Abstract
Purpose
Up to half of unique genetic variants in genomic evaluations of familial cancer risk will be rare variants of uncertain significance. Classification of rare variants will be an ongoing issue as genomic testing becomes more common.
Methods
We modified standard power calculations to explore sample sizes necessary to classify and estimate relative disease risk for rare variant frequencies (0.001 to 0.00001) and varying relative risk (20 to 1.5) and using population-based and family-based designs focusing on breast and colon cancer. We required 80% power and tolerated a 10% false positive rate, since variants tested will be in known genes with high pretest probability.
Results
Using population-based strategies, hundreds to millions of cases are necessary to classify rare cancer variants. Larger samples are necessary for less frequent and less penetrant variants. Family-based strategies are robust to changes in variant frequency and require between 8 and 1175 individuals, depending on risk.
Conclusion
It is unlikely that most rare missense variants will be classifiable in the near future and accurate relative risk estimates may never be available for very rare variants. This knowledge may alter strategies for communicating information about variants of uncertain significance to patients.
Keywords: SAMPLE SIZE CALCULATION, POWER, VARIANT OF UNCERTAIN SIGNIFICANCE, STUDY DESIGN, ODDS RATIO, RELATIVE RISK, VUS, CANCER RISK
Introduction
Up to half of unique genetic variants in evaluations of familial cancer risk are variants of uncertain significance (1–3). The number of rare missense variants identified increases linearly proportionate with the length of DNA sequenced at a rate of approximately 0.008 rare variants identified per kilobase of exonic DNA sequence (4). New next-generation sequencing based clinical assays aimed at comprehensive evaluation of cancer risk genes are predicted to identify at least one rare missense variant in over half of the individuals sequenced (5). These rare variants of uncertain significance can cause confusion and patient anxiety, so definitive classification of these variants is a high priority (6–8). There have been several frameworks proposed for classifying novel variants in known cancer genes with ongoing debate about the level of evidence necessary to classify a novel variant in each category (9–13). For example, the framework outlined by Plon et al 2008 suggests a variant could be considered pathogenic if combined evidence from multiple sources indicates a 99% or greater probability that the variant causes the phenotype in question and the Partners Laboratory for Molecular Medicine has multiple criteria for pathogenic including LOD > 3 (≥10 meiosis) (9, 10). Similarly, a variant could be considered likely pathogenic if there is greater than 95% probability that variant is pathogenic or segregation is seen across more than 3 meiosis with other supporting evidence, depending on the classification framework (9, 10). Several groups have detailed specific mechanisms that might be used to combine evidence from multiple sources to classify variants (14–17).
Classification is important, but the information about risk of disease is what drives clinical decisions. This risk information intuitively appears implicit in classification; however, classification may or may not facilitate accurate risk prediction. For genes associated with familial cancer risk, understanding novel variants could be seen as a two-step process: 1) categorizing the variant in a broad class, and 2) estimating actual cancer risk conferred by the novel variant. This second step can be more challenging than simply classifying a variant, particularly for missense and splice site mutations.
In practice, cancer risk is usually inferred from literature based on other variants with the same classification, and many methods for categorizing uncertain variants explicitly assume that risk for novel variants will be identical to that for previously described, highly penetrant variants (6, 14). Grouping variants that clearly completely abrogate gene function, such as premature stop codons, early frameshift mutations, and large deletions, appears appropriate for many genes, particularly for highly penetrant cancer risk genes such as BRCA1 or MLH1. However, all variants that alter activity may not confer similar risk, such as missense variants, leaky splice site variants, or variants that occur near the 3’ end of a gene (18–20). In this paper we will first illustrate the level of risk implied by classification groups, acknowledging that there is large uncertainty surrounding this implied risk. Then we will describe the magnitude of effort that would be necessary to define better cancer risk estimates for novel, rare variants in known cancer genes through power calculations of sample sizes necessary to generate minimally useful mutation specific relative risk estimates for rare missense variants.
Current Practice in Risk Estimation: Implied Risk from Classification
Variant classification may imply different levels of risk to patients (see Figure 1). In the Plon framework (9) as implemented by current classification schemes, the “Definitely Pathogenic” classification implies risk similar to that reported in the literature for pathogenic mutations. For example, pathogenic variants in the most studied breast and colon cancer risk genes eliminate one functional copy of the gene; risk estimates from well defined cases that completely eliminate one functional copy of the gene represent a theoretical upper limit of risk conferred by a heterozygous variant in a specific gene. “Likely Pathogenic” implies that there is enough evidence to conclude the relative risk of the variant is greater than one and suggests relative risk may be similar to the upper limit defined by definitely pathogenic variants. “Uncertain Significance” implies the relative risk may be anywhere from slightly less than one to the upper limit of risk seen in pathogenic variants. “Likely Benign” implies there is enough evidence to conclude the relative risk is unlikely to be as great as the risk for pathogenic variants, and that evidence suggests similar risk to that of the general population, but that there is not enough evidence to definitively conclude that risk is similar to the general population risk. “Benign” implies relative risk near one (or very slightly less than one as these individuals lack risk conferred by reported variants). Explicitly illustrating this implied risk framework may be a useful to genetic counselors for helping patients visualize and understand variant categories (Figure 1).
Figure 1.
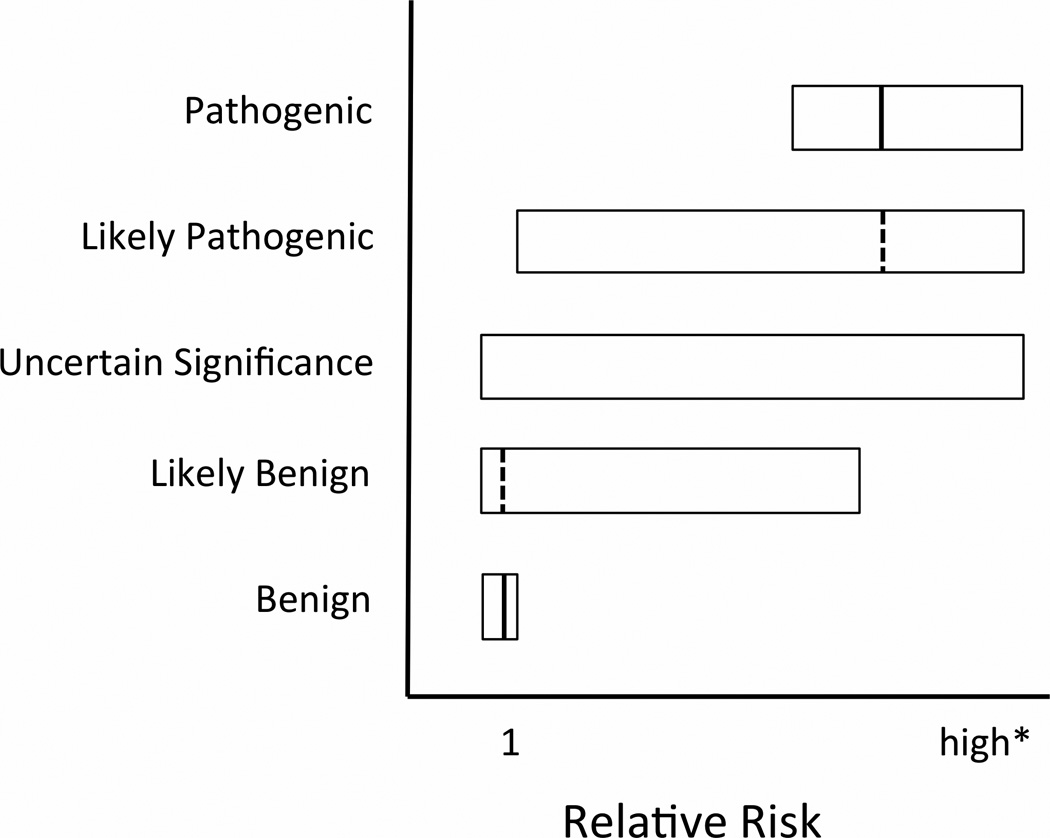
Visualization of current standard of care – Implied cancer relative risk from variant classification for dominant diseases with incomplete penetrance. Boxes indicate confidence intervals for relative risk. Solid vertical lines represent point estimates for relative risk for which data exists. Dotted vertical lines represent assumed point estimates not supported by independent, variant-specific studies. * High risk is specific to disease and gene and is defined by variants that completely eliminate one functional copy of the gene; this is the theoretical upper limit of risk conferred by a heterozygous variant in a specific gene.
This display of implied risk illustrates how simple classification can be suboptimal for patient management because of the high degree of uncertainty in implied risk for missense and splice variants, even those classified as likely pathogenic or likely benign. Variant specific relative risk estimates, beyond classification, allow quantitative estimates of outcome probabilities that are necessary for rational medical planning. Current studies indicate that risk conferred by different missense mutations can vary substantially (18–20). However, current classification systems often draw from many sources, including sources that provide no information about clinical outcomes or risk, such as in-silico protein predictions, in-vitro protein function studies, cross-species sequence conservation (9–11). This is likely to lead to over-estimates of risk for many rare variants and creates a genetic counseling dilemma, as low frequency missense variants may be grouped into risk categories before population or family based relative risk data is available (9, 11). Furthermore, in the setting of novel genes that have been linked with cancer and less common cancers relative risk estimates may be unavailable for any variant, even those classified as known pathogenic. In order to ascertain the magnitude of this problem, we evaluated the samples sizes that might be necessary to generate a minimally accurate relative risk estimate for hypothetical rare variants of uncertain significance using the examples of breast and colon cancer risk.
Materials and Methods
Calculation of Sample Size Needed for Minimally Useful Risk Estimates
Risk estimates often come from odds ratios generated by case-control studies since odds ratio and relative risk converge for rare diseases. Another strategy to evaluate variants is to use families with the mutation. We modified standard power calculations to explore sample sizes necessary to determine if relative risk for a novel variant is greater than 1.
We used standard formulas for calculating sample size from allele frequency modified as described in Fleiss et. al. to include continuity correction, and in the case of family data, to permit unequal numbers of affected and unaffected individuals (21, 22). The R-script that we used for calculations of population and family-based sample size is included as an online appendix to facilitate additional power calculations across a wider spectrum of allele frequency, relative risk, desired power, and ascertainment parameters.
We specifically examined variant population frequencies of 0.1%, 0.01% and 0.001%. We performed power calculations for population based case-control studies and family based linkage studies across several levels of cancer relative risk. We used relative risk of 12, 6, 3, and 1.5 for breast cancer and 20, 10, 5, and 2.5 for colon cancer. From literature we identified 12 as relative risk for established breast cancer genes (i.e. BRCA1, BRCA2) and 20 as relative risk for established colon cancer genes (i.e. MLH1), then used regular fractions of these to explore sample size over the spectrum of possible risk (23–25). We assumed breast cancer cumulative incidence of 0.08 and colon cancer cumulative incidence of 0.03 for individuals between 40 and 70 likely to be included in this type of study.
Population-based sample size calculation
Because variants of clinical interest will be in known genes and will presumably have in-silico data available, we assume in-silico data in known cancer genes is equivalent to a pretest probability of 0.9, and we use a Bayesian approach to define thresholds for power calculations similar to approaches used for variant classification in previous studies (14). Hence we used desired power of 0.8 and an alpha of 0.1, which would be consistent with a post-test probability of pathogenicity 99% for a pathogenic variant. We used a one-tailed test to calculate sample size because we are assuming that alleles increase cancer risk. We purposefully used these liberal assumptions, which result in low samples size estimates, because we are considering the situation where we desire definitive classification and a reasonable independent estimate of relative risk for rare variants in established cancer genes. The estimated relative risk will have some degree of error. More conservative assumptions would obviously result in larger sample requirements and more precise relative risk estimates, which may be desirable in certain clinical or research scenarios. If the measured relative risk is extremely high, this is not a major concern because the practical upper limit of risk is defined by well-studied, highly pathogenic variants. Similarly the statistical lower bound for relative risk is zero, but the practical lower limit is one because only elevated cancer risk is clinically actionable.
Family-based sample size calculation
For family based variant classification analysis, there have been several strategies proposed to generate likelihood ratios that can be used for multifactorial classification of rare variants (26–29). Variant classification strategies usually favor genotyping individuals with extreme phenotypes such as distant relatives with cancer at a young age. This strategy takes advantage of the fact that identifying a shared rare allele in an unlikely clinical situation can generate very large likelihood ratios with minimal genotyping. Although this strategy may work well for classifying a variant as pathogenic, it does not create information that can be used to define the relative risk conferred by the variant. Likelihood based classification studies may dramatically over-estimate risk (the winner’s curse). However, for extremely rare variants, it is unlikely that unrelated carriers can be identified. Despite its drawbacks, a family based approach may be the only way to estimate relative risk. However, in order to mitigate the probability of dramatically overestimating risk, studies of families with novel mutations should recruit individuals related to identified variant carriers without regard for disease status. As noted above for population-based studies, extreme overestimates can be avoided by capping risk at the level defined by common, highly pathogenic variants.
One efficient way to gather the most informative individuals for relative risk estimates in a family would be to iteratively genotype close relatives of individuals carrying the rare allele starting with the proband (but excluding the proband in calculations to avoid ascertainment bias). Case-family and case-family-control methods have been described previously (29–32). One would recruit all available family members of appropriate age and gender that are likely to carry the variant of interest regardless of personal cancer history. The strategy would be to ascertain genotype data for all available first- and second-degree relatives of the proband, and repeat this process for newly identified carriers branching to new first- and second-degree relatives while gathering data on disease status, but not skewing recruitment based on this data. When a variant is very rare, first-degree relatives have 50% chance and second-degree relatives have a 25% chance of being carriers. Alternatively one could only recruit first-degree relatives of identified carriers, which would require additional iterations of testing, or recruit both near and distant relatives, resulting in a lower variant frequency but potentially fewer stages of iterative recruiting. Regardless of strategy, it should be emphasized that for accurate relative risk estimates relatives must be recruited without regard for disease status. In order to calculate relative risk one must phenotype enough individuals with and without the variant and with and without the disease to generate a meaningful risk ratio.
Confidence intervals for the risk estimates could be computed using linear mixed models to account for within-family genotype and environmental cancer risk correlation (29, 33, 34). For simplicity in our calculations we assumed that non-genetic factors influencing cancer risk are uncorrelated and genetic cancer risk beyond the variant of interest is negligible. We also assumed that that the baseline cancer risk in a family is independent and identical to the population risk. This allows definition of the lower bound of sample size for risk estimates with confidence intervals small enough to classify the variant without knowing clinical details about specific families. For our analysis we assumed that equal numbers of first and second degree relatives would be genotyped, resulting in an overall rare variant frequency of 37.5% in the genotyped cohort. We used similar assumptions as we did for population-based studies, one-sided alpha of 0.1 and 80% power. As with population-based studies these are low estimates of sample size because correlation between family members would widen confidence intervals. More accurate power estimates would require more specific disease models, and sample sizes required for adequate power may be substantially higher.
Results
Population-based case-control study size necessary to define risk for a low frequency VUS
Population-based studies to categorize additional variants that may confer cancer risk will need to be increasingly large as variant frequency decreases and as relative risk decreases (Tables 1 and 2). When there is very high relative risk of disease case-control studies will yield sufficient cases to prove the variant is pathogenic, but insufficient controls to accurately calculate odds ratios because of the extremely low frequency of the mutation in controls, so using case-only analysis and known population disease frequencies from larger samples in the denominator might yield more accurate odds ratios.
Table 1.
Case/control study number consists of 50% cancer cases and 50% cancer free controls. Assuming cumulative incidence of breast cancer of 0.08 for individuals in study, α= 0.1, β = 0.2, MAF = minor allele frequency
| Subjects necessary to characterize as pathogenic | ||||
|---|---|---|---|---|
| RR | Tumor Type | MAF = 0.001 | MAF = 0.0001 | MAF = 0.00001 |
| RR = 12 | Breast | 663 | 6,544 | 65,358 |
| RR = 6 | Breast | 1,652 | 16,392 | 163,792 |
| RR = 3 | Breast | 5,491 | 54,650 | 546,238 |
| RR = 1.5 | Breast | 49,162 | 490,135 | 4,899,864 |
Table 2.
Case/control study number consists of 50% cancer cases and 50% cancer free controls. Assuming cumulative incidence of colon cancer of 0.03 for individuals in study, α= 0.1, β = 0.2, MAF = minor allele frequency
| Subjects necessary to characterize as pathogenic | ||||
|---|---|---|---|---|
| RR | Tumor Type | MAF = 0.001 | MAF = 0.0001 | MAF = 0.00001 |
| RR = 20 | Colon | 368 | 3,606 | 35,988 |
| RR = 10 | Colon | 830 | 8,204 | 81,944 |
| RR = 5 | Colon | 2,178 | 21,632 | 216,170 |
| RR = 2.5 | Colon | 8,331 | 82,959 | 829,236 |
Family-based study size necessary to define risk for a low frequency VUS
Family-based studies to categorize additional variants that may confer cancer risk will need to be increasingly large as relative risk decreases, but are robust to changes in variant frequency since rare variant carrier frequency in families is a direct function of relationship to identified carriers (Tables 3 and 4). Note that in the unrelated and familial study designs, the two groups being compared are orthogonal: in population-based studies the carrier frequency is compared in cases and controls. In families the affected frequency is compared in carriers and non-carriers.
Table 3.
| Total number of family members to be tested* |
Expected cancer cases with mutation |
Expected cancer cases not carrying mutation |
Expected number of mutation carriers without cancer |
||
|---|---|---|---|---|---|
| RR = 12 | Breast | 8 | 3 | 0 | 0 |
| RR = 6 | Breast | 32 | 6 | 2 | 6 |
| RR = 3 | Breast | 122 | 11 | 6 | 35 |
| RR = 1.5 | Breast | 1174 | 53 | 59 | 387 |
Assuming family members tested are first-degree relatives of a known carrier old enough to be at risk for cancer (total carrier frequency 0.375). Cumulative incidence of breast cancer of 0.08 for individuals in study, α= 0.1, β = 0.2
Table 4.
| Total number of family members to be tested* |
Expected cancer cases with mutation |
Expected cancer cases not carrying mutation |
Expected number of mutation carriers without cancer |
||
|---|---|---|---|---|---|
| RR = 20 | Colon | 17 | 4 | 0 | 3 |
| RR = 10 | Colon | 48 | 5 | 1 | 13 |
| RR = 5 | Colon | 137 | 8 | 3 | 44 |
| RR = 2.5 | Colon | 550 | 15 | 10 | 191 |
Assuming family members tested are first-degree relatives of a known carrier old enough to be at risk for cancer (total carrier frequency 0.375). Cumulative incidence of colon cancer of 0.03 for individuals in study, α= 0.1, β = 0.2
GALNT12 example
GALNT12 has recently been identified as a colorectal cancer risk gene, but relative risk has not been established for GALNT12 variants. There are 69 exonic missense variants identified by the exome variant server project in approximately 6,500 individuals sequenced; 33 of these were missense variants at a frequency < 0.001 of which 28 had a frequency < 0.0002 (35). Some of these rare variants may have been oversampled due to chance and have actual population frequencies that are much lower.
We recently identified an individual with a GALNT12 D303N mutation. This is present at a frequency of approximately 0.001 in both the 1000 genomes and exome variant server databases. There is limited literature that suggests this variant may be pathogenic (36, 37). However, the literature does not indicate what the relative risk or odds ratio might be for this variant, but suggests risk may be lower than that for known pathogenic mutations in well-defined hereditary non-polyposis colon cancer genes (36, 37). If the relative risk of colon cancer conferred by this variant is 5, we would expect a case-control study with approximately 1,089 cases and 1,089 cancer-free controls would have a reasonable likelihood of definitively categorizing the variant and generating a reasonable relative risk estimate (see Table 2). We would expect such a study to identify approximately 5 individuals carrying the variant among cases and 1 with the variant among controls. As noted above, using a larger control dataset, such as the exome variant server database may allow more accurate estimates of odds ratio (35). If we were to calculate risk from family based studies, and if we can successfully sample relatives of the proband such that 37.5% of genotyped individuals carry the variant in question and are old enough to be at risk of cancer, we would need to genotype 137 relatives of the proband to classify the variant and define a reasonable independent relative risk estimate. In this process we would to identify approximately 11 individuals who have had colon cancer of which 8 would be expected to carry the variant of interest and 3 would be incidental cancer cases. Despite the substantial relative risk, only a portion of the approximately 52 (8 + 44) related individuals carrying the risk variant would have developed cancer (see Table 4).
Discussion
Some patients, physicians, and genetic counselors may have the hope that many VUS will be classified in the near future (6). However, despite the liberal assumptions resulting in lower bounds on sample size estimates that we report, it appears unlikely that most very rare missense variants will be classifiable in the near future. Furthermore, accurate relative risk estimates are more challenging from an epidemiological perspective. Unfortunately, based on samples sizes necessary, independent relative risk estimates may not be available for most rare variants anytime in the foreseeable future. Functional studies will probably improve and may help with Bayesian classification of some variants; however, since functional assays are usually targeted at specific domains and typically generate likelihood ratios between 1.5 and 10, there will not be functional assays for many variants, and even when these exist some epidemiological would likely be required as additional support (17, 38, 39).
Efforts to build large shared databases of cases and population-based controls are promising and may make it possible to classify and estimate risk for the highest risk variants near 0.1% frequency in the population, such as the GALNT12 variant described above. However, the use of population based relative risk calculations may not be feasible for most rare variants. It is unlikely that the enormous research funds required could be made available to do adequate population based surveys to classify extremely rare variants, but some data may become available from pooled clinical testing institutions that are early adopters of genomic methodologies for cancer risk testing.
Family based analysis requires the same sample size regardless of variant frequency, so despite substantial limitations this may be the best strategy for classifying extremely rare missense variants, particularly if relative risk is predicted to be high. However, it will be necessary to identify many distant relatives or multiple apparently unrelated families to classify and estimate relative risk for most rare variants using families. This may be challenging as average family size has been decreasing in much of the world, knowledge of family medical history is often limited, and obtaining additional family history can be difficult due to geography, family communication, and limited availability of older records. Unfortunately, the probability of finding more than one independent family carrying a rare variant is directly proportional to variant frequency. Although this type of family based analysis might be feasible in a research setting for highly penetrant genes, in the current funding environment it is highly unlikely that grant funding will become available for classification of private mutations in already well-characterized genes. From a clinical perspective, identifying enough family members to classify and estimate risk for most rare variants will constitute a heroic genetic counseling effort and insurance coverage for such testing would be difficult to justify.
The GALNT12 D303N mutation example presented above is illustrative. Although this specific variant is common enough that it may be definitively classified relatively soon, the risk conferred by this variant may remain unclear even after the variant is definitively classified. Dozens of other rare GALNT12 missense variants have already been identified in fewer than 0.5% of individuals sequenced for this gene (35). It is clear from recent population-based exome and genome sequencing projects that the number of rare variants with potential clinical implications identified in the future will increase with the number of individuals receiving genomic testing (4, 40).
We demonstrate that generating clinically actionable estimates of relative risk for rare missense will be very challenging even after extensive efforts to categorize these as likely pathogenic or pathogenic. This demonstrates a significant limitation to personalized cancer risk estimates based on genetic information.
Supplementary Material
Acknowledgements
This manuscript was supported in-part by the following grants: U01HG006507 (GPJ), U01HG006375 (GPJ), U01HG007307 (GPJ), and HG004960 (BLB)
References
- 1.Peltomaki P, Vasen H. Mutations associated with HNPCC predisposition -- Update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20:269–276. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frank TS, Deffenbaugh AM, Reid JE, Hulick M, Ward BE, Lingenfelter B, Gumpper KL, Scholl T, Tavtigian SV, Pruss DR, Critchfield GC. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol. 2002;20:1480–1490. doi: 10.1200/JCO.2002.20.6.1480. [DOI] [PubMed] [Google Scholar]
- 3.Goldgar DE, Easton DF, Byrnes GB, Spurdle AB, Iversen ES, Greenblatt MS. Genetic evidence and integration of various data sources for classifying uncertain variants into a single model. Hum Mutat. 2008;29:1265–1272. doi: 10.1002/humu.20897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tennessen JA, Bigham AW, O'Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walsh T, Lee MK, Casadei S, Thornton AM, Stray SM, Pennil C, Nord AS, Mandell JB, Swisher EM, King MC. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107:12629–12633. doi: 10.1073/pnas.1007983107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller-Samuel S, MacDonald DJ, Weitzel JN, Santiago F, Martino MA, Namey T, Augustyn A, Mueller R, Forman A, Bradbury AR, Morris GJ. Variants of uncertain significance in breast cancer-related genes: real-world implications for a clinical conundrum. Part one: clinical genetics recommendations. Semin Oncol. 2011;38:469–480. doi: 10.1053/j.seminoncol.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 7.O'Neill SC, Rini C, Goldsmith RE, Valdimarsdottir H, Cohen LH, Schwartz MD. Distress among women receiving uninformative BRCA1/2 results: 12-month outcomes. Psychooncology. 2009;18:1088–1096. doi: 10.1002/pon.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Culver J, Brinkerhoff C, Clague J, Yang K, Singh K, Sand S, Weitzel J. Variants of uncertain significance in BRCA testing: evaluation of surgical decisions, risk perception, and cancer distress. Clin Genet. 2013;17:12097. doi: 10.1111/cge.12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plon SE, Eccles DM, Easton D, Foulkes WD, Genuardi M, Greenblatt MS, Hogervorst FB, Hoogerbrugge N, Spurdle AB, Tavtigian SV. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat. 2008;29:1282–1291. doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rhem HL. LABORATORY FOR MOLECULAR MEDICINE VARIANT CLASSIFICATION RULES. Cambridge, MA: Partners Laboratory for Molecular Medicine; 2012. [cited 2013 July 17]; Available from: http://pcpgm.partners.org/sites/default/files/LMM/Resources/LMM_VariantClassification_05.26.11.pdf. [Google Scholar]
- 11.Bell J, Bodmer D, Sistermans E, Ramsden SC. Practice guidelines for the Interpretation and Reporting of Unclassified Variants (UVs) in Clinical Molecular Genetics. UK clinical Molecular Genetics Society; January 11, 2008: UK Clinical Molecular Genetics Society and Dutch Society of Clinical Genetics Laboratory Specialists. 2008 [Google Scholar]
- 12.Lindor NM, Goldgar DE, Tavtigian SV, Plon SE, Couch FJ. BRCA1/2 sequence variants of uncertain significance: a primer for providers to assist in discussions and in medical management. Oncologist. 2013;18:518–524. doi: 10.1634/theoncologist.2012-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorschner MO, Amendola LM, Turner EH, Robertson PD, Shirts BH, Gallego CJ, Bennett RL, Jones KL, Tokita MJ, Bennett JT, Kim JH, Rosenthal EA, Kim DS, Tabor HK, Bamshad MJ, Motulsky AG, Scott CR, Pritchard CC, Walsh T, Burke W, Raskind WH, Byers P, Hisama FM, Nickerson DA, Jarvik GP. Actionable, pathogenic incidental findings in 1,000 participants' exomes. Am J Hum Genet. 2013;93:631–640. doi: 10.1016/j.ajhg.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson BA, Goldgar DE, Paterson C, Clendenning M, Walters R, Arnold S, Parsons MT, Michael DW, Gallinger S, Haile RW, Hopper JL, Jenkins MA, Lemarchand L, Lindor NM, Newcomb PA, Thibodeau SN, Young JP, Buchanan DD, Tavtigian SV, Spurdle AB. A multifactorial likelihood model for MMR gene variant classification incorporating probabilities based on sequence bioinformatics and tumor characteristics: a report from the Colon Cancer Family Registry. Hum Mutat. 2013;34:200–209. doi: 10.1002/humu.22213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bayrak-Toydemir P, McDonald J, Mao R, Phansalkar A, Gedge F, Robles J, Goldgar D, Lyon E. Likelihood ratios to assess genetic evidence for clinical significance of uncertain variants: hereditary hemorrhagic telangiectasia as a model. Exp Mol Pathol. 2008;85:45–49. doi: 10.1016/j.yexmp.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Spurdle AB. Clinical relevance of rare germline sequence variants in cancer genes: evolution and application of classification models. Curr Opin Genet Dev. 2010;20:315–323. doi: 10.1016/j.gde.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 17.Lindor NM, Guidugli L, Wang X, Vallee MP, Monteiro AN, Tavtigian S, Goldgar DE, Couch FJ. A review of a multifactorial probability-based model for classification of BRCA1 and BRCA2 variants of uncertain significance (VUS) Hum Mutat. 2012;33:8–21. doi: 10.1002/humu.21627. Epub 2011 Nov 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang B, Beeghly-Fadiel A, Long J, Zheng W. Genetic variants associated with breastcancer risk: comprehensive research synopsis, meta-analysis, and epidemiological evidence. Lancet Oncol. 2011;12:477–488. doi: 10.1016/S1470-2045(11)70076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vink GR, van Asperen CJ, Devilee P, Breuning MH, Bakker E. Unclassified variants in disease-causing genes: nonuniformity of genetic testing and counselling, a proposal for guidelines. Eur J Hum Genet. 2005;13:525–527. doi: 10.1038/sj.ejhg.5201379. [DOI] [PubMed] [Google Scholar]
- 20.Nieuwenhuis MH, Vasen HF. Correlations between mutation site in APC and phenotype of familial adenomatous polyposis (FAP): a review of the literature. Crit Rev Oncol Hematol. 2007;61:153–61. doi: 10.1016/j.critrevonc.2006.07.004. Epub 2006 Oct 24. [DOI] [PubMed] [Google Scholar]
- 21.Ziegler A, Konig IR. A Statistical Approach to Genetic Epidemiology. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co; 2006. p. 335. [Google Scholar]
- 22.Fleiss JL, Levin B, Paik MC. Statistical Methods for Rates and Proportions. 3rd ed. Hoboken, New Jersey: Wiley-Interscience; 2003. p. 760. [Google Scholar]
- 23.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton- Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tulinius H, Thorlacius S, Eerola H, Nevanlinna H, Syrjakoski K, Kallioniemi OP, Thompson D, Evans C, Peto J, Lalloo F, Evans DG, Easton DF. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. Epub 2003 Apr 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol. 2007;25:1329–1333. doi: 10.1200/JCO.2006.09.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quehenberger F, Vasen HF, van Houwelingen HC. Risk of colorectal and endometrial cancer for carriers of mutations of the hMLH1 and hMSH2 gene: correction for ascertainment. J Med Genet. 2005;42:491–496. doi: 10.1136/jmg.2004.024299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petersen GM, Parmigiani G, Thomas D. Missense mutations in disease genes: a Bayesian approach to evaluate causality. Am J Hum Genet. 1998;62:1516–1524. doi: 10.1086/301871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson D, Easton DF, Goldgar DE. A full-likelihood method for the evaluation of causality of sequence variants from family data. Am J Hum Genet. 2003;73:652–655. doi: 10.1086/378100. Epub 2003 Jul 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohammadi L, Vreeswijk MP, Oldenburg R, van den Ouweland A, Oosterwijk JC, van der Hout AH, Hoogerbrugge N, Ligtenberg M, Ausems MG, van der Luijt RB, Dommering CJ, Gille JJ, Verhoef S, Hogervorst FB, van Os TA, Gomez Garcia E, Blok MJ, Wijnen JT, Helmer Q, Devilee P, van Asperen CJ, van Houwelingen HC. A simple method for co-segregation analysis to evaluate the pathogenicity of unclassified variants; BRCA1 and BRCA2 as an example. BMC Cancer. 2009:9. doi: 10.1186/1471-2407-9-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao LP, Aragaki C, Hsu L, Potter J, Elston R, Malone KE, Daling JR, Prentice R. Integrated designs for gene discovery and characterization. J Natl Cancer Inst Monogr. 1999:71–80. doi: 10.1093/oxfordjournals.jncimonographs.a024229. [DOI] [PubMed] [Google Scholar]
- 30.Cui JS, Spurdle AB, Southey MC, Dite GS, Venter DJ, McCredie MR, Giles GG, Chenevix- Trench G, Hopper JL. Regressive logistic and proportional hazards disease models for within-family analyses of measured genotypes, with application to a CYP17 polymorphism and breast cancer. Genet Epidemiol. 2003;24:161–172. doi: 10.1002/gepi.10222. [DOI] [PubMed] [Google Scholar]
- 31.Jenkins MA, Croitoru ME, Monga N, Cleary SP, Cotterchio M, Hopper JL, Gallinger S. Risk of colorectal cancer in monoallelic and biallelic carriers of MYH mutations: a population-based case-family study. Cancer Epidemiol Biomarkers Prev. 2006;15:312–314. doi: 10.1158/1055-9965.EPI-05-0793. [DOI] [PubMed] [Google Scholar]
- 32.Jenkins MA, Baglietto L, Dowty JG, Van Vliet CM, Smith L, Mead LJ, Macrae FA, St John DJ, Jass JR, Giles GG, Hopper JL, Southey MC. Cancer risks for mismatch repair gene mutation carriers: a population-based early onset case-family study. Clin Gastroenterol Hepatol. 2006;4:489–498. doi: 10.1016/j.cgh.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, Buckler ES. Mixed linear model approach adapted for genome-wide association studies. Nat Genet. 2010;42:355–360. doi: 10.1038/ng.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Project NGES. NHLBI Exome Sequencing Project (ESP) Exome Variant Server. Seattle, Wa: University of Washington; 2013. Seattle GO; [updated June 7, 2013; cited 2013 August 20]; v.0.0.20:[Exome Variant Server]. Available from: http://evs.gs.washington.edu/EVS/ [Google Scholar]
- 36.Clarke E, Green RC, Green JS, Mahoney K, Parfrey PS, Younghusband HB, Woods MO. Inherited deleterious variants in GALNT12 are associated with CRC susceptibility. Hum Mutat. 2012;33:1056–1058. doi: 10.1002/humu.22088. [DOI] [PubMed] [Google Scholar]
- 37.Guda K, Moinova H, He J, Jamison O, Ravi L, Natale L, Lutterbaugh J, Lawrence E, Lewis S, Willson JK, Lowe JB, Wiesner GL, Parmigiani G, Barnholtz-Sloan J, Dawson DW, Velculescu VE, Kinzler KW, Papadopoulos N, Vogelstein B, Willis J, Gerken TA, Markowitz SD. Inactivating germ-line and somatic mutations in polypeptide N19 acetylgalactosaminyltransferase 12 in human colon cancers. Proc Natl Acad Sci U S A. 2009;106:12921–12925. doi: 10.1073/pnas.0901454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bouwman P, van der Gulden H, van der Heijden I, Drost R, Klijn CN, Prasetyanti P, Pieterse M, Wientjens E, Seibler J, Hogervorst FB, Jonkers J. A high-throughput functional complementation assay for classification of BRCA1 missense variants. Cancer Discov. 2013;23:23. doi: 10.1158/2159-8290.CD-13-0094. [DOI] [PubMed] [Google Scholar]
- 39.Tram E, Savas S, Ozcelik H. Missense variants of uncertain significance (VUS) altering the phosphorylation patterns of BRCA1 and BRCA2. PLoS One. 2013;8:e62468. doi: 10.1371/journal.pone.0062468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Consortium GP. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


