Abstract
The microbiota that populates the intestinal tract affects many physiological processes, such as cell proliferation, epithelial barrier function, and immune responses. However, the molecular mechanisms by which the microbiota influences these events remain unknown. It was recently reported by our research group that specific taxa of intestinal bacteria induce the rapid and transient enzymatic production of reactive oxygen species (ROS) within enterocytes. Whereas NADPH oxidase 2 (Nox2) catalyzed ROS generation in response to microbial perception by bone marrow-derived phagocytes is well-studied, the function of ROS generated by Nox1 in enterocytes in response to microbial signals is not fully understood. It is established that ROS can act as signaling molecules in diverse transduction pathways by the rapid and transient oxidation of oxidant-sensitive thiol groups harbored within sensor regulatory proteins. Because commensal-bacterial-stimulated ROS generation in enterocytes has been shown to induce a wide range of physiological processes, in our recent manuscript, we proposed a paradigm wherein the influence of the microbiota on intestinal physiology is mediated in part by redox-dependant signaling.
Keywords: lactobacilli, ROS, proliferation, symbiosis, microbiota
Host-Microbe Interactions and Influences on Intestinal Physiology
Symbiotic microbes within the metazoan gut lumen have co-evolved within their hosts over millennia, with the inhabitants of the human and murine intestine being the topic of intensive research efforts over the past 10 y. The mammalian intestinal microbiota is composed of up to 100 trillion microbes from over 500 genera of bacteria from two main phyla, namely Bacteroidetes and Firmicutes.1 Deep sequencing approaches, coupled with computational taxonomy, have expanded our knowledge of the community composition of the gut microbiota.2-4 Population numbers differ considerably along the intestinal tract with approximately 103 bacteria per gram in the jejunum, 108 bacteria per gram in the ileum, and 1012 bacteria per gram within the colon.5 As well as absolute numbers, it is important to consider the ecological niche within the intestine, with many bacteria residing in the luminal fecal stream and others bound to the mucous layer or even tightly adhered to the surface of epithelial cells.6
The use of germ-free animals has enabled discovery of diverse and expanding roles for the microbiota in the modulation of host metabolic and immune functions.7 In addition, the microbiota has been shown to modulate epithelial cell homeostasis, including proliferation and survival, regulation of barrier function, and epithelial restitution.8,9 For example, small intestinal villi of germ-free mice have impaired angiogenesis10 and have a slower crypt to tip transit time.11 Colonization of germ-free mice with well-characterized symbiotic bacteria elicits host transcriptional responses, indicating the ability of the intestine to sense the presence of non-pathogenic bacteria.10 These observations are evidence for a dynamic symbiotic host-microbe relationship between the intestine and its luminal occupants. It has also become apparent that alterations in this relationship resulting from changes in the composition of the microbiota may induce or aggravate intestinal immunity, resulting in pathological conditions as is seen in inflammatory bowel disease (IBD). In addition, changes in the microbiota have been shown to be associated with conditions including celiac disease, metabolic syndrome, and the onset of obesity, autoimmune disorders such as multiple sclerosis, infectious disease such as pseudomembranous colitis, and allergic conditions such as asthma.12-14
Thus, increasing evidence shows that the microbiota beneficially contributes to intestinal health, and increased efforts have been made to exploit symbiotic bacteria by enhancing the native microbiota with exogenous viable bacteria. Such “probiotic” microorganisms may potentially be therapeutic in inflammatory and developmental disorders of the intestinal tract.15 However, little is known about how the native microbiota and probiotic organisms mechanistically influence gut biology and how the intestine perceives these microbes. This addendum summarizes recent findings by our research group showing redox-based responses within enterocytes following contact with bacteria that represents, at least in part, a plausible mechanism by which symbiotic bacteria transmits their beneficial influences on the host.
Deliberate Generation of Physiological ROS by Tissues
In macrophages and neutrophils (“professional phagocytes”), deliberate generation of high levels of ROS function as a microbicidal response. In this context, perception of prokaryotic translation products, tagged with a bacterial-specific N-formyl group—of which N-formyl methionyl-leucyl-phenylalanine (fMLF) is the archetypical example—by a distinct class of pattern recognition receptors (PRRs) called formyl peptide receptors (FPRs), triggers a signaling cascade that eventuates in catalyzed generation of ROS and bacterial killing. This so-called “oxidative burst” in phagocytes is catalyzed by an NADPH oxidase, Nox2 (formerly designated gp120phox)—a basally inactive multi-subunit complex comprised of a membrane-bound dimer of gp91phox and gp22phox.16 The NADPH oxidase family, or “Nox” enzymes, are also present in many non-phagocytic cell types, with Nox1 and Duox2 functionally expressed in intestinal epithelia where they likely mediate ROS induction in response to bacterial contact.17-19 Non-phagocyte Nox-dependent generation of ROS functions in controlling cell signal transduction and is observed after receptor activation by various growth factors in a wide range of tissues.18 Interestingly, orthologs of the Nox/NAPDH family mediate ROS generation throughout multicellular life, with recent studies demonstrating a role for ROS in the control of cellular proliferation and differentiation of Drosophila hematopoietic progenitors,20 in the control of the transition from proliferation to differentiation in the plant root,21 and for regeneration of an amputated Xenopus tadpole tail.22
Symbiotic Lactobacilli Stimulate Gut Epithelial Proliferation via Nox-Mediated Generation of Reactive Oxygen Species
In the research article featured in this commentary,23 we report that commensal bacteria, particularly members of the genus Lactobacillus, can stimulate NADPH oxidase 1 (Nox1)-dependent ROS generation and consequent cell proliferation in intestinal stem cells rapidly upon initial ingestion into the Drosophila or murine intestine. In Drosophila, six distinct bacteria were isolated and cultured (3 Gram-negative and 3 Gram-positive) from the luminal content of adult fly and gnotobiotically fed to germ-free larvae. Only Lactobacillus plantarum induced the dNox-dependent generation of cellular ROS, and ROS-dependent epithelial cell proliferation at time points up to 4 h after ingestion. This was in contrast to ingestion of the fly pathogen Erwinia carotovora, which is reported to induce the generation of ROS in enterocytes at 24 h following ingestion.24 In addition, depletion of dDuox levels in fly enterocytes did not inhibit the ability of L. plantarum to elicit this response. We recapitulated these data showing specificity of lactobacilli to induce ROS generation in cultured cells, where the well-studied mammalian probiotic Lactobacillus rhamnosus GG was the most potent inducer of ROS in cultured CaCo-2 cells. Finally, using an epithelial cell-specific Nox1-deficient (B6.Nox1ΔIEC) mouse, we showed that ingestion of L. rhamnosus GG induces Nox1-dependent ROS-generation and cell proliferation in both the small intestine and in the colon. Together, the data from both Drosophila and mouse identify a highly conserved mechanism by which symbiotic microorganisms promote epithelial growth and development, and thus homeostasis. Additionally, the work suggests specific redox-mediated functions may be assigned to specific bacterial taxa and may contribute to the identification of microbes with probiotic potential.
In a complementary study, we also showed that FPR1-mediated sensing of fMLF by the intestine activates redox signaling cascades that promote restitution of an injured mucosa.25 Injury to the intestinal mucosa can occur with infection, surgical trauma, and in IBD. Restitution of mucosa following injury involves induced and coordinated proliferation and migration of intestinal epithelial cells. Our studies showed that L. rhamnosus GG or purified fMLF stimulate FPR1 on intestinal epithelial cells to activate ROS generation via enterocyte Nox1, causing rapid phosphorylation of focal adhesion kinase (FAK) and the promotion of the migration and proliferation of cells adjacent to colonic wounds.26 Our findings thus demonstrated a novel role for FPR1 in perceiving the enteric microbiota and facilitation of mucosal wound restitution.
Redox Signaling and Reactive Cysteines
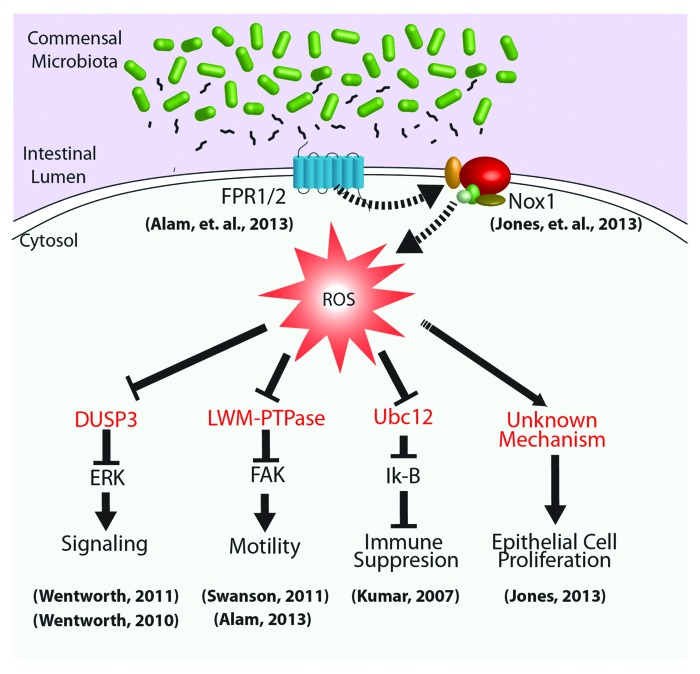
As mentioned, non-radical ROS, such as H2O2, produced by Nox enzymes are now recognized as key regulators of numerous intracellular signaling pathways.27 The physiological outputs of generated ROS depend on the intensity and/or duration of the production and the subcellular sites of generation.28-30 As short-lived molecules, ROS can have very small functional radii, which contribute to their selectivity of action. Some receptors physically interact with a ROS-generating Nox enzyme, probably to limit ROS-mediated influences to the immediate vicinity of target effector proteins.31 The mechanism by which ROS exert their effects on cell signaling circuitry is by the rapid and reversible oxidation of regulatory cysteine residues in sensor proteins.32-34 These proteins serve as ROS-sensitive signal transducers via the reversible H2O2-mediated oxidation of their active site cysteines, allowing a graded response to intracellular H2O2 concentrations. In redox-insensitive enzymes, cysteine residues are protonated at physiological pH (Cys-SH) (pKa ˜8.5), whereas so-called low-pKa cysteines exist as thiolate anions (Cys-S−) and are more readily oxidized by H2O2.34 These redox-sensitive thiolates are present under physiological conditions in a limited but increasingly recognized subset of sensor enzymes, including protein tyrosine phosphatases (PTPs),35 the lipid phosphatase (PTEN),26 MAPKP such as DUSP3,36,37 and low-molecular weight (LMW)-PTP,38 as well as enzymes involved in sumoylation and neddylation reactions39 (Fig. 1). The next logical step in our investigations will be to expand the characterization of regulatory proteins and pathways that are sensitive to cellular ROS generated in response to symbiotic bacteria.
Figure 1. Cellular signaling pathways regulated by microbial-elicited ROS generation. Commensal microbiota and/or their products within the intestinal lumen influence the activity of homeostatic processes through the regulation of cellular redox processes. For example, luminal bacteria produce and shed small formylated peptides, which are perceived via formyl peptide receptors localized to the apical surface of gut epithelia.25 These, and likely other receptors, activate NADPH oxidases that transduce microbial signals via highly localized ROS production,23 affecting the oxidation status and thus the activity of redox sensor regulatory proteins (in red), such as DUSP3, LMW-PTPase, and the Nedd8 ligase, Ubc12. Downstream basic cellular processes, including proliferation, motility, and inflammation, can thus be modulated by changes in microbial-dependent cellular redox balance.
Concluding Remarks
Epithelial generation of ROS through microbial contact is a highly conserved process with many known, well-characterized downstream responses. This mechanism is an attractive means by which a complex microbial community could influence a wide range of host signaling and homeostatic processes.40 A comprehensive understanding of this mechanism will advance understanding of the physiological importance of the microbiota in health and disease. As has been discussed, oxidative modulation of a wide range of sensor proteins is an increasingly recognized mechanism of signal transduction.30,35 Newly developed, high throughput proteomic approaches have been developed to identify reactive cysteines that can be used to screen for microbial-specific, oxidant-sensitive regulatory proteins.41 Functional analyses of microbial and ROS-dependent outcomes on multiple pathways in vivo will be challenging future work.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
R.M.J. is supported by NIH Grant R01DK098391, and A.S.N. is supported, in part, by National Institutes of Health Grant R01DK071604 and RO1AI064462.
References
- 1.Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–8. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–9. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 5.Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, Henrissat B, Coutinho PM, Minx P, Latreille P, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Ismail AS, Hooper LV. Epithelial cells and their neighbors. IV. Bacterial contributions to intestinal epithelial barrier integrity. Am J Physiol Gastrointest Liver Physiol. 2005;289:G779–84. doi: 10.1152/ajpgi.00203.2005. [DOI] [PubMed] [Google Scholar]
- 9.Madsen K, Cornish A, Soper P, McKaigney C, Jijon H, Yachimec C, Doyle J, Jewell L, De Simone C. Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology. 2001;121:580–91. doi: 10.1053/gast.2001.27224. [DOI] [PubMed] [Google Scholar]
- 10.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci U S A. 2002;99:15451–5. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc Natl Acad Sci U S A. 2005;102:99–104. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–94. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 13.Noverr MC, Huffnagle GB. Does the microbiota regulate immune responses outside the gut? Trends Microbiol. 2004;12:562–8. doi: 10.1016/j.tim.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Wills-Karp M, Santeliz J, Karp CL. The germless theory of allergic disease: revisiting the hygiene hypothesis. Nat Rev Immunol. 2001;1:69–75. doi: 10.1038/35095579. [DOI] [PubMed] [Google Scholar]
- 15.Sansonetti PJ. War and peace at mucosal surfaces. Nat Rev Immunol. 2004;4:953–64. doi: 10.1038/nri1499. [DOI] [PubMed] [Google Scholar]
- 16.Cheng G, Lambeth JD. NOXO1, regulation of lipid binding, localization, and activation of Nox1 by the Phox homology (PX) domain. J Biol Chem. 2004;279:4737–42. doi: 10.1074/jbc.M305968200. [DOI] [PubMed] [Google Scholar]
- 17.Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol. 2004;4:181–9. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- 18.Lambeth JD, Neish AS. Nox Enzymes and New Thinking on Reactive Oxygen: A Double-Edged Sword Revisited. Annu Rev Pathol. 2013 doi: 10.1146/annurev-pathol-012513-104651. [DOI] [PubMed] [Google Scholar]
- 19.Ogier-Denis E, Mkaddem SB, Vandewalle A. NOX enzymes and Toll-like receptor signaling. Semin Immunopathol. 2008;30:291–300. doi: 10.1007/s00281-008-0120-9. [DOI] [PubMed] [Google Scholar]
- 20.Owusu-Ansah E, Banerjee U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature. 2009;461:537–41. doi: 10.1038/nature08313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsukagoshi H, Busch W, Benfey PN. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell. 2010;143:606–16. doi: 10.1016/j.cell.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 22.Love NR, Chen Y, Ishibashi S, Kritsiligkou P, Lea R, Koh Y, Gallop JL, Dorey K, Amaya E. Amputation-induced reactive oxygen species are required for successful Xenopus tadpole tail regeneration. Nat Cell Biol. 2013;15:222–8. doi: 10.1038/ncb2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones RM, Luo L, Ardita CS, Richardson AN, Kwon YM, Mercante JW, Alam A, Gates CL, Wu H, Swanson PA, et al. Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J. 2013;32:3017–28. doi: 10.1038/emboj.2013.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe. 2009;5:200–11. doi: 10.1016/j.chom.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Alam A, Leoni G, Wentworth CC, Kwal JM, Wu H, Ardita CS, Swanson PA, Lambeth JD, Jones RM, Nusrat A, et al. Redox signaling regulates commensal-mediated mucosal homeostasis and restitution and requires formyl peptide receptor 1. Mucosal Immunol. 2013;2013 doi: 10.1038/mi.2013.84. Accepted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swanson PA, 2nd, Kumar A, Samarin S, Vijay-Kumar M, Kundu K, Murthy N, Hansen J, Nusrat A, Neish AS. Enteric commensal bacteria potentiate epithelial restitution via reactive oxygen species-mediated inactivation of focal adhesion kinase phosphatases. Proc Natl Acad Sci U S A. 2011;108:8803–8. doi: 10.1073/pnas.1010042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernández-García D, Wood CD, Castro-Obregón S, Covarrubias L. Reactive oxygen species: A radical role in development? Free Radic Biol Med. 2010;49:130–43. doi: 10.1016/j.freeradbiomed.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 28.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 29.Bedard K, Lardy B, Krause KH. NOX family NADPH oxidases: not just in mammals. Biochimie. 2007;89:1107–12. doi: 10.1016/j.biochi.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 30.Terada LS. Specificity in reactive oxidant signaling: think globally, act locally. J Cell Biol. 2006;174:615–23. doi: 10.1083/jcb.200605036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim SC, Tonkonogy SL, Karrasch T, Jobin C, Sartor RB. Dual-association of gnotobiotic IL-10-/- mice with 2 nonpathogenic commensal bacteria induces aggressive pancolitis. Inflamm Bowel Dis. 2007;13:1457–66. doi: 10.1002/ibd.20246. [DOI] [PubMed] [Google Scholar]
- 32.Barford D. The role of cysteine residues as redox-sensitive regulatory switches. Curr Opin Struct Biol. 2004;14:679–86. doi: 10.1016/j.sbi.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 33.Kamata H, Honda S, Maeda S, Chang L, Hirata H, Karin M. Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell. 2005;120:649–61. doi: 10.1016/j.cell.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 34.Rhee SG, Kang SW, Jeong W, Chang TS, Yang KS, Woo HA. Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr Opin Cell Biol. 2005;17:183–9. doi: 10.1016/j.ceb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 35.Chiarugi P, Buricchi F. Protein tyrosine phosphorylation and reversible oxidation: two cross-talking posttranslation modifications. Antioxid Redox Signal. 2007;9:1–24. doi: 10.1089/ars.2007.9.1. [DOI] [PubMed] [Google Scholar]
- 36.Wentworth CC, Alam A, Jones RM, Nusrat A, Neish AS. Enteric commensal bacteria induce extracellular signal-regulated kinase pathway signaling via formyl peptide receptor-dependent redox modulation of dual specific phosphatase 3. J Biol Chem. 2011;286:38448–55. doi: 10.1074/jbc.M111.268938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wentworth CC, Jones RM, Kwon YM, Nusrat A, Neish AS. Commensal-epithelial signaling mediated via formyl peptide receptors. Am J Pathol. 2010;177:2782–90. doi: 10.2353/ajpath.2010.100529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tonks NK. Redox redux: revisiting PTPs and the control of cell signaling. Cell. 2005;121:667–70. doi: 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 39.Kumar A, Wu H, Collier-Hyams LS, Hansen JM, Li T, Yamoah K, Pan ZQ, Jones DP, Neish AS. Commensal bacteria modulate cullin-dependent signaling via generation of reactive oxygen species. EMBO J. 2007;26:4457–66. doi: 10.1038/sj.emboj.7601867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee WJ. Bacterial-modulated signaling pathways in gut homeostasis. Sci Signal. 2008;1:pe24. doi: 10.1126/stke.121pe24. [DOI] [PubMed] [Google Scholar]
- 41.Sethuraman M, McComb ME, Huang H, Huang S, Heibeck T, Costello CE, Cohen RA. Isotope-coded affinity tag (ICAT) approach to redox proteomics: identification and quantitation of oxidant-sensitive cysteine thiols in complex protein mixtures. J Proteome Res. 2004;3:1228–33. doi: 10.1021/pr049887e. [DOI] [PubMed] [Google Scholar]