Abstract
Epithelial ovarian cancer (EOC) is a heterogeneous cancer with both genetic and environmental risk factors. Variants influencing the risk of developing the less-common EOC subtypes have not been fully investigated. We performed a genome-wide association study (GWAS) of EOC according to subtype by pooling genomic DNA from 545 cases and 398 controls of European descent, and testing for allelic associations. We evaluated for replication 188 variants from the GWAS (56 variants for mucinous, 55 for endometrioid and clear cell, 53 for low malignant potential (LMP) serous, and 24 for invasive serous EOC), selected using pre-defined criteria. Genotypes from 13,188 cases and 23,164 controls of European descent were used to perform unconditional logistic regression under the log-additive genetic model; odds ratios (OR) and 95% confidence intervals are reported. Nine variants tagging 6 loci were associated with subtype-specific EOC risk at P<0.05, and had an OR that agreed in direction of effect with the GWAS results. Several of these variants are in or near genes with a biological rationale for conferring EOC risk, including ZFP36L1 and RAD51B for mucinous EOC (rs17106154, OR=1.17, P=0.029, n=1,483 cases), GRB10 for endometrioid and clear cell EOC (rs2190503, P=0.014, n=2,903 cases), and C22orf26/BPIL2 for LMP serous EOC (rs9609538, OR=0.86, P=0.0043, n=892 cases). In analyses that included the 75 GWAS samples, the association between rs9609538 (OR=0.84, P=0.0007) and LMP serous EOC risk remained statistically significant at P<0.0012 adjusted for multiple testing. Replication in additional samples will be important to verify these results for the less-common EOC subtypes.
Keywords: histological subtype, serous, endometrioid, clear cell, mucinous, BPIL2
Background
Epithelial ovarian cancer (EOC) is a heterogeneous cancer with distinct and clinically relevant subtypes that are characterized by differences in morphology, gene expression profile, and molecular genetic features (Gilks et al. 2008; Kalloger et al. 2010; Kobel et al. 2010). It has become apparent that the main histological subtypes, comprising ∼70% serous, 11% endometrioid, 12% clear cell, and 3% mucinous EOC (Kobel et al. 2010), have different genetic (Gayther and Pharoah 2010; Lynch et al. 1991; Lynch et al. 1985; Shulman 2010) and epidemiologic (Faber et al. 2013) risk factors, precursor lesions (Pearce et al. 2012; Piek et al. 2001), pattern of spread, response to platinum-taxane based treatment, and patient outcome (Vaughan et al. 2011), compelling many to assert that they are different diseases (Gomez-Raposo et al. 2010; Kobel et al. 2010; Kurman and Shih 2010). Gene expression profiling has further classified serous EOC tumours into those of a less-common low grade (3% of all EOC) and more-common high grade (68% of all EOC) type (Kobel et al. 2010; Tothill et al. 2008), a finding supported by differences in the clinical behaviour of these tumours (Matsuno et al. 2013). Much of the excess familial risk observed for EOC remains unexplained and may be improved by investigations that stratify by histological subtype.
Genome-wide association studies (GWAS) have identified several common susceptibility variants for EOC (Bolton et al. 2010; Goode et al. 2010; Permuth-Wey et al. 2013; Pharoah et al. 2013; Song et al. 2009). The majority of these has been most strongly associated with serous EOC, unsurprising given the GWAS design carries forward for genotyping in subsequent stages those single nucleotide polymorphisms (SNPs) with the smallest P-values associated with the most prevalent serous EOC subtype. Fewer genome-wide significant associations have been reported for the less-common EOC subtypes. In our most recent analyses of data from over 40 international studies of EOC within the Ovarian Cancer Association Consortium (OCAC), we reported that common susceptibility variants in the candidate HNF1B gene (Shen et al. 2013), and the candidate TERT locus (Bojesen et al. 2013) differentially associate with risk by subtype and imply that distinct mechanisms are involved in pathogenesis. Examining risk factors separately by histological subtype, together with assembling studies of large numbers of women with these cancers, is critical to understanding this disease.
To extend the findings of the existing EOC GWAS (Bolton et al. 2010; Goode et al. 2010; Pharoah et al. 2013; Song et al. 2009), we performed a GWAS according to EOC histological subtype using a DNA pooling strategy. The focus was to discover genetic variants associated with the less-common EOC subtypes (endometrioid, clear cell, mucinous, and low-malignant potential [LMP] serous).The DNA pooling strategy is an efficient approach to assess genetic associations and has been successfully performed for various disease subtypes (Pearson et al. 2007; Schrauwen et al. 2009; Skibola et al. 2009). In the first stage, individual DNA samples were physically combined to create subtype-specific case pools, and a control pool, and DNA pools (not individual samples) were assayed using commercially available SNP arrays. Data from arrays were used to estimate SNP allele frequencies or allelotypes for each DNA pool, and not to determine genotypes. Pool allelotypes were then used in allele-based tests to evaluate SNP associations with EOC subtypes. In the second stage, replication of allelic associations was performed by individual genotyping (IG) for a large number of women contributing samples to the OCAC.
Methods
Discovery stage study population
Participants were from the Ovarian Cancer in Alberta and British Columbia (OVAL-BC) population-based case-control study (abbreviated as “OVA”). Eligible cases had incident, histologically-confirmed EOC, and were identified from the provincial cancer registries of British Columbia (BC) and Alberta (AB) between 2002 and 2011. Eligible control women were identified from provincial health care enrollment rosters or from a province-wide mammography program (BC after 2005). Participants provided blood or saliva samples for DNA. Of 1,578 cases and 2,222 controls (response 64.9% and 55.6%, respectively) in OVA, 545 cases and 398 controls were recruited in BC before June 30, 2008 and comprised the discovery stage sample. The study was approved by the Research Ethics Boards of the BC Cancer Agency, the University of British Columbia and the University of Calgary. All subjects gave written informed consent.
Discovery stage pool construction and quality control (QC)
Histologic review of cases at our centre adhered to contemporary diagnostic criteria (Gilks et al. 2008). Tumour histology codes were abstracted from pathology reports and four case pools were constructed. These pools contained DNA from: 1) 24 LMP and 60 invasive mucinous cases combined, 2) 72 invasive endometrioid and 42 invasive clear cell cases combined, 3) 75 LMP serous cases, and 4) 272 invasive serous cases (Table 1). We based our decision to combine LMP and invasive mucinous cases on the recent evidence that mucinous tumours develop along a continuum from benign to borderline to invasive (Kurman and Ronnett 2010), and therefore reflect the same disease (i.e. pooling these samples should not introduce genetic heterogeneity). Although the morphologies of the endometrioid and clear cell subtypes differ (Tavassoulu and Devilee 2003), both are associated with endometriosis (Pearce et al. 2012) and share similar somatic alterations, including mutations in ARID1A (Jones et al. 2010; Wiegand et al. 2010), KRAS (Kuo et al. 2009; Stewart et al. 2012), PTEN (Catasus et al. 2004; Obata et al. 1998), and defects in mismatch repair genes (Albarracin et al. 2004; Cai et al. 2004). To explore possible shared heritable risk factors, these two subtypes were combined and analyzed as a single pool. LMP and invasive serous samples were analyzed as separate pools, consistent with these being distinct molecular subtypes (Tothill et al. 2008). Controls in OVA were frequency-matched by age to all cases used in pools (by 10 year age bins), and one control pool was constructed using 398 samples (Table 1). No exclusions were made for family history of cancer or BRCA1 and BRCA2 status (unknown), and only women of self-reported European descent were included.
Table 1.
Characteristics of samples in the discovery stage case-control DNA pools.
| DNA pool | ICD-O-3 codea | Pool Size, N | Ageb (standard deviation) | Minimum Age | Maximum Age |
|---|---|---|---|---|---|
| Mucinous | 84721, 84803, 84703, 90151 | 84 | 53 ± 13.2 | 23 | 78 |
| Endometrioid/Clear cell | 83803, 83103 | 114 | 56 ± 10.5 | 25 | 79 |
| LMP Serous | 84421, 84621 | 75 | 53 ± 12.8 | 21 | 80 |
| Invasive Serous | 84603, 84413, 84613 | 272 | 62 ± 10.1 | 36 | 80 |
| Control pool | 398 | 57 ± 10.5 | 31 | 80 |
International Classification of Diseases for Oncology, 3rd Edition
Median age at diagnosis or control selection, years
DNA was extracted from peripheral venous blood (90% of subjects) using a modified salting out protocol (Sambrook J 2000), and from saliva (10% of subjects) using OraGene kits (DNA Genotek, PA, USA). Genotyping call rates were compared previously between OVA blood (99.7%) and saliva (98.1%) (unpublished). DNA samples were adjusted to between 50-100ng/uL and then precisely quantified in duplicate by fluorometry using PicoGreen™ (Molecular Probes, Eugene, OR, USA). For each EOC subtype, individual samples of 2-4uL were manually pipetted into a single pool of 200ng of DNA. Pools were assayed on Human660W-Quad v1 (660-Quad) genotyping beadchips (Illumina, San Diego, CA, USA), and imaged at The Centre for Applied Genomics (Toronto, ON, Canada). The red and green channel intensities for each 660-Quad array were extracted and used to estimate the relative allele frequency (RAF) of SNPs for each DNA pool as red intensity/(red intensity + green intensity), following a previously described approach (Pearson et al. 2007). Each DNA pool was assayed using 12 replicate 660-Quad beadchips. Replicate arrays were used to reduce the error in RAF estimation(Earp et al. 2011). Details of the 660-Quad array QC are described in the online Supplementary Methods.
Discovery stage association analysis
The RAF of SNPs for each EOC DNA pool was compared with the RAF of SNPs for the control DNA pool, and allele-based tests were used to evaluate SNP associations according to subtypes using the SingleMarker test, implemented in the program GENEPOOL (Homer et al. 2008; Pearson et al. 2007). The SingleMarker test is a modified two-tailed Student's t-test that divides the difference in RAF of SNPs between cases and controls by the variance components specific to pooling (for example, variance due to pool construction, and variance due to SNP arrays) (Earp et al. 2011; Homer et al. 2008; Pearson et al. 2007).
SNP selection for replication stage
One hundred and ninety-eight SNPs from the pool-based GWAS were individually genotyped in samples from OCAC using a custom Illumina Infinium iSelect array as part of the Collaborative Oncological Gene-environment Study (COGS) (Pharoah et al. 2013). These 198 SNPs were selected roughly equally from among the mucinous (61 SNPs), endometrioid/clear cell (59 SNPs), and LMP serous (54 SNPs) GWAS results (Table 2). Fewer invasive serous SNPs (24 SNPs) were selected for replication as the previous EOC GWAS had greater power to detect associations for this subtype (Song et al. 2009). During SNP selection, two categories of SNPs were considered: (1) SNPs in high linkage disequilibrium (LD) r2>0.8 (based on HapMap European ancestry data) with at least one other SNP analyzed by the pool-based GWAS (referred to as ‘cluster’ SNPs), and (2) SNPs from the pool-based GWAS that had no other SNPs in high LD (referred to as ‘singleton’ SNPs). In the first category, consistency in the strength of P-values between SNPs in high LD was used to prioritize SNPs for selection. If a top-ranked SNP (ranked by smallest P-value) was in high LD with a SNP(s) that failed to also rank highly, then the top-ranked SNP was not chosen for replication. We defined a top-ranked SNP as one within the top ∼0.5% (n=3,300) of GWAS P-values. We reasoned that multiple highly-ranked SNPs in LD with each other were more likely to reflect true positive association results, and SNPs in high LD should yield very similar results assuming experimental error is not an issue. In the second category, singleton SNPs were chosen based on rank alone (by smallest SingleMarker P-value). We selected 30 clusters (48 SNPs) and 13 singleton SNPs to tag a total of 43 loci for replication in mucinous EOC, 30 clusters (46 SNPs) and 13 singleton SNPs tagged 42 loci for endometrioid/clear cell EOC, 30 clusters (41 SNPs) and 13 singleton SNPs tagged 43 loci for LMP serous EOC, and 15 clusters (24 SNPS) and 0 singletons tagged 15 loci for invasive serous EOC, for a total of 159 cluster SNPs and 39 singletons SNPs chosen for replication (Table 2). Some clusters were composed of 2 - 5 SNPs depending on the number of highly-ranked SNPs in the region, and the potential for biological relevance given proximity to a gene. Genome-wide significance (or other P-value thresholds) were not used as criteria in choosing SNPs for replication because pool-based GWAS often do not achieve these stringent thresholds; but, this strategy has been successful in selecting SNPs that attain stringent P-value thresholds in the replication stage (Pearson et al. 2007; Schrauwen et al. 2009; Skibola et al. 2009).
Table 2. Number of pool-based GWAS SNPs selected for replication by EOC subtype.
| Subtype | SNP selection methoda | # SNPs chosen | # SNPs successfully genotyped | # Independent tests of association |
|---|---|---|---|---|
| Mucinous | Cluster | 48 | 43 | 43 |
| Singleton | 13 | 13 | ||
|
| ||||
| Endometrioid/Clear cell | Cluster | 46 | 43 | 42 |
| Singleton | 13 | 12 | ||
|
| ||||
| LMP Serous | Cluster | 41 | 40 | 43 |
| Singleton | 13 | 13 | ||
|
| ||||
| Invasive Serous | Cluster | 24 | 24 | 15 |
| Singleton | 0 | 0 | ||
|
| ||||
| Total | 198 | 188 | -- | |
Cluster refers to SNPs chosen based on P-value and their being in linkage disequilibrium (LD) with other GWAS SNPs having a small P-value. Singleton refers to SNPs not in LD with other GWAS SNPs, and chosen based on P-value alone. See Methods for details.
Replication stage study populations
Forty-three studies participating in OCAC contributed samples and data to the COGS effort. The OCAC studies have been described previously (Pharoah et al. 2013). All studies had data on disease status, age at diagnosis or interview, and histological subtype. Most studies frequency-matched controls to cases on age group and race. Nine studies were case-only and were combined with case-control studies from the same geographical regions. Two Australian studies were also combined, creating 34 case-control sets.
Replication stage genotyping and QC
The COGS genotyping and QC process has been described (Pharoah et al. 2013). Briefly, OCAC samples were genotyped at two centers: McGill University and Genome Quebec Innovation Centre (Montreal, PQ) and the Mayo Clinic Medical Genome Facility (Rochester, MN) and genotype calling and QC were performed centrally at the University of Cambridge (Cambridge, UK). Of 47,630 OCAC samples genotyped, 44,308 passed QC. Concordance was > 99.6% among duplicates. Samples were excluded as follows: 1) a call rate of < 95%; 2) heterozygosity >5 standard deviations from the ancestry-specific mean; 3) ambiguous sex; 4) lowest call rate from a first-degree relative pair; 5) duplicate samples that were non-concordant for genotype or genotypic duplicates not concordant for phenotype. Of the 198 SNPs chosen by the current investigation, 188 (94.9%) passed QC. SNPs were excluded if: (1) the call rate was < 95% with MAF > 5% or < 99% with MAF < 5%; (2) they were monomorphic; (3) P-values of HWE in controls were < 10-7; (4) there was > 2% discordance in duplicate pairs; or (5) no genotypes were called.
As an additional QC check, SNP-EOC associations detected in the pool-based GWAS data were compared with SNP-EOC associations evaluated using genotyped data for 915 of the 943 discovery samples (97%) with sufficient DNA for genotyping. These samples were genotyped on the custom Illumina Infinium iSelect array as part of the COGS initiative, and evaluated with an allelic χ2 test (PLINK v1.07), the test most comparable to the SingleMarker test.
Replication stage association analysis
Analyses were further restricted to 36,352 eligible subjects (13,188 cases and 23,164 controls) of European descent. For each EOC subtype, SNP associations were estimated using unconditional logistic regression treating the number of minor alleles as an ordinal variable (log-additive model), and adjusting for population substructure by including the first 5 eigenvalues from principal components analysis (see (Pharoah et al. 2013)). Minor allele frequency for each SNP was calculated using genotypes for control subjects of European descent in OCAC. Separate analyses were carried out for each study within EOC subtype, and odds ratios (ORs) and 95% confidence intervals (CIs) were then combined across studies using fixed-effects meta-analysis. Analyses were performed including and excluding the OVA study (i.e. the source of the discovery stage samples). The I2 test of heterogeneity was estimated to quantify the proportion of total variation due to heterogeneity across studies, and the heterogeneity of ORs between studies was tested with Cochran's Q statistic. The R statistical package rmeta was used to generate forest plots. Statistical analysis was conducted in PLINK (v1.07) (Purcell et al. 2007). Adjustment for multiple testing was performed using a Bonferroni correction of the Type I error. Because unique SNPs were selected for each EOC subtype, we treated each set of SNPs independently, and treated correlated (cluster) SNPs as one independent test (Table 2).
Results
Descriptive characteristics of the discovery stage cases and controls are shown in Table 1 and of the pool-based GWAS SNP selection in Table 2. Pool-based allelotype associations were highly concordant with their values determined from individual genotypes (online Supplementary Table 1). Of the 188 SNPs successfully genotyped, 89% were significant at P<0.05. This level of concordance is consistent with previous pool-based GWAS (Skibola et al. 2009). Associations between SNPs and each EOC subtype are reported in Table 3 and in the Figures. There was no heterogeneity in ORs between studies for any of the associations.
Table 3. Associations between SNPs and subtype-specific ovarian cancer risk in discovery and replication stages.
| Subtype(s) | SNP (Maj>Min) | Location | MAF | Sample description | # Cases | # Controls | OR (95% CI)a | P | Phet |
|---|---|---|---|---|---|---|---|---|---|
| Mucinous | rs11108890 C>A | Chr12:96137530 | 0.04 | Discovery | 78 | 392 | 3.05 (1.32-7.06) | 0.0091 | |
| Replication | 1483 | 21530 | 1.33 (1.11-1.61) | 0.0026 | 0.42 | ||||
| Rep+OVA | 1578 | 22278 | 1.38 (1.16-1.66) | 0.0004 | 0.38 | ||||
|
| |||||||||
| rs933518 G>A | Chr16: 53079622 | 0.08 | Discovery | 78 | 392 | 2.29 (1.32-4.00) | 0.0034 | ||
| Replication | 1483 | 21530 | 1.20 (1.04-1.38) | 0.0141 | 0.81 | ||||
| Rep+OVA | 1578 | 22278 | 1.26 (1.11-1.45) | 0.0008 | 0.41 | ||||
|
| |||||||||
| rs17106154 A>G | Chr14: 68230927 | 0.07 | Discovery | 78 | 392 | 1.78 (0.95-3.37) | 0.0738 | ||
| Replication | 1483 | 21530 | 1.17 (1.02-1.35) | 0.0287 | 0.96 | ||||
| Rep+OVA | 1578 | 22278 | 1.21 (1.06-1.39) | 0.0058 | 0.83 | ||||
|
| |||||||||
| rs970651 G>A | Chr13: 47351705 | 0.16 | Discovery | 78 | 392 | 2.54 (1.54-4.19) | 0.0003 | ||
| Replication | 1483 | 21530 | 1.12 (1.01-1.24) | 0.0334 | 0.81 | ||||
| Rep+OVA | 1578 | 22278 | 1.16 (1.05-1.28) | 0.0042 | 0.44 | ||||
|
| |||||||||
| rs7981902 A>G | Chr13: 47368792 | 0.13 | Discovery | 78 | 392 | 2.26 (1.32-3.86) | 0.0029 | ||
| Replication | 1483 | 21530 | 1.13 (1.01-1.27) | 0.0308 | 0.94 | ||||
| Rep+OVA | 1578 | 22278 | 1.15 (1.03-1.28) | 0.0114 | 0.84 | ||||
|
| |||||||||
| Endometrioid/Clear Cell | rs2190503 G>A | Chr7: 50710111 | 0.13 | Discovery | 114 | 392 | 1.85 (1.21-2.85) | 0.0049 | |
| Replication | 2903 | 21528 | 1.11 (1.02-1.21) | 0.0137 | 0.12 | ||||
| Rep+OVA | 3060 | 22276 | 1.12 (1.04-1.22) | 0.0050 | 0.11 | ||||
|
| |||||||||
| rs6593140 A>G | Chr7: 50765627 | 0.12 | Discovery | 114 | 392 | 2.03 (1.30-3.17) | 0.0018 | ||
| Replication | 2903 | 21528 | 1.11 (1.02-1.21) | 0.0195 | 0.28 | ||||
| Rep+OVA | 3060 | 22276 | 1.09 (1.03-1.17) | 0.0060 | 0.46 | ||||
|
| |||||||||
| rs2329554 G>A | Chr7: 50842524 | 0.22 | Discovery | 114 | 392 | 1.91 (1.33-2.74) | 0.0004 | ||
| Replication | 2903 | 21528 | 1.08 (1.01-1.16) | 0.0195 | 0.59 | ||||
| Rep+OVA | 3060 | 22276 | 1.12 (1.03-1.22) | 0.0060 | 0.25 | ||||
|
| |||||||||
| LMP Serous | rs9609538 A>G | Chr22: 31139832 | 0.24 | Discovery | 68 | 392 | 0.48 (0.30-0.77) | 0.0023 | |
| Replication | 892 | 21529 | 0.86 (0.77-0.95) | 0.0043 | 1.00 | ||||
| Rep+OVA | 970 | 22277 | 0.84 (0.76-0.93) | 0.0007 | 0.99 | ||||
Abbreviations used: SNP, single nucleotide polymorphism; MAF, minor allele frequency; OR, odds ratio; CI, confidence interval. Within subtype, table is ordered by replication sample P-value. SNP location given for human genome build NBCI36.3. MAF is based on all European controls in OCAC and genotyped as part of COGS. Discovery samples are OVA samples used in the pool-based GWAS and subsequently genotyped by COGS (28 samples with insufficient DNA were not genotyped; hence, the number of cases and controls in pools may be smaller than in Table 1). Replication samples are from OCAC studies participating in COGS, excluding all OVA samples. Rep+OVA includes OCAC studies participating in COGS and all OVA samples.
Discovery sample OR and P-value are from logistic regression assuming an additive genetic model, implemented in PLINK (v1.07). Replication sample OR and P-value are from fixed effects meta-analysis carried out using the rmeta library implemented in the R project, excluding the OVA site. Rep+OVA sample OR and P-value are from fixed effects meta-analysis carried out using the rmeta library implemented in the R project, including the OVA site. Phet is the P-value for Cochran's-Q measure of between study heterogeneity, generated in rmeta. All values are based on individual genotyping data.
Mucinous subtype
Fifty-six SNPs representing 43 loci were tested for association with 1,483 LMP and invasive mucinous EOC cases and 21,530 controls. Five SNPs were associated with mucinous EOC risk at P<0.05 with an OR that agreed in direction of effect with the OVA discovery samples (Table 3). Replication results including the OVA study are also presented in Table 3 with the corresponding Forest plots shown in Figure 1. SNP rs11108890 on chromosome 12 had the lowest P-value and was associated with increasing mucinous EOC risk (OR= 1.33, 95%CI: 1.11–1.61, P=0.0026). With the exception of SNPs rs970651 and rs7981902 on chromosome 13 (r2=0.76), which tagged a cluster of 8 highly ranked SNPs, these 5 SNPs are not in LD with other SNPs chosen for replication by the pool-based GWAS. These SNPs were statistically significant at the Bonferroni adjusted critical value of 0.0012. In exploratory analyses, we examined the associations of these 5 SNPS in the other EOC subtypes (see online Supplementary Table 2). rs11108890 was associated with endometrioid (but not clear cell) EOC risk (OR=1.23, 95%CI: 1.02-1.49, P= 0.03). No other associations at P<0.05 were evident.
Figure 1.
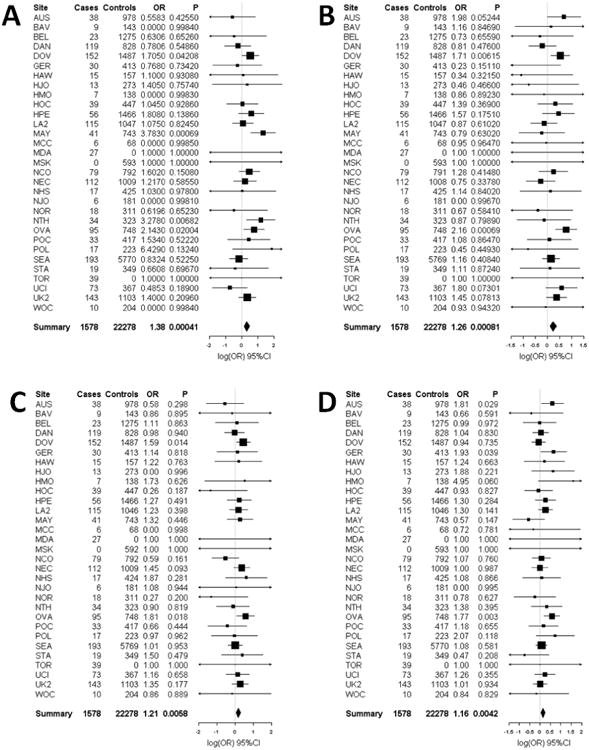
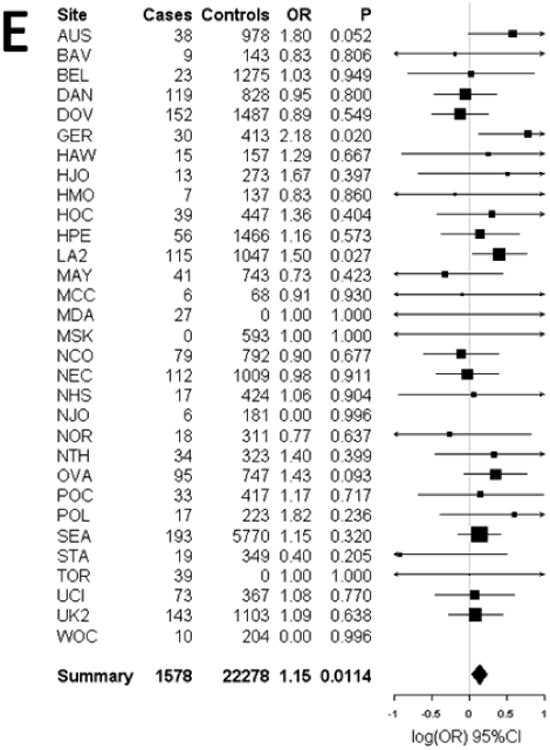
Forest plots of the study specific and summary odds ratios and 95% confidence intervals for the association between mucinous ovarian cancer risk and five SNPs nominated by our pool-based GWAS: (A) rs11108890 (B) rs933518 (C) rs17106154 (D) rs970651, and (E) rs7981902. Study-specific odds ratios, 95% confidence intervals, and P-values are based on logistic regression assuming an additive genetic mode, adjusting for the first 5 eigenvalues from principal components analysis. Summary odds ratios and 95% confidence intervals are from fixed effects meta-analysis and include the OVA study site. Forest plots were generated using the rmeta library implemented in the R project; association analyses were performed using PLINK
Endometrioid/Clear Cell subtype
Fifty-five SNPs representing 42 loci were tested for association with 2,903 invasive endometrioid/clear cell EOC cases (these subtypes were combined to be consistent with the DNA pool constructed in the discovery stage) and 21,528 controls. Three SNPs (rs2190503, rs6593140, rs2329554) were associated with endometrioid/clear cell EOC risk at P<0.05 with an OR that agreed in direction of effect with the OVA discovery samples (Table 3). rs2190503 and rs6593140 are in complete LD, and rs2329554 is in moderate LD with them (r2=0.6). These SNPs were selected to tag a cluster of 13 highly-ranked SNPs on chromosome 7. SNP rs2190503 had the lowest P-value (P= 0.014) and was associated with increased risk (OR=1.11, 95%CI: 1.02-1.21); but, these SNPs were not significant at the Bonferroni adjusted critical value of 0.0012. In exploratory analyses, we examined the associations of these 3 SNPS in the other EOC subtypes and found them to also be associated with all subtype and invasive serous EOC risk (see online Supplementary Table 2). When endometrioid and clear cell EOC were stratified, effect estimates were consistent with those reported in the endometrioid/clear cell combined analysis for all three SNPs, though the associations were marginally or not statistically significant. This result likely relates to power limitations given the small number of endometrioid and clear cell cases.
LMP serous subtype
Fifty-three SNPs representing 43 loci were tested for association with 892 LMP serous EOC and 21,528 controls. SNP rs9609538 was associated with risk at P<0.05 with an OR that agreed in direction of effect with the OVA discovery samples (OR= 0.86, 95%CI: 0.77-0.95, p=0.0043; Table 3). In analyses that included the discovery stage samples (2.6% of total samples), the association between rs9609538 and LMP serous EOC was statistically significant at the Bonferroni adjusted critical value of 0.0012 (OR=0.84, 95%CI: 0.76-0.93, P=0.0007). rs9609538 was the only SNP chosen to tag a cluster of 4 highly-ranked SNPs on chromosome 22. rs9609538 was not associated with risk in any of the other EOC subtypes (see online Supplementary Table 2).
Invasive serous subtype
Twenty-four SNPs representing 15 loci were tested for association with 6,881 invasive serous EOC and 21,530 controls. None of these SNPs was associated with risk (data not shown).
Discussion
Our objective was to discover risk alleles for the less-common EOC subtypes by performing a pool-based GWAS, followed by replication of associations using genotypes from 13,188 cases and 23,164 controls from OCAC. Nine SNPs tagging 6 loci were found to be associated with risk at P<0.05 with ORs that agreed in direction of effect with the discovery stage samples. Only one of these, rs9609538, remained statistically significant with LMP serous EOC following correction for multiple testing in analyses that included the 75 discovery samples.
SNP rs9609538 was associated with decreased risk for LMP serous EOC and, in exploratory analyses, was not associated with any other subtype. This SNP lies on chromosome 22 within a 1Mb region of 11 genes (YWHAH, LOC402057, SLC5A1, LOC150297, RFPL2, SLC5A4, RFPL3, C22orf28, BPIL2, FBXO7, and SYN3). The minor allele of rs9609538 is predicted to alter transcription factor (TF) binding site activity (multiple TFs including AIRE, AP-4, and CDP CR3) and miRNA binding site activity (hsa-miR-516a-5p and hsa-miR-548d-3p) based on FuncPred algorithms (Xu and Taylor 2009). SNP rs9609538 is positioned between C22orf28 (∼500 bp upstream) and BPIL2 (5bp downstream). BPIL2 is reported to be a rarely expressed lipid transfer/lipopolysaccharide binding protein, involved in recognizing the outer membrane of Gram-negative bacteria (Mulero et al. 2002). It was reported to be abnormally highly-expressed in the inflamed skin of psoriasis patients, and implicated in the inflammation and/or immune response (Mulero et al. 2002). The relevance of inflammation processes to risk of LMP tumours was also recently suggested by the association of TNFSF10 (or TRAIL) with this EOC subtype (Charbonneau et al., in submission). C22orf28 encodes a tRNA-splicing ligase protein. Although BPIL2 seems a plausible candidate gene, fine mapping of the association in a larger sample following by functional assays are needed to determine the gene targeted by this association, followed by further work to determine how it exerts its effects.
Although the other 8 SNPs were not significantly associated with subtype-specific EOC risk following adjustments for multiple testing, several loci tagged by these SNPs are in or near genes that have a plausible biological rationale for influencing ovarian cancer pathogenesis. These include rs17106154, which lies within a ∼150kb LD region of ZFP36L1 (also known as BRF1, TIS11B, and Berg36). ZFP36L1 is highly expressed in the ovary (Hacker et al. 2010) and was identified as a VEGF mRNA-destabilizing protein (Planel et al. 2010). That ZFP36L1 is altered in 7% of adenoid cystic carcinomas (Ho et al. 2013) and only 1% of invasive serous EOCs in The Cancer Genome Atlas is consistent with our finding that the SNP is associated with mucinous (a cystic tumor) but not invasive serous EOC. Three non-coding SNPs (rs2190503, rs6593140, rs2329554) tagging one locus upstream/intronic to GRB10 were associated with risk of endometrioid/clear cell EOC. GRB10 functions in the feedback inhibition of the PI3K/AKT and RAS/MAPK pathways (Hsu et al. 2011; Yu et al. 2011), and genes in these pathway are frequently mutated in endometrioid and clear cell tumours, and occasionally in serous tumours (Gilks 2010). GRB10 may be a tumour suppressor gene that acts in parallel with PTEN to ensure proper levels of activation of the PI3K/AKT pathway (Hsu et al. 2011; Yu et al. 2011). No SNPs were found to be associated with invasive serous risk in the replication samples. However, a previously reported GWAS SNP for invasive serous EOC with a moderately large OR (rs10088218, OR=0.76) ranked highly in our pool-based data (ranked 3389 and in perfect LD with SNPs that ranked 295 and 445), but did not meet our stringent criteria for selection in replication.
There are several limitations to our study. First, the discovery stage sample size was small, reflecting the low incidence of the less-common EOC subtypes. We, therefore, combined endometrioid and clear cell samples for analysis and primarily investigated associations shared between these subtypes. Thus, associations unique to one subtype could not be evaluated. Second, despite rapid progress in recent years, robust histological subtyping remains a challenge for studies of EOC (Gilks et al. 2008; Gilks and Prat 2009; Han et al. 2008; Kobel et al. 2010; Kobel et al. 2009). For example, many tumours that have previously been designated high-grade endometrioid are likely to be high-grade serous EOC (Kobel et al. 2009), and metastatic carcinoma from other organ sites is still difficult to correctly identify from primary mucinous EOC (Kelemen and Kobel 2011). Samples used in the pool-based stage of this study were reviewed using contemporary diagnostic criteria (Gilks et al. 2008); however, many of the OCAC replication studies including samples in our previous GWAS (Song et al. 2009) were not centrally-reviewed, and subtype misclassification may have introduced genetic heterogeneity and reduced statistical power. Third, the number of SNPs (maximum 200) chosen for replication was low. This was a factor restricted by cost and assay design across OCAC investigators and the three other consortia participating in the COGS initiative. The low coverage of genotyped SNPs per locus was also insufficient to allow imputation. Thus, additional genotyping of loci of interest will be needed to narrow down the regions of association.
This study also has several strengths. The DNA pooling design approach has successfully been applied in the GWAS context, including cancer GWAS (Brown et al. 2008; Skibola et al. 2009). The lack of identified risk alleles for EOC subtypes other than invasive serous prompted the current study, and we report a promising candidate for further interrogation for LMP serous EOC. Finally, the large number of EOC samples in OCAC - the largest assembled to date – and specifically of the less-common EOC subtypes, together with the coordinated genotyping and QC success rates achieved for over 200,000 samples and SNPs across 4 consortia, is a major strength of this study.
In conclusion, our pool-based GWAS of EOC risk according to subtype identified 9 SNPs tagging 6 loci with suggestive associations in the mucinous (5 SNPs), endometrioid/clear cell (3 SNPs), and LMP serous (1 SNP) subtypes. Several tagged loci harbor genes that have a plausible biological rationale for conferring EOC risk. Further evaluation in additional samples will be important to verify these results for the less-common EOC subtypes.
Supplementary Material
Figure 2.
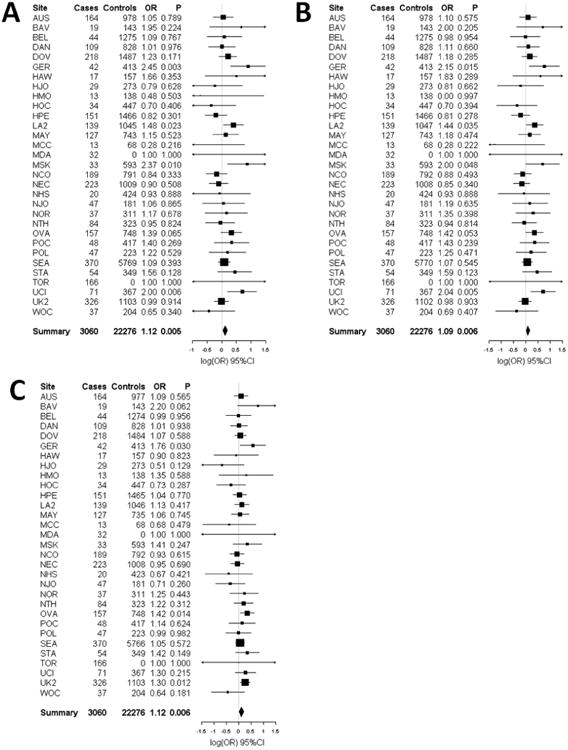
Forest plots of the study specific and summary odds ratios and 95% confidence intervals for the association between endometrioid/clear cell ovarian cancer risk and three SNPs nominated by our pool-based GWAS: (A) rs2190503 (B) rs6593140 and (C) rs2329554. Study-specific odds ratios, 95% confidence intervals, and P-values are based on logistic regression assuming an additive genetic mode, adjusting for the first 5 eigenvalues from principal components analysis. Summary odds ratios and 95% confidence intervals are from fixed effects meta-analysis and include the OVA study site. Forest plots were generated using the rmeta library implemented in the R project; association analyses were performed using PLINK
Figure 3.
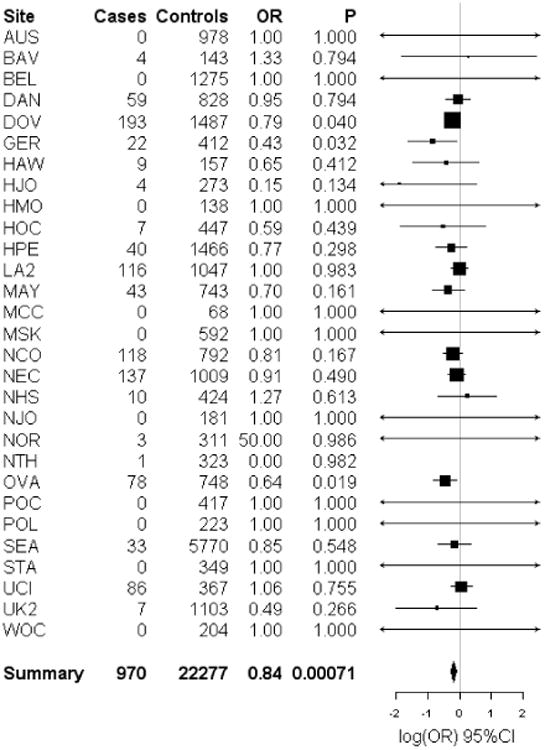
Forest plot of the study specific and summary odds ratios and 95% confidence intervals for the association between low malignant potential serous ovarian cancer risk and one SNP nominated by our pool-based GWAS, rs9609538. Study-specific odds ratios, 95% confidence intervals, and P-values are based on logistic regression assuming an additive genetic model, adjusting for the first 5 eigenvalues from principal components analysis. Summary odds ratios and 95% confidence intervals are from fixed effects meta-analysis and include the OVA study site. Forest plots were generated using the rmeta library implemented in the R project; association analyses were performed using PLINK. The MDA and TOR study sites are not plotted; neither study had any LMP serous ovarian cancer cases or controls
Acknowledgments
Named individuals: This study would not have been possible without the contributions of the following: P. Hall (COGS); A. M. Dunning and A. Lee (Cambridge); J. Benitez, A. Gonzalez-Neira and the staff of the CNIO genotyping unit; D. C. Tessier, F. Bacot, D. Vincent, S. LaBoissière and F. Robidoux and the staff of the Genome Quebec genotyping unit; S. E. Bojesen, S. F. Nielsen, B. G. Nordestgaard, and the staff of the Copenhagen DNA laboratory; and J.M. Cunningham, S. A. Windebank, C. A. Hilker, J. Meyer and the staff of Mayo Clinic Genotyping Core Facility. We thank all the individuals who took part in this study and all the researchers, clinicians and technical and administrative staff who have made possible the many studies contributing to this work. In particular, we thank: D. Bowtell, A. deFazio, D. Gertig, A. Green, P. Parsons, N. Hayward, P. Webb, and D. Whiteman (AUS); G. Peuteman, T. Van Brussel, and D. Smeets (BEL); U. Eilber (GER); L. Gacucova (HMO); P. Schürmann, F. Kramer, W. Zheng, T.W. Park-Simon, K. Beer-Grondke, and D. Schmidt (HJO); J. Vollenweider (MAY); the MD Anderson Center for Translational and Public Health Genomics (MDA); the state cancer registries of AL, AZ, AR, CA, CO, CT, DE, FL, GA, HI, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, and WY (NHS); L. Paddock, M. King, L. Rodriguez-Rodriguez, A. Samoila, and Y. Bensman (NJO); M. Sherman, A. Hutchinson, N. Szeszenia-Dabrowska, B. Peplonska, W. Zatonski, A. Soni, P. Chao, and M. Stagner (POL); C. Luccarini, P. Harrington, the SEARCH team and ECRIC (SEA); the Scottish Gynaecological Clinical Trails group and SCOTROC1 investigators (SRO); I. Jacobs, M. Widschwendter, E. Wozniak, N. Balogun, A. Ryan, and J. Ford (UKO); and Carole Pye (UKR).
Higher level funding: The COGS project is funded through a European Commission's Seventh Framework Programme grant (agreement number 223175 - HEALTH-F2-2009-223175). The Ovarian Cancer Association Consortium is supported by a grant from the Ovarian Cancer Research Fund thanks to donations by the family and friends of Kathryn Sladek Smith (PPD/RPCI.07). The scientific development and funding for this project were in part supported by the US National Cancer Institute GAME-ON Post-GWAS Initiative (U19-CA148112).
Investigator support: L.E.K. is supported by a Canadian Institutes of Health Research Investigator award (MSH-87734). G.C.-T. is supported by the National Health and Medical Research Council. B.Y. K. holds an American Cancer Society Early Detection Professorship (SIOP-06-258-01- COUN). F.M. is supported by a K-award from the National Cancer Institute (K07-CA080668). W.S. is supported by a K-award from the National Cancer Institute (K07-CA143047). D.F.E. is a Principal Research Fellow of Cancer Research UK.
Funding of constituent studies: This project was funded through grants from the Canadian Institutes of Health Research (MOP-86727, MOP-84340); WorkSafeBC 14, and OvCaRe: BC's Ovarian Cancer Research Team. Funding of the constituent studies was provided by the American Cancer Society (CRTG-00-196-01-CCE); the California Cancer Research Program (00-01389V-20170, N01-CN25403, 2II0200); Cancer Council Victoria; Cancer Council Queensland; Cancer Council New South Wales; Cancer Council South Australia; Cancer Council Tasmania; Cancer Foundation of Western Australia; the Cancer Institute of New Jersey; Cancer Research UK (C490/A6187, C490/A10119, C490/A10124, C536/A13086, C536/A6689); the Celma Mastry Ovarian Cancer Foundation ; the Danish Cancer Society (94-222-52); the ELAN Program of the University of Erlangen-Nuremberg; the Eve Appeal (Oak Foundation); the Fred C. and Katherine B. Andersen Foundation; the German Cancer Research Center; the German Federal Ministry of Education and Research of Germany, Program of Clinical Biomedical Research (01GB 9401); the Helsinki University Central Hospital Research Fund; Helse Vest; Imperial Experimental Cancer Research Centre (C1312/A15589); the L & S Milken Foundation; the Lon V. Smith Foundation (LVS-39420); the Mayo Foundation; the Mermaid I project; the Minnesota Ovarian Cancer Alliance; the National Health and Medical Research Council of Australia (199600, 209057, 251533, 396414, 400281, and 504715); Nationaal Kankerplan of Belgium; the Norwegian Cancer Society; the Norwegian Research Council; the OHSU Foundation; the Polish Ministry of Science and Higher Education (4 PO5C 028 14, 2 PO5A 068 27); Pomeranian Medical University; Radboud University Medical Center; the Roswell Park Cancer Institute Alliance Foundation; the Royal Marsden Hospital; the Rudolf-Bartling Foundation; the Sigrid Juselius Foundation; the state of Baden-Württemberg through Medical Faculty of the University of Ulm (P.685); the UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge and the University College London Hospitals; the US Army Medical Research and Material Command (DAMD17-98-1- 8659, DAMD17-01-1-0729, DAMD17-02-1-0666, DAMD17-02-1-0669, W81XWH-10-1-0280); the Department of Defense Ovarian Cancer Research Program (W81XWH-07-1-0449); the US National Cancer Institute (K07-CA095666, K22-CA138563, N01-CN55424, N01-PC067010, N01-PC035137, P01-CA017054, P01-CA087696, P30-CA15083, P50-CA105009, P50- CA136393, R01-CA014089, R01-CA016056, R01-CA017054, R01-CA049449, R01-CA050385, R01-CA054419, R01-CA058598, R01-CA058860, R01-CA061107, R01-CA061132, R01-CA063678, R01-CA063682, R01-CA064277, R01-CA067262, R01- CA071766, R01-CA074850, R01-CA076016, R01-CA080742, R01-CA080978, R01-CA083918, R01-CA087538, R01- CA092044, R01-095023, R01-CA106414, R01-CA122443, R01-CA112523, R01-CA114343, R01-CA126841, R01- CA136924, R01-CA149429, R03-CA113148, R03-CA115195, R37-CA070867, R37-CA70867, U01-CA069417, U01- CA071966 and Intramural research funds); the US National Institutes of Health/National Center for Research Resources/General Clinical Research Center (MO1-RR000056); and the US Public Health Service (PSA-042205).
References
- Albarracin CT, Silva EG, Malpica A, Luthra R, Liu J. The role of hMSH3 and hMSH6 in ovarian endometrioid carcinoma and relationship with microsatellite instability phenotype. Oncol Rep. 2004;12:1217–9. [PubMed] [Google Scholar]
- Bojesen SE, Pooley KA, Johnatty SE, Beesley J, Michailidou K, Tyrer JP, Edwards SL, Pickett HA, Shen HC, Smart CE, Hillman KM, Mai PL, Lawrenson K, Stutz MD, Lu Y, Karevan R, Woods N, Johnston RL, French JD, Chen X, Weischer M, Nielsen SF, Maranian MJ, Ghoussaini M, Ahmed S, Baynes C, Bolla MK, Wang Q, Dennis J, McGuffog L, Barrowdale D, Lee A, Healey S, Lush M, Tessier DC, Vincent D, Bacot F, Vergote I, Lambrechts S, Despierre E, Risch HA, Gonzalez-Neira A, Rossing MA, Pita G, Doherty JA, Alvarez N, Larson MC, Fridley BL, Schoof N, Chang-Claude J, Cicek MS, Peto J, Kalli KR, Broeks A, Armasu SM, Schmidt MK, Braaf LM, Winterhoff B, Nevanlinna H, Konecny GE, Lambrechts D, Rogmann L, Guenel P, Teoman A, Milne RL, Garcia JJ, Cox A, Shridhar V, Burwinkel B, Marme F, Hein R, Sawyer EJ, Haiman CA, Wang-Gohrke S, Andrulis IL, Moysich KB, Hopper JL, Odunsi K, Lindblom A, Giles GG, Brenner H, Simard J, Lurie G, Fasching PA, Carney ME, Radice P, Wilkens LR, Swerdlow A, Goodman MT, Brauch H, Garcia-Closas M, Hillemanns P, Winqvist R, Durst M, Devilee P, Runnebaum I, Jakubowska A, Lubinski J, Mannermaa A, Butzow R, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet. 2013;45:371–84. 384e1–2. doi: 10.1038/ng.2566ng.2566[pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton KL, Tyrer J, Song H, Ramus SJ, Notaridou M, Jones C, Sher T, Gentry-Maharaj A, Wozniak E, Tsai YY, Weidhaas J, Paik D, Van Den Berg DJ, Stram DO, Pearce CL, Wu AH, Brewster W, Anton-Culver H, Ziogas A, Narod SA, Levine DA, Kaye SB, Brown R, Paul J, Flanagan J, Sieh W, McGuire V, Whittemore AS, Campbell I, Gore ME, Lissowska J, Yang HP, Medrek K, Gronwald J, Lubinski J, Jakubowska A, Le ND, Cook LS, Kelemen LE, Brook-Wilson A, Massuger LF, Kiemeney LA, Aben KK, van Altena AM, Houlston R, Tomlinson I, Palmieri RT, Moorman PG, Schildkraut J, Iversen ES, Phelan C, Vierkant RA, Cunningham JM, Goode EL, Fridley BL, Kruger-Kjaer S, Blaeker J, Hogdall E, Hogdall C, Gross J, Karlan BY, Ness RB, Edwards RP, Odunsi K, Moyisch KB, Baker JA, Modugno F, Heikkinenen T, Butzow R, Nevanlinna H, Leminen A, Bogdanova N, Antonenkova N, Doerk T, Hillemanns P, Durst M, Runnebaum I, Thompson PJ, Carney ME, Goodman MT, Lurie G, Wang-Gohrke S, Hein R, Chang-Claude J, Rossing MA, Cushing-Haugen KL, Doherty J, Chen C, Rafnar T, Besenbacher S, Sulem P, Stefansson K, Birrer MJ, Terry KL, Hernandez D, Cramer DW, Vergote I, Amant F, Lambrechts D, Despierre E, et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nature genetics. 2010;42:880–884. doi: 10.1038/ng.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KM, Macgregor S, Montgomery GW, Craig DW, Zhao ZZ, Iyadurai K, Henders AK, Homer N, Campbell MJ, Stark M, Thomas S, Schmid H, Holland EA, Gillanders EM, Duffy DL, Maskiell JA, Jetann J, Ferguson M, Stephan DA, Cust AE, Whiteman D, Green A, Olsson H, Puig S, Ghiorzo P, Hansson J, Demenais F, Goldstein AM, Gruis NA, Elder DE, Bishop JN, Kefford RF, Giles GG, Armstrong BK, Aitken JF, Hopper JL, Martin NG, Trent JM, Mann GJ, Hayward NK. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nature genetics. 2008;40:838–840. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai KQ, Albarracin C, Rosen D, Zhong R, Zheng W, Luthra R, Broaddus R, Liu J. Microsatellite instability and alteration of the expression of hMLH1 and hMSH2 in ovarian clear cell carcinoma. Hum Pathol. 2004;35:552–9. doi: 10.1016/j.humpath.2003.12.009. doi:S0046817704000450[pii] [DOI] [PubMed] [Google Scholar]
- Catasus L, Bussaglia E, Rodrguez I, Gallardo A, Pons C, Irving JA, Prat J. Molecular genetic alterations in endometrioid carcinomas of the ovary: similar frequency of beta-catenin abnormalities but lower rate of microsatellite instability and PTEN alterations than in uterine endometrioid carcinomas. Hum Pathol. 2004;35:1360–8. doi: 10.1016/j.humpath.2004.07.019. doi:S0046817704004368[pii]10.1016/j.humpath.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Earp MA, Rahmani M, Chew K, Brooks-Wilson A. Estimates of array and pool-construction variance for planning efficient DNA-pooling genome wide association studies. BMC medical genomics. 2011;4:81. doi: 10.1186/1755-8794-4-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber MT, Kjaer SK, Dehlendorff C, Chang-Claude J, Andersen KK, Hogdall E, Webb PM, Jordan SJ, Rossing MA, Doherty JA, Lurie G, Thompson PJ, Carney ME, Goodman MT, Ness RB, Modugno F, Edwards RP, Bunker CH, Goode EL, Fridley BL, Vierkant RA, Larson MC, Schildkraut J, Cramer DW, Terry KL, Vitonis AF, Bandera EV, Olson SH, King M, Chandran U, Kiemeney LA, Massuger LF, van Altena AM, Vermeulen SH, Brinton L, Wentzensen N, Lissowska J, Yang HP, Moysich KB, Odunsi K, Kasza K, Odunsi-Akanji O, Song H, Pharaoh P, Shah M, Whittemore AS, McGuire V, Sieh W, Sutphen R, Menon U, Gayther SA, Ramus SJ, Gentry-Maharaj A, Pearce CL, Wu AH, Pike MC, Risch HA, Jensen A. Cigarette smoking and risk of ovarian cancer: a pooled analysis of 21 case-control studies. Cancer Causes Control. 2013;24:989–1004. doi: 10.1007/s10552-013-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayther SA, Pharoah PD. The inherited genetics of ovarian and endometrial cancer. Current opinion in genetics & development. 2010;20:231–238. doi: 10.1016/j.gde.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks CB. Molecular abnormalities in ovarian cancer subtypes other than high-grade serous carcinoma. J Oncol. 2010;2010:740968. doi: 10.1155/2010/740968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks CB, Ionescu DN, Kalloger SE, Kobel M, Irving J, Clarke B, Santos J, Le N, Moravan V, Swenerton K Cheryl Brown Ovarian Cancer Outcomes Unit of the British Columbia Cancer A. Tumor cell type can be reproducibly diagnosed and is of independent prognostic significance in patients with maximally debulked ovarian carcinoma. Human pathology. 2008;39:1239–1251. doi: 10.1016/j.humpath.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Gilks CB, Prat J. Ovarian carcinoma pathology and genetics: recent advances. Human pathology. 2009;40:1213–1223. doi: 10.1016/j.humpath.2009.04.017. [DOI] [PubMed] [Google Scholar]
- Gomez-Raposo C, Mendiola M, Barriuso J, Hardisson D, Redondo A. Molecular characterization of ovarian cancer by gene-expression profiling. Gynecologic oncology. 2010;118:88–92. doi: 10.1016/j.ygyno.2010.03.012. [DOI] [PubMed] [Google Scholar]
- Goode EL, Chenevix-Trench G, Song H, Ramus SJ, Notaridou M, Lawrenson K, Widschwendter M, Vierkant RA, Larson MC, Kjaer SK, Birrer MJ, Berchuck A, Schildkraut J, Tomlinson I, Kiemeney LA, Cook LS, Gronwald J, Garcia-Closas M, Gore ME, Campbell I, Whittemore AS, Sutphen R, Phelan C, Anton-Culver H, Pearce CL, Lambrechts D, Rossing MA, Chang-Claude J, Moysich KB, Goodman MT, Dork T, Nevanlinna H, Ness RB, Rafnar T, Hogdall C, Hogdall E, Fridley BL, Cunningham JM, Sieh W, McGuire V, Godwin AK, Cramer DW, Hernandez D, Levine D, Lu K, Iversen ES, Palmieri RT, Houlston R, van Altena AM, Aben KK, Massuger LF, Brooks-Wilson A, Kelemen LE, Le ND, Jakubowska A, Lubinski J, Medrek K, Stafford A, Easton DF, Tyrer J, Bolton KL, Harrington P, Eccles D, Chen A, Molina AN, Davila BN, Arango H, Tsai YY, Chen Z, Risch HA, McLaughlin J, Narod SA, Ziogas A, Brewster W, Gentry-Maharaj A, Menon U, Wu AH, Stram DO, Pike MC, Wellcome Trust Case-Control C. Beesley J, Webb PM, Australian Cancer S. Australian Ovarian Cancer Study G. Ovarian Cancer Association C. Chen X, Ekici AB, Thiel FC, Beckmann MW, Yang H, Wentzensen N, Lissowska J, Fasching PA, Despierre E, Amant F, Vergote I, Doherty J, Hein R, Wang-Gohrke S, Lurie G, et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nature genetics. 2010;42:874–879. doi: 10.1038/ng.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker C, Valchanova R, Adams S, Munz B. ZFP36L1 is regulated by growth factors and cytokines in keratinocytes and influences their VEGF production. Growth Factors. 2010;28:178–90. doi: 10.3109/08977190903578660. [DOI] [PubMed] [Google Scholar]
- Han G, Gilks CB, Leung S, Ewanowich CA, Irving JA, Longacre TA, Soslow RA. Mixed ovarian epithelial carcinomas with clear cell and serous components are variants of high-grade serous carcinoma: an interobserver correlative and immunohistochemical study of 32 cases. Am J Surg Pathol. 2008;32:955–64. doi: 10.1097/PAS.0b013e318164edf7. [DOI] [PubMed] [Google Scholar]
- Ho AS, Kannan K, Roy DM, Morris LG, Ganly I, Katabi N, Ramaswami D, Walsh LA, Eng S, Huse JT, Zhang J, Dolgalev I, Huberman K, Heguy A, Viale A, Drobnjak M, Leversha MA, Rice CE, Singh B, Iyer NG, Leemans CR, Bloemena E, Ferris RL, Seethala RR, Gross BE, Liang Y, Sinha R, Peng L, Raphael BJ, Turcan S, Gong Y, Schultz N, Kim S, Chiosea S, Shah JP, Sander C, Lee W, Chan TA. The mutational landscape of adenoid cystic carcinoma. Nat Genet. 2013 doi: 10.1038/ng.2643ng.2643[pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer N, Tembe WD, Szelinger S, Redman M, Stephan DA, Pearson JV, Nelson SF, Craig D. Bioinformatics. Vol. 24. Oxford, England: 2008. Multimarker analysis and imputation of multiple platform pooling-based genome-wide association studies; pp. 1896–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, Marto JA, Sabatini DM. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–22. doi: 10.1126/science.1199498332/6035/1317[pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Wang TL, Shih le M, Mao TL, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA, Jr, Vogelstein B, Kinzler KW, Velculescu VE, Papadopoulos N. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–31. doi: 10.1126/science.1196333science.1196333[pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalloger SE, Kobel M, Leung S, Mehl E, Gao D, Marcon KM, Chow C, Clarke BA, Huntsman DG, Gilks CB. Calculator for ovarian carcinoma subtype prediction. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc.; 2010. [DOI] [PubMed] [Google Scholar]
- Kelemen LE, Kobel M. Mucinous carcinomas of the ovary and colorectum: different organ, same dilemma. Lancet Oncol. 2011;12:1071–80. doi: 10.1016/S1470-2045(11)70058-4S1470-2045(11)70058-4[pii]. [DOI] [PubMed] [Google Scholar]
- Kobel M, Kalloger SE, Baker PM, Ewanowich CA, Arseneau J, Zherebitskiy V, Abdulkarim S, Leung S, Duggan MA, Fontaine D, Parker R, Huntsman DG, Gilks CB. Diagnosis of ovarian carcinoma cell type is highly reproducible: a transcanadian study. The American Journal of Surgical Pathology. 2010;34:984–993. doi: 10.1097/PAS.0b013e3181e1a3bb. [DOI] [PubMed] [Google Scholar]
- Kobel M, Kalloger SE, Carrick J, Huntsman D, Asad H, Oliva E, Ewanowich CA, Soslow RA, Gilks CB. A limited panel of immunomarkers can reliably distinguish between clear cell and high-grade serous carcinoma of the ovary. Am J Surg Pathol. 2009;33:14–21. doi: 10.1097/PAS.0b013e3181788546. [DOI] [PubMed] [Google Scholar]
- Kuo KT, Mao TL, Jones S, Veras E, Ayhan A, Wang TL, Glas R, Slamon D, Velculescu VE, Kuman RJ, Shih Ie M. Frequent activating mutations of PIK3CA in ovarian clear cell carcinoma. Am J Pathol. 2009;174:1597–601. doi: 10.2353/ajpath.2009.081000. doi:S0002-9440(10)61017-6[pii]10.2353/ajpath.2009.081000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurman RJ, Ronnett BM. Ovarian intestinal-type mucinous borderline tumors: a nomenclature change is long overdue. Int J Gynecol Pathol. 2010;29:552–3. doi: 10.1097/PGP.0b013e3181eb2fb5. author reply 553-4. [DOI] [PubMed] [Google Scholar]
- Kurman RJ, Shih I. The origin and pathogenesis of epithelial ovarian cancer: a proposed unifying theory. The American Journal of Surgical Pathology. 2010;34:433–443. doi: 10.1097/PAS.0b013e3181cf3d79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch HT, Conway T, Lynch J. Hereditary ovarian cancer. Pedigree studies. Part II Cancer Genet Cytogenet. 1991;53:161–83. doi: 10.1016/0165-4608(91)90094-b. doi:0165-4608(91)90094-B[pii] [DOI] [PubMed] [Google Scholar]
- Lynch HT, Schuelke GS, Kimberling WJ, Albano WA, Lynch JF, Biscone KA, Lipkin ML, Deschner EE, Mikol YB, Sandberg AA, et al. Hereditary nonpolyposis colorectal cancer (Lynch syndromes I and II). II. Biomarker studies. Cancer. 1985;56:939–51. doi: 10.1002/1097-0142(19850815)56:4<939::aid-cncr2820560440>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Matsuno RK, Sherman ME, Visvanathan K, Goodman MT, Hernandez BY, Lynch CF, Ioffe OB, Horio D, Platz C, Altekruse SF, Pfeiffer RM, Anderson WF. Agreement for tumor grade of ovarian carcinoma: analysis of archival tissues from the surveillance, epidemiology, and end results residual tissue repository. Cancer Causes Control. 2013;24:749–57. doi: 10.1007/s10552-013-0157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero JJ, Boyle BJ, Bradley S, Bright JM, Nelken ST, Ho TT, Mize NK, Childs JD, Ballinger DG, Ford JE, Rupp F. Three new human members of the lipid transfer/lipopolysaccharide binding protein family (LT/LBP) Immunogenetics. 2002;54:293–300. doi: 10.1007/s00251-002-0467-3. [DOI] [PubMed] [Google Scholar]
- Obata K, Morland SJ, Watson RH, Hitchcock A, Chenevix-Trench G, Thomas EJ, Campbell IG. Frequent PTEN/MMAC mutations in endometrioid but not serous or mucinous epithelial ovarian tumors. Cancer Res. 1998;58:2095–7. [PubMed] [Google Scholar]
- Pearce CL, Templeman C, Rossing MA, Lee A, Near AM, Webb PM, Nagle CM, Doherty JA, Cushing-Haugen KL, Wicklund KG, Chang-Claude J, Hein R, Lurie G, Wilkens LR, Carney ME, Goodman MT, Moysich K, Kjaer SK, Hogdall E, Jensen A, Goode EL, Fridley BL, Larson MC, Schildkraut JM, Palmieri RT, Cramer DW, Terry KL, Vitonis AF, Titus LJ, Ziogas A, Brewster W, Anton-Culver H, Gentry-Maharaj A, Ramus SJ, Anderson AR, Brueggmann D, Fasching PA, Gayther SA, Huntsman DG, Menon U, Ness RB, Pike MC, Risch H, Wu AH, Berchuck A on behalf of the Ovarian Cancer Association C. Association between endometriosis and risk of histological subtypes of ovarian cancer: a pooled analysis of case-control studies. The lancet oncology. 2012 doi: 10.1016/s1470-2045(11)70404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JV, Huentelman MJ, Halperin RF, Tembe WD, Melquist S, Homer N, Brun M, Szelinger S, Coon KD, Zismann VL, Webster JA, Beach T, Sando SB, Aasly JO, Heun R, Jessen F, Kolsch H, Tsolaki M, Daniilidou M, Reiman EM, Papassotiropoulos A, Hutton ML, Stephan DA, Craig DW. Identification of the genetic basis for complex disorders by use of pooling-based genomewide single-nucleotide-polymorphism association studies. American Journal of Human Genetics. 2007;80:126–139. doi: 10.1086/510686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permuth-Wey J, Lawrenson K, Shen HC, Velkova A, Tyrer JP, Chen Z, Lin HY, Chen YA, Tsai YY, Qu X, Ramus SJ, Karevan R, Lee J, Lee N, Larson MC, Aben KK, Anton-Culver H, Antonenkova N, Antoniou AC, Armasu SM, Bacot F, Baglietto L, Bandera EV, Barnholtz-Sloan J, Beckmann MW, Birrer MJ, Bloom G, Bogdanova N, Brinton LA, Brooks-Wilson A, Brown R, Butzow R, Cai Q, Campbell I, Chang-Claude J, Chanock S, Chenevix-Trench G, Cheng JQ, Cicek MS, Coetzee GA, Cook LS, Couch FJ, Cramer DW, Cunningham JM, Dansonka-Mieszkowska A, Despierre E, Doherty JA, Dork T, du Bois A, Durst M, Easton DF, Eccles D, Edwards R, Ekici AB, Fasching PA, Fenstermacher DA, Flanagan JM, Garcia-Closas M, Gentry-Maharaj A, Giles GG, Glasspool RM, Gonzalez-Bosquet J, Goodman MT, Gore M, Gorski B, Gronwald J, Hall P, Halle MK, Harter P, Heitz F, Hillemanns P, Hoatlin M, Hogdall CK, Hogdall E, Hosono S, Jakubowska A, Jensen A, Jim H, Kalli KR, Karlan BY, Kaye SB, Kelemen LE, Kiemeney LA, Kikkawa F, Konecny GE, Krakstad C, Kjaer SK, Kupryjanczyk J, Lambrechts D, Lambrechts S, Lancaster JM, Le ND, Leminen A, Levine DA, Liang D, Lim BK, Lin J, Lissowska J, Lu KH, Lubinski J, et al. Identification and molecular characterization of a new ovarian cancer susceptibility locus at 17q21.31. Nat Commun. 2013;4:1627. doi: 10.1038/ncomms2613ncomms2613[pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharoah PD, Tsai YY, Ramus SJ, Phelan CM, Goode EL, Lawrenson K, Buckley M, Fridley BL, Tyrer JP, Shen H, Weber R, Karevan R, Larson MC, Song H, Tessier DC, Bacot F, Vincent D, Cunningham JM, Dennis J, Dicks E, Aben KK, Anton-Culver H, Antonenkova N, Armasu SM, Baglietto L, Bandera EV, Beckmann MW, Birrer MJ, Bloom G, Bogdanova N, Brenton JD, Brinton LA, Brooks-Wilson A, Brown R, Butzow R, Campbell I, Carney ME, Carvalho RS, Chang-Claude J, Chen YA, Chen Z, Chow WH, Cicek MS, Coetzee G, Cook LS, Cramer DW, Cybulski C, Dansonka-Mieszkowska A, Despierre E, Doherty JA, Dork T, du Bois A, Durst M, Eccles D, Edwards R, Ekici AB, Fasching PA, Fenstermacher D, Flanagan J, Gao YT, Garcia-Closas M, Gentry-Maharaj A, Giles G, Gjyshi A, Gore M, Gronwald J, Guo Q, Halle MK, Harter P, Hein A, Heitz F, Hillemanns P, Hoatlin M, Hogdall E, Hogdall CK, Hosono S, Jakubowska A, Jensen A, Kalli KR, Karlan BY, Kelemen LE, Kiemeney LA, Kjaer SK, Konecny GE, Krakstad C, Kupryjanczyk J, Lambrechts D, Lambrechts S, Le ND, Lee N, Lee J, Leminen A, Lim BK, Lissowska J, Lubinski J, Lundvall L, Lurie G, Massuger LF, Matsuo K, McGuire V, et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat Genet. 2013;45:362–70. doi: 10.1038/ng.2564ng.2564[pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piek JM, van Diest PJ, Zweemer RP, Jansen JW, Poort-Keesom RJ, Menko FH, Gille JJ, Jongsma AP, Pals G, Kenemans P, Verheijen RH. Dysplastic changes in prophylactically removed Fallopian tubes of women predisposed to developing ovarian cancer. J Pathol. 2001;195:451–6. doi: 10.1002/path.1000[pii]10.1002/path.1000. [DOI] [PubMed] [Google Scholar]
- Planel S, Salomon A, Jalinot P, Feige JJ, Cherradi N. A novel concept in antiangiogenic and antitumoral therapy: multitarget destabilization of short-lived mRNAs by the zinc finger protein ZFP36L1. Oncogene. 2010;29:5989–6003. doi: 10.1038/onc.2010.341onc2010341[pii]. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. doi:S0002-9297(07)61352-4[pii]10.1086/519795[doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook JRD. Molecular Cloning: A Laboratory Manual. Third. Cold Spring Harbor Laboratory Press; USA: 2000. [Google Scholar]
- Schrauwen I, Ealy M, Huentelman MJ, Thys M, Homer N, Vanderstraeten K, Fransen E, Corneveaux JJ, Craig DW, Claustres M, Cremers CW, Dhooge I, Van de Heyning P, Vincent R, Offeciers E, Smith RJ, Van Camp G. A genome-wide analysis identifies genetic variants in the RELN gene associated with otosclerosis. American Journal of Human Genetics. 2009;84:328–338. doi: 10.1016/j.ajhg.2009.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Fridley BL, Song H, Lawrenson K, Cunningham JM, Ramus SJ, Cicek MS, Tyrer J, Stram D, Larson MC, Kobel M, Ziogas A, Zheng W, Yang HP, Wu AH, Wozniak EL, Woo YL, Winterhoff B, Wik E, Whittemore AS, Wentzensen N, Weber RP, Vitonis AF, Vincent D, Vierkant RA, Vergote I, Van Den Berg D, Van Altena AM, Tworoger SS, Thompson PJ, Tessier DC, Terry KL, Teo SH, Templeman C, Stram DO, Southey MC, Sieh W, Siddiqui N, Shvetsov YB, Shu XO, Shridhar V, Wang-Gohrke S, Severi G, Schwaab I, Salvesen HB, Rzepecka IK, Runnebaum IB, Rossing MA, Rodriguez-Rodriguez L, Risch HA, Renner SP, Poole EM, Pike MC, Phelan CM, Pelttari LM, Pejovic T, Paul J, Orlow I, Omar SZ, Olson SH, Odunsi K, Nickels S, Nevanlinna H, Ness RB, Narod SA, Nakanishi T, Moysich KB, Monteiro AN, Moes-Sosnowska J, Modugno F, Menon U, McLaughlin JR, McGuire V, Matsuo K, Adenan NA, Massuger LF, Lurie G, Lundvall L, Lubinski J, Lissowska J, Levine DA, Leminen A, Lee AW, Le ND, Lambrechts S, Lambrechts D, Kupryjanczyk J, Krakstad C, Konecny GE, Kjaer SK, Kiemeney LA, Kelemen LE, Keeney GL, Karlan BY, Karevan R, Kalli KR, Kajiyama H, Ji BT, Jensen A, Jakubowska A, et al. Epigenetic analysis leads to identification of HNF1B as a subtype-specific susceptibility gene for ovarian cancer. Nat Commun. 2013;4:1628. doi: 10.1038/ncomms2629ncomms2629[pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman LP. Hereditary breast and ovarian cancer (HBOC): clinical features and counseling for BRCA1 and BRCA2, Lynch syndrome, Cowden syndrome, and Li-Fraumeni syndrome. Obstetrics and gynecology clinics of North America. 2010;37:109–33. doi: 10.1016/j.ogc.2010.03.003. Table of Contents. [DOI] [PubMed] [Google Scholar]
- Skibola CF, Bracci PM, Halperin E, Conde L, Craig DW, Agana L, Iyadurai K, Becker N, Brooks-Wilson A, Curry JD, Spinelli JJ, Holly EA, Riby J, Zhang L, Nieters A, Smith MT, Brown KM. Genetic variants at 6p21.33 are associated with susceptibility to follicular lymphoma. Nature genetics. 2009;41:873–875. doi: 10.1038/ng.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Ramus SJ, Tyrer J, Bolton KL, Gentry-Maharaj A, Wozniak E, Anton-Culver H, Chang-Claude J, Cramer DW, DiCioccio R, Dork T, Goode EL, Goodman MT, Schildkraut JM, Sellers T, Baglietto L, Beckmann MW, Beesley J, Blaakaer J, Carney ME, Chanock S, Chen Z, Cunningham JM, Dicks E, Doherty JA, Durst M, Ekici AB, Fenstermacher D, Fridley BL, Giles G, Gore ME, De Vivo I, Hillemanns P, Hogdall C, Hogdall E, Iversen ES, Jacobs IJ, Jakubowska A, Li D, Lissowska J, Lubinski J, Lurie G, McGuire V, McLaughlin J, Medrek K, Moorman PG, Moysich K, Narod S, Phelan C, Pye C, Risch H, Runnebaum IB, Severi G, Southey M, Stram DO, Thiel FC, Terry KL, Tsai YY, Tworoger SS, Van Den Berg DJ, Vierkant RA, Wang-Gohrke S, Webb PM, Wilkens LR, Wu AH, Yang H, Brewster W, Ziogas A, Australian Cancer S. Australian Ovarian Cancer Study G. Ovarian Cancer Association C. Houlston R, Tomlinson I, Whittemore AS, Rossing MA, Ponder BA, Pearce CL, Ness RB, Menon U, Kjaer SK, Gronwald J, Garcia-Closas M, Fasching PA, Easton DF, Chenevix-Trench G, Berchuck A, Pharoah PD, Gayther SA. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nature genetics. 2009;41:996–1000. doi: 10.1038/ng.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CJ, Leung Y, Walsh MD, Walters RJ, Young JP, Buchanan DD. KRAS mutations in ovarian low-grade endometrioid adenocarcinoma: association with concurrent endometriosis. Hum Pathol. 2012;43:1177–83. doi: 10.1016/j.humpath.2011.10.009S0046-8177(11)00435-7[pii]. [DOI] [PubMed] [Google Scholar]
- Tavassoulu FA, Devilee P. World Health Organization classification of tumors. Pathology and genetics of tumors of the breast and female genital organs. IARC Breast 2003 [Google Scholar]
- Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro B, Traficante N, Fereday S, Hung JA, Chiew YE, Haviv I, Gertig D, DeFazio A, Bowtell DD. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–208. doi: 10.1158/1078-0432.CCR-08-019614/16/5198[pii]. [DOI] [PubMed] [Google Scholar]
- Vaughan S, Coward JI, Bast RC, Jr, Berchuck A, Berek JS, Brenton JD, Coukos G, Crum CC, Drapkin R, Etemadmoghadam D, Friedlander M, Gabra H, Kaye SB, Lord CJ, Lengyel E, Levine DA, McNeish IA, Menon U, Mills GB, Nephew KP, Oza AM, Sood AK, Stronach EA, Walczak H, Bowtell DD, Balkwill FR. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer. 2011;11:719–25. doi: 10.1038/nrc3144nrc3144[pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, Yang W, Heravi-Moussavi A, Giuliany R, Chow C, Fee J, Zayed A, Prentice L, Melnyk N, Turashvili G, Delaney AD, Madore J, Yip S, McPherson AW, Ha G, Bell L, Fereday S, Tam A, Galletta L, Tonin PN, Provencher D, Miller D, Jones SJ, Moore RA, Morin GB, Oloumi A, Boyd N, Aparicio SA, Shih I, Mes-Masson AM, Bowtell DD, Hirst M, Gilks B, Marra MA, Huntsman DG. ARID1A mutations in endometriosis-associated ovarian carcinomas. The New England journal of medicine. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37:W600–5. doi: 10.1093/nar/gkp290gkp290[pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, Blenis J. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–6. doi: 10.1126/science.1199484332/6035/1322[pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


