Abstract
Several hierarchical levels of DNA packaging are believed to exist in chromatin, starting from a 10-nm chromatin fiber that is further packed into a 30-nm fiber. Transitions between the 30-nm and 10-nm fibers are thought to be essential for the control of chromatin transcriptional status. However, recent studies demonstrate that in the nuclei, DNA is packed in tightly associated 10-nm fibers that are not compacted into 30-nm fibers. Additionally, the accessibility of DNA in chromatin depends on the local mobility of nucleosomes rather than on decompaction of chromosome regions. These findings argue for reconsidering the hierarchical model of chromatin packaging and some of the basic definitions of chromatin. In particular, chromatin domains should be considered as three-dimensional objects, which may include genomic regions that do not necessarily constitute a continuous domain on the DNA chain.
Keywords: chromatin fiber, DNA packaging, histone modifications, nucleosome mobility, chromatin domain, 3D genome organization
Introduction
In eukaryotic cells, a very long DNA chain (the total length of DNA in a haploid set of human chromosomes is ~2 meters) is packaged within a relatively small volume of the cell nucleus, which typically has a diameter of ~10 μm. In spite of its tight packaging, DNA remains accessible for the transcription and replication machinery. The mechanisms of DNA packaging in chromatin have been intensively studied over the past 40 y. It is commonly believed that DNA is packed at several hierarchical levels, the first of them being the wrapping of DNA over histone octamers to form nucleosomes.1 The nucleosomal chain has been thought to be packed into the so-called 30-nm fiber and further folded to form loops or several hierarchical coiled structures. Of all these structures, only the nucleosomal particle has been characterized in detail (including the high-resolution structure of a histone octamer and an octamer wrapped with DNA). The structure of the 30-nm chromatin fiber remains unclear,2 and the very existence of this fiber in living cells has been questioned by some authors.3-5 As for the higher level of chromatin packaging, the existing experimental data are so controversial that no comprehensive picture can be drawn.6,7 The principal difficulty was the absence of an experimental tool allowing for the study of the DNA path in chromatin within an undisturbed cell nucleus. For this reason, the existing models of chromatin folding at supranucleosomal scales are mostly based on observations made in experiments with chromatin fibers either folded or both assembled and folded in vitro.2,8-12 Although experiments of this type allow for elucidation of the basic mechanisms of chromatin fiber folding and intranucleosomal interactions, the relevance of models based on observations made in vitro to the actual status of chromatin folding in vivo is not obvious. One aspect of the problem that is usually overlooked is the dynamics of nucleosomes and folded nucleosomal fibers. In the case of X-ray analysis, the method of study already modifies the object being studied. Indeed, only static objects can be crystallized. Therefore, specific (and frequently “unnatural”) conditions are used to stabilize a particular configuration of either a nucleosomal particle or a fragment of a chromatin fiber (e.g., see ref. 8). For this reason, the existence of a variety of other configurations may have escaped the attention of researchers.
30-nm Chromatin Fiber
The 30-nm chromatin fiber was first seen when chromatin released from nuclei was inspected under an electron microscope.13,14 Based on the results of X-ray analysis, it was proposed that this fiber forms from the folding of a nucleosomal chain into a helical structure (solenoid or “one-start” helix) where the nucleosomes coil around a central cavity with ∼six nucleosomes per turn, so that each nucleosome in the fiber interacts with its fifth and sixth neighbor nucleosomes.15 Later, electron microscopy (EM) studies of chromatin that had been frozen immediately after release from nuclei demonstrated that in a 30-nm fiber, nucleosomes are arranged not linearly along the same helical path but rather zig-zag back and forth to form a “two-start” helix, in which each nucleosome binds to the second neighbor nucleosome.16-18 A solenoid has been proposed to represent a structure that is the most stable thermodynamically and, thus, any other type of supranucleosomal fiber is easily converted into a solenoid, as long as this conversion was not prevented by freezing. Crystallization of a tetranucleosomal array provided additional evidence for the zig-zag model of the 30-nm fiber.8 However, the tetramer that was crystallized was assembled without H1, and the nucleosomal repeat length was only 167 bp (in contrast to the 200 bp typical for most eukaryotic organisms).8 Thus, the biological relevance of this structure is unclear. Nevertheless, some conclusions that followed from the analysis of the crystallized tetramer appear highly important. In particular, it was observed that the fiber was stabilized by interaction of a charged H4 tail with the acidic patch on the globule of a neighboring nucleosome.8 The acidic patch is composed of several negatively charged amino acid residues present in histones H2A and H2B. Variant forms of histone H2A (H2A.Z, H2A.Bbd and macro-H2A) have substitutions in the region involved in the formation of the acidic patch. The presence of these substitutions correlates well with the stability of the 30-nm fiber and the biological properties of chromatin domains containing these variant histones.19,20 Also of note is that acetylation of the tail of H4 (in particular at K1621,22) results in destabilization of the 30-nm fiber, as one would expect based on analysis of the crystallized tetramer structure.
Cryo-electron microscopy of long regular nucleosomal arrays assembled in vitro on regularly spaced positioning sequences demonstrated that spacer length and the presence of a linker histone are of crucial importance for the parameters of the 30-nm fibers.9,23 Short-to-medium repeat length, including the most typical repeat length for metazoan cells (200 bp), favors formation of zig-zag structures, while longer repeat lengths allow for the assembly of solenoid-like structures.10 Most importantly, results of several recent studies strongly suggest that the 30-nm fiber is not uniform. In this heteromorphic 30-nm fiber, the regions with two-start organization (zig-zag) are interspersed by regions possessing properties of one-start solenoids.10 If it exists in nature (see below), the 30-nm fiber most likely represents a delicate balance of different configurations that can be modified by a number of factors, including but not limited to, deposition of linker histones and high mobility group (HMG) proteins, incorporation of variant histones, modifications of histone tails, activity of chromatin remodeling factors, presence of phased nucleosome arrays, and interruptions in the continuity of nucleosomal fibers due to the presence of nucleosome-free regions.24-26
Importantly, all of the above observations were made in experiments performed in vitro. Meanwhile, estimation of the contour length of active segments of the genome made using fluorescence in situ hybridization (FISH) and other more sophisticated approaches suggests that these regions are compacted far beyond the level that can be achieved by packaging in a 30-nm fiber.27-31 It is thus likely that in both active and repressed genome regions, chromatin is folded by side condensation of 10-nm fibers into a compact structure that textbooks usually show to illustrate a possible way of forming heterochromatin (Fig. 1). Fiber association can be stabilized by interaction between nucleosomes that belong to different fibers.32 This interaction may be either direct (e.g., interaction of the H4 tail with the acidic patch of another nucleosome) or assisted by architectural proteins, such as Hp1 (Fig. 1).33 One can imagine the side-to-side association of both 10-nm and 30-nm fibers. Until recently, the existing experimental approaches did not allow visualization of the path of DNA within the nucleus due to the overlapping of multiple chromatin fibers projected onto a single 2D image. This problem was solved by combining electron spectroscopic imaging (ESI) with electron tomography. Surprisingly, 10-nm fibers were the only regular chromatin structures that were observed in eukaryotic cell nuclei using this technique. Both euchromatic and heterochromatic regions appeared to be composed of tightly packed 10-nm fibers with no traces of 30-nm fibers seen anywhere.3 The compaction level (i.e., the density of nucleosomal particles per a volume unit) was higher in heterochromatic regions, but this was achieved by more densely packaged 10-nm fibers and not by their folding into a regular structure. In heterochromatic regions, the bending of individual fibers was more pronounced.3 Experiments performed using electron cryotomography also demonstrated that there is no regularity in the higher-order packaging of a nucleosomal fiber. Interphase chromatin appeared to be a disordered assemblage of nucleosomes and could best be described by the polymer melt model.4 Using Hi-C analysis (a biochemical approach based on the proximity ligation procedure), Dekker and collaborators also came to the conclusion that both open (active) and closed (repressed) chromatin domains were composed of a single type of fiber.5 No evidence for the hierarchical folding of chromatin fibers was found.
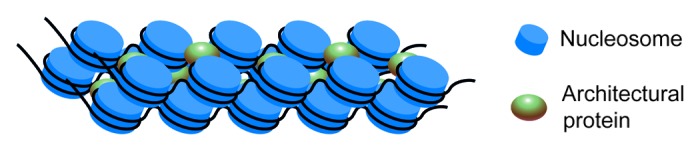
Figure 1. Schematic illustrating a possible pathway for the condensation of several 10-nm fibers into a compact heterochromatin-like structure. See description in the text.
Active Chromatin and the Histone Code
Pioneering observations made by Weintraub and Groudine34 that were confirmed in a number of subsequent studies35 demonstrated that actively transcribed genes were preferentially sensitive to nucleases in permeabilized cells. The proposed explanation for this preferential sensitivity to nucleases was that in active chromatin regions, DNA is more accessible to trans-acting factors.35 Additional accessibility can be gained both by reorganization of core particles, allowing for nucleases to attack DNA wrapped around the octamer, and by reorganization of higher-order chromatin structures, allowing for more easy diffusion. Although nucleosomal particles isolated from active chromatin possess some special characteristics,36,37 preferential sensitivity of active genes to nucleases was commonly considered to be a consequence of partial chromatin unfolding. In particular, the unfolding of 30 nm-fibers is believed to be the cause of preferential sensitivity to nucleases.38 As discussed above, current results show that this is an oversimplification. Still, less dense packaging of chromatin in active (euchromatic) regions is an established fact confirmed by numerous studies, including those that demonstrated the absence of 30-nm chromatin fibers in undisturbed nuclei.3,39 Active and repressed genomic regions are characterized by different profiles of histone modifications. Since the histone code hypothesis was first proposed,40 the significance of different histone modifications has been intensively studied.41-45 Development of modern genomic approaches (including chromatin immunoprecipitation [ChIP] followed by high-throughput sequencing) allowed for comprehensive analysis of profiles of histone modifications in different organisms, different genomic regions and under different conditions (e.g., differentiation and transcriptional activation in response to hormones).46-48 The histone code hypothesis gradually transformed into the histone context hypothesis,41,49 as several different modifications should be considered simultaneously to predict positions of genome regulatory elements50-53 and to determine the type of chromatin domain (reviewed in refs. 54 and 55). Histone modifications essential for activation of chromatin domains have been discussed in terms of transitions between 30-nm and 10-nm chromatin fibers.38 For example, acetylation of histone H4 at position K16 was shown to prevent interaction of the H4 N-terminal tail with the acidic patch on the neighboring nucleosomal globule, therefore destabilizing the 30-nm chromatin fiber.21,22 Substitution of histone H2A by H2A.Bbd had similar consequences20 due to a decrease in the negative charge of the acidic patch. In view of the new concepts of chromatin folding (see the previous section), one may question the biological relevance of all the above-mentioned conclusions, although such reasoning appears to be too hasty. Irrespectively of whether 30-nm fibers exist or not, the generic mechanisms of higher-order hierarchical folding that depend on histone substitutions or modifications are still likely to play major roles in higher order folding structures. For example, in the absence of intra-fiber interactions resulting in the formation of a 30-nm fiber, these mechanisms may stabilize inter-fiber associations necessary to keep chromatin in a folded state.32 Consequently, loss of interactions between neighboring nucleosomes will lead to local decompaction of a chromatin domain.
Is Decompaction of Chromatin Necessary to Grant Accessibility of DNA?
The answer to this question seems obvious. Indeed, it is known that active chromatin is preferentially digested by nucleases and is packed in a less dense fashion. However, to what extent dense packaging of nucleosomal arrays interferes with accessibility of DNA has remained unknown until recently. In a recent study,56 Maeshima and collaborators directly approached this question using fluorescence correlation spectroscopy. Unexpectedly, a relatively large object (in this case, a pentamer of enhanced green fluorescent protein [EGFP] that is slightly larger than a nucleosome) moved with comparable speed through the cytoplasm, interphase chromatin, and metaphase chromatin, although the estimated density of nucleosomal particles in metaphase chromatin was 5 to 10 times higher than in interphase chromatin.57-59 Computer simulations demonstrated that an object of this size should not move at all through metaphase chromatin unless nucleosomes are mobile (i.e., can make short non-directional movements at distances of up to 10–20 nm). Single nucleosome imaging studies showed that nucleosomes are indeed mobile in living cells. For both interphase and mitotic chromatin, the fluctuation of individual nucleosomes was ~50 nm/30 ms.56 If confirmed by other researchers, these observations may open a new chapter in the study of chromatin domains. Modifications of histones, collectively known as the histone code, may control the local mobility of nucleosomes. In the computer simulations mentioned above, the nucleosomes and the EGFP pentamer were considered as solid spheres. In reality, all cellular constituents, including nucleosomes,60 are relatively soft, i.e., they can change their shape to some extent. This can further enhance the mobility of different objects in chromatin.
In Conclusion
During the past 35 y, the hierarchical model of chromatin folding constituted the basis for the study of the functionally relevant spatial organization of the eukaryotic genome. However, this model was mostly based on results obtained from experiments performed in vitro. The development of methods allowing for the study of chromatin folding in living cells permitted researchers to make observations that have questioned this model. To account for the new data, many current concepts of chromatin need to be revised. Although some established results (such as the relation of histone acetylation to chromatin activation) remain clear, their interpretation of particular molecular events should be reconsidered in view of a new model of interphase chromatin folding. There is no need to revise the basic principles of signals (either activation or repressive) that spread in chromatin via modification of histones in neighboring nucleosomes and attraction of “histone code writers” to these modified nucleosomes. However, in a mesh of stochastically folded and tightly packed chromatin, the activating/repressive signal will spread in all directions in 3D space, as nucleosomes from different fibers will be positioned close to a nucleation center (Fig. 2). In this situation, it is not clear how the spreading of this signal can be restricted to a particular genome area. Perhaps organization of chromosomes into topologically associating domains (TADs) or smaller globular units61 may put restrictions upon chromatin domain expansion. In classical molecular biology, distances were calculated in kilobases rather than in micrometers. This measurement principle also applies to the mapping of chromatin modifications (see, for example, the ENCODE project). Recognition of the importance of long-range intrachromosomal and interchromosomal interactions has made modern genomics three-dimensional;62 chromatin studies will expand in the same direction. Chromatin domains are not necessarily continuous. These domains may be composed of a mosaic of chromosome regions that become united in a 3D nuclear space. The Hi-C data strongly support the idea of spatial segregations of active and repressive genomic regions.
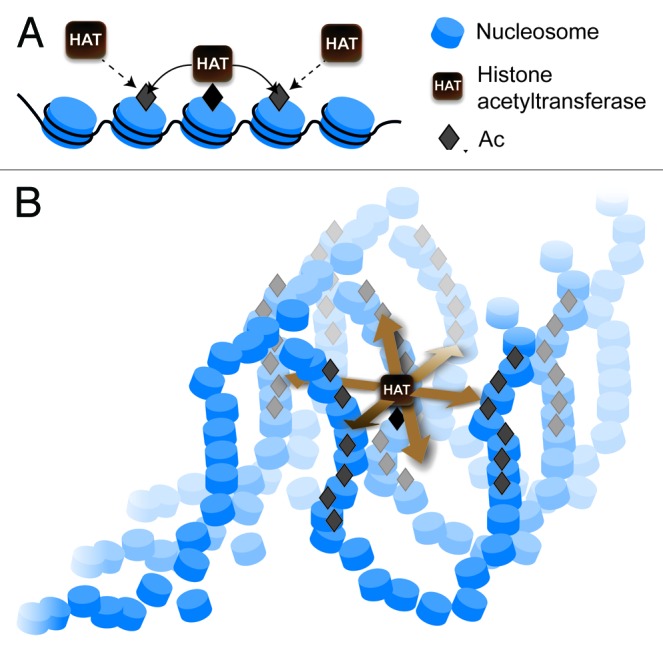
Figure 2. Mechanisms of signal spreading in chromatin (exemplified by the spreading of histone acetylation). (A) A classical view of the linear spreading of a signal in two directions along the chromatin fiber. (B) Three-dimensional spreading of a signal in all directions from a nucleation center resulting in modification of multiple chromatin regions both in cis and in trans.
Disclosure of the mechanisms controlling the assembly of 3D chromatin domains that incorporate segments of different chromosomal regions constitutes an important task for the future studies. By solving this problem it will become possible to reinterpret the existing data on the distribution of histone modifications and to provide new insights into the principles of histone code functioning. The current progress in the development of high-resolution microscopic techniques, allowing the study of living cells, promises exciting new discoveries in the chromatin field.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the Presidium of the Russian Academy of Sciences (grants MCB to SVR and AAG), by RFBR grants 12-04-93109, 12-04-0036, 14-04-00010 and by Dmitri Zimin’s foundation “Dynasty.”
References
- 1.Getzenberg RH, Pienta KJ, Ward WS, Coffey DS. Nuclear structure and the three-dimensional organization of DNA. J Cell Biochem. 1991;47:289–99. doi: 10.1002/jcb.240470402. [DOI] [PubMed] [Google Scholar]
- 2.Grigoryev SA, Woodcock CL. Chromatin organization - the 30 nm fiber. Exp Cell Res. 2012;318:1448–55. doi: 10.1016/j.yexcr.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 3.Fussner E, Strauss M, Djuric U, Li R, Ahmed K, Hart M, Ellis J, Bazett-Jones DP. Open and closed domains in the mouse genome are configured as 10-nm chromatin fibres. EMBO Rep. 2012;13:992–6. doi: 10.1038/embor.2012.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gan L, Ladinsky MS, Jensen GJ. Chromatin in a marine picoeukaryote is a disordered assemblage of nucleosomes. Chromosoma. 2013;122:377–86. doi: 10.1007/s00412-013-0423-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sajan SA, Hawkins RD. Methods for identifying higher-order chromatin structure. Annu Rev Genomics Hum Genet. 2012;13:59–82. doi: 10.1146/annurev-genom-090711-163818. [DOI] [PubMed] [Google Scholar]
- 7.Maeshima K, Eltsov M. Packaging the genome: the structure of mitotic chromosomes. J Biochem. 2008;143:145–53. doi: 10.1093/jb/mvm214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–41. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 9.Robinson PJ, Fairall L, Huynh VA, Rhodes D. EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc Natl Acad Sci U S A. 2006;103:6506–11. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grigoryev SA, Arya G, Correll S, Woodcock CL, Schlick T. Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc Natl Acad Sci U S A. 2009;106:13317–22. doi: 10.1073/pnas.0903280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blacketer MJ, Feely SJ, Shogren-Knaak MA. Nucleosome interactions and stability in an ordered nucleosome array model system. J Biol Chem. 2010;285:34597–607. doi: 10.1074/jbc.M110.140061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghirlando R, Felsenfeld G. Chromatin structure outside and inside the nucleus. Biopolymers. 2013;99:225–32. doi: 10.1002/bip.22157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGhee JD, Rau DC, Charney E, Felsenfeld G. Orientation of the nucleosome within the higher order structure of chromatin. Cell. 1980;22:87–96. doi: 10.1016/0092-8674(80)90157-9. [DOI] [PubMed] [Google Scholar]
- 14.Tremethick DJ. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell. 2007;128:651–4. doi: 10.1016/j.cell.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976;73:1897–901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodcock CL, Frado LL, Rattner JB. The higher-order structure of chromatin: evidence for a helical ribbon arrangement. J Cell Biol. 1984;99:42–52. doi: 10.1083/jcb.99.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bednar J, Horowitz RA, Dubochet J, Woodcock CL. Chromatin conformation and salt-induced compaction: three-dimensional structural information from cryoelectron microscopy. J Cell Biol. 1995;131:1365–76. doi: 10.1083/jcb.131.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–3. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 19.Fan JY, Rangasamy D, Luger K, Tremethick DJ. H2A.Z alters the nucleosome surface to promote HP1alpha-mediated chromatin fiber folding. Mol Cell. 2004;16:655–61. doi: 10.1016/j.molcel.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 20.Zhou J, Fan JY, Rangasamy D, Tremethick DJ. The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat Struct Mol Biol. 2007;14:1070–6. doi: 10.1038/nsmb1323. [DOI] [PubMed] [Google Scholar]
- 21.Allahverdi A, Yang R, Korolev N, Fan Y, Davey CA, Liu CF, Nordenskiöld L. The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic Acids Res. 2011;39:1680–91. doi: 10.1093/nar/gkq900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang D, Arya G. Structure and binding of the H4 histone tail and the effects of lysine 16 acetylation. Phys Chem Chem Phys. 2011;13:2911–21. doi: 10.1039/c0cp01487g. [DOI] [PubMed] [Google Scholar]
- 23.Arya G, Schlick T. Role of histone tails in chromatin folding revealed by a mesoscopic oligonucleosome model. Proc Natl Acad Sci U S A. 2006;103:16236–41. doi: 10.1073/pnas.0604817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong H, Victor JM, Mozziconacci J. An all-atom model of the chromatin fiber containing linker histones reveals a versatile structure tuned by the nucleosomal repeat length. PLoS One. 2007;2:e877. doi: 10.1371/journal.pone.0000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Depken M, Schiessel H. Nucleosome shape dictates chromatin fiber structure. Biophys J. 2009;96:777–84. doi: 10.1016/j.bpj.2008.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perišić O, Collepardo-Guevara R, Schlick T. Modeling studies of chromatin fiber structure as a function of DNA linker length. J Mol Biol. 2010;403:777–802. doi: 10.1016/j.jmb.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawrence JB, Singer RH. Spatial organization of nucleic acid sequences within cells. Semin Cell Biol. 1991;2:83–101. [PubMed] [Google Scholar]
- 28.Lawrence JB, Singer RH, McNeil JA. Interphase and metaphase resolution of different distances within the human dystrophin gene. Science. 1990;249:928–32. doi: 10.1126/science.2203143. [DOI] [PubMed] [Google Scholar]
- 29.Mahy NL, Perry PE, Bickmore WA. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J Cell Biol. 2002;159:753–63. doi: 10.1083/jcb.200207115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Y, Kireev I, Plutz M, Ashourian N, Belmont AS. Large-scale chromatin structure of inducible genes: transcription on a condensed, linear template. J Cell Biol. 2009;185:87–100. doi: 10.1083/jcb.200809196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belmont AS, Hu Y, Sinclair PB, Wu W, Bian Q, Kireev I. Insights into interphase large-scale chromatin structure from analysis of engineered chromosome regions. Cold Spring Harb Symp Quant Biol. 2010;75:453–60. doi: 10.1101/sqb.2010.75.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pepenella S, Murphy KJ, Hayes JJ. Intra- and inter-nucleosome interactions of the core histone tail domains in higher-order chromatin structure. Chromosoma. 2013 doi: 10.1007/s00412-013-0435-8. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McBryant SJ, Adams VH, Hansen JC. Chromatin architectural proteins. Chromosome Res. 2006;14:39–51. doi: 10.1007/s10577-006-1025-x. [DOI] [PubMed] [Google Scholar]
- 34.Weintraub H, Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976;193:848–56. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- 35.Yaniv M, Cereghini S. Structure of transcriptionally active chromatin. CRC Crit Rev Biochem. 1986;21:1–26. doi: 10.3109/10409238609113607. [DOI] [PubMed] [Google Scholar]
- 36.Bazett-Jones DP, Mendez E, Czarnota GJ, Ottensmeyer FP, Allfrey VG. Visualization and analysis of unfolded nucleosomes associated with transcribing chromatin. Nucleic Acids Res. 1996;24:321–9. doi: 10.1093/nar/24.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Czarnota GJ, Bazett-Jones DP, Mendez E, Allfrey VG, Ottensmeyer FP. High resolution microanalysis and three-dimensional nucleosome structure associated with transcribing chromatin. Micron. 1997;28:419–31. doi: 10.1016/S0968-4328(97)00050-4. [DOI] [PubMed] [Google Scholar]
- 38.Razin SV, Iarovaia OV, Sjakste N, Sjakste T, Bagdoniene L, Rynditch AV, Eivazova ER, Lipinski M, Vassetzky YS. Chromatin domains and regulation of transcription. J Mol Biol. 2007;369:597–607. doi: 10.1016/j.jmb.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 39.Bian Q, Belmont AS. Revisiting higher-order and large-scale chromatin organization. Curr Opin Cell Biol. 2012;24:359–66. doi: 10.1016/j.ceb.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 41.Gardner KE, Allis CD, Strahl BD. Operating on chromatin, a colorful language where context matters. J Mol Biol. 2011;409:36–46. doi: 10.1016/j.jmb.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–95. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–99. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 44.Rando OJ. Combinatorial complexity in chromatin structure and function: revisiting the histone code. Curr Opin Genet Dev. 2012;22:148–55. doi: 10.1016/j.gde.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karch KR, Denizio JE, Black BE, Garcia BA. Identification and interrogation of combinatorial histone modifications. Front Genet. 2013;4:264. doi: 10.3389/fgene.2013.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boyle AP, Furey TS. High-resolution mapping studies of chromatin and gene regulatory elements. Epigenomics. 2009;1:319–29. doi: 10.2217/epi.09.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ku CS, Naidoo N, Wu M, Soong R. Studying the epigenome using next generation sequencing. J Med Genet. 2011;48:721–30. doi: 10.1136/jmedgenet-2011-100242. [DOI] [PubMed] [Google Scholar]
- 48.Yavartanoo M, Choi JK. ENCODE: A Sourcebook of Epigenomes and Chromatin Language. Genomics Inform. 2013;11:2–6. doi: 10.5808/GI.2013.11.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horikoshi M. Histone acetylation: from code to web and router via intrinsically disordered regions. Curr Pharm Des. 2013;19:5019–42. doi: 10.2174/1381612811319280002. [DOI] [PubMed] [Google Scholar]
- 50.Hon GC, Hawkins RD, Ren B. Predictive chromatin signatures in the mammalian genome. Hum Mol Genet. 2009;18(R2):R195–201. doi: 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hwang YC, Zheng Q, Gregory BD, Wang LS. High-throughput identification of long-range regulatory elements and their target promoters in the human genome. Nucleic Acids Res. 2013;41:4835–46. doi: 10.1093/nar/gkt188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49:825–37. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murr R. Interplay between different epigenetic modifications and mechanisms. Adv Genet. 2010;70:101–41. doi: 10.1016/B978-0-12-380866-0.60005-8. [DOI] [PubMed] [Google Scholar]
- 54.Boros IM. Histone modification in Drosophila. Brief Funct Genomics. 2012;11:319–31. doi: 10.1093/bfgp/els029. [DOI] [PubMed] [Google Scholar]
- 55.White R. Packaging the fly genome: domains and dynamics. Brief Funct Genomics. 2012;11:347–55. doi: 10.1093/bfgp/els020. [DOI] [PubMed] [Google Scholar]
- 56.Hihara S, Pack CG, Kaizu K, Tani T, Hanafusa T, Nozaki T, Takemoto S, Yoshimi T, Yokota H, Imamoto N, et al. Local nucleosome dynamics facilitate chromatin accessibility in living mammalian cells. Cell Rep. 2012;2:1645–56. doi: 10.1016/j.celrep.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 57.Weidemann T, Wachsmuth M, Knoch TA, Müller G, Waldeck W, Langowski J. Counting nucleosomes in living cells with a combination of fluorescence correlation spectroscopy and confocal imaging. J Mol Biol. 2003;334:229–40. doi: 10.1016/j.jmb.2003.08.063. [DOI] [PubMed] [Google Scholar]
- 58.Wachsmuth M, Caudron-Herger M, Rippe K. Genome organization: balancing stability and plasticity. Biochim Biophys Acta. 2008;1783:2061–79. doi: 10.1016/j.bbamcr.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 59.Nozaki T, Kaizu K, Pack CG, Tamura S, Tani T, Hihara S, Nagai T, Takahashi K, Maeshima K. Flexible and dynamic nucleosome fiber in living mammalian cells. Nucleus. 2013;4:349–56. doi: 10.4161/nucl.26053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miyagi A, Ando T, Lyubchenko YL. Dynamics of nucleosomes assessed with time-lapse high-speed atomic force microscopy. Biochemistry. 2011;50:7901–8. doi: 10.1021/bi200946z. [DOI] [PubMed] [Google Scholar]
- 61.Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49:773–82. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cavalli G. Chromosomes: now in 3D! Nat Rev Mol Cell Biol. 2014;15:6. doi: 10.1038/nrm3717. [DOI] [PubMed] [Google Scholar]


