal formation by EPEC, as mentioned above. Other tyrosines of EPEC Tir at positions 483 and 511 also become phosphorylated and are essential for recruiting SHIP2, a host inositol phosphatase controlling pedestal formation by generating a phosphatidylinositol 3,4-bisphosphate-enriched lipid platform (Smith et al., 2010). It appears that tyrosine-mediated interactions of Tir with specific peptide recognition domains of the host cell are critical for pedestal formation of both EHEC and EPEC.
The almost parallel orientation and the distance of the two Tir-derived peptides in the complex suggest that one IRSp53 homodimer can link two Tir molecules in vivo. An affinity of 60 μM was found for the binding reaction under in vitro conditions, which is comparable to the relatively weak interactions between SH3 domains and proline-rich motifs (Ball et al., 2005; Kuriyan and Cowburn, 1997). The affinity and specificity of ligand binding by SH3 can be greatly enhanced by additional contacts between the SH3 loop regions and residues of the ligand outside the proline-rich motif (Mayer and Saksela, 2004; Arold et al., 1998). Regions outside the Tir-derived peptide might likewise contribute to binding to IRSp53. This notion is supported by the fact that weak binding of the I-BAR mutant F196A to a longer C-terminal Tir fragment was observed (Figure 5C) in pull-down experiments, whereas binding to short Tir-derived peptides was not detectable (Figure 5B). Tir forms homodimers and is clustered beneath the attached bacteria upon interaction with intimin (Luo et al., 2000; Touzé et al., 2004). Together with the presence of two binding sites on the I-BAR homodimer, this is expected to lead to a higher apparent affinity due to the avidity effect and, consequently, efficient recruitment of IRSp53 at physiological concentrations.
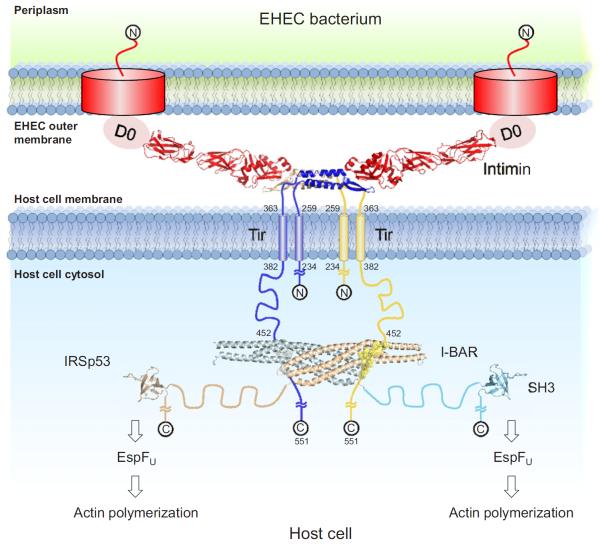
The EHEC bacterium utilizes a large protein scaffold including bacterial and host proteins that immobilizes the bacterium and leads to N-WASP mediated Arp2/3-complex dependent actin polymerization (Weiss et al., 2009). Figure 6 shows a model of this scaffold consisting of Tir, intimin, and IRSp53. An alternative model with one IRSp53 homodimer linking two distinct Tir homodimers appears also possible. I-BAR domains recognize and even induce convex deformation of lipid membranes (Scita et al., 2008; Suetsugu et al., 2006). Membrane binding by human IRSp53 is short lived and highly dynamic (Saarikangas et al., 2009), and it is dispensable for pedestal formation (Weiss et al., 2009). The orientation of the Tir-derived peptides on the I-BAR domain suggests that the flat I-BAR surface, which is assumed to contact the plasma membrane, is oriented toward it in the complex, thus supporting the possibility of Tir-bound IRSp53 contacting the plasma membrane.
Figure 6. Model of the Intimin:Tir:IRSp53 Complex.
A model of the scaffold formed by EHEC Tir, intimin, and human IRSp53 was drawn to scale based on the available structures. Whether the Tir-bound I-BAR domain interacts with the plasma membrane directly is currently unknown. The extracellular Tir and intimin domains are represented by the structure of the complex of the homologous EPEC proteins by Luo et al. (2000) (PDB entry 1FO2). The model excludes the N terminus of Tir, which modulates pedestal length (Campellone et al., 2006). The IRSp53 SH3 domain is represented by the similar PDB entry 2KXC. Regions of unknown structure are represented by ropes with end-to-end distances corresponding to root-mean-square end-to-end distances of random coils of the respective chain length (Tanford et al., 1966). See also Table S3.
EXPERIMENTAL PROCEDURES
Sequence Analysis
Protein sequences were aligned with EMBOSS emma/ClustalW2 (Larkin et al., 2007) and displayed with ESPript (Gouet et al., 2003). Disordered protein regions were predicted by DISOPRED2 (Ward et al., 2004). The proportion of human protein sequences in SwissProt containing the NPY motif was determined with Scansite (Obenauer et al., 2003).
Protein Purification and Crystallization
E. coli SCS1 cells bearing the I-BARIRSp53 expression plasmid (see Tables S1 and S2 and Supplement Experimental Procedures for construction of plasmids) and pRARE (Novy et al., 2001) were grown in SB medium (12 g/l bactotryptone, 24 g/l yeast extract, 0.4% [v/v] glycerol, 17 mM KH2PO4, 72 mM K2HPO4) supplemented with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol at 37°C to an OD of 1.5. Protein expression was induced with 1 mM IPTG at 37°C for 4 hr. Cells were lysed by three thaw and freeze cycles in 20 mM Tris-HCl (pH 7.5), 500 mM NaCl, 20 mM imidazole, 5 mM 2-mercaptoethanol, and 1 mM PMSF, and centrifugation (35,000 × g, 45 min). The soluble protein was purified by nickel affinity chromatography and gel filtration, followed by cleavage with TEV protease and nickel rechromatography. The protein was supplemented with 10% (v/v) glycerol, flash frozen in liquid nitrogen, and stored at −80°C. A peptide with the sequence 452GTVQNPYADVKT463, derived from EHEC Tir (SwissProt Q7DB77), was synthesized in house with a C-terminal amide and HPLC purified for complex formation and crystallization. The buffers of I-BAR and peptide were exchanged against 20 mM HEPES-NaOH, 100 mM NaCl, 0.1 mM EDTA, and 1 mM DTT (pH 7.5). The complex was prepared by adding a 2-fold molar excess of peptide to purified I-BAR with a final concentration of 11 mg/ml.
Crystallization was performed at 19°C using the hanging drop-vapor phase diffusion method. Equal volumes of concentrated protein and reservoir solution (16% [w/v] PEG 3350, 0.3 M (NH4)2SO4) were mixed. Crystals appeared after 1 day and were improved by microseeding. Crystals were transferred into reservoir solution with 20% (v/v) PEG 400 before freezing.
Structure Determination and Refinement
The data sets were collected at beamline 14.2 of the synchrotron storage ring BESSY II in Berlin. Images were indexed and processed with XDS (Kabsch, 1993), and the structure was solved by molecular replacement using CCP4 MOLREP (Vagin and Teplyakov, 1997) with PDB entry 1Y2O as the search model. REFMAC5 (Murshudov et al., 1997) was used for refinement and Coot (Emsley and Cowtan, 2004) for model building. Figures were prepared with PyMOL. Figure 3 was calculated with LIGPLOT (Wallace et al., 1995). Buried surface areas were calculated with PISA (Krissinel and Henrick, 2007). Rmsds between common Cα positions were calculated with ProFit using the McLachlan algorithm (McLachlan, 1982).
ITC
Binding reactions were measured by ITC using a MCS-ITC calorimeter (MicroCal, Northampton, MA, USA). Peptides and I-BAR were dialyzed against 20 mM HEPES-NaOH, 100 mM NaCl, 0.1 mM EDTA, and 0.8 mM tris(2-carboxyethyl)phosphine (TCEP) (pH 7.5). During titration, 2 mM peptide was injected into 0.1 mM of His-tagged I-BAR protomers at 24°C in 10 μl steps up to a 3-fold molar excess. The mixing heat of the peptide, measured by injecting peptide into buffer, was subtracted from the signals obtained from the binding reactions. ITC data of three independent measurements were analyzed using Origin V7.0 with MicroCal ITC add-on. Peptides 452GTVQNPYADVKT463, derived from EHEC Tir, GTVQNPFADVKT, 448SEVVNPYAEVGG459, derived from EPEC Tir (UniProt B7UM99), and a sequence of the human insulin receptor 995YSNPEYLSASDV1005 (Swissprot P06213) were synthesized with C-terminal amide and HPLC purified.
PIP Assays
In this assay, GFP-fusion proteins can be recruited to inclusions (platforms) by a potential interaction partner fused to the μNS reoviral scaffolding protein upon coexpression in yeast. Physical interaction of the two fusion proteins is observed as fluorescent foci (Schmitz et al., 2009). Yeast cells were transformed with either of the μNS-TirC and GFP-I-BARIRTKS expression plasmids (see Supplement Experimental Procedures) and were mated to obtain diploids that carry both expression plasmids. The resulting diploids were grown overnight in synthetic-complete medium lacking leucine and uracil (SC-LU), containing 2% raffinose as a carbon source. The cultures were back diluted to OD 0.5–1.0 in fresh media (SC-HU 2% raffinose) and grown for another 2 hr. Expression of μNS-TirC and GFP-I-BARIRTKS fusions was induced by addition of galactose (4%). Fluorescence was visualized 4–5 hr post-induction using a Nikon TE300 microscope with Chroma Technology filters; images were captured digitally using a black and white SenSys charge-coupled device (CCD) camera and IPLab software (Scanalytics). The experiment was performed three times with similar results, and the percentage of cells containing GFP inclusions was determined visually from one representative experiment.
Cells, Transfections, and Pull-Down Assays
COS7 cells (ATCC: CRL-1651) were grown in DMEM 4.5 g/l glucose (Invitrogen) supplemented with 10% FBS (PAA, Germany), 2 mM glutamine, and 50 U/ml Penicillin/Streptomycin (Invitrogen). Transfections were carried out with FuGENE 6 (Roche, Mannheim, Germany) or SuperFect (QIAGEN, Germany), according to manufacturer's protocols. For immunoprecipitations, cells grown in 10 cm diameter dishes were washed with PBS, lysed in 500 ml of ice-cold lysis buffer (8 mM Tris base, 12 mM HEPES [pH 7.5], 50 mM NaCl, 15 mM KCl, 3 mM MgCl2, 1% Triton X-100, and EDTA-free protease inhibitor cocktail Complete Mini [Roche]) for 20 min on ice. Homogenates were centrifuged for 45 min at 15,000 × g, and cleared lysates were snap frozen in liquid nitrogen for storage. For pull-downs with immobilized I-BARs, recombinant His-tagged proteins were expressed and purified following standard protocols and immobilized on Ni-NTA Sepharose beads (QIAGEN) at 1 mg protein/ml slurry in binding buffer (50 mM Na2HPO4, 20 mM imidazole, 300 mM NaCl [pH 8.0]). The buffer was supplemented with protease inhibitor cocktail, and 10% glycerol and beads were snap frozen, and stored at −70°C. For pull-downs using immobilized peptides, peptides were synthesized carrying an N-terminal biotin residue with an alanine spacer and were coupled to Streptavidin-agarose (Fluka, Germany) at 2 mg/ml slurry. Peptides used were EHEC-NPY (biotin-A-450SIGTVQNPYADVKTS464), EPEC-NPY (biotin-A-446SSSEVVNPYAEVGGA460), and EPEC Y474 (biotin-A-470EEHI(phospho-Tyr)DEVAADP481). For pull-downs, 400 μl of the respective lysate was incubated with 30 μl of slurry for 1 hr at 4°C on a rotary wheel. Samples were washed three times with washing buffer (as above “binding buffer” for His-I-BARs; lysis buffer with 0.1% Triton X-100 for biotinylated peptides), resolved on SDS-PAGE, and analyzed by immunoblotting using monoclonal anti-GFP antibodies.
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to Victor Wray for careful reading and helpful comments. The authors thank Uwe Müller and Nora Darowski for operation of the BESSY II beamlines, Manfred Nimtz and Undine Felgenträger for mass spectrometry, and Werner Tegge for peptide synthesis. This work was funded by the German Federal Ministry of Education and Research (BMBF) through the National Genome Network (NGFN-Plus Grant 01GS08181), the Helmholtz Association of German Research Centres through the Protein Sample Production Facility, and NIH Grant R01 AI46454 (to J.M.L.). Support by the Fonds der Chemischen Industrie to D.W.H. and by the DFG to T.E.B.S. (SPP1150 and FOR629) is gratefully acknowledged.
Footnotes
ACCESSION NUMBERS Coordinates have been deposited in the Protein Data Bank with accession code 2YKT.
SUPPLEMENTAL INFORMATION Supplemental Information includes Supplemental Experimental Procedures, two figures, three tables, and one movie and can be found with this article online at doi:10.1016/j.str.2011.06.015.
REFERENCES
- Arold S, O'Brien R, Franken P, Strub MP, Hoh F, Dumas C, Ladbury JE. RT loop flexibility enhances the specificity of Src family SH3 domains for HIV-1 Nef. Biochemistry. 1998;37:14683–14691. doi: 10.1021/bi980989q. [DOI] [PubMed] [Google Scholar]
- Ball LJ, Kühne R, Schneider-Mergener J, Oschkinat H. Recognition of proline-rich motifs by protein-protein-interaction domains. Angew. Chem. Int. Ed. Engl. 2005;44:2852–2869. doi: 10.1002/anie.200400618. [DOI] [PubMed] [Google Scholar]
- Brady MJ, Campellone KG, Ghildiyal M, Leong JM. Enterohaemorrhagic and enteropathogenic Escherichia coli Tir proteins trigger a common Nck-independent actin assembly pathway. Cell. Microbiol. 2007;9:2242–2253. doi: 10.1111/j.1462-5822.2007.00954.x. [DOI] [PubMed] [Google Scholar]
- Campellone KG, Leong JM. Tails of two Tirs: actin pedestal formation by enteropathogenic E. coli and enterohemorrhagic E. coli O157:H7. Curr. Opin. Microbiol. 2003;6:82–90. doi: 10.1016/s1369-5274(03)00005-5. [DOI] [PubMed] [Google Scholar]
- Campellone KG, Robbins D, Leong JM. EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev. Cell. 2004;7:217–228. doi: 10.1016/j.devcel.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Campellone KG, Brady MJ, Alamares JG, Rowe DC, Skehan BM, Tipper DJ, Leong JM. Enterohaemorrhagic Escherichia coli Tir requires a C-terminal 12-residue peptide to initiate EspF-mediated actin assembly and harbours N-terminal sequences that influence pedestal length. Cell. Microbiol. 2006;8:1488–1503. doi: 10.1111/j.1462-5822.2006.00728.x. [DOI] [PubMed] [Google Scholar]
- Cantarelli VV, Kodama T, Nijstad N, Abolghait SK, Iida T, Honda T. Cortactin is essential for F-actin assembly in enteropathogenic Escherichia coli (EPEC)- and enterohaemorrhagic E. coli (EHEC)-induced pedestals and the α-helical region is involved in the localization of cortactin to bacterial attachment sites. Cell. Microbiol. 2006;8:769–780. doi: 10.1111/j.1462-5822.2005.00664.x. [DOI] [PubMed] [Google Scholar]
- Cheng HC, Skehan BM, Campellone KG, Leong JM, Rosen MK. Structural mechanism of WASP activation by the enterohaemorrhagic E. coli effector EspF(U) Nature. 2008;454:1009–1013. doi: 10.1038/nature07160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer T, Hoofnagle AN, Enmon JL, Bowers RC, Yamabhai M, Kay BK, Overduin M. Molecular mechanism of NPF recognition by EH domains. Nat. Struct. Biol. 2000;7:1018–1022. doi: 10.1038/80924. [DOI] [PubMed] [Google Scholar]
- Disanza A, Mantoani S, Hertzog M, Gerboth S, Frittoli E, Steffen A, Berhoerster K, Kreienkamp HJ, Milanesi F, Di Fiore PP, et al. Regulation of cell shape by Cdc42 is mediated by the synergic actin-bundling activity of the Eps8-IRSp53 complex. Nat. Cell Biol. 2006;8:1337–1347. doi: 10.1038/ncb1502. [DOI] [PubMed] [Google Scholar]
- Eck MJ, Dhe-Paganon S, Trüb T, Nolte RT, Shoelson SE. Structure of the IRS-1 PTB domain bound to the juxtamembrane region of the insulin receptor. Cell. 1996;85:695–705. doi: 10.1016/s0092-8674(00)81236-2. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Frankel G, Phillips AD. Attaching effacing Escherichia coli and paradigms of Tir-triggered actin polymerization: getting off the pedestal. Cell. Microbiol. 2008;10:549–556. doi: 10.1111/j.1462-5822.2007.01103.x. [DOI] [PubMed] [Google Scholar]
- Frese S, Schubert W-D, Findeis AC, Marquardt T, Roske YS, Stradal TEB, Heinz DW. The phosphotyrosine peptide binding specificity of Nck1 and Nck2 Src homology 2 domains. J. Biol. Chem. 2006;281:18236–18245. doi: 10.1074/jbc.M512917200. [DOI] [PubMed] [Google Scholar]
- Frost A, Unger VM, De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosney DL, DeVinney R, Pfuetzner RA, Frey EA, Strynadka NC, Finlay BB. Enteropathogenic E. coli translocated intimin receptor, Tir, interacts directly with α-actinin. Curr. Biol. 2000;10:735–738. doi: 10.1016/s0960-9822(00)00543-1. [DOI] [PubMed] [Google Scholar]
- Gouet P, Robert X, Courcelle E. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenheid S, DeVinney R, Bladt F, Goosney D, Gelkop S, Gish GD, Pawson T, Finlay BB. Enteropathogenic E. coli Tir binds Nck to initiate actin pedestal formation in host cells. Nat. Cell Biol. 2001;3:856–859. doi: 10.1038/ncb0901-856. [DOI] [PubMed] [Google Scholar]
- Hayward RD, Leong JM, Koronakis V, Campellone KG. Exploiting pathogenic Escherichia coli to model transmembrane receptor signalling. Nat. Rev. Microbiol. 2006;4:358–370. doi: 10.1038/nrmicro1391. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Krugmann S, Jordens I, Gevaert K, Driessens M, Vandekerckhove J, Hall A. Cdc42 induces filopodia by promoting the formation of an IRSp53:Mena complex. Curr. Biol. 2001;11:1645–1655. doi: 10.1016/s0960-9822(01)00506-1. [DOI] [PubMed] [Google Scholar]
- Kuriyan J, Cowburn D. Modular peptide recognition domains in eukaryotic signaling. Annu. Rev. Biophys. Biomol. Struct. 1997;26:259–288. doi: 10.1146/annurev.biophys.26.1.259. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee SH, Kerff F, Chereau D, Ferron F, Klug A, Dominguez R. Structural basis for the actin-binding function of missing-in-metastasis. Structure. 2007;15:145–155. doi: 10.1016/j.str.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KB, Bu W, Goh WI, Koh E, Ong SH, Pawson T, Sudhaharan T, Ahmed S. The Cdc42 effector IRSp53 generates filopodia by coupling membrane protrusion with actin dynamics. J. Biol. Chem. 2008;283:20454–20472. doi: 10.1074/jbc.M710185200. [DOI] [PubMed] [Google Scholar]
- London N, Movshovitz-Attias D, Schueler-Furman O. The structural basis of peptide-protein binding strategies. Structure. 2010;18:188–199. doi: 10.1016/j.str.2009.11.012. [DOI] [PubMed] [Google Scholar]
- Luo Y, Frey EA, Pfuetzner RA, Creagh AL, Knoechel DG, Haynes CA, Finlay BB, Strynadka NCJ. Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature. 2000;405:1073–1077. doi: 10.1038/35016618. [DOI] [PubMed] [Google Scholar]
- Mattila PK, Pykäläinen A, Saarikangas J, Paavilainen VO, Vihinen H, Jokitalo E, Lappalainen P. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J. Cell Biol. 2007;176:953–964. doi: 10.1083/jcb.200609176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer BJ, Saksela K. SH3 domains. In: Cesareni G, Gimona M, Sudol M, Yaffe M, editors. Modular Protein Domains. Willey-VCH Verlag GmbH; Weinheim, Germany: 2004. pp. 37–58. [Google Scholar]
- McLachlan AD. Rapid comparison of protein structures. Acta Crystallogr. A. 1982;38:871–873. [Google Scholar]
- Miki H, Yamaguchi H, Suetsugu S, Takenawa T. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature. 2000;408:732–735. doi: 10.1038/35047107. [DOI] [PubMed] [Google Scholar]
- Millard TH, Dawson J, Machesky LM. Characterisation of IRTKS, a novel IRSp53/MIM family actin regulator with distinct filament bundling properties. J. Cell Sci. 2007;120:1663–1672. doi: 10.1242/jcs.001776. [DOI] [PubMed] [Google Scholar]
- Millard TH, Bompard G, Heung MY, Dafforn TR, Scott DJ, Machesky LM, Fütterer K. Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. EMBO J. 2005;24:240–250. doi: 10.1038/sj.emboj.7600535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novy R, Drott D, Yaeger K, Mierendorf R. Overcoming the codon bias of E. coli for enhanced protein expression. Innovations. 2001;12:1–3. [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips N, Hayward RD, Koronakis V. Phosphorylation of the enteropathogenic E. coli receptor by the Src-family kinase c-Fyn triggers actin pedestal formation. Nat. Cell Biol. 2004;6:618–625. doi: 10.1038/ncb1148. [DOI] [PubMed] [Google Scholar]
- Saarikangas J, Zhao H, Pykäläinen A, Laurinmäki P, Mattila PK, Kinnunen PK, Butcher SJ, Lappalainen P. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr. Biol. 2009;19:95–107. doi: 10.1016/j.cub.2008.12.029. [DOI] [PubMed] [Google Scholar]
- Sawallisch C, Berhörster K, Disanza A, Mantoani S, Kintscher M, Stoenica L, Dityatev A, Sieber S, Kindler S, Morellini F, et al. The insulin receptor substrate of 53 kDa (IRSp53) limits hippocampal synaptic plasticity. J. Biol. Chem. 2009;284:9225–9236. doi: 10.1074/jbc.M808425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz AM, Morrison MF, Agunwamba AO, Nibert ML, Lesser CF. Protein interaction platforms: visualization of interacting proteins in yeast. Nat. Methods. 2009;6:500–502. doi: 10.1038/nmeth.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scita G, Confalonieri S, Lappalainen P, Suetsugu S. IRSp53: crossing the road of membrane and actin dynamics in the formation of membrane protrusions. Trends Cell Biol. 2008;18:52–60. doi: 10.1016/j.tcb.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Smith K, Humphreys D, Hume PJ, Koronakis V. Enteropathogenic Escherichia coli recruits the cellular inositol phosphatase SHIP2 to regulate actin-pedestal formation. Cell Host Microbe. 2010;7:13–24. doi: 10.1016/j.chom.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Murayama K, Sakamoto A, Hanawa-Suetsugu K, Seto A, Oikawa T, Mishima C, Shirouzu M, Takenawa T, Yokoyama S. The RAC binding domain/IRSp53-MIM homology domain of IRSp53 induces RAC-dependent membrane deformation. J. Biol. Chem. 2006;281:35347–35358. doi: 10.1074/jbc.M606814200. [DOI] [PubMed] [Google Scholar]
- Swimm A, Bommarius B, Reeves P, Sherman M, Kalman D. Complex kinase requirements for EPEC pedestal formation. Nat. Cell Biol. 2004;6:795. doi: 10.1038/ncb0904-795a. [DOI] [PubMed] [Google Scholar]
- Tanford C, Kawahara K, Lapanje S. Proteins in 6-M guanidine hydrochloride. Demonstration of random coil behavior. J. Biol. Chem. 1966;241:1921–1923. [PubMed] [Google Scholar]
- Touzé T, Hayward RD, Eswaran J, Leong JM, Koronakis V. Self-association of EPEC intimin mediated by the β-barrel-containing anchor domain: a role in clustering of the Tir receptor. Mol. Microbiol. 2004;51:73–87. doi: 10.1046/j.1365-2958.2003.03830.x. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- Vingadassalom D, Kazlauskas A, Skehan B, Cheng HC, Magoun L, Robbins D, Rosen MK, Saksela K, Leong JM. Insulin receptor tyrosine kinase substrate links the E. coli O157:H7 actin assembly effectors Tir and EspF(U) during pedestal formation. Proc. Natl. Acad. Sci. USA. 2009;106:6754–6759. doi: 10.1073/pnas.0809131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Weiss SM, Ladwein M, Schmidt D, Ehinger J, Lommel S, Städing K, Beutling U, Disanza A, Frank R, Jänsch L, et al. IRSp53 links the enterohemorrhagic E. coli effectors Tir and EspFU for actin pedestal formation. Cell Host Microbe. 2009;5:244–258. doi: 10.1016/j.chom.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Wilson DR, Finlay BB. The `Asx-Pro turn' as a local structural motif stabilized by alternative patterns of hydrogen bonds and a consensus-derived model of the sequence Asn-Pro-Asn. Protein Eng. 1997;10:519–529. doi: 10.1093/protein/10.5.519. [DOI] [PubMed] [Google Scholar]
- Yamagishi A, Masuda M, Ohki T, Onishi H, Mochizuki N. A novel actin bundling/filopodium-forming domain conserved in insulin receptor tyrosine kinase substrate p53 and missing in metastasis protein. J. Biol. Chem. 2004;279:14929–14936. doi: 10.1074/jbc.M309408200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.