Abstract
Constitutional epigenetic changes detected in blood or non-disease involving tissues have been associated with disease susceptibility. We measured promoter methylation of CDKN2A (p16 and p14ARF) and 13 melanoma-related genes using bisulfite pyrosequencing of blood DNA from 114 cases and 122 controls in 64 melanoma-prone families (26 segregating CDKN2A germline mutations). We also obtained gene expression data for these genes using microarrays from the same blood samples. We observed that CDKN2A epimutation is rare in melanoma families, and therefore is unlikely to cause major susceptibility in families without CDKN2A mutations. Although methylation levels for most gene promoters were very low (<5%), we observed a significantly reduced promoter methylation (odds ratio = 0.63, 95% confidence interval = 0.50, 0.80, P < 0.001) and increased expression (fold change = 1.27, P = 0.048) for TNFRSF10C in melanoma cases. Future research in large prospective studies using both normal and melanoma tissues is required to assess the significance of TNFRSF10C methylation and expression changes in melanoma susceptibility.
Keywords: familial melanoma, CDKN2A, promoter methylation, peripheral blood mononuclear cells, TNFRSF10C
Introduction
Cutaneous malignant melanoma (CMM) is a potentially fatal form of skin cancer with a heterogeneous etiology.1 The cyclin-dependent kinase inhibitor 2A (CDKN2A) gene is one of the most established major melanoma susceptibility genes identified to date. However, it only occurs in 20–40% of melanoma-prone families,2 suggesting the existence of additional high-risk genes or other susceptibility mechanisms. Further, the incomplete penetrance of CDKN2A as well as variations in phenotypic manifestations among mutation carriers1,2 suggest that other factors may modify melanoma risk even in families with known genetic causes. Recently, constitutional epigenetic changes including gene-specific promoter hypermethylation in blood or non-diseased tissues have been associated with disease susceptibility. The most striking example is the identification of epimutations in MLH1 and MSH2 as major susceptibility mechanisms for familial cancers.3,4 The goal of this study was to evaluate whether constitutional promoter methylation of CDKN2A and other melanoma-related genes was related to melanoma susceptibility in families with and without CDKN2A mutations.
Results and Discussion
Our study population was comprised of families with at least two living first degree relatives with a history of invasive melanoma ascertained from the United States.5 The current study was based on 114 CMM cases (45 CDKN2A-carriers and 69 non-carriers) and 122 controls (32 CDKN2A-carriers and 90 non-carriers) from 64 families (26 families segregating CDKN2A mutations and 38 families without known mutations) (Table 1). The study was approved by the National Cancer Institute Clinical Center Institutional Review Board and conducted according to the Declaration of Helsinki. Informed consent was obtained from all participants.
Table 1. Distribution of age, gender, CDKN2A, pigmentation phenotype, and sun exposure variables in 64 melanoma-prone families by CMM status.
| Unaffected Individuals (n = 122) |
CMM Cases (n = 114) |
P value |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | |||||||
|
Age at blood draw |
||||||||||
| < 50 | 73 | 59.84 | 70 | 61.40 | ||||||
| 50+ | 49 | 40.16 | 44 | 38.60 | 0.79 | |||||
| Gender | ||||||||
|---|---|---|---|---|---|---|---|---|
| Female | 77 | 63.11 | 64 | 56.14 | ||||
| Male | 45 | 36.89 | 50 | 43.86 | 0.15 |
| CDKN2A | ||||||||
|---|---|---|---|---|---|---|---|---|
| Non-Carrier | 90 | 73.77 | 69 | 60.53 | ||||
| Carrier | 32 | 26.23 | 45 | 39.47 | 0.003 |
| Dysplastic nevi | ||||||||
|---|---|---|---|---|---|---|---|---|
| Unaffected | 71 | 69.61 | 2 | 2.17 | ||||
| Affected | 31 | 30.39 | 90 | 97.83 | < 0.0001 |
| Moles | ||||||||
|---|---|---|---|---|---|---|---|---|
| 0–24 | 41 | 36.94 | 9 | 9.18 | ||||
| 25–99 | 46 | 41.44 | 23 | 23.47 | ||||
| 100+ | 24 | 21.62 | 66 | 67.35 | < 0.0001 |
| Freckles | ||||||||
|---|---|---|---|---|---|---|---|---|
| None/few | 33 | 45.83 | 12 | 19.35 | ||||
| Moderate | 21 | 29.17 | 15 | 24.19 | ||||
| Many | 18 | 25.00 | 35 | 56.45 | < 0.0001 |
| Solar injury | ||||||||
|---|---|---|---|---|---|---|---|---|
| None/mild | 77 | 68.75 | 48 | 48.98 | ||||
| Moderate | 22 | 19.64 | 27 | 27.55 | ||||
| Severe | 13 | 11.61 | 23 | 23.47 | 0.006 |
| Tanning ability | ||||||||
|---|---|---|---|---|---|---|---|---|
| Tan/Little burn | 59 | 60.82 | 40 | 46.51 | ||||
| Burn/Little tan | 38 | 39.18 | 46 | 53.49 | 0.07 |
| Skin type | ||||||||
|---|---|---|---|---|---|---|---|---|
| Dark/medium | 33 | 32.04 | 13 | 14.94 | ||||
| Pale/fair | 70 | 67.96 | 74 | 85.06 | 0.006 |
| Eye color | ||||||||
|---|---|---|---|---|---|---|---|---|
| Black/brown | 27 | 26.47 | 18 | 20.69 | ||||
| Hazel | 25 | 24.51 | 18 | 20.69 | ||||
| Green/gray | 14 | 13.73 | 12 | 13.79 | ||||
| Blue | 36 | 35.29 | 39 | 44.83 | 0.51 |
| Hair color | ||||||||
|---|---|---|---|---|---|---|---|---|
| Black/brown | 45 | 44.12 | 35 | 40.23 | ||||
| Blonde brown/light brown | 29 | 28.43 | 27 | 31.03 | ||||
| Blonde | 17 | 16.67 | 12 | 13.79 | ||||
| Red | 11 | 10.78 | 13 | 14.94 | 0.71 |
| MC1R | ||||||||
|---|---|---|---|---|---|---|---|---|
| Wild type | 25 | 27.17 | 3 | 3.85 | ||||
| 1 nonsynonymous variant | 39 | 42.39 | 40 | 51.28 | ||||
| 2 nonsynonymous variants | 28 | 30.43 | 35 | 44.87 | 0.0001 |
P values were obtained by comparing CMM cases to unaffected individuals using a generalized estimating equation and adjusting for familial correlation in the variance.
We investigated the constitutional methylation of the p16 and p14ARF promoters of the CDKN2A locus as well 13 melanoma-related genes known to be involved in important cellular pathways relevant to melanoma including CDH1, COL1A2, DAPK1, DDIT4L, HSPB6, LOX, MAGE-A3, MT1G, NPM2, PTEN, RASSF1, TNFRSF10C, and TNFRSF10D (Table 2) using DNA extracted from peripheral blood mononuclear cells (PBMCs). The methylation status of each promoter region was measured across multiple CpG sites (range: 7–27 CpG sites) for each gene using bisulfite pyrosequencing (Table 2). Each CpG was analyzed individually as a T/C SNP and then averaged together to provide an overall percent 5-MeC for each gene promoter (Supplementary Methods).
Table 2. Overall gene-specific methylation levels in cutaneous malignant melanoma (CMM) cases and unaffected individuals.
| Gene Symbol (Ca/Co) |
Chromosomal Location | No. CpGs | Genomic Location (hg19) of Promoter CpGs | Average % methylation Non-carrier controls (n = 90) |
Average % methylation CDKN2A-carrier controls (n = 32) |
Average % methylation Non-carrier cases (n = 69) |
Average % methylation CDKN2A-carrier cases (n = 45) |
P value All cases vs. controls |
|---|---|---|---|---|---|---|---|---|
|
CDH1 (114/122) |
16q22.1 | 7 | 21405–21555 | 4.36 | 4.16 | 3.67 | 4.15 | 0.002 |
|
COL1A2 (114/121) |
7q22.1 | 9 | 68771138–68771203 | 58.9 | 58.19 | 59.05 | 57.09 | 0.722 |
|
DAPK1 (113/116) |
9q34.1 | 25 | 90112806–90113020 | 3.17 | 3.96 | 2.66 | 3.39 | 0.021 |
|
DDIT4L (114/119) |
4q24 | 8 | 101111643–101111547 | 2.53 | 2.45 | 2.33 | 2.43 | 0.170 |
|
HSPB6 (114/121) |
19q13.12 | 11 | 36248078–36247921 | 24.12 | 23.35 | 23.04 | 22.9 | 0.130 |
|
LOX (114/120) |
5q23.2 | 16 | 121414112–121413916 | 3.34 | 2.96 | 3.05 | 3.05 | 0.073 |
|
MAGE-A3 (Females) (64/72) |
8q28 | 13 | 151938243–151938137 | 91.41 | 90.66 | 91.78 | 88.93 | 0.0003 |
|
MAGE-A3 (Males) (50/42) |
8q28 | 13 | 151938243–151938137 | 94.22 | 91.84 | 93.03 | 90.96 | 0.317 |
|
MT1G (114/120) |
16q13 | 5 | 56701919–56701865 | 10.29 | 11.64 | 8.96 | 11.16 | 0.006 |
|
NPM2 (113/122) |
8p21.3 | 19 | 21881609–21881783 | 1.61 | 1.41 | 1.4 | 1.6 | 0.232 |
|
p14ARF (114/121) |
9p21 | 19 | 21994866–21994723 | 0.82 | 0.80 | 0.65 | 0.82 | 0.017 |
|
p16 (114/119) |
9p21 | 7 | 21974890–21974866 | 1.74 | 1.57 | 1.7 | 1.5 | 0.209 |
|
PTEN (112/116) |
10q23.3 | 27 | 89623432–89623620 | 1.11 | 0.96 | 0.82 | 1.09 | 0.025 |
|
RASSF1 (111/117) |
3p21.3 | 9 | 50378294–50378232 | 0.6 | 0.53 | 0.47 | 0.58 | 0.022 |
|
TNFRSF10C 9113/120) |
8p21 | 10 | 22960386–22960481 | 2.26 | 2.55 | 1.48 | 2.63 | 0.0002 |
|
TNFRSF10D (114/120) |
8p21 | 12 | 23021611–23021470 | 1.38 | 1.63 | 1.0 | 1.69 | 0.042 |
P values were obtained by comparing all CMM cases to all unaffected control individuals using a generalized estimating equation and adjusting for familial correlation in the variance. No, number; Ca, case; Co, unaffected individual/control. Cases (Ca), control (Co) numbers for each gene promoter region are shown in parentheses under gene symbol.
In agreement with other studies examining blood PBMCs from healthy subjects,6 we found that the overall methylation levels at 11 out of the 15 genes (including p16/CDKN2A and p14ARF/CDKN2A, Fig. 1) were low (<5%) among non-carrier controls. Higher methylation levels (hypermethylation) (10.8–92.3%) were only observed for COL1A, HSPB6, MAGE-A3 and MT1G (Table 2). Several CMM risk factors such as eye color and hair color were also related to promoter methylation levels for a number of genes among unaffected individuals (Table S1).
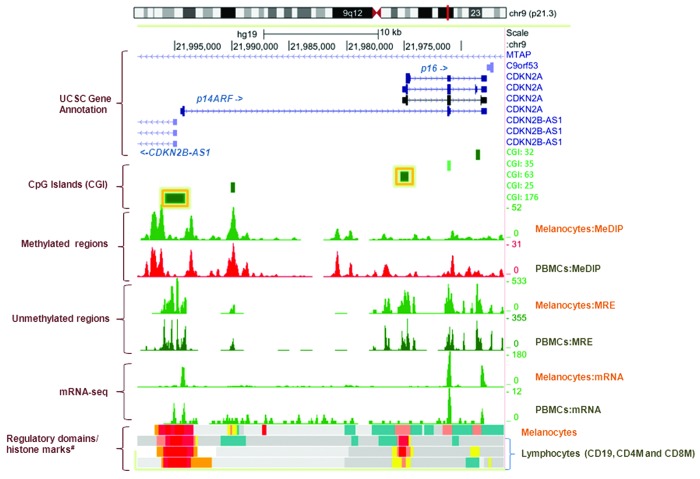
Figure 1. Genome Browser (http://genome.ucsc.edu/) image of the CDKN2A promoter region on human assembly hg19 based on NIH Epigenomics Roadmap data.10 The promoter CpG islands (CGIs) of p16/CDKN2A (CGI:63) and p14ARF/CDKN2A (CGI: 176) analyzed in this study are highlighted with yellow boxes. The CGIs (CGI: 176, 63, 35 and 32) and/or differentially methylated regions technically validated using the DMH-array based method7 in CDKN2A-negative families are annotated to the right of the figure. MeDIP, methylated DNA immunoprecipitation; MRE, methylation-sensitive restrictive enzyme; Melanocytes, normal primary penile foreskin melanocytes (UCSF-UBC-USC and UCSF-UBC); PBMCs, peripheral blood mononuclear cells (UCSF-UBC-UCD and UCSF UBC); lymphocytes, CD19, CD4, and CD8 cells (NIH Epigenomics Roadmap data). Regulatory domains (chromatin state segmentation using a hidden Markov Model [ChromHMM]) and core histone marks: red, active transcriptional start site (TSS); dark salmon, poised TSS; crimson, flanking TSS; orange, active to weak enhancer; yellow, poised enhancer; cadet blue, H3K9me3_K27me3.
We measured methylation at 7 CpG sites in CpG island 63 (CGI: 63, UCSC Browser) for p16/CDKN2A and 19 CpG sites in CGI:176 for p14ARF/CDKN2A (Fig. 1) and we found no evidence for promoter hypermethylation in either region among 114 CMM cases regardless of their CDKN2A germline mutation status. This finding was validated using differential methylation hybridization (DMH)7 of the same DNA samples, in which methylation levels corresponding to four CGIs (CGIs: 176, 63, 35 and 32, Fig. 1) spanning the CDKN2A gene were shown to be similar in cases and controls. We then measured gene expression levels of p16/CDKN2A and p14ARF/CDKN2A performed on total RNA co-extracted from the same PBMC samples (Hyland et al., in preparation). We observed no case-control differences in CDKN2A mRNA (data not shown), further indicating that germline epimutations of CDKN2A do not explain melanoma susceptibility in our melanoma-prone families. Our findings are consistent with a previous report that hypermethylation of CDKN2A was absent in Dutch patients with familial melanoma.8
Among the other 13 genes we evaluated, we observed reduced promoter methylation in CMM cases for a number of genes, however, the overall promoter methylation levels were very low for most genes (Table 2). To determine the functional relevance of the observed methylation changes, we examined the mRNA levels of these genes, and the expected negative correlation between promoter methylation and gene expression was only observed for TNFRS10C (r = –0.26, P = 0.011 among all unaffected individuals). We further observed the expected negative correlation between TNFRSF10C promoter methylation and gene expression in a small number of fibroblasts (n = 25) and a strong positive correlation between DNA methylation in matched fibroblasts and PBMC pairs (n = 8) (data not shown).
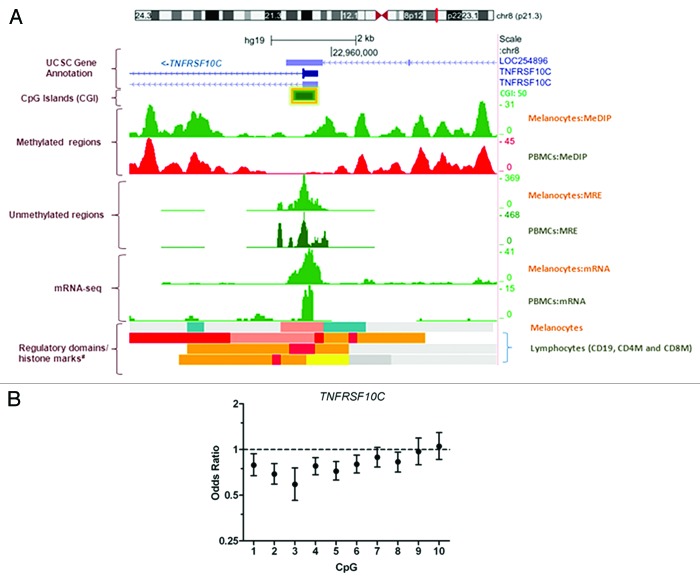
Compared with unaffected individuals, the overall promoter methylation of TNFRSF10C (Fig. 2A) was significantly reduced in CMM cases (odds ratio [OR] = 0.63, 95% confidence interval [CI] = 0.50–0.80, P < 0.001) after adjusting for age, sex, CDKN2A mutation status and familial correlation using a generalized estimating equation (GEE) with the independence working correlation matrix (Table S2 and Supplemental Materials).9 The association remained significant (OR = 0.64, 95% CI = 0.47–0.87, P = 0.004) with the additional adjustment of the number of moles and hair color (Table S2). Although based on a small number of spouses, similar results were obtained when comparing cases to spouses only (data not shown). Reduced methylation of TNFRSF10C in CMM cases was observed for 7 of 10 individual CpG sites (Fig. 2B). In addition, we observed a significant increased expression of TNFRSF10C (fold change [fc] = 1.27, P = 0.048) in cases compared with controls after controlling for age, sex, CDKN2A mutation status and familial correction in the variance computation, and this finding was further technically validated using qRT-PCR of RNAs from 112 cases and 110 controls (fc 3.4, P = 0.007). We also found a significant interaction between CDKN2A germline mutation and TNFRSF10C methylation levels (P = 0.009) and data from CDKN2A-stratified analyses showed that reduced promoter methylation levels for TNFRSF10C were only seen in CDKN2A mutation negative cases (OR = 0.47, 95% CI = 0.31–0.72, P = 0.0005) but not in mutation positive cases (OR = 0.97, 95% CI = 0.62–1.52, P = 0.88). Interestingly, in a previous analysis of genetic variants in these families, we showed that rs10866820 in the 3′ gene region of TNFRSF10C was associated with CMM risk,5 and the association was also stronger in CDKN2A-negative families. This SNP is located in a DNaseI site in melanocytes10 and is strongly predicted to alter transcription factor binding in this region (http://www.regulomedb.org).
Figure 2. (A) Genome Browser (http://genome.ucsc.edu/) image of the TNFRSF10C promoter region on human assembly hg19 based on NIH Epigenomics Roadmap data.10 The promoter of TNFRF10C (CGI:50) analyzed in this study is highlighted with a yellow box. MeDIP, methylated DNA immunoprecipitation; MRE, methylation-sensitive restrictive enzyme; Melanocytes, normal primary penile foreskin melanocytes (UCSF-UBC-USC and UCSF-UBC); PBMCs, peripheral blood mononuclear cells (UCSF-UBC-UCD and UCSF UBC); lymphocytes, CD19, CD4, and CD8 cells (NIH Epigenomics Roadmap data). Regulatory domains (chromatin state segmentation using a hidden Markov Model [ChromHMM]) and core histone marks: red, active transcriptional start site (TSS); dark salmon, poised TSS; crimson, flanking TSS; orange, active to weak enhancer; yellow, poised enhancer; cadet blue, H3K9me3_K27me3. (B) Odds ratios showing association between methylation at each CpG site in the TNFRSF10C promoter (CGI 50) and CMM status adjusting for age at blood draw, sex and CDKN2A status, and accounting for family correlation in the variance.
TNFRSF10C (and TNFRSF10D) are truncated “decoy” receptors that bind tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), but do not induce apoptosis, and thus protect cells from TRAIL-induced apoptosis.11,12 In our study, TNFRSF10C mRNA was significantly differentially expressed in PBMCs in CMM cases compared with unaffected individuals, whereas TNFRSF10D and the TRAIL receptor mRNAs TNFRSF10A and TNFRSF10B were similar in cases and controls (data not shown). Previous studies showed that melanoma cell lines overexpressing TNFRSF10C and TNFRSF10D exhibited increased resistance to TRAIL-induced apoptosis.11 Based on NIH Epigenomics Roadmap data,10 methylation marks (and chromatin marks) at the TNFRSF10C promoter, as well as TNFRSF10C mRNA levels, are similar in normal melanocytes and PBMCs (Fig. 2A). Thus, collectively this data might suggest that reduced methylation and increased expression of TNFRSF10C in blood may characterize reduced apoptosis or prolonged survival of PBMCs and/or melanocytes in our melanoma-prone families. Arguably, the observed difference in TNFRSF10C promoter methylation and gene expression between cases and controls could be attributable to a general immune response to melanoma in our cases or differences in the cell composition of PBMCs. However, with the exception of CD4, CD6, and CD14 mRNAs, which are positive markers for CD4+, CD8+, T lymphocytes and mixed PBMC-monocyte abundance, respectively, there were no significant differences in expression levels of 57 other mRNAs used to examine PBMC cell composition13 between CMM cases and controls (data not shown). Also, we evaluated gene-specific promoter methylation and expression directly from the same PBMCs, thereby circumventing any potential artifacts caused by cell culture.
In conclusion, our data suggest that constitutional epimutation of the CDKN2A gene is rare in our melanoma-prone families. Reduced methylation of the TNFRSF10C promoter in blood was significantly associated with the risk of CMM. The associated promoter demethylation of TNFRSF10C and concomitant increase in mRNA levels among CMM cases may cause reduced apoptosis and prolonged cell survival particularly in CDKN2A mutation negative cases. Our study is limited by the small sample size, low methylation levels for most genes examined, and the lack of pre-diagnostic collection of DNA. Future research in large prospective studies are required to validate these findings and to investigate the functional significance of the TNFRSF10C demethylation and its expression in both blood and melanoma tissues.
Materials and Methods
Study population
The study population of this family study has been previously described in detail.14,15 In brief, US families with at least two living first degree relatives with a history of invasive melanoma were ascertained through health care professionals or self-referrals. All participants in the study underwent a full-body skin examination to characterize phenotypes and completed risk factor questionnaires for sun-related exposures such as tanning ability. All diagnoses of melanoma were confirmed by histologic review of pathologic material and pathology reports. The current study was based on 64 families (26 families segregating CDKN2A mutations and 38 families without known mutations). All study participants were Caucasian and CMM cases and controls with and without CDKN2A mutations were selected from families based on the availability of primary frozen PBMCs. The study was approved by the National Cancer Institute Clinical Center Institutional Review Board and conducted according to the Declaration of Helsinki. Informed consent was obtained from all participants.
Gene selection
We investigated the constitutional methylation status of the p16 and p14ARF promoters of the CDKN2A locus as well a number of specific melanoma-related gene promoters in blood known to be involved in melanoma (melanoma-associated antigen 3 [MAGE-A3]) and different cellular pathways such as cell adhesion, migration and response to stress (cadherin 1 [CDH]; collagen, type I, α 2 [COL1A2]; lysyl oxidase [LOX]; heat shock protein, α-crystallin-related, B6 [HSPB6]), metal detoxification and protection against oxidative stress (metallothionein-1G [MT1G]), chromatin organization (nucleoplasmin 2 [NPM2]), cell cycle and DNA damage (DNA-damage-inducible transcript 4-like [DDIT4], phosphatase and tensin homolog [PTEN]) and apoptosis (death-associated protein kinase 1 [DAPK1], Ras association [RalGDS/AF-6] domain family member 1 [RASSF1], tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain [TNFRSF10C] and tumor necrosis factor receptor superfamily, member 10d, decoy without an intracellular domain [TNFRSF10D]). In addition to p16/CDKN2A and p14ARF/CDKN2A, the 13 melanoma-associated genes were selected based on current literature at the time of the study. Altered mRNA and protein expression of these genes have been shown to be associated with melanoma development, progression and prognosis.16-19 In addition, aberrant promoter methylation of these genes in melanoma tissues, cultured melanocytes or serum from melanoma patients has also been described.17,18,20-27
Total genomic DNA extraction
Deterioration of DNA methylation levels in cultured PBMC samples has previously been reported.28 To avoid this problem, we extracted total genomic DNA directly from cryopreserved primary PBMC cells (3–5 × 106 cells) using TRIzol® as per manufacturers’ guidelines. All extracted DNA samples were run on a 0.8% agarose gel to assess integrity and purity, and concentration was determined using NanoDrop method.
Methylation pyrosequencing analyses
The Zymo Research EZ Methylation Kit was used for bisulfite modification of 500 ng-1000 ng of PBMC DNA and promoter methylation assays for each of the selected genes were performed by EpigenDx (Hopkinton, MA, USA) using a PSQ96 HS system (Biotage AB). The methylation status of each promoter region (and/or selected CpG dinucleotides) was measured across multiple CpG sites (range: 7–27 CpG sites) for each gene using commercially available assays (Qiagen, Valencia CA). Validation of each assay was previously performed using bisulfite-modified methylated control DNA and non-methylated control DNA (EpigenDx, Hopkinton, MA, USA). The methylation status at each CpG was analyzed individually as a T/C SNP using QCpG software (Pyrosequencing Qiagen) and then averaged together to provide a mean or overall percent 5-MeC for each gene promoter. Methylated and unmethylated controls were included with each batch. Percent DNA methylation within each promoter was measured for all samples and a coefficient of variation (CV) among blinded replicates (n = 12) was used to determine intra- and inter-batch variation. Individual “pyrograms” and percent of methylated DNA at each CpG site were returned for each of the 15 genes. Pyrosequencing promoter methylation values less than or equal to 5% has been reported previously for CDKN2A, CDH1 and RASSF1 in normal healthy cell/tissue types.29,30 The median CV for intra- and inter-overall methylation levels among 12 replicate samples for each gene promoter was below 5% and 9%, respectively.
Gene expression
We extracted RNA expression levels for 14 of 15 selected genes from an independently conducted gene expression microarray analysis of 93 cases and 98 unaffected individuals (Hyland et al., in preparation). Gene expression data was not available for MAGE-A3. In brief, total RNA was simultaneously co-extracted with total genomic DNA TRIzol® from all cryopreserved primary PBMC cells (3–5 × 106 cells). Each microarray experiment was performed using the SurePrint G3 Agilent expression array (GE 8x60K, Design 028004) by Oxford Gene Technology (OGT), UK according to manual instructions (G4140–90050 version 5.0.01). Agilent feature extraction software (Agilent Technologies) was used to assess fluorescent hybridization signals and to normalize signals (intra-normalization) using Linear Lowess. Gene expression analysis was performed using Agilent GeneSpring v12.6 (Agilent Technologies) and inter-array normalization was performed by baseline transformation to the median of all samples. For a number of our genes we had two probes for targetting mRNA levels. To validate TNFRSF10C gene expression, we modeled and analyzed qRT-PCR PBMC-based data using the 2−ΔΔCt method.31 All reactions were performed in triplicate using commercially available kits for TNFRSF10C (Hs00182570_m1, Applied Biosystems Inc.) and GAPDH (Hs02758991_g1) as an internal control. The N-fold differential expression of TNFRSF10C in cases (n = 112) compared with controls (n = 110) was expressed as the mean gene expression at the group level in cases normalized to GAPDH and relative to all controls.
Statistical analysis
We examined overall promoter methylation (the average methylation levels across all CpG sites) for each gene (as well as CpG-site specific methylation within each promoter) as a continuous variable and demographic or cutaneous malignant melanoma (CMM) risk factors among unaffected individuals using multivariable linear regression. Normality of residuals was assessed using the Cramer von Mises test statistic. To account for familial correlations among family members, we computed variances and P values based on a generalized estimating equations (GEEs) with the independence working correlation matrix.9 To assess our main hypothesis, we estimated odds ratios (ORs) and 95% confidence intervals (95% CIs) for the association between overall methylation at each gene promoter and CMM status using unconditional logistic regression adjusting for age at blood draw (categorical variable), sex, CDKN2A mutation status using GEE to account for familial correlation in the variance computation. We examined the association between CMM status and MAGE-A3 (X-linked) methylation in gender-stratified analyses. Associations were also evaluated separately for individuals with and without CDKN2A mutation. We used Spearman’s rank correlation coefficient to compare the overall (and CpG site-specific) methylation values for each gene promoter to mRNA expression. To examine whether gene expression levels differed between cases and controls, we used gene expression levels for each probe as a continuous variable in a linear regression model that included CMM status and was additionally adjusted for age, sex and CDKN2A status. In these models we accounted for familial correlation using the GEE approach. For qRT-PCR technical validation, a 2-sample t test was conducted to test whether mean 2-∆∆Ct TNFRSF10C levels differed between cases and controls. All tests were two-sided. Statistical analyses were performed using SAS software version 9.1 (SAS Institute) and R program language (http://www.r-project.org).
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are indebted to the participating families, whose generosity and cooperation have made our study possible. We also acknowledge Virginia Pichler, Deborah Zametkin, and Mary Fraser for their contributions to this work. This research was supported by the Intramural Research Program of the NIH, NCI, DCEG; Cancer Prevention Fellowship Program, Division of Cancer Prevention, NCI (to P.L.H); and Health and Social Care (HSC), Northern Ireland, UK (to P.L.H).
References
- 1.Goldstein AM, Tucker MA. Genetic epidemiology of cutaneous melanoma: a global perspective. Arch Dermatol. 2001;137:1493–6. doi: 10.1001/archderm.137.11.1493. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein AM. Familial melanoma, pancreatic cancer and germline CDKN2A mutations. Hum Mutat. 2004;23:630. doi: 10.1002/humu.9247. [DOI] [PubMed] [Google Scholar]
- 3.Chan TL, Yuen ST, Kong CK, Chan YW, Chan AS, Ng WF, Tsui WY, Lo MW, Tam WY, Li VS, et al. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat Genet. 2006;38:1178–83. doi: 10.1038/ng1866. [DOI] [PubMed] [Google Scholar]
- 4.Hitchins MP, Wong JJ, Suthers G, Suter CM, Martin DI, Hawkins NJ, Ward RL. Inheritance of a cancer-associated MLH1 germ-line epimutation. N Engl J Med. 2007;356:697–705. doi: 10.1056/NEJMoa064522. [DOI] [PubMed] [Google Scholar]
- 5.Yang XR, Pfeiffer RM, Wheeler W, Yeager M, Chanock S, Tucker MA, Goldstein AM. Identification of modifier genes for cutaneous malignant melanoma in melanoma-prone families with and without CDKN2A mutations. Int J Cancer. 2009;125:2912–7. doi: 10.1002/ijc.24622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browne EP, Punska EC, Lenington S, Otis CN, Anderton DL, Arcaro KF. Increased promoter methylation in exfoliated breast epithelial cells in women with a previous breast biopsy. Epigenetics. 2011;6:1425–35. doi: 10.4161/epi.6.12.18280. [DOI] [PubMed] [Google Scholar]
- 7.Tran A, Escovedo C, Migdall-Wilson J, Chou AP, Chen W, Cloughesy T, Nelson S, Lai A. In Silico Enhanced Restriction Enzyme Based Methylation Analysis of the Human Glioblastoma Genome Using Agilent 244K CpG Island Microarrays. Front Neurosci. 2009;3:57. doi: 10.3389/neuro.15.005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erlandson A, Appelqvist F, Enerbäck C. Epigenetic mutations in CDKN2A in western Swedish families with hereditary malignant melanoma. Mol Med Rep. 2008;1:89–91. [PubMed] [Google Scholar]
- 9.Liang KY, Zeger SL. Longitudinal Data-Analysis Using Generalized Linear-Models. Biometrika. 1986;73:13–22. doi: 10.1093/biomet/73.1.13. [DOI] [Google Scholar]
- 10.Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–9. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang XD, Franco AV, Nguyen T, Gray CP, Hersey P. Differential localization and regulation of death and decoy receptors for TNF-related apoptosis-inducing ligand (TRAIL) in human melanoma cells. J Immunol. 2000;164:3961–70. doi: 10.4049/jimmunol.164.8.3961. [DOI] [PubMed] [Google Scholar]
- 12.Hersey P, Zhang XD. How melanoma cells evade trail-induced apoptosis. Nat Rev Cancer. 2001;1:142–50. doi: 10.1038/35101078. [DOI] [PubMed] [Google Scholar]
- 13.Palmer C, Diehn M, Alizadeh AA, Brown PO. Cell-type specific gene expression profiles of leukocytes in human peripheral blood. BMC Genomics. 2006;7:115. doi: 10.1186/1471-2164-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldstein AM, Landi MT, Tsang S, Fraser MC, Munroe DJ, Tucker MA. Association of MC1R variants and risk of melanoma in melanoma-prone families with CDKN2A mutations. Cancer Epidemiol Biomarkers Prev. 2005;14:2208–12. doi: 10.1158/1055-9965.EPI-05-0321A. [DOI] [PubMed] [Google Scholar]
- 15.Goldstein AM, Struewing JP, Chidambaram A, Fraser MC, Tucker MA. Genotype-phenotype relationships in U.S. melanoma-prone families with CDKN2A and CDK4 mutations. J Natl Cancer Inst. 2000;92:1006–10. doi: 10.1093/jnci/92.12.1006. [DOI] [PubMed] [Google Scholar]
- 16.Roeder C, Schuler-Thurner B, Berchtold S, Vieth G, Driesch Pv, Schuler G, Lüftl M. MAGE-A3 is a frequent tumor antigen of metastasized melanoma. Arch Dermatol Res. 2005;296:314–9. doi: 10.1007/s00403-004-0527-7. [DOI] [PubMed] [Google Scholar]
- 17.Marini A, Mirmohammadsadegh A, Nambiar S, Gustrau A, Ruzicka T, Hengge UR. Epigenetic inactivation of tumor suppressor genes in serum of patients with cutaneous melanoma. J Invest Dermatol. 2006;126:422–31. doi: 10.1038/sj.jid.5700073. [DOI] [PubMed] [Google Scholar]
- 18.Lahtz C, Stranzenbach R, Fiedler E, Helmbold P, Dammann RH. Methylation of PTEN as a prognostic factor in malignant melanoma of the skin. J Invest Dermatol. 2010;130:620–2. doi: 10.1038/jid.2009.226. [DOI] [PubMed] [Google Scholar]
- 19.Zhuang L, Lee CS, Scolyer RA, McCarthy SW, Zhang XD, Thompson JF, Screaton G, Hersey P. Progression in melanoma is associated with decreased expression of death receptors for tumor necrosis factor-related apoptosis-inducing ligand. Hum Pathol. 2006;37:1286–94. doi: 10.1016/j.humpath.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 20.Sigalotti L, Fratta E, Coral S, Tanzarella S, Danielli R, Colizzi F, Fonsatti E, Traversari C, Altomonte M, Maio M. Intratumor heterogeneity of cancer/testis antigens expression in human cutaneous melanoma is methylation-regulated and functionally reverted by 5-aza-2′-deoxycytidine. Cancer Res. 2004;64:9167–71. doi: 10.1158/0008-5472.CAN-04-1442. [DOI] [PubMed] [Google Scholar]
- 21.Liu S, Ren S, Howell P, Fodstad O, Riker AI. Identification of novel epigenetically modified genes in human melanoma via promoter methylation gene profiling. Pigment Cell Melanoma Res. 2008;21:545–58. doi: 10.1111/j.1755-148X.2008.00484.x. [DOI] [PubMed] [Google Scholar]
- 22.Bonazzi VF, Nancarrow DJ, Stark MS, Moser RJ, Boyle GM, Aoude LG, Schmidt C, Hayward NK. Cross-platform array screening identifies COL1A2, THBS1, TNFRSF10D and UCHL1 as genes frequently silenced by methylation in melanoma. PLoS One. 2011;6:e26121. doi: 10.1371/journal.pone.0026121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koga Y, Pelizzola M, Cheng E, Krauthammer M, Sznol M, Ariyan S, Narayan D, Molinaro AM, Halaban R, Weissman SM. Genome-wide screen of promoter methylation identifies novel markers in melanoma. Genome Res. 2009;19:1462–70. doi: 10.1101/gr.091447.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furuta J, Umebayashi Y, Miyamoto K, Kikuchi K, Otsuka F, Sugimura T, Ushijima T. Promoter methylation profiling of 30 genes in human malignant melanoma. Cancer Sci. 2004;95:962–8. doi: 10.1111/j.1349-7006.2004.tb03184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mirmohammadsadegh A, Marini A, Nambiar S, Hassan M, Tannapfel A, Ruzicka T, Hengge UR. Epigenetic silencing of the PTEN gene in melanoma. Cancer Res. 2006;66:6546–52. doi: 10.1158/0008-5472.CAN-06-0384. [DOI] [PubMed] [Google Scholar]
- 26.Hoon DS, Spugnardi M, Kuo C, Huang SK, Morton DL, Taback B. Profiling epigenetic inactivation of tumor suppressor genes in tumors and plasma from cutaneous melanoma patients. Oncogene. 2004;23:4014–22. doi: 10.1038/sj.onc.1207505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spugnardi M, Tommasi S, Dammann R, Pfeifer GP, Hoon DS. Epigenetic inactivation of RAS association domain family protein 1 (RASSF1A) in malignant cutaneous melanoma. Cancer Res. 2003;63:1639–43. [PubMed] [Google Scholar]
- 28.Saferali A, Grundberg E, Berlivet S, Beauchemin H, Morcos L, Polychronakos C, Pastinen T, Graham J, McNeney B, Naumova AK. Cell culture-induced aberrant methylation of the imprinted IG DMR in human lymphoblastoid cell lines. Epigenetics. 2010;5:50–60. doi: 10.4161/epi.5.1.10436. [DOI] [PubMed] [Google Scholar]
- 29.Wong CM, Anderton DL, Smith-Schneider S, Wing MA, Greven MC, Arcaro KF. Quantitative analysis of promoter methylation in exfoliated epithelial cells isolated from breast milk of healthy women. Epigenetics. 2010;5:645–55. doi: 10.4161/epi.5.7.12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaw RJ, Liloglou T, Rogers SN, Brown JS, Vaughan ED, Lowe D, Field JK, Risk JM. Promoter methylation of P16, RARbeta, E-cadherin, cyclin A1 and cytoglobin in oral cancer: quantitative evaluation using pyrosequencing. Br J Cancer. 2006;94:561–8. doi: 10.1038/sj.bjc.6602972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.