Abstract
The capacity for Borrelia burgdorferi to cause disseminated infection in humans or mice is associated with the genotype of the infecting strain. The cytokine profiles elicited by B. burgdorferi clinical isolates of different genotype (ribosomal spacer type) groups were assessed in a human PBMC co-incubation model. RST1 isolates, which are more frequently associated with disseminated Lyme disease in humans and mice, induced significantly higher levels of IFN-α and IFN-λ1/IL29 relative to RST3 isolates, which are less frequently associated with disseminated infection. No differences in the protein concentrations of IFN-γ, IL-1β, IL-6, IL-8, IL-10 or TNF-α were observed between isolates of differing genotype. The ability of B. burgdorferi to induce type I and type III IFNs was completely dependent on the presence of linear plasmid (lp) 36. An lp36-deficient B. burgdorferi mutant adhered to, and was internalized by, PBMCs and specific dendritic cell (DC) subsets less efficiently than its isogenic B31 parent strain. The association defect with mDC1s and pDCs could be restored by complementation of the mutant with the complete lp36. The RST1 clinical isolates studied were found to contain a 2.5-kB region, located in the distal one-third of lp36, which was not present in any of the RST3 isolates tested. This divergent region of lp36 may encode one or more factors required for optimal spirochetal recognition and the production of type I and type III IFNs by human DCs, thus suggesting a potential role for DCs in the pathogenesis of B. burgdorferi infection.
Introduction
Borrelia burgdorferi, a tick-transmitted spirochete, is the infectious agent of Lyme disease [1], [2]. This disease is a multisystemic disorder with possible neurologic, rheumatologic, and cardiac symptoms which develop following hematogenous dissemination of the bacterium from the skin to distal target tissues [3]. The potential for Lyme disease to develop from the bite of an infected tick is highly variable due to the existence in nature of distinct B. burgdorferi genotypes with diverse capacities to cause infection or to disseminate. The genome of B. burgdorferi type strain B31-MI consists of a single linear chromosome and 21 linear and circular plasmids [4], [5]. Linear plasmid 54 (lp54), circular plasmid 26 (cp26) and the chromosome are highly conserved and have been present in all tested natural isolates of B. burgdorferi. However, the presence and content of other plasmids vary among isolates [4]–[7]. A genotyping method for B. burgdorferi isolates based on restriction-fragment length polymorphism of the 16S–23S ribosomal DNA spacer region has been developed [8], [9]. A relationship between ribosomal spacer type (RST) and the capacity for disseminated infection was observed; RST1 strains are more frequently associated with disseminated infection in both Lyme disease patients and mice, whereas RST3 isolates are less frequently detected in the blood of patients and some do not disseminate in mice [10]–[13]. The molecular mechanisms underlying the differential risk for hematogenous dissemination of different B. burgdorferi genotypes, however, have not been elucidated.
B. burgdorferi induces the production of both pro- and anti-inflammatory cytokines through the nuclear factor-kappa B (NF-κB) signaling pathway [14]–[18]. Internalization of intact B. burgdorferi and subsequent degradation and release of pathogen associated molecular patterns (PAMPs) within the phagolysosome are critical events leading to full activation of the innate immune response to this extracellular pathogen [16], [17], [19]–[21]. Phagocytic uptake of intact spirochetes induces secretion of IFN-γ by NK cells and triggers both apoptosis and the production of high levels of pro-inflammatory cytokines by human monocytes [16], [17]. Recent findings have identified the endosomal receptors, TLR7 and TLR9 in dendritic cells, and TLR8 in monocytes, as pathogen recognition receptors (PRRs) essential for the production of type I IFN protein and the transcription of IFN-responsive genes by B. burgdorferi-stimulated human immune cells [20]–[23]. Moreover, Cervantes et al. provided evidence using human monocytes that coordinated interactions between TLR8 and TLR2 facilitate the development of a diverse pro- and anti-inflammatory cytokine response by human monocytes [21]. However, despite the contribution of TLR2 to the initiation of B. burgdorferi-triggered signaling cascades both at the cell surface and within phagolysosomes, TLR2 is not required for spirochetal uptake [19]. Hawley et al. recently identified CR3 (CD11b) as a MyD88-independent phagocytic receptor for B. burgdorferi that also modulates TNF production by murine bone marrow-derived macrophages [24]. The B. burgdorferi ligand for CR3, as well as spirochetal components that mediate internalization by other phagocytic cell populations, remain unknown.
Type I interferons (IFN-α/β), an innate defense classically associated with an antiviral immune state and produced at high levels by plasmacytoid dendritic cells (pDCs), are now known to be induced in response to a variety of intracellular and extracellular bacterial pathogens [20], [25]–[29]. Previously, using an ex vivo co-incubation model, we demonstrated that human pDCs and CD11c+CD14+ monocytoid cells produce IFN-α protein following phagocytosis of B. burgdorferi [20]. It has also been shown that IFN-α is present at higher levels in the serum of Lyme disease patients with multiple erythema migrans (EM) lesions, an indicator of disseminated infection, compared to those of patients with a single EM [26]. The expression of IFN-α in the EM lesion correlates with the presence of monocytoid (mDC) and plasmacytoid (pDC) dendritic cells, both of which are enriched approximately 5-fold in the EM lesion infiltrate as compared with the skin of uninfected controls [26]. The interactions of B. burgdorferi with a specific innate immune cell population might facilitate dissemination and promote Lyme disease pathogenesis.
The type III IFNs, or IFN-λs, discovered in 2003, are a family of novel cytokines that includes three members: IFN-λ1, IFN-λ2, and IFN-λ3, also known as IL-29, IL-28A, and IL-28B, respectively [30]. Similar to type I IFNs, IFN-λs are expressed primarily by monocytes, macrophages, and dendritic cells, and can be produced by these cells simultaneously with IFN-α and IFN-β [30]–[33]. Although initially identified as anti-viral proteins, recent studies demonstrate that type III IFNs are an important component of the innate immune response to non-viral pathogens, including Salmonella enterica serovar typhimurium [31], [34] and Listeria monocytogenes [35]. However, the expression pattern of the IFN-λ receptor (IFNλR) is far more restricted than that of the type I IFN receptor; IFNλR is expressed solely in epithelial tissues and epithelial-like cell types, suggesting that the effects exerted on innate immunity likely are more important for pathogens interacting with the epithelium [36], [37]. Notably, L. monocytogenes pathogenesis is promoted by type I IFNs but can be further enhanced by cooperative interactions between type I IFNs and IL-28B [35].
Two reports from our laboratory were the first to describe the induction of IFN-λ1 (IL-29) by B. burgdorferi [23], [38]. Love et al. demonstrated the production of IL-29 by TLR7-dependent signaling in human PBMCs in response to live B. burgdorferi or isolated B. burgdorferi RNA. Those results were generated using B. burgdorferi B515, an RST1 clinical isolate that disseminates in mice [11]. In the current study, the cytokine profiles induced by B. burgdorferi clinical isolates of varying genotypes were analyzed using a human PBMC ex vivo co-incubation model. Intriguingly, differential recognition by human dendritic cells was found to be dependent on a highly divergent region of lp36 which is conserved among RST1 isolates.
Results
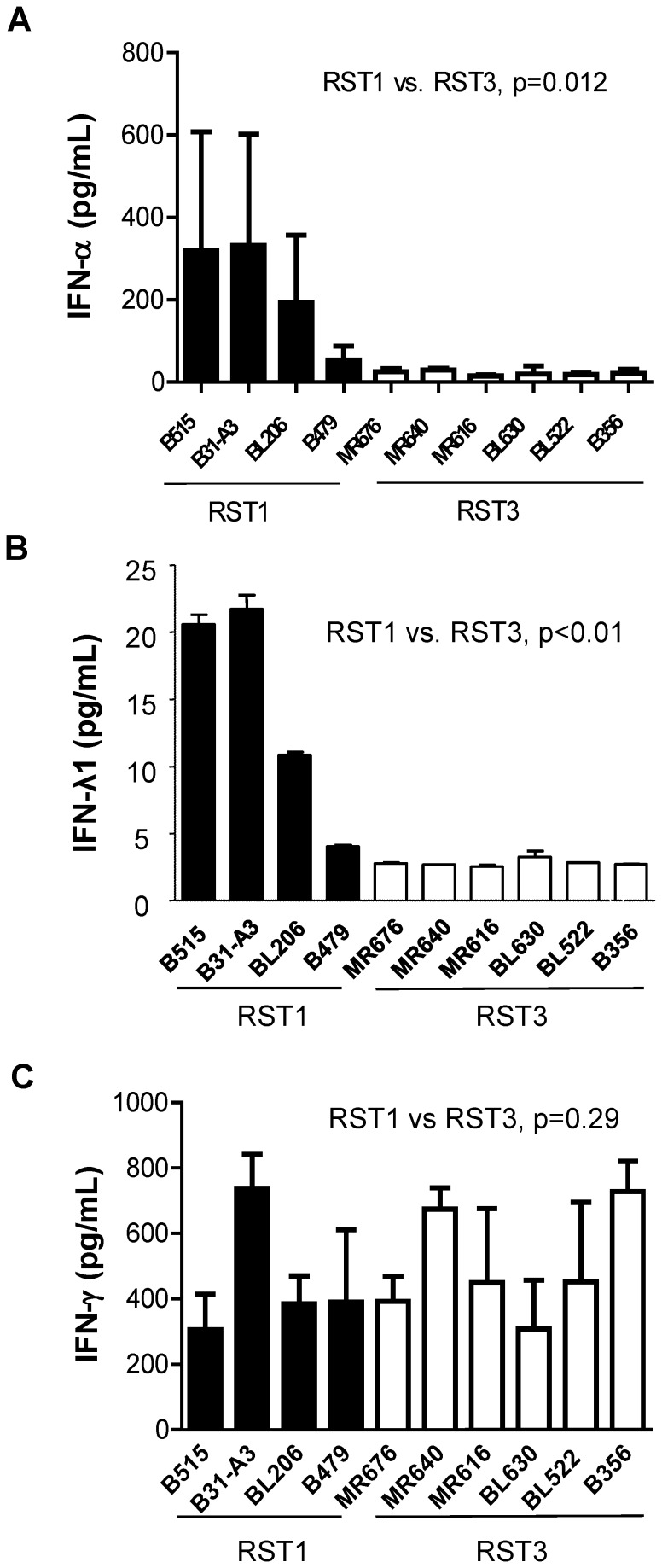
B. burgdorferi Clinical Isolates Induce Type I and Type III IFNs in a Genotype-dependent Manner
B. burgdorferi clinical isolates obtained from the blood or skin biopsies of early Lyme disease patients were classified into distinct genotypes [9]. The interferon response of human PBMCs to B. burgdorferi strains with varying genotypes was assessed using an ex vivo co-incubation model as previously described [20,20] (Table 1). IFN proteins in the cell-free culture supernatants were quantitated by ELISA (IFN-α, IFN-λ) or by cytometric bead array (IFN-γ). Representative results are shown in Figure 1. RST1 isolates induced production of 224.70±337.46 pg/mL of IFN-α, whereas RST3 isolates induced 21.57±14.86 pg/mL IFN-α (p = 0.012) (Fig. 1A). In addition, RST1 isolates were also found to elicit significantly higher production of IL-29/IFN-λ1, a type III IFN, as compared to RST3 isolates [RST1, 14.28±8.41 pg/mL; RST3, 2.78±0.24 pg/mL (p<0.01)] (Fig. 1B). In contrast, both RST1 and RST3 isolates induced equivalent amounts of IFN-γ (Fig. 1C). These results demonstrate that there is a significant correlation between the induction of IFN-α and IFN–λ1 and B. burgdorferi genotype.
Table 1. B. burgdorferi clinical isolates and strains used in this study.
| Identifier | Genotypea | Biological source | Reference |
| B31-A3 | RST1 | B31 derivative | [101] |
| B31-4 | RST1 | B31 derivative | [39] |
| A3-M9 lp36− | RST1 | B31 derivative | [43] |
| A3-M9 lp36−/lp36+ | RST1 | B31 derivative | [43] |
| B479 | RST1 | Human EM skin biopsy | [11] |
| B515 | RST1 | Human EM skin biopsy | [11] |
| BL206 | RST1 | Lyme disease patient blood | [10], [11] |
| B331 | RST3A | Human EM skin biopsy | [11] |
| B356 | RST3A | Human EM skin biopsy | [10], [11] |
| BL522 | RST3A | Lyme disease patient blood | |
| BL630 | RST3A | Lyme disease patient blood | |
| MR616 | RST3A | Human EM skin biopsy | [7] |
| MR640 | RST3A | Human EM skin biopsy | |
| MR676 | RST3A | Human EM skin biopsy |
Genotype was determined by PCR-RFLP analysis of 16S-23S ribosomal RNA gene spacer [9].
Figure 1. RST1 isolates induce expression of multiple interferons.
Human PBMCs (4×106) were co-incubated for 20 hrs with RST1 (black bars) or RST3 (white bars) B. burgdorferi clinical isolates (4×107) (MOI = 10∶1). Concentrations of human IFN-α (A), IFN-λ1/IL29 (B), or IFN-γ (C) proteins in cell-free culture supernatants were quantitated by ELISA (A , B) or cytometric bead array (C). Columns represent the mean ± SD of values from three blood donors assessed in independent experiments, with the exception of (B) which represents results from a single donor. Statistical analysis was performed using a non-parametric, two-tailed Mann-Whitney U-test.
B. burgdorferi Isolates of Varying Genotypes Induce Comparable Levels of Pro- and Anti-inflammatory Cytokines
In order to better understand the role that genotype plays in the B. burgdorferi-elicited cytokine response of human PBMCs, a human FlowCytomix bead array was employed to simultaneously measure various cytokine levels in cell-free culture supernatants from 20-hour co-incubations of PBMCs with B. burgdorferi isolates (Table 2). Protein levels for IL-1β, IL-6, IL-8 and TNF-α were the highest of the cytokines measured. Concentrations of IL-10 and IL-12p70 were markedly lower. IL-2, IL-4, IL-5 and TNF-β proteins were below the level of detection in most of the samples tested. No significant differences were detected when comparing RST1- and RST3-induced levels of any of the cytokines (Table 2). While it appeared that higher levels of IL-12p70 were induced by RST1 isolates relative to RST3 isolates, this difference did not achieve statistical significance (Table 2). In summary, of all cytokine proteins assayed, only IFN-α and IFN-λ1 levels differed significantly in PBMCs incubated in the presence of B. burgdorferi isolates of varying genotype, with higher induction by RST1 isolates.
Table 2. Pro- and anti-inflammatory cytokine production by RST1 and RST3 B. burgdorferi clinical isolates.
| Cytokine (pg/mL) | ||||||
| Genotype | IL-1β | IL-6 | IL-8 | IL-10 | IL-12p70 | TNF-α |
| RST1 * (n = 4) | 24841±7187 | 79738±17430 | 73678±21296 | 9468±1876 | 3441±3243 | 45748±21388 |
| RST3 (n = 6) | 21898±8148 | 71340±10956 | 69671±10190 | 9807±2641 | 2392±2728 | 43941±18476 |
| P value | 0.317 | 0.479 | 0.826 | 0.788 | 0.137 | 0.981 |
*B31-4 and A3-M9 lp36− were not included in the RST1 group when calculating averages and performing statistical analysis. Statistical significance was determined using a non-parametric Mann-Whitney U-test. Data shown are from three independent donors assessed in separate experiments.
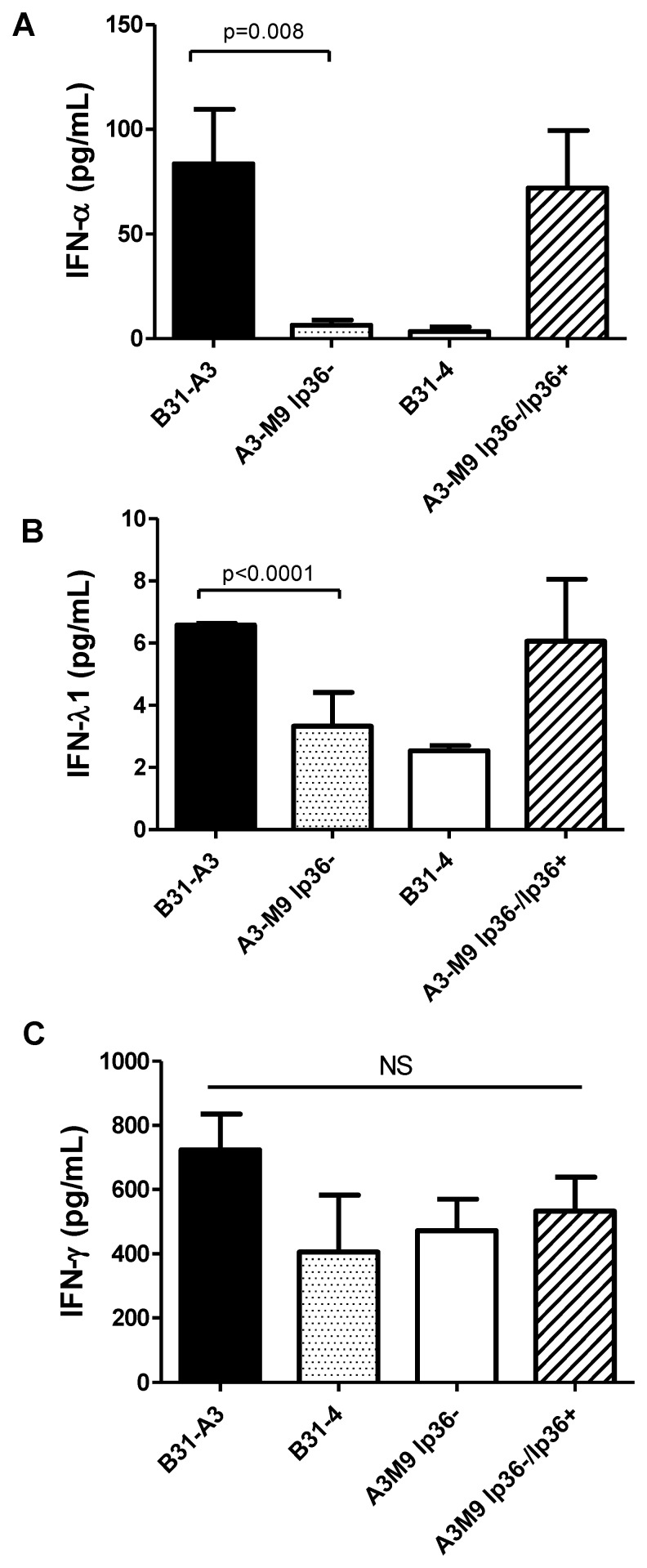
B. burgdorferi Mutants Lacking Linear Plasmid 36 (lp36) Elicit Virtually no IFN-α or IFN-λ1
The sequenced B. burgdorferi genome of the B. burgdorferi type strain B31-MI consists of a single linear chromosome and 12 linear and 9 circular plasmids [4], [5]. B31 is an RST1 strain [9]. B31-4 is a derivative of strain B31 that lacks all linear plasmids [39]. This strain elicited approximately 3.46±4.17 pg/ml IFN-α protein, whereas B31-A3, a strain that contains all plasmids, elicited 95.91±58.91 pg/ml IFN-α (p = 0.02) (Figure 2A).
Figure 2. The presence of lp36 is required for IFN-α and IFN-λ induction.
Human PBMCs (4×106) were co-incubated for 20 hrs with 4×107 wt B31-A3 (black bars), B31-4 (gray bars), A3-M9 lp36− (white bars) or A3-M9 lp36−/lp36+ (cross-hatched bar)(MOI = 10). Protein concentrations of human IFN-α (A), IFN-λ1 (B), or IFN-γ (C) in cell-free culture supernatants were quantitated by ELISA (A,B) or cytometric bead array (C). The results are shown as the mean ± SD of values from three to five blood donors assessed in two or three independent experiments. Statistics were obtained using a non-parametric Mann-Whitney U-test.
To identify the linear plasmid that may encode the factor(s) responsible for type I IFN induction, B. burgdorferi mutants that lacked single individual linear plasmids were assessed for their ability to induce IFN-α. These included B31 strains which lacked: lp28-1 [40], lp25 [40], two independent mutants lacking lp28-4 [41], [42], and two independent mutants lacking lp36 [43]. The absence of lp28-1, lp25, and lp28-4 had no effect on IFN-α production by human PBMCs (data not shown). However, the B. burgdorferi strains which lacked only lp36 elicited virtually no IFN-α protein. Results are shown in Figure 2A for A3-M9 lp36−, a B31 derivative that lacks lp36 but contains all other plasmids known to be required for infectivity [43]; an independently derived B31 strain lacking lp36 produced essentially identical results (data not shown). A3-M9 lp36− elicited only 4.93±4.95 pg/ml IFN-α protein (p = 0.008) (Figure 2A). Furthermore, strain A3-M9 lp36− complemented by the re-introduction of lp36 (designated as A3-M9 lp36−/lp36+) [43] produced IFN-α at levels equivalent to those induced by the parent strain B31-A3 (Figure 2A). B. burgdorferi strains B31-4 and A3-M9 lp36− also elicited significantly lower levels of IFN-λ1 as compared to B31-A3 (2.53±0.24 pg/mL and 2.97±1.25 pg/mL vs. 11.62±8.73 pg/mL, respectively; p<0.001 for both) (Figure 2B). In contrast, IFN-γ, IL-1β, IL-6, IL-8, IL-10, and TNF-α production by PBMCs was not dependent on lp36; B31-A3 and A3-M9 lp36− induced comparable levels of these cytokines (Figure 2C and Table 3). This finding suggests that a B. burgdorferi factor(s) required for induction of type I and type III IFN is likely encoded on lp36.
Table 3. Pro- and anti-inflammatory cytokine profiles elicited by B. burgdorferi B31-A3 and A3-M9 lp36−.
| Cytokine (pg/mL) | ||||||
| Genotype | IL-1β | IL-6 | IL-8 | IL-10 | IL-12p70 | TNF-α |
| B31-A3 (wt) | 21120±3259 | 112152±71306 | 54369±4144 | 8594±4851 | 916.9±842.6 | 6688±10922 |
| A3-M9 lp36− | 16535±9775 | 81210±56966 | 50509±12690 | 4475±1581 | 483.6±687.8 | 2973±4324 |
| P value | 0.151 | 0.421 | 0.691 | 0.095 | 0.222 | 0.486 |
Statistical analysis was performed using a non-parametric Mann Whitney U-test. Data shown are from three to five independent donors assessed in separate experiments.
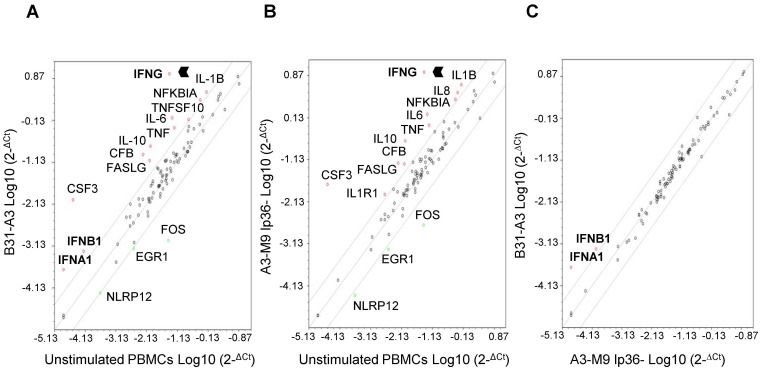
Strains B31-A3 and A3-M9 lp36− elicit Virtually Identical NF-κB-mediated Expression Profiles, but Differentially Induce Type I IFN Gene Transcripts
In order to obtain a global picture of the NF-κB response induced in human PBMCs exposed to B31-A3 or A3-M9 lp36−, PBMC total RNA was isolated following co-incubation and analyzed using an NF-κB RT2 Profiler PCR Array. This allows for the simultaneous quantitative measurement of 84 NF-κB-related gene transcripts. Similar transcriptional response profiles were generated by B31-A3 and A3-M9 lp36− (Figure 3). Genes which were commonly induced by either strain ≥4-fold relative to unstimulated PBMCs included those for IFN-γ, IL-10, TNF-α, IL-6, IL-1β, colony stimulating factor 3 (CSF3), complement factor B (CFB), Fas ligand (FASLG) and nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor alpha (NFKBIA). Genes which were commonly repressed by either isolate ≥4-fold relative to unstimulated PBMCs consisted of those encoding NLR family pyrin domain containing 12 (NLRP12), early growth response 1 (EGR1) and FBJ murine osteosarcoma viral oncogene homolog (FOS) (Figure 3A and 3B, respectively). Notably, genes for the type I IFNs, IFNα1 and IFNβ1, were uniquely induced following PBMC stimulation with B31-A3 but not with A3-M9 lp36− (Figure 3A and 3B). A direct comparison of the expression profiles elicited by B31-A3 and A3-M9 lp36− revealed that these were the only two of the 84 genes on the NF-kB PCR array that were differentially regulated by the two strains (Figure 3C). These findings corroborate the IFN protein data shown in Figure 2 and further validate the importance of lp36 for induction of the type I IFN response.
Figure 3. The absence of lp36 does not affect NF-κB-mediated signaling, but is required for transcription of ifna1 and ifnb1.
Human PBMCs (5×106) were co-incubated for 12 hours with 5×107 B31-A3 or A3-M9 lp36−. Equal amounts of PBMC total RNA from each of three biological replicates was pooled, converted to cDNA and used as a template for the Human NF-κB Signaling Pathway RT2 Profiler PCR Array. Genes that were transcriptionally induced or repressed ≥4-fold are indicated by red or green circles, respectively. Scatter plots depict transcriptional regulation in PBMCs stimulated with B31-A3 ( A ) or A3-M9 lp36− (B) versus unstimulated PBMCs. C , Comparison of transcript profiles of PBMCs stimulated with either B31-A3 or A3-M9 lp36− B. burgdorferi.
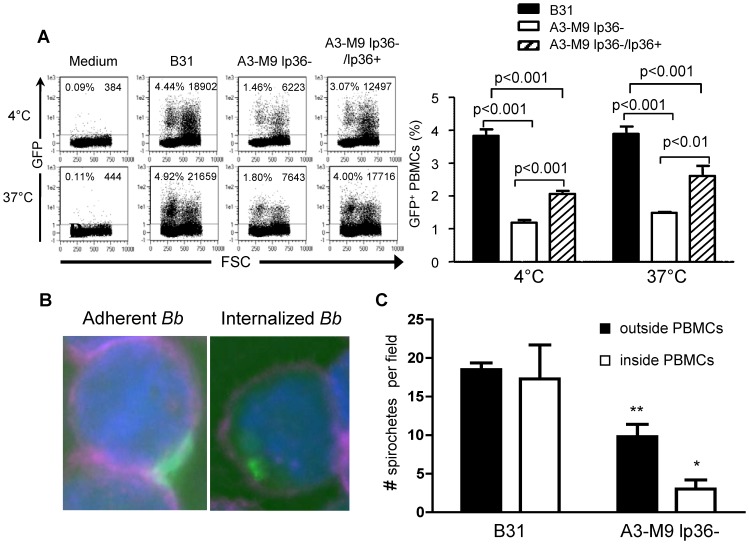
Spirochetes that Lack lp36 have Reduced Adherence to Human PBMCs and are Phagocytosed Less Efficiently
The production of IFN-α by B. burgdorferi-stimulated human PBMCs is dependent on phagocytosis of the bacterium, as it is blocked by the addition of cytochalasin D [20]. This suggested the possibility that lp36-deficient B. burgdorferi failed to induce IFN-α as a result of a defect in uptake of the mutant by phagocytic cells. To test this hypothesis, B31, A3-M9 lp36− and A3-M9 lp36−/lp36+ transformants containing green fluorescent protein (GFP) were produced and their association with PBMCs was evaluated by flow cytometry or microscopy. To verify that introduction of the GFP reporter plasmid did not affect the ability to elicit type I IFN, the GFP-transformed strains were assayed for IFN-α induction in human PBMCs and were found not to differ significantly from the non-GFP-transformed strains (data not shown). Adhesion and association of GFP-tagged B31, A3-M9 lp36− and A3-M9 lp36−/lp36+ that had been co-incubated with human PBMCs for 6 hr at either 4°C (prevents phagocytosis) or 37°C (allows phagocytosis) were assessed by flow cytometry. A significantly higher percentage of B31 adhered to PBMCs (3.9±0.2%GFP+ PBMCs vs. 1.50±0.04%, p<0.001; 4°C), and associated with PBMCs (3.88±0.23% vs. 1.48±0.04%GFP+ PBMCs, p<0.001; 37°C), than did A3-M9 lp36− (Figure 4A). A partial restoration in adhesion and association was observed with the lp36-complemented B. burgdorferi strain (Figure 4A).
Figure 4. The adhesion and uptake of B. burgdorferi is significantly reduced when lp36 is absent.
Human PBMCs (4×106) were co-incubated with 4×107 GFP-tagged B31-A3 (black bars), A3-M9 lp36− (white bars) or A3-M9 lp36−/lp36+ (cross-hatched bars) for 6 hours at 4°C or 37°C. A, Flow cytometry was performed to compare the ability of the GFP-tagged B. burgdorferi strains to associate with PBMCs. Representative dot plots are shown on the left. Graphs (right) depict the mean percentage ± SD of PBMCs identified as FITC+, indicating either adhesion (4°C) or adhesion and uptake (37°C) of GFP-tagged B. burgdorferi. Data are representative of results from three blood donors assessed in triplicate. A one-way ANOVA with a Tukey’s post-test was used to determine statistical significance. B and C, Three-dimensional fluorescence deconvolution microscopy was performed to quantify spirochetes adherent to, or internalized by, PBMCs. Samples were co-incubated with GFP-tagged B. burgdorferi for 6 hours at 37°C. PBMC outer cell membranes were labeled with APC-conjugated anti-CD45 to label the outer cell membrane (cyan) and DAPI (blue) was used to label the cell nucleus. Single optical sections of images acquired under 100X magnification were examined for adherent (B, left) or internalized (B, right) GFP-tagged B. burgdorferi. C, Columns show the mean (± SD) number of spirochetes outside (black bars) or inside (white bars) PBMCs. Data were collected from single optical sections taken from five 100X fields from each of three independent biological replicates. An unpaired, two-tailed Student’s t-test was used for statistical analysis.
The same co-incubation conditions were used to acquire samples for 3D-fluorescence deconvolution microscopy (optical sectioning) to further differentiate adherent from internalized spirochetes. Cytocentrifugation was used to deposit cells from co-incubations onto slides. Cells were fixed and stained with anti-CD45, a pan-leukocyte marker, which labels the PBMC outer membrane and allows the identification of spirochetes inside PBMCs, as opposed to spirochetes adherent to the outside of the cell membrane. Five 100X fields from three separate co-incubations (B31 or A3-M9 lp36−) at 37°C were analyzed. Figure 4B shows a representative single optical section from a deconvolved Z-stack for an adherent GFP-tagged B31 (left panel) versus an internalized B31 B. burgdorferi (right panel). As shown in Figure 4C, significant more wild type B. burgdorferi per field were adherent to the outside of PBMCs as compared to lp36− mutants (18.5±0.87 vs. 9.8±1.6 A3-M9 lp36−; p<0.01). Similarly, significantly more wild type spirochetes were internalized relative to lp36− B. burgdorferi (17.3±4.4 vs. 3.0±1.2; p<0.05) (Figure 4C). These data demonstrate that the absence of lp36 results in defects in both B. burgdorferi spirochetal adhesion to, and internalization by, PBMCs.
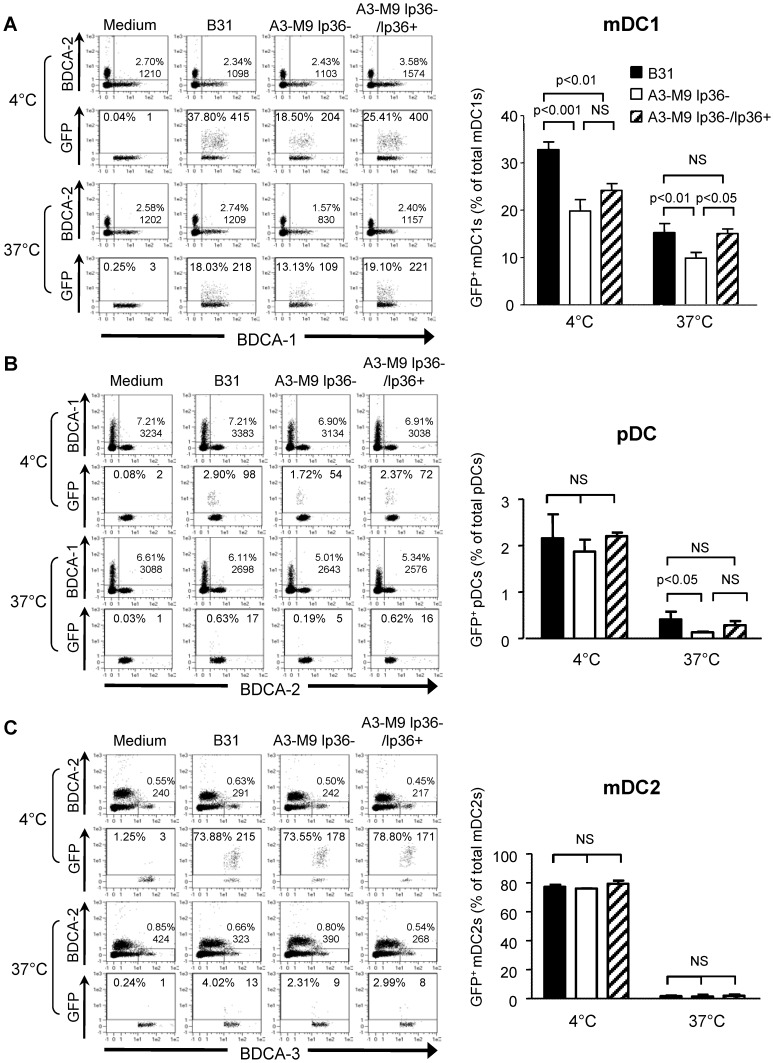
The Association of B. burgdorferi with mDC1s and pDCs is Significantly Reduced when lp36 is Absent
PBMCs are a heterogeneous population of innate immune cells consisting of T-cells, B- cells, monocytes, NK cells and dendritic cells [44]. Human blood dendritic cells are classified as plasmacytoid dendritic cells (pDCs), myeloid BDCA1+ dendritic cells (mDC1s) and myeloid BDCA3+ dendritic cells (mDC2s) [45]. Although pDCs constitute only ∼0.4% of the total PBMC population, they specialize in the production of large amounts of type I IFNs [46], [47]. Previous findings from this laboratory demonstrated that pDCs and CD11c+CD14+ monocytoid cells are the primary producers of IFN-α on co-incubation with B. burgdorferi [20]. The cell population(s) involved in the differential association and phagocytosis of B. burgdorferi based on the presence of lp36 was further examined using flow cytometry (Figure 5). A significantly higher percentage of BDCA1+ mDC1s (15.3±1.9 vs. 9.9±1.2; p<0.01) and BDCA2+ pDCs (0.4±0.2 vs. 0.1±0.02; p<0.05) were associated with GFP-tagged B31 B. burgdorferi than with the lp36− strain. Re-introduction of lp36 restored spirochetal association with mDC1s and pDCs to levels observed with the wild type (Figure 5A and 5B, respectively). However, it should be noted that the percentages of pDCs associated with lp36− mutant or complemented strains at 37°C were not significantly different. Interestingly, both wild type and lp36−deficient B. burgdorferi strongly adhered to approximately 80% of BDCA3+ mDC2s at 4°C, but failed to associate with these cells at 37°C (Figure 5C), although the total numbers of mDC2 cells remained unchanged (Figure 5C, left). Together, these results demonstrate that lp36 is required for optimal recognition and phagocytosis of spirochetes by pDC and mDC1 populations.
Figure 5. lp36 contributes to the association of B. burgdorferi with specific populations of dendritic cells.
Human PBMCs (4×106) were co-incubated with 4×107 GFP-tagged B31 (black bars), A3-M9 lp36− (white bars) or A3-M9 lp36−/lp36+ (cross-hatched bars) B. burgdorferi for 6 hours at 4°C or 37°C. The percentages of GFP+ mDC1s (CD19−CD3−BDCA2−BDCA1+) (A), pDCs (CD19−CD3−BDCA2+BDCA1−) or (B) mDC2s (CD19−CD3−BDCA3+BDCA2−) (C) were determined by multiparameter flow cytometry. Dot plots representing 500,000 collected events are provided to illustrate gating strategies (left). Column graphs represent the mean and standard deviation of three biological replicates (right). Statistical analysis was performed using a one-way ANOVA with a Tukey’s post-test for multiple comparisons.
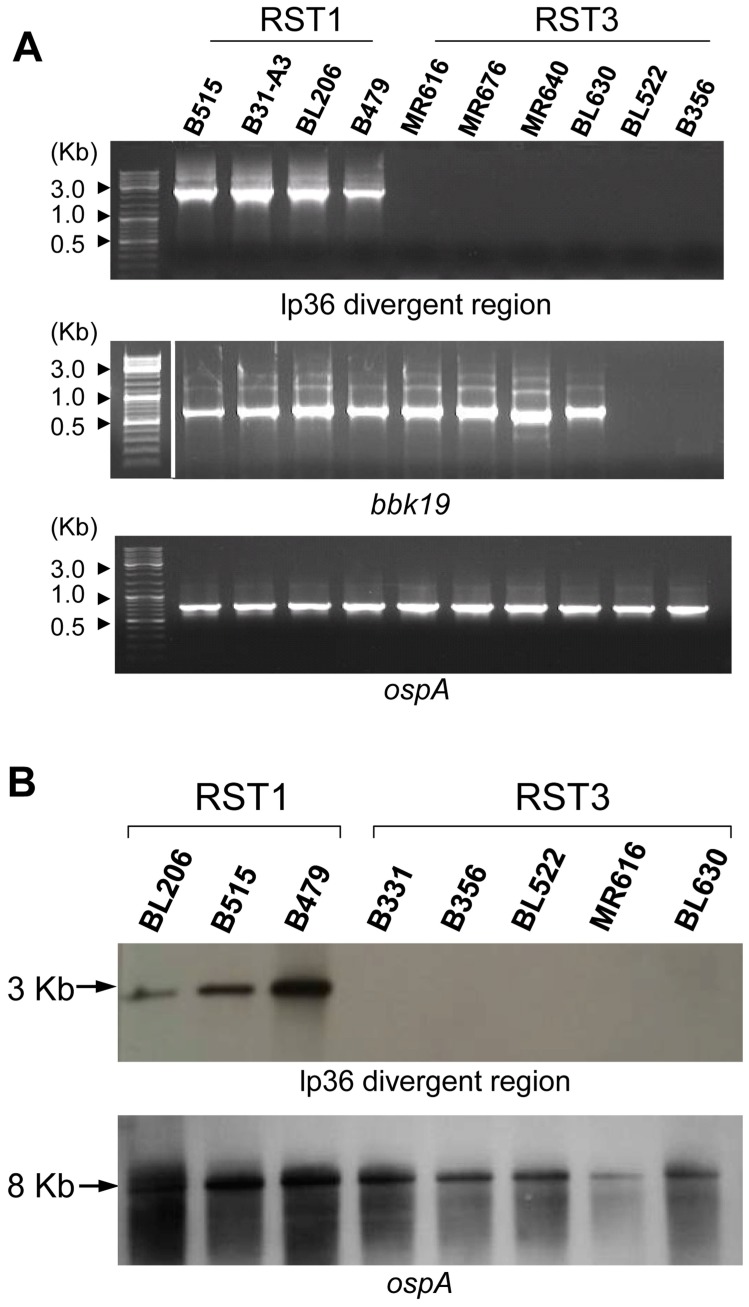
RST1 Isolates contain a Region of lp36 which is Absent in RST3A Isolates
It has been previously observed that plasmids constitute the most divergent portion of the B. burgdorferi genome and that lp36 is highly variable among clinical isolates of B. burgdorferi: the divergent region of lp36 was shown to encompass ORFs BBK35-BBK50 [7]. It was, therefore, of interest to explore whether the presence or absence of this region of lp36 correlated with the differential induction of IFN-α described earlier (Figure 1). PCR primers (primer set lp36A; Table 4) were designed to amplify a 2.5-kb sequence (bbk42-bbk45) within the divergent region of lp36. The same four RST1 isolates and six RST3 isolates that were tested for IFN-α production (Figure 1) were scored for the presence of the lp36-divergent sequence. All RST1 isolates yielded a PCR product of expected size, whereas no PCR product was obtained for RST3 isolates (Figure 6A, top panel). The presence of bbk19, which is located in a conserved area of lp36 adjacent to the plasmid partition locus, was assessed to determine if the RST3 isolates lack the entire plasmid. All RST1 and four of the six RST3 isolates tested were PCR positive for bbk19; a bbk19 PCR product was not detected for B356 or BL522 (Figure 6A, middle panel). B356 was previously shown to lack lp36 [7]; the absence of bbk19 in BL522 suggests that this isolate may also lack lp36. All isolates yielded a PCR product for ospA, which is located on the conserved lp54 plasmid [7], [48] (Figure 6A, bottom panel).
Table 4. Oligonucleotides used for plasmid content analysis.
| Primer name | Primer sequence | Predicted fragment size (bp) | Plasmid nucleotide coordinates |
| lp36A-Forward | 5′-GGG AAA TGC TAT AAG CCT CAA GCC CAG CC-3′ | 2480 | 26244–28724 |
| lp36A-Reverse | 5′-GCT TAG CCT GCA CCA AGC TAT TGC TAA GG-3′ | ||
| lp36B-Forward | 5′-GCATCTCCTATAAGTATAC-3′ | 184 | 27069–27253 |
| lp36B-Reverse | 5′-TCCCTGGTAAAGTAATTCATTTATCC-3′ | ||
| BBK19-Forward | 5′-CTA TTG TTT GTC CAA GTC TTA ATC-3′ | 1196 | 12269–13465 |
| BBK19-Reverse | 5′-ATT GTA CGA AGT TGA ATT TTA GCA TC-3′ | ||
| OspA-Forward | 5′-GGG AAT AGG TCT AAT ATT AGC C-3′ | 770 | 9410–10160 |
| OspA-Reverse | 5′-TTT CAA CTG CTG ACC CCT C-3′ |
Figure 6. RST1 isolates contain a region of lp36 that is lacking in RST3 isolates.
RST1 and RST3 B. burgdorferi isolates were grown to late log phase prior to isolation of plasmid DNA. (A) PCR primers were designed to amplify a ∼2.5 kilobase region on the distal end of lp36. The presence of lp36 was assessed by amplification of bbk19, an ORF located adjacent to the plasmid partition locus. PCR products were analyzed by gel electrophoresis in 1% agarose gels prepared in 1X TBE buffer and stained with ethidium bromide. (B) Southern blotting of SpeI digests of total genomic DNAs of B. burgdorferi isolates was performed using a probe specific for the variable region of lp36 (upper panel). As a control, membranes were stripped and re-probed for B. burgdorferi ospA (BBA15), located on lp54 (lower panel).
One limitation of PCR is the requirement that primer and target DNA contain nucleotide sequences that are nearly 100% complementary; even single nucleotide substitutions in the primer binding sequence can substantially reduce primer annealing and subsequent PCR product yield. Therefore, Southern hybridization of SpeI-digested genomic DNA was performed for eight clinical isolates using a gene probe specific for a 184-bp sequence located within the 2.5-kB lp36 divergent region. These included three RST1 and four RST3 isolates that had been assessed by PCR. Due to insufficient yields of DNA, RST3 isolates MR676 and MR640 could not be included; B331, an RST3 clinical isolate that does not disseminate in mice [11], was substituted. The predicted SpeI digestion fragment size of 3085 bp (lp36 nucleotide coordinates 24828–27913) corresponding to the lp36 variable region was observed in all three of the RST1 isolates but none of the RST3 isolates, confirming the PCR results (Figure 6B). These findings suggest that the RST3 isolates studied possess an ‘lp36-like’ plasmid that lacks the divergent region conserved among RST1 isolates, and that this region of lp36 encodes the factor(s) required for B. burgdorferi-elicited type I IFN production by human dendritic cells.
Discussion
In this study, we used an established human PBMC ex vivo co-incubation model [20] to explore the effect of B. burgdorferi genotype on the induction of cytokines, including three major classes of IFNs, and to delineate the host cell-pathogen interactions that contribute to the differential induction of IFNs. We observed that RST1 clinical isolates induced significantly higher levels of IFN-α and IFN-λ1 (IL-29) than did RST3 clinical isolates. In contrast, there were no significant differences between these two B. burgdorferi genotypes with regard to levels of induced IFN-γ, IL-1β, IL-6, IL-8, IL-10, IL-12p70 and TNF-α. Strle and colleagues reported that B. burgdorferi RST1 isolates, OspC type A genotype in particular, are associated with greater inflammation and more severe disease [49]. After a 24-hour co-incubation of human PBMCs with B. burgdorferi clinical isolates in that study, significantly higher amounts of IL-6, IL-8, CCL3, and CCL4 were produced in response to RST1 isolates as compared to those incubated with RST2 or RST3 isolates. In addition, RST1 isolates induced significantly higher levels of IFN-α than did RST2 isolates [49]. There are several differences between the experiments performed by Strle et al. and those described here that may contribute to the contrasting results with regard to IL-6 and IL-8 production. Strle et al. used an MOI of 25∶1 and co-incubation of 24 hours or 5 days, whereas an MOI of 10∶1 and co-incubation for 20 hours were employed in this study. Importantly, despite some differences, both studies demonstrate that there is a correlation between B. burgdorferi genotype, the capacity to cause a disseminated infection and the ability to induce a type I IFN response.
The finding that the capacity of B. burgdorferi to induce a type I and type III IFN response is genotype-dependent, and correlates with pathogenic potential, raises a number of questions. One important inquiry relates to the genomic component of B. burgdorferi responsible for the differential IFN induction. The genome of B. burgdorferi is comprised of a single linear chromosome (∼910 kb) and twelve linear and nine circular plasmids which total an additional 610 kb [4], [5]. There have been extensive studies correlating plasmid content and the pathogenesis of B. burgdorferi infection [40], [41], [43], [50]–[53]. To identify the B. burgdorferi-encoded factor(s) required for the induction of type I and type III IFNs, various B. burgdorferi mutants lacking specific linear plasmids were screened for their ability to induce a type I IFN response. Strikingly, B. burgdorferi strains that selectively lacked only lp36 had a significantly attenuated capacity to induce the production of type I and type III IFNs. In contrast, both wild-type and lp36-deficient B. burgdorferi induced comparable levels of IL-1β, -6, -8, -10, IL12-p70, IFN-γ and TNF-α. In support of these data, out of 84 different NF-κB-related genes, only the type I IFN gene transcripts ifna1 and ifnb1 were found to be induced by wild-type B31-A3 relative to A3-M9 lp36−.
The lp36 of B. burgdorferi type strain B31-MI is a 36-kilobase plasmid that encodes 54 putative ORFs, many of which are annotated as encoding hypothetical proteins [4]. There is a limited body of literature regarding lp36 and its encoded ORFs. Using whole cell DNA from a group of 15 North American B. burgdorferi isolates, Palmer et al. determined that lp36-derived DNA probes hybridized to targets of significantly different size in B31 as compared to B31-MI [6]. Terekhova and colleagues performed comparative genome hybridization and Southern blot analysis with various clinical isolates of B. burgdorferi and determined that RST1 isolates contain an lp36 plasmid virtually identical to that of B31-MI, whereas the non-RST1 isolates contain an “lp36-like” plasmid that is considerably different from the lp36 of B31-MI, yet contains the lp36 partition locus [7]. Jewett et al. investigated the role of lp36 in the infectious cycle of B. burgdorferi and showed that while lp36 was not required for spirochete survival in the tick, B. burgdorferi lacking lp36 exhibited an approximately 4-log increase in ID50 relative to wild-type B. burgdorferi in a murine infection model [43]. The lower infectivity was partially attributed to the loss of bbk17 (adeC), which encodes an adenine deaminase [43]. In addition to bbk17, the functions of several other lp36-encoded genes have been examined: bbk32 encodes a fibronectin-binding protein that may promote full infectivity in mice [54]–[56], and bbk07 has been investigated as a potential marker for serodiagnosis of Lyme disease [57], [58]. We tested mutant strains lacking bbk07, bbk17 or bbk32 and found that these mutants induced IFN-α at levels that were indistinguishable from those elicited by the wild-type strain (data not shown). It is worthy to note that none of these three genes is located within the divergent region (bbk35-bbk50) of lp36.
The current investigation was designed to explore the relationship between B. burgdorferi genotype and induction of a type I IFN response. Rather than a systematic analysis of all OspC genotypes, the study focused on RST1 (OspC type A) genotypes that have a high probability of causing disseminated infection and RST3 strains with OspC genotypes (E, G, M and U) that have a low probability of causing disseminated infection [59]. A limitation of the study is that several RST2 and RST3 genotypes (OspC types H, K and I) that have a high likelihood of producing disseminated infection in humans were not analyzed. It would be of interest in a future investigation to explore whether these B. burgdorferi genotypes induce high levels of type I IFN by human PBMCs. Recently, whole-genome sequences for other B. burgdorferi isolates have become available [60] and provide further evidence for a correlation between genotype, disseminating capacity and lp36 content. Strain N40, a B. burgdorferi OspC type E tick isolate that disseminates in mice [61], contains sequences that are highly homologous to the variable region of B31 lp36. However, the genetic information is divided between lp17 and lp36: the last ∼1400 nucleotides of the 2.5 kb fragment are present on N40 lp17 whereas the first ∼800 nucleotides of the fragment remain on N40 lp36 [60]. In contrast, B. burgdorferi isolates 118a (OspC type J), 72a (OspC type G) and 94a (OspC type U) [62], [63], which based on OspC type are likely to be RST3 isolates, do not encode the 2.5-kb variable region of B31 lp36. Taken together, these data suggest that this highly variable region may encode a factor(s) that is responsible for the induction of type I and type III IFNs by human PBMCs and that may contribute to the differential pathogenic potential of RST1 and RST3 B. burgdorferi isolates.
We have previously demonstrated that B. burgdorferi induces production of type I IFNs by human PBMCs in a TLR7- and TLR9-dependent manner [20]. While generally host protective in viral infections, the biological effects of type I IFNs on the outcome of non-viral infections can be variable. A potent type I IFN response contributes to the control of infection by some bacteria [64]–[73] but promotes the pathogenesis of other bacteria, including Listeria monocytogenes [25], [74], Mycobacterium bovis [75], Mycobacterium tuberculosis [76] and uropathogenic E. coli (UPEC) [77]. In the context of B. burgdorferi infection, type I IFN signaling has been shown to be crucial for the development of arthritis in a susceptible mouse model [78], [79]. In addition, significantly higher levels of serum IFN-α have been detected in human patients with multiple EM lesions, a symptom of disseminated B. burgdorferi infection, as compared to patients with a single EM [26]. Similar to type I IFNs, IFN-λs are expressed primarily by monocytes, macrophages and dendritic cells and can be produced by these cells simultaneously with IFN-α and IFN-β [30]–[33]. Type I and type III IFNs possess overlapping biological activities: both can induce MHC class I antigen expression, regulate transcription of a common subset of interferon-stimulated genes in various cell lines [30], increase NK and T cell cytotoxicity [80], promote Th1 responses [81] and mediate cellular apoptosis [82]. Recent studies demonstrate that type III IFNs are an important component of the innate immune response to non-viral pathogens, particularly those that are associated with epithelial tissues, including Salmonella enterica serovar typhimurium [31], [34] and L. monocytogenes [35]. We have previously shown that IFN-λ1 production can be elicited from human PBMCs by a disseminating isolate of B. burgdorferi, as well as by purified RNA from this same isolate [23], [23,38,38].
We have previously established that phagocytosis is required for B. burgdorferi-induced IFN-α production by pDCs and mDC precursors [20]. The finding that lp36 is required for the induction of both type I and type III IFN suggests that lp36 may mediate the association of B. burgdorferi with DC populations by encoding a phagocytic ligand. This hypothesis is supported by multi-parameter flow cytometry and 3-D fluorescence deconvolution microscopy. B. burgdorferi that lacked lp36 were attenuated not only in the ability to adhere to human PBMCs, but also in promoting phagocytic uptake by these cells. The absence of lp36 did not affect the ability of B. burgdorferi to either adhere to or associate with mDC2s; it is therefore unlikely that this cell type has a critical role in mediating spirochete dissemination. In contrast, optimal association of B. burgdorferi with mDC1s and pDCs was found to be dependent on the presence of lp36; thus these particular cells, while comprising an extremely small percentage (∼1%) of total PBMCs, may preferentially associate with B. burgdorferi and aid the spirochetes in hematogenous dissemination.
The data reported here support a conclusion that appears to be paradoxical: B. burgdorferi with greater capacity for dissemination are preferentially phagocytosed by DCs. However, a body of literature has emerged that describes a crucial role for DCs in immune suppression. Tolerogenic DCs have been shown to promote the generation of regulatory T-cells (Tregs) through expression of the immunosuppressive enzyme 2,3-indoleamine dioxygenase (IDO) [83]–[86]. IDO expression has been implicated in the generation of both central and peripheral tolerance, but a particularly crucial function appears to be the establishment of localized immune privilege in epithelial tissues. IDO-expressing DCs and Foxp3+ Tregs have been implicated in the development of various infectious and autoimmune pathological conditions of the epithelium, including lichen planus, lupus vulgaris, sarcoidosis and infections caused by Mycobacterium leprae, Haemophilus ducreyi, UPEC and Leishmania sp. [77], [87]–[91]. Of particular relevance to the current study, Tregs have been found in the peripheral blood of patients with secondary syphilis, a disease that evolves following initial skin infection with Treponema pallidum, another spirochetal pathogen [92]. Although it is known that human monocytoid and plasmacytoid DCs are present in the EM lesion infiltrate of Lyme disease patients [26], [93], [94], the potential role of these innate immune cells in determining disease outcome remains to be established. We hypothesize that recognition of B. burgdorferi by DCs may direct the differentiation of these cells towards a tolerogenic phenotype, resulting in localized immune suppression that may be exploited by B. burgdorferi to establish infection and enable dissemination. These questions are the topics of ongoing investigation in our laboratory.
In summary, this study demonstrates that B. burgdorferi lp36 is indispensable for the induction of IFN-α and IFN-λ1 by human PBMCs and for full recognition of B. burgdorferi by human dendritic cell populations. In contrast, B. burgdorferi genotype did not affect induction of IFN-γ or other NF-κB-dependent cytokines. The ability of B. burgdorferi to induce IFN-α and IFN-λ was associated with the presence of a variable 2.5-kb segment of lp36 (bbk42–bbk45) that was conserved among the RST1 isolates, but absent in all RST3 isolates evaluated. These results provide new insight into the connection between B. burgdorferi genotype, lp36 content and pathogenic potential. Although the presence of DCs in EM lesions has been reported, the potential role of these innate immune cells in determining disease outcome remains to be defined. The ability of tolerogenic DCs to generate localized immune suppression suggests a mechanism that may be exploited by B. burgdorferi in order to establish infection and enable subsequent dissemination.
Materials and Methods
Ethics Statement
Written informed consent was obtained from all human subjects in accordance with protocols approved by the Institutional Review Board of New York Medical College.
Culture of Borrelia burgdorferi
Low-passage B. burgdorferi clinical isolates were obtained from the blood or skin biopsies of the EM lesions of early Lyme disease patients enrolled in a prospective study at the Lyme Disease Diagnostic Center at New York Medical College, Valhalla, NY [12], [95]. B31-4, a derivative of B. burgdorferi strain B31 that lacks all linear plasmids, has been described [39]. A3-M9 lp36− and A3-M9 lp36−/lp36+, both B31 derivative strains, have been previously described [42]. Spirochetes were cultured in modified Barbour-Stoenner-Kelly (BSK-S) medium [96]. To simulate the temperature shifts that occur during tick-to-mammal transmission and that may enhance the expression of virulence factors, spirochetes were grown at 25°C, subcultured and incubated at 37°C until cultures reached the mid- to late-log phase of growth (4 to 8×107/ml). Spirochetes were enumerated and assessed for motility by dark-field microscopy [95]. The genotypes and biological sources of the B. burgdorferi clinical isolates for which data are reported in this study are provided in Table 1.
Isolation of Human PBMCs
Venous blood was obtained from five healthy adult volunteers who had no prior history of Lyme disease, had not been vaccinated for Lyme disease and were seronegative for B. burgdorferi antibodies as confirmed by Western immunoblotting. Written informed consent was obtained from all subjects before blood collection, in accordance with the protocol approved by the Institutional Review Board of New York Medical College. Blood was collected directly into BD-Vacutainer CPT tubes (BD Biosciences) and PBMCs were isolated by centrifugation according to the manufacturer’s instructions. PBMCs were washed twice with Hank’s Balanced Salt Solution (HBSS) and resuspended in RPMI 1640 medium without phenol red containing 10% (v/v) heat-inactivated and endotoxin-free fetal bovine serum (FBS) (Hyclone). The viability of PBMCs was determined by trypan blue staining and was found to be >90%.
Co-incubation of PBMCs with B. burgdorferi
Ex vivo stimulation of freshly isolated PBMCs by B. burgdorferi was performed essentially as previously described [20]. Spirochetes were centrifuged at 8000×g for 10 min, washed twice with HBSS and resuspended in RPMI 1640 without phenol red containing 10% (v/v) heat-inactivated and endotoxin-free FBS (Hyclone). Live B. burgdorferi were added to 4×106 (or 5×106 for experiments involving RNA isolation) viable PBMCs at a multiplicity of infection (MOI) of 10∶1 (spirochetes:PBMC) in 24-well tissue culture plates, in a final volume of 1.1 ml. Co-cultures were maintained in a humidified 37°C incubator containing 5% CO2 for the specified time period.
Measurement of Secreted Cytokine Proteins
PBMCs were co-incubated with B. burgdorferi for 20 hrs. Cell-free culture supernatants were prepared by removal of PBMCs at 300×g for 10 min followed by centrifugation of the supernatant at 9000×g for 10 min to remove non-adherent spirochetes and cellular debris. Cell-free supernatants were aliquoted and stored at −20°C until further use. Concentrations of human IFN-α and IFN–λ1 were quantitated using the Human Verikine IFN-α (PBL Biomedical Laboratories) or the Human IL-29 ELISA Ready-Set Go! Kit (eBioscience), respectively. Protein levels of IFN-γ, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12 p70, TNF-α and TNF-β in cell-free culture supernatants were measured using the FlowCytomix Human Th1/Th2 11plex Kit (BMS810FF;Bender MedSystems) according to the manufacturer’s instructions. Data were acquired with a MACSQuant analyzer (Miltenyi Biotec) and analyzed using FlowCytomix Pro Software (Bender MedSystems).
RNA Isolation
PBMCs were co-incubated with B. burgdorferi as described above for 12 hours. The contents of each well were transferred to a 1.5-ml microcentrifuge tube and centrifuged at 300×g for 5 min at 4°C. PBMCs were washed with HBSS and RNA was isolated using the RNeasy Plus Mini Kit with QIAshredder and gDNA eliminator columns (Qiagen). RNA was eluted in 20 µl of RNase/DNase-free water and stored at −80°C after the addition of 0.8 µl of RNase inhibitor (40 U/µl; Promega).
NF-κB-mediated Gene Expression PCR Array
PBMCs were co-incubated in triplicate with B. burgdorferi for 12 hr and RNA was isolated as described above. The RT2 First Strand Kit (SABiosciences) was used for cDNA synthesis, according to the manufacturer’s instructions. cDNA from biological triplicates was pooled and used as a template for the RT2 Profiler PCR Array Human NFκB Signaling Pathway (PAHS-025A;SABiosciences), according to the manufacturer’s instructions. This array contains primer pairs for 84 well-characterized NF-κB-related genes, 5 housekeeping genes and various quality controls in a 96-well plate format. Transcript levels were quantified based on the 2−ΔΔCT method [97] through the measurement of SYBR green fluorescence using the RT2 Real-Time SYBR Green/Rox PCR Master Mix (PA-012; SABiosciences).
Generation of Green Fluorescent Protein-expressing B. burgdorferi
A GFP reporter plasmid, based on plasmid pCE320 [17], [98], was used to electrotransform B. burgdorferi A3-M9 lp36− B. burgdorferi as previously described [99]. This plasmid contains the GFP structural gene fused to the constitutive flaB promoter of B. burgdorferi and encodes for gentamicin resistance. Transformants were selected by growth in BSK medium containing 40 µg/mL of gentamicin and plated by limiting dilution to obtain single clones. The GFP-expressing lp36 complement, A3-M9 lp36−/lp36+, was constructed using the GFP reporter plasmid pSPC-S [100]. This plasmid contains the GFP structural gene fused to the constitutive flaB promoter of B. burgdorferi and encodes for streptomycin resistance. Transformants were selected by growth in BSK-S medium containing 100 µg/mL streptomycin. Wild-type B31 B. burgdorferi expressing GFP was kindly provided by Drs. Justin Radolf and Melissa Caimano, University of Connecticut Health Center [98].
Multi-parameter Flow Cytometric Analysis
Human PBMCs were co-incubated for 6 hours at 4°C or 37°C with GFP-tagged B. burgdorferi at MOI of 10. Following co-incubation, the contents of each well were transferred to a 5 mL FACS tube and pelleted at 300×g for 10 min. Cells were washed twice with 1 mL HBSS to remove unattached spirochetes, fixed with 3.2% formaldehyde in HBSS for 30 min, pelleted and resuspended in 0.5 mL FACS stain buffer (1X PBS, 0.2% bovine serum albumin, pH 7.4). Total numbers of GFP+ PBMCs were determined by flow cytometric analysis of 100,000 to 300,000 events using a MACSQuant Analyzer and MACSQuantify software (Miltenyi Biotec).
Dendritic cell populations present within PBMCs were identified by labeling of surface antigens. Following co-incubation, the contents of each well were transferred to two, 5 mL FACS tubes (∼2×106 cells each) and centrifuged at 300×g for 10 min. Cells were washed once with 1 ml HBSS and once with 1 ml FACS stain buffer. To block Fc receptors, cells were resuspended in 70 µl stain buffer containing 1 µg/mL human IgG1, kappa, purified myeloma protein (Sigma 5154). 30 µL of the appropriate antibody staining panel (mDC1/pDC or mDC2) was then added to each sample. Antibody fluorochrome conjugates consisted of anti-CD19-eFluor450 (mouse IgG1,k, clone HIB19 eBioscience 48-0199), anti-CD3-eFluor450 (mouse IgG1,k, clone UCHTI eBioscience 48-0038), anti-BDCA1 (CD1c)-APC (mouse IgG2a, clone AD5-8E7 Miltenyi 130-090-903), anti-BDCA2 (CD303)-PE (mouse IgG1, clone AC144 Miltenyi 130-090-511) and anti-BDCA3 (CD141)-APC (mouse IgG1, clone AD5-14H12 Miltenyi 130-090-907). Cell populations were defined as follows: mDC1s (CD19−CD3−BDCA2−BDCA1+), pDCs (CD19−CD3−BDCA2+BDCA1−) or mDC2s (CD19−CD3−BDCA3+BDCA2−). Samples were mixed well, covered and refrigerated at 4–8°C for 10 min. Cells were washed by adding 2 mL stain buffer followed by centrifugation at 300×g for 10 min at 4°C. Cells were then fixed in 800 µL of 3.2% formaldehyde in HBSS, covered and stored overnight at 4–8°C. Following fixation, cells were pelleted at 300×g for 10 min at 4–8°C and resuspended in 500 µL stain buffer. A minimum of 500,000 events per sample were collected on a MACSQuant analyzer and analyzed using MACSQuantify software (Miltenyi).
Quantitation of Internalized versus Extracellular Spirochetes using 3-D-fluorescence Deconvolution Microscopy
Human PBMCs were co-incubated with GFP-tagged B. burgdorferi for 6 hours at 4°C or 37°C. Cells were pelleted at 300×g for 5 minutes, washed twice with tissue culture grade 1X PBS to remove non-adherent spirochetes and deposited onto glass microscope slides by cytocentrifugation. Cells were fixed in 3.7% formaldehyde for 20 minutes and stored in 1X PBS overnight at 4°C. Cells were protected from light and stained for 1 hour at room temperature with APC-conjugated mouse anti-CD45 (cyan) (Invitrogen; MHCD4505), a pan-leukocyte cell membrane marker, to define the PBMC outer membrane. Coverslips were mounted using Vectashield containing DAPI (Vector Laboratories, Inc.). Counts were obtained from deconvolved images using an inverse filter algorithm. Five 100X fields (∼250 total PBMCs) from each of three independent co-incubations were examined in a blinded manner for adherent or internalized spirochetes. Samples were imaged on a Zeiss Axiovert 200 fluorescence microscope and data were analyzed using Axiovision 4.6.3 software (both from Carl Zeiss, Inc.).
PCR Analysis of B. burgdorferi Linear Plasmid Sequences
Plasmid DNA was isolated from 4-ml B. burgdorferi cultures (1×107 to 1×108 cells/ml) with the Qiaprep Spin Miniprep kit (Qiagen) and resuspended in 30 µl of nuclease-free water. PCR primers (Table 4) were designed to amplify a 2.5-kB sequence on lp36, encompassing bbk42–bbk45 [7], [62]. Each PCR mixture contained approximately 10 ng DNA, 250 µM deoxynucleoside triphosphates (Roche), 10 ng of each primer and 1 U Taq DNA polymerase (Denville Scientific, Inc.). PCR conditions were as follows: initial denaturation at 95°C for 3 minutes; 35 cycles of 95°C for 1 minute, 60°C for 40 seconds, 72°C for 3 minutes; and final extension at 72°C for 2 minutes. PCR of bbk19 was performed to assess the presence or absence of lp36. PCR parameters were as follows: initial denaturation at 95°C for 3 minutes, followed by 35 cycles of 95°C for 30 seconds, 48°C for 30 seconds, 72°C for 3 minutes, and final extension at 72°C for 2 minutes. ospA (bba15) was amplified as a control for the presence of plasmid DNA using the following conditions: initial denaturation at 95°C for 3 minutes; followed by 35 cycles of 95°C for 30 seconds, 52°C for 30 seconds, 72°C for 3 minutes; and final extension at 72°C for 2 minutes. PCR products were analyzed by electrophoresis in 1% agarose gels prepared in 1X TBE buffer and stained with ethidium bromide.
Southern Hybridization
Total DNA was isolated from 10-ml cultures of B. burgdorferi B31-MI using a commercial DNA extraction kit (DNeasy Blood and Tissue Kit; Qiagen) and suspended in 100 µl nuclease-free water. DNA (3 µg) was digested overnight at 37°C with SpeI (10 U) in 50 µL 1X Buffer 2 (New England Biolabs) supplemented with 100 µg/ml BSA. Digested DNA was separated by electrophoresis on a 1% agarose gel prepared in 1X TBE buffer.
Probes to ospA and lp36 were separately generated by PCR using 10 ng B31-MI DNA template and 10 ng of each primer (Table 4). PCR parameters for bba15 (ospA) were as previously reported [48]. A 184-bp sequence located in the divergent region of lp36 was amplified using the lp36-B primer set and the following thermocycler conditions: initial denaturation at 95°C for 2 minutes; 35 cycles of 94°C for 30 seconds, 42°C for 30 seconds and 72°C for 30 seconds; and final extension at 72°C for 2 minutes. PCR products were purified using a commercial kit (Qiagen) and labeled with digoxygenin-11-dUTP according to the manufacturer’s protocol (Roche Molecular Biochemicals). Southern blotting was performed as previously described [48]. Hybridization was first performed for the lp36 sequence. Hybridization patterns were visualized by addition of CDP-Star chemiluminescent substrate (Sigma-Aldrich) followed by exposure of the membrane to radiographic film for 2 to 4 minutes. The membrane was then stripped and re-probed for ospA.
Statistical Analysis
Statistical significance was determined by one-way Analysis of Variance (ANOVA), a non-parametric Mann-Whitney U-test, a Kruskal-Wallis test, or a two-tailed, unpaired Student’s t-test using GraphPad Prism 5 software (GraphPad Software, Inc.), as appropriate.
Acknowledgments
The authors gratefully acknowledge Diane Holmgren, R.N., and the staff of the Division of Infectious Diseases, for performing phlebotomy; Drs. Patricia Rosa (National Institutes of Allergy and Infectious Diseases, NIH), Mollie Jewett (University of Central Florida), Utpal Pal (University of Maryland), and Jon T. Skare (Texas A&M Health Sciences Center) for B. burgdorferi strains and mutants; Drs. Justin Radolf and Melissa Caimano (University of Connecticut Health Center), and Darrin Akins (University of Oklahoma Health Sciences Center) for GFP-expressing plasmid constructs; Dr. Radha Iyer for technical assistance and advice; and Nicholas Camacho for assisting with plasmid isolation and PCR analysis.
Funding Statement
This work was supported by a cooperative agreement (U01-CI000160) from the Centers for Disease Control and Prevention (to IS), The William and Sylvia Silberstein Foundation (to IS), ARRA supplement (3R01-AR41511) from the National Institutes of Health (to IS), and a predoctoral fellowship to MAK-G from the American Heart Association. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Steere AC (1989) Lyme disease. N Engl J Med 321: 586–596. [DOI] [PubMed] [Google Scholar]
- 2. Steere AC (2001) Lyme disease. N Engl J Med 345: 115–125. [DOI] [PubMed] [Google Scholar]
- 3. Steere AC, Bartenhagen NH, Craft JE, Hutchinson GJ, Newman JH, et al. (1986) Clinical manifestations of Lyme disease. Zentralbl Bakteriol Mikrobiol Hyg A 263: 201–205. [DOI] [PubMed] [Google Scholar]
- 4. Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, et al. (1997) Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi . Nature 390: 580–586. [DOI] [PubMed] [Google Scholar]
- 5. Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, et al. (2000) A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi . Mol Microbiol 35: 490–516. [DOI] [PubMed] [Google Scholar]
- 6. Palmer N, Fraser C, Casjens S (2000) Distribution of twelve linear extrachromosomal DNAs in natural isolates of Lyme disease spirochetes. J Bacteriol 182: 2476–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Terekhova D, Iyer R, Wormser GP, Schwartz I (2006) Comparative genome hybridization reveals substantial variation among clinical isolates of Borrelia burgdorferi sensu stricto with different pathogenic properties. J Bacteriol 188: 6124–6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liveris D, Gazumyan A, Schwartz I (1995) Molecular typing of Borrelia burgdorferi sensu lato by PCR-restriction fragment length polymorphism analysis. J Clin Microbiol 33: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liveris D, Wormser GP, Nowakowski J, Nadelman R, Bittker S, et al. (1996) Molecular typing of Borrelia burgdorferi from Lyme disease patients by PCR-restriction fragment length polymorphism analysis. J Clin Microbiol 34: 1306–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang G, Ojaimi C, Iyer R, Saksenberg V, McClain SA, et al. (2001) Impact of genotypic variation of Borrelia burgdorferi sensu stricto on kinetics of dissemination and severity of disease in C3H/HeJ mice. Infect Immun 69: 4303–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang G, Ojaimi C, Wu H, Saksenberg V, Iyer R, et al. (2002) Disease severity in a murine model of lyme borreliosis is associated with the genotype of the infecting Borrelia burgdorferi sensu stricto strain. J Infect Dis 186: 782–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wormser GP, Brisson D, Liveris D, Hanincova K, Sandigursky S, et al. (2008) Borrelia burgdorferi genotype predicts the capacity for hematogenous dissemination during early Lyme disease. J Infect Dis 198: 1358–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wormser GP, Liveris D, Nowakowski J, Nadelman RB, Cavaliere LF, et al. (1999) Association of specific subtypes of Borrelia burgdorferi with hematogenous dissemination in early Lyme disease. J Infect Dis 180: 720–725. [DOI] [PubMed] [Google Scholar]
- 14. Wooten RM, Ma Y, Yoder RA, Brown JP, Weis JH, et al. (2002) Toll-like receptor 2 plays a pivotal role in host defense and inflammatory response to Borrelia burgdorferi . Vector Borne Zoonotic Dis 2: 275–278. [DOI] [PubMed] [Google Scholar]
- 15. Akira S (2003) Mammalian Toll-like receptors. Curr Opin Immunol 15: 5–11. [DOI] [PubMed] [Google Scholar]
- 16. Moore MW, Cruz AR, LaVake CJ, Marzo AL, Eggers CH, et al. (2007) Phagocytosis of Borrelia burgdorferi and Treponema pallidum potentiates innate immune activation and induces gamma interferon production. Infect Immun 75: 2046–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cruz AR, Moore MW, La Vake CJ, Eggers CH, Salazar JC, et al. (2008) Phagocytosis of Borrelia burgdorferi, the Lyme disease spirochete, potentiates innate immune activation and induces apoptosis in human monocytes. Infect Immun 76: 56–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sjowall J, Fryland L, Nordberg M, Sjogren F, Garpmo U, et al. (2011) Decreased Th1-type inflammatory cytokine expression in the skin is associated with persisting symptoms after treatment of erythema migrans. PLoS One 6: e18220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Salazar JC, Duhnam-Ems S, La Vake C, Cruz AR, Moore MW, et al. (2009) Activation of human monocytes by live Borrelia burgdorferi generates TLR2-dependent and -independent responses which include induction of IFN-beta. PLoS Pathog 5: e1000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Petzke MM, Brooks A, Krupna MA, Mordue D, Schwartz I (2009) Recognition of Borrelia burgdorferi, the Lyme disease spirochete, by TLR7 and TLR9 induces a type I IFN response by human immune cells. J Immunol 183: 5279–5292. [DOI] [PubMed] [Google Scholar]
- 21. Cervantes JL, Dunham-Ems SM, La Vake CJ, Petzke MM, Sahay B, et al. (2011) Phagosomal signaling by Borrelia burgdorferi in human monocytes involves Toll-like receptor (TLR) 2 and TLR8 cooperativity and TLR8-mediated induction of IFN-{beta}. Proc Natl Acad Sci U S A 108: 3683–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cervantes JL, La Vake CJ, Weinerman B, Luu S, O’Connell C, et al. (2013) Human TLR8 is activated upon recognition of Borrelia burgdorferi RNA in the phagosome of human monocytes. J Leukoc Biol 94: 1231–1241. jlb.0413206 [pii]; 10.1189/jlb.0413206 [doi]. [DOI] [PMC free article] [PubMed]
- 23.Love AC, Schwartz I, Petzke MM (2014) Borrelia burgdorferi RNA induces type I and type III interferons via TLR7 and contributes to the production of NF-kappaB-dependent cytokines. Infect Immun. IAI.01617-14 [pii]; 10.1128/IAI.01617-14 [doi]. [DOI] [PMC free article] [PubMed]
- 24. Hawley KL, Olson CM Jr, Iglesias-Pedraz JM, Navasa N, Cervantes JL, et al. (2012) CD14 cooperates with complement receptor 3 to mediate MyD88-independent phagocytosis of Borrelia burgdorferi . Proc Natl Acad Sci U S A 109: 1228–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Auerbuch V, Brockstedt DG, Meyer-Morse N, O’Riordan M, Portnoy DA (2004) Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes . J Exp Med 200: 527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Salazar JC, Pope CD, Sellati TJ, Feder HM Jr, Kiely TG, et al. (2003) Coevolution of markers of innate and adaptive immunity in skin and peripheral blood of patients with erythema migrans. J Immunol 171: 2660–2670. [DOI] [PubMed] [Google Scholar]
- 27. Mancuso G, Gambuzza M, Midiri A, Biondo C, Papasergi S, et al. (2009) Bacterial recognition by TLR7 in the lysosomes of conventional dendritic cells. Nat Immunol 10: 587–594. [DOI] [PubMed] [Google Scholar]
- 28. Carrero JA, Calderon B, Unanue ER (2004) Type I interferon sensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. J Exp Med 200: 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carrero JA, Unanue ER (2007) Impact of lymphocyte apoptosis on the innate immune stages of infection. Immunol Res 38: 333–341. [DOI] [PubMed] [Google Scholar]
- 30. Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, et al. (2003) IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol 4: 69–77. [DOI] [PubMed] [Google Scholar]
- 31. Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, et al. (2003) IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol 4: 63–68. [DOI] [PubMed] [Google Scholar]
- 32. Coccia EM, Severa M, Giacomini E, Monneron D, Remoli ME, et al. (2004) Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur J Immunol 34: 796–805. [DOI] [PubMed] [Google Scholar]
- 33. Wolk K, Witte K, Witte E, Proesch S, Schulze-Tanzil G, et al. (2008) Maturing dendritic cells are an important source of IL-29 and IL-20 that may cooperatively increase the innate immunity of keratinocytes. J Leukoc Biol 83: 1181–1193. [DOI] [PubMed] [Google Scholar]
- 34. Pietila TE, Latvala S, Osterlund P, Julkunen I (2010) Inhibition of dynamin-dependent endocytosis interferes with type III IFN expression in bacteria-infected human monocyte-derived DCs. J Leukoc Biol 88: 665–674. [DOI] [PubMed] [Google Scholar]
- 35. Lebreton A, Lakisic G, Job V, Fritsch L, Tham TN, et al. (2011) A bacterial protein targets the BAHD1 chromatin complex to stimulate type III interferon response. Science 331: 1319–1321. [DOI] [PubMed] [Google Scholar]
- 36. Bierne H, Travier L, Mahlakoiv T, Tailleux L, Subtil A, et al. (2012) Activation of type III interferon genes by pathogenic bacteria in infected epithelial cells and mouse placenta. PLoS One 7: e39080 10.1371/journal.pone.0039080 [doi];PONE-D-12-04118 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jewell NA, Cline T, Mertz SE, Smirnov SV, Flano E, et al. (2010) Lambda interferon is the predominant interferon induced by influenza A virus infection in vivo. J Virol 84: 11515–11522 JVI.01703-09 [pii];10.1128/JVI.01703-09 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krupna MA, Wormser GP, Schwartz I, Petzke MM (2011) Borrelia burgdorferi pathogenic potential correlates with type I and type III IFN production by human immune cells. J Immunol.
- 39. Sadziene A, Wilske B, Ferdows MS, Barbour AG (1993) The cryptic ospC gene of Borrelia burgdorferi B31 is located on a circular plasmid. Infect Immun 61: 2192–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Labandeira-Rey M, Skare JT (2001) Decreased infectivity in Borrelia burgdorferi strain B31 is associated with loss of linear plasmid 25 or 28-1. Infect Immun 69: 446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Labandeira-Rey M, Seshu J, Skare JT (2003) The absence of linear plasmid 25 or 28–1 of Borrelia burgdorferi dramatically alters the kinetics of experimental infection via distinct mechanisms. Infect Immun 71: 4608–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grimm D, Tilly K, Bueschel DM, Fisher MA, Policastro PF, et al. (2005) Defining plasmids required by Borrelia burgdorferi for colonization of tick vector Ixodes scapularis (Acari: Ixodidae). J Med Entomol 42: 676–684. [DOI] [PubMed] [Google Scholar]
- 43. Jewett MW, Lawrence K, Bestor AC, Tilly K, Grimm D, et al. (2007) The critical role of the linear plasmid lp36 in the infectious cycle of Borrelia burgdorferi . Mol Microbiol 64: 1358–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Teetson W, Cartwright C, Dreiling BJ, Steinberg MH (1983) The leukocyte composition of peripheral blood buffy coat. Am J Clin Pathol 79: 500–501. [DOI] [PubMed] [Google Scholar]
- 45. Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, et al. (2010) Nomenclature of monocytes and dendritic cells in blood. Blood 116: e74–e80. [DOI] [PubMed] [Google Scholar]
- 46. Siegal FP, Kadowaki N, Shodell M, Fitzgerald-Bocarsly PA, Shah K, et al. (1999) The nature of the principal type 1 interferon-producing cells in human blood. Science 284: 1835–1837. [DOI] [PubMed] [Google Scholar]
- 47. Gregorio J, Meller S, Conrad C, Di NA, Homey B, et al. (2010) Plasmacytoid dendritic cells sense skin injury and promote wound healing through type I interferons. J Exp Med 207: 2921–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Iyer R, Kalu O, Purser J, Norris S, Stevenson B, et al. (2003) Linear and circular plasmid content in Borrelia burgdorferi clinical isolates. Infect Immun 71: 3699–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Strle K, Jones KL, Drouin EE, Li X, Steere AC (2011) Borrelia burgdorferi RST1 (OspC type A) genotype is associated with greater inflammation and more severe Lyme disease. Am J Pathol 178: 2726–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Anguita J, Samanta S, Revilla B, Suk K, Das S, et al. (2000) Borrelia burgdorferi gene expression in vivo and spirochete pathogenicity. Infect Immun 68: 1222–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Purser JE, Norris SJ (2000) Correlation between plasmid content and infectivity in Borrelia burgdorferi . Proc Natl Acad Sci U S A 97: 13865–13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schwan TG, Burgdorfer W, Garon CF (1988) Changes in infectivity and plasmid profile of the Lyme disease spirochete, Borrelia burgdorferi, as a result of in vitro cultivation. Infect Immun 56: 1831–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xu Y, Kodner C, Coleman L, Johnson RC (1996) Correlation of plasmids with infectivity of Borrelia burgdorferi sensu stricto type strain B31. Infect Immun 64: 3870–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. He M, Boardman BK, Yan D, Yang XF (2007) Regulation of expression of the fibronectin-binding protein BBK32 in Borrelia burgdorferi . J Bacteriol 189: 8377–8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li X, Liu X, Beck DS, Kantor FS, Fikrig E (2006) Borrelia burgdorferi lacking BBK32, a fibronectin-binding protein, retains full pathogenicity. Infect Immun 74: 3305–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Seshu J, Esteve-Gassent MD, Labandeira-Rey M, Kim JH, Trzeciakowski JP, et al. (2006) Inactivation of the fibronectin-binding adhesin gene bbk32 significantly attenuates the infectivity potential of Borrelia burgdorferi . Mol Microbiol 59: 1591–1601. [DOI] [PubMed] [Google Scholar]
- 57. Coleman AS, Rossmann E, Yang X, Song H, Lamichhane CM, et al. (2011) BBK07 Immunodominant Peptides as Serodiagnostic Markers of Lyme Disease. Clin Vaccine Immunol 18: 406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Coleman AS, Pal U (2009) BBK07, a dominant in vivo antigen of Borrelia burgdorferi, is a potential marker for serodiagnosis of Lyme disease. Clin Vaccine Immunol 16: 1569–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pal U, Wang P, Bao F, Yang X, Samanta S, et al. (2008) Borrelia burgdorferi basic membrane proteins A and B participate in the genesis of Lyme arthritis. J Exp Med 205: 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schutzer SE, Fraser-Liggett CM, Casjens SR, Qiu WG, Dunn JJ, et al. (2011) Whole-genome sequences of thirteen isolates of Borrelia burgdorferi . J Bacteriol 193: 1018–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Barthold SW, Moody KD, Terwilliger GA, Duray PH, Jacoby RO, et al. (1988) Experimental Lyme arthritis in rats infected with Borrelia burgdorferi . J Infect Dis 157: 842–846. [DOI] [PubMed] [Google Scholar]
- 62. Casjens SR, Mongodin EF, Qiu WG, Luft BJ, Schutzer SE, et al. (2012) Genome stability of Lyme disease spirochetes: comparative genomics of Borrelia burgdorferi plasmids. PLoS One 7: e33280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Qiu WG, Schutzer SE, Bruno JF, Attie O, Xu Y, et al. (2004) Genetic exchange and plasmid transfers in Borrelia burgdorferi sensu stricto revealed by three-way genome comparisons and multilocus sequence typing. Proc Natl Acad Sci U S A 101: 14150–14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bogdan C, Mattner J, Schleicher U (2004) The role of type I interferons in non-viral infections. Immunol Rev 202: 33–48. [DOI] [PubMed] [Google Scholar]
- 65. Rothfuchs AG, Gigliotti D, Palmblad K, Andersson U, Wigzell H, et al. (2001) IFN-alpha beta-dependent, IFN-gamma secretion by bone marrow-derived macrophages controls an intracellular bacterial infection. J Immunol 167: 6453–6461. [DOI] [PubMed] [Google Scholar]
- 66. de la Maza LM, Peterson EM, Goebel JM, Fennie CW, Czarniecki CW (1985) Interferon-induced inhibition of Chlamydia trachomatis: dissociation from antiviral and antiproliferative effects. Infect Immun 47: 719–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kazar J, Gillmore JD, Gordon FB (1971) Effect of Interferon and Interferon Inducers on Infections with a Nonviral Intracellular Microorganism, Chlamydia trachomatis . Infect Immun 3: 825–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hess CB, Niesel DW, Klimpel GR (1989) The induction of interferon production in fibroblasts by invasive bacteria: a comparison of Salmonella and Shigella species. Microb Pathog 7: 111–120. [DOI] [PubMed] [Google Scholar]
- 69. Hess CB, Niesel DW, Cho YJ, Klimpel GR (1987) Bacterial invasion of fibroblasts induces interferon production. J Immunol 138: 3949–3953. [PubMed] [Google Scholar]
- 70. Niesel DW, Hess CB, Cho YJ, Klimpel KD, Klimpel GR (1986) Natural and recombinant interferons inhibit epithelial cell invasion by Shigella spp. Infect Immun 52: 828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bukholm G, Berdal BP, Haug C, Degre M (1984) Mouse fibroblast interferon modifies Salmonella typhimurium infection in infant mice. Infect Immun 45: 62–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lindahl G, Stalhammar-Carlemalm M, Areschoug T (2005) Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin Microbiol Rev 18: 102–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mancuso G, Midiri A, Biondo C, Beninati C, Zummo S, et al. (2007) Type I IFN signaling is crucial for host resistance against different species of pathogenic bacteria. J Immunol 178: 3126–3133. [DOI] [PubMed] [Google Scholar]
- 74. O’connell RM, Saha SK, Vaidya SA, Bruhn KW, Miranda GA, et al. (2004) Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J Exp Med 200: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bouchonnet F, Boechat N, Bonay M, Hance AJ (2002) Alpha/beta interferon impairs the ability of human macrophages to control growth of Mycobacterium bovis BCG. Infect Immun 70: 3020–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Manca C, Tsenova L, Bergtold A, Freeman S, Tovey M, et al. (2001) Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha/beta. Proc Natl Acad Sci U S A 98: 5752–5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Loughman JA, Hunstad DA (2012) Induction of indoleamine 2,3-dioxygenase by uropathogenic bacteria attenuates innate responses to epithelial infection. J Infect Dis. [DOI] [PMC free article] [PubMed]
- 78.Crandall H, Dunn DM, Ma Y, Wooten RM, Zachary JF, et al.. (2006) Gene expression profiling reveals unique pathways associated with differential severity of lyme arthritis. J Immunol 177: 7930–7942. 177/11/7930 [pii]. [DOI] [PubMed]
- 79.Miller JC, Ma Y, Bian J, Sheehan KC, Zachary JF, et al. (2008) A critical role for type I IFN in arthritis development following Borrelia burgdorferi infection of mice. J Immunol 181: 8492–8503. 181/12/8492 [pii]. [DOI] [PMC free article] [PubMed]
- 80. Numasaki M, Tagawa M, Iwata F, Suzuki T, Nakamura A, et al. (2007) IL-28 elicits antitumor responses against murine fibrosarcoma. J Immunol 178: 5086–5098. [DOI] [PubMed] [Google Scholar]
- 81. Li Q, Kawamura K, Ma G, Iwata F, Numasaki M, et al. (2010) Interferon-lambda induces G1 phase arrest or apoptosis in oesophageal carcinoma cells and produces anti-tumour effects in combination with anti-cancer agents. Eur J Cancer 46: 180–190. [DOI] [PubMed] [Google Scholar]
- 82. Li W, Lewis-Antes A, Huang J, Balan M, Kotenko SV (2008) Regulation of apoptosis by type III interferons. Cell Prolif 41: 960–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Baban B, Chandler PR, Sharma MD, Pihkala J, Koni PA, et al. (2009) IDO activates regulatory T cells and blocks their conversion into Th17-like T cells. J Immunol 183: 2475–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mellor A (2005) Indoleamine 2,3 dioxygenase and regulation of T cell immunity. Biochem Biophys Res Commun 338: 20–24. [DOI] [PubMed] [Google Scholar]
- 85. Mellor AL, Munn DH (2004) IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol 4: 762–774. [DOI] [PubMed] [Google Scholar]
- 86. Le BH, Gougeon ML, Mathian A, Lebon P, Dupont JM, et al. (2011) IFN-alpha and CD46 stimulation are associated with active lupus and skew natural T regulatory cell differentiation to type 1 regulatory T (Tr1) cells. Proc Natl Acad Sci U S A 108: 18995–19000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Li W, Katz BP, Spinola SM (2011) Haemophilus ducreyi lipooligosaccharides induce expression of the immunosuppressive enzyme indoleamine 2,3-dioxygenase via type I interferons and tumor necrosis factor alpha in human dendritic cells. Infect Immun 79: 3338–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Makala LH, Baban B, Lemos H, El-Awady AR, Chandler PR, et al. (2011) Leishmania major attenuates host immunity by stimulating local indoleamine 2,3-dioxygenase expression. J Infect Dis 203: 715–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. von Bubnoff D, Scheler M, Wilms H, Wenzel J, von Bubnoff N, et al. (2011) Indoleamine 2,3-dioxygenase-expressing myeloid dendritic cells and macrophages in infectious and noninfectious cutaneous granulomas. J Am Acad Dermatol 65: 819–832. [DOI] [PubMed] [Google Scholar]
- 90. Scheler M, Wenzel J, Tuting T, Takikawa O, Bieber T, et al. (2007) Indoleamine 2,3-dioxygenase (IDO): the antagonist of type I interferon-driven skin inflammation? Am J Pathol 171: 1936–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Bourreau E, Ronet C, Darcissac E, Lise MC, Sainte MD, et al. (2009) Intralesional regulatory T-cell suppressive function during human acute and chronic cutaneous leishmaniasis due to Leishmania guyanensis . Infect Immun 77: 1465–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Babolin C, Amedei A, Ozolins D, Zilevica A, D’Elios MM, et al. (2011) TpF1 from Treponema pallidum activates inflammasome and promotes the development of regulatory T cells. J Immunol 187: 1377–1384. [DOI] [PubMed] [Google Scholar]
- 93. Filgueira L, Nestle FO, Rittig M, Joller HI, Groscurth P (1996) Human dendritic cells phagocytose and process Borrelia burgdorferi . J Immunol 157: 2998–3005. [PubMed] [Google Scholar]
- 94. Hovius JW, de Jong MA, den Dunnen J, Litjens M, Fikrig E, et al. (2008) Salp15 binding to DC-SIGN inhibits cytokine expression by impairing both nucleosome remodeling and mRNA stabilization. PLoS Pathog 4: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Schwartz I, Wormser GP, Schwartz JJ, Cooper D, Weissensee P, et al. (1992) Diagnosis of early Lyme disease by polymerase chain reaction amplification and culture of skin biopsies from erythema migrans lesions. J Clin Microbiol 30: 3082–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wang G, Iyer R, Bittker S, Cooper D, Small J, et al. (2004) Variations in Barbour-Stoenner-Kelly culture medium modulate infectivity and pathogenicity of Borrelia burgdorferi clinical isolates. Infect Immun 72: 6702–6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 98. Eggers CH, Caimano MJ, Clawson ML, Miller WG, Samuels DS, et al. (2002) Identification of loci critical for replication and compatibility of a Borrelia burgdorferi cp32 plasmid and use of a cp32-based shuttle vector for the expression of fluorescent reporters in the lyme disease spirochaete. Mol Microbiol 43: 281–295. [DOI] [PubMed] [Google Scholar]
- 99. Samuels DS (1995) Electrotransformation of the spirochete Borrelia burgdorferi . Methods Mol Biol 47: 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kenedy MR, Vuppala SR, Siegel C, Kraiczy P, Akins DR (2009) CspA-mediated binding of human factor H inhibits complement deposition and confers serum resistance in Borrelia burgdorferi . Infect Immun 77: 2773–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Elias AF, Stewart PE, Grimm D, Caimano MJ, Eggers CH, et al. (2002) Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect Immun 70: 2139–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]