Abstract
Apomixis (asexual seed formation) is the result of a plant gaining the ability to bypass the most fundamental aspects of sexual reproduction: meiosis and fertilization. Without the need for male fertilization, the resulting seed germinates a plant that develops as a maternal clone. This dramatic shift in reproductive process has been documented in many flowering plant species, although no major seed crops have been shown to be capable of apomixis. The ability to generate maternal clones and therefore rapidly fix desirable genotypes in crop species could accelerate agricultural breeding strategies. The potential of apomixis as a next-generation breeding technology has contributed to increasing interest in the mechanisms controlling apomixis. In this review, we discuss the progress made toward understanding the genetic and molecular control of apomixis. Research is currently focused on two fronts. One aims to identify and characterize genes causing apomixis in apomictic species that have been developed as model species. The other aims to engineer or switch the sexual seed formation pathway in non-apomictic species, to one that mimics apomixis. Here we describe the major apomictic mechanisms and update knowledge concerning the loci that control them, in addition to presenting candidate genes that may be used as tools for switching the sexual pathway to an apomictic mode of reproduction in crops.
Keywords: apomixis, sexual reproduction, epigenetic, apomeiosis, diplospory, apospory
WITHIN flowering plants (angiosperms), reproduction through seeds occurs by sexual and surprisingly, by asexual pathways. The former generates variation, while asexual seed reproduction (apomixis) produces genetically identical progeny. (See Table 1 for definitions.) The ability to produce genetically identical progeny via seed is of significant value to agriculture for its power to fix complex favorable genotypes, including the higher yields often found in F1 hybrids. Apomixis is documented in more than 120 angiosperm genera (Carman 1997); however, no major seed crop species are apomictic, and attempts to introduce the apomixis trait to crops from apomictic relatives by cross-pollination have been largely unsuccessful (Savidan 2000). Apomixis is also of interest from developmental and evolutionary perspectives. Most natural apomicts are classified as facultative because they have the ability to reproduce both sexually or asexually. To understand the different pathways of apomictic seed formation, it is instructive to first examine the sequence of events required for sexual seed formation.
Table 1. Definitions.
| Term | Definition |
|---|---|
| Apomixis | Plant asexual reproduction through seed. Progeny of an apomictic plant are genetically identical to the maternal plant. |
| Embryo sac | Multicellular female gamete-producing structure of a flowering plant. Also known as the female gametophyte. |
| Apomeiosis | Avoidance or failure of meiosis during the development of an embryo sac. |
| Diplospory | Apomeiosis pathway where a diploid embryo sac develops from the megaspore mother cell. |
| Apospory | Apomeiosis pathway where a diploid embryo sac develops from a somatic ovule cell that is not the megaspore mother cell, called the aposporous initial (AI) cell. |
| Parthenogenesis | Development of an egg found in a diplosporous or aposporous embryo sac into an embryo without fertilization. |
Sexual Seed Formation in Flowering Plants
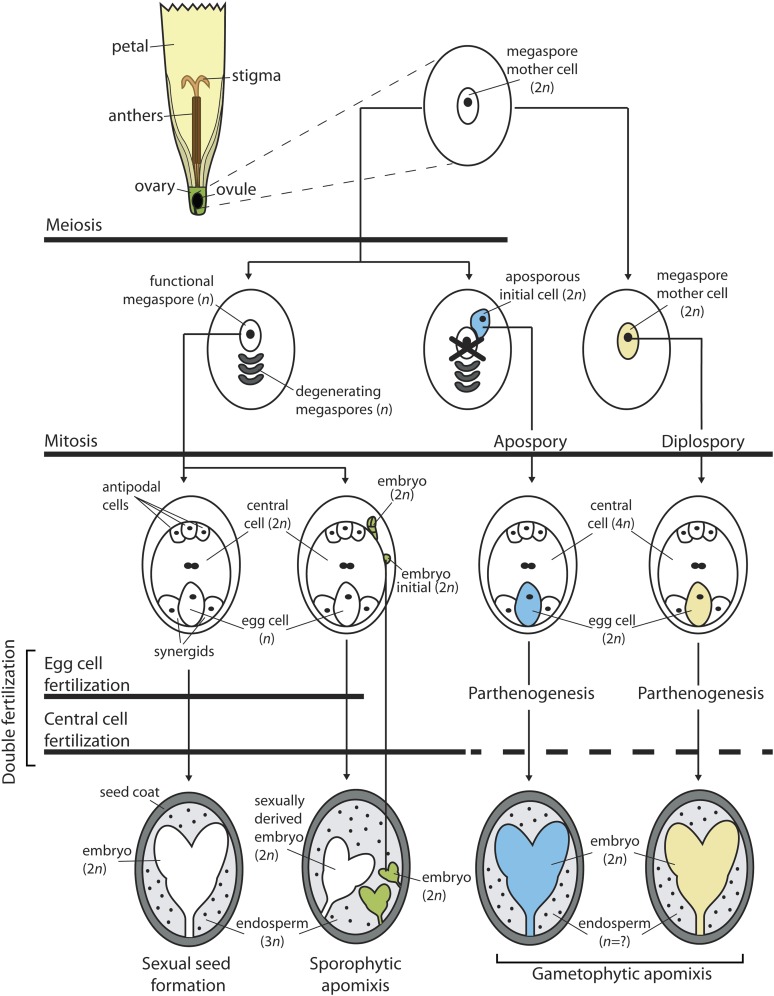
Seed formation in angiosperms begins with the developmental decision to switch from a vegetative to a reproductive mode of growth. This manifests with the formation of a flower containing male (anther) and female (ovule) reproductive organs, and within these organs, diploid gamete precursor cells form (Figure 1). Many diploid male gamete precursor cells differentiate in the anther of the flower; however within the ovule, which is the progenitor of the seed, a single female gamete precursor cell differentiates. This cell is called a megaspore mother cell. Both male and female gamete precursor cells undergo meiosis followed by mitosis to form the mature multicellular male and female gamete-containing structures, known as the pollen grain and embryo sac, respectively. Figure 1 illustrates the events leading to the formation of the most common form of embryo sac, called the Polygonum-type, which is found in >70% of flowering plants. The diploid megaspore mother cell undergoes meiosis, forming four haploid megaspores. One of these megaspores is selected to become the functional megaspore and the others degenerate. The haploid functional megaspore then undergoes three mitotic divisions without cytokinesis, to produce a coenocyte containing eight haploid nuclei surrounded by a cell wall (stage not shown in Figure 1) (reviewed by Drews and Koltunow 2011). The nuclei migrate and cellularize, producing a seven-celled mature embryo sac. It contains three antipodal cells whose function is unknown, two synergid cells that have roles in attracting the male gametes, a single egg cell, which is the progenitor of the embryo in the seed, and a large central cell containing two nuclei that fuse to form a diploid precursor nucleus of the endosperm (Figure 1). Seed development initiates following double fertilization, where the male pollen tube containing two sperm cells enters the ovule. One sperm cell fuses with the egg cell stimulating divisions and pattern-forming events that give rise to the diploid embryo. The other sperm cell fuses with the diploid central cell nucleus to initiate divisions to form the nutritive triploid endosperm, which provides essential resources to the developing embryo (Berger et al. 2008). The ovule tissues surrounding the developing embryo and endosperm contribute to the seed coat in the mature seed.
Figure 1.
Mechanisms of sexual and apomictic seed development. Seed developmental processes occur within the ovule of the flower, which is depicted as a single floret typical of Hieracium species in this figure. This diagram compares the major differences in the seed development pathway for sexual seed formation and the apomictic mechanisms of sporophytic and gametophytic apomixis. Meiosis, mitosis, and double fertilization constitute the major components of the seed formation pathway. Arrows passing through each of these components represents the involvement of a given component within a particular pathway. In the process of gametophytic apomixis, embryo sac formation can occur via either apospory or diplospory, which are distinguished by different embyro sac precursor cells. In gametophytic apomixis, embryo formation is initiated in the absence of fertilization (parthenogenesis); however, endosperm formation can occur either with or without fertilization, which is represented by a dashed line. The relative ploidy level of cells (n) is tracked for various components throughout each pathway. The ploidy level of endosperm formed through gametophytic apomixis is variable, depends on a number of factors, and is therefore represented by a question mark (?). In the depicted apospory pathway, the sexual pathway is shown to terminate once the aposporous initial cell undergoes mitosis. Different colors track the precursor cells that form the embryo for each pathway: sexual (white), sporophytic apomixis (green), diplospory (yellow), and apospory (blue).
Apomixis Mechanisms
In contrast to sexual seed formation, apomixis can occur by various mechanisms that share three common developmental components: (i) a bypass of meiosis during embryo sac formation (apomeiosis), (ii) development of an embryo independent of fertilization (a process known as parthenogenesis), and (iii) formation of viable endosperm either via fertilization-independent means or following fertilization with a sperm cell (Koltunow and Grossniklaus 2003). Derivation of the egg from a diploid maternal cell without meiotic reduction, and its subsequent fertilization-independent development into an embryo, means that the progeny derived from apomictic development are clonal and therefore genetically identical to the maternal parent.
Apomixis mechanisms are historically subdivided into two categories and classified as either gametophytic or sporophytic, based on whether the embryo develops via a gametophyte (embryo sac) or directly from diploid somatic (sporophytic) cells within the ovule (Figure 1; Nogler 1984; Koltunow 1993). During sporophytic apomixis, development of an embryo sac following the typical angiosperm sexual pathway still occurs. However, during mitosis of the functional megaspore, diploid somatic ovule cells surrounding the embryo sac differentiate and have an embryogenic cell fate. These embryo initial cells begin mitosis forming multiple globular-shaped embryos that can develop to maturity only if the sexually derived embryo sac is fertilized, as the sexual and asexual embryos share the nutrititive endosperm. Sporophytic apomixis can therefore lead to a seed containing multiple embryos (Figure 1) and is common in citrus. The sexually derived embryo may or may not mature or germinate (Koltunow et al. 1995; Koltunow et al. 1996). Sporophytic apomixis has not been extensively studied at the molecular level; however, it appears genetically complex (García et al. 1999). The remainder of this review focuses on gametophytic apomixis.
Gametophytic apomixis relates to mechanisms where an embryo sac is mitotically formed from a diploid cell in the ovule, bypassing meiosis. Another term for such mitotic embryo sac development is apomeiosis. Embryo development in gametophytic apomixis is fertilization independent whereas endosperm formation may or may not require fertilization (Figure 1). Apomeiotic embryo sac development is further subdivided into two types (diplospory and apospory) based upon the origin of the diploid precursor cell that ultimately gives rise to the mitotically derived embryo sac. In diplospory, the precursor is the megaspore mother cell (or a cell with an altered program that differentiates in the megaspore mother cell position) (Figure 1). This cell may enter meiosis and abort the process or it may immediately begin mitosis. Diplospory has been observed in species including Taraxacum officinale (dandelion), Boechera spp., Erigeron annuus, and Tripsacum dactyloides. By contrast, apospory involves development of the embryo sac via mitosis not from the megaspore mother cell, but from a diploid somatic cell positioned adjacent to the megaspore mother cell (Figure 1). This cell, termed the aposporous initial cell, undergoes mitosis and the nuclei cellularize. The mitotic events of diplospory and apospory may or may not make a seven-nucleate Polygonum-type embryo sac; however, an egg, a central cell, and synergids are typically formed (Figure 1). Depending on the species, both sexually derived and aposporous embryo sacs can coexist within the one ovule, as occurs in Brachiaria species (Araujo et al. 2000). Alternatively, development of the aposporous embryo sac may lead to the demise of the sexually derived embryo sac or pathway as occurs in aposporous Hieracium and Pennisetum species (Peel et al. 1997; Koltunow et al. 2011). Embryo development from the diploid egg formed in aposporous and diplosporous embryo sacs occurs without fertilization and the term parthenogenesis is used to describe this process (Figure 1). Endosperm development can occur without fertilization of the central cell, although this is rare, occurring predominantly in members of the daisy family (Asteraceae). Apomicts that require fertilization to produce endosperm have disturbed maternal and paternal genome contributions (m:p) in the endosperm. For example, in such apomicts, fertilization of a tetraploid central cell may lead to a 4m:1p endosperm genome ratio, in contrast to the typical 2m:1p ratio of fully sexually reproducing species. Those apomicts that require fertilization to develop endosperm have therefore developed multiple strategies to ensure seed viability and these have been discussed in previous reviews (Koltunow and Grossniklaus 2003; Curtis and Grossniklaus 2008).
Genetics and Inheritance of Apomixis
The presence of varied apomictic mechanisms and the phylogenetic positioning of apomictic species throughout many angiosperm families together imply that apomixis has evolved independently multiple times (Carman 1997; Van Dijk and Vijverberg 2005). Genetic analyses using apomicts as pollen donors in crosses with sexual individuals as maternal parents have shown apomixis to be inherited as a dominant trait. Early genetic studies proposed that a single dominant locus controlled apomixis in most studied apomictic species. For some species, it has since been revealed that many apomixis loci exhibit suppressed recombination. Studies involving genetic analyses searching for rare recombinants in suppressed regions and gamma deletion mutagenesis have revealed that in some species, the developmental components of apomixis (meiotic avoidance, parthenogenesis, and fertilization-independent endosperm development) are controlled by independent loci. For instance, in Taraxacum and Erigeron species, two independent loci have been identified that control diplospory and parthenogenesis (Van Dijk et al. 1999; Noyes and Rieseberg 2000). Similarly, apospory and parthenogenesis are controlled by two independent loci in Hypericum, Poa, Hieracium, and Cenchrus species (Albertini et al. 2001; Catanach et al. 2006; Schallau et al. 2010; Conner et al. 2013). Genetic studies in Hieracium, which is also capable of fertilization-independent endosperm formation, have revealed that this trait can also segregate independently of the other two apomictic components (Ogawa et al. 2013).
Interestingly, characterized gamma deletion mutants have revealed that sexual reproduction is the default pathway in apomictic Hieracium praealtum. A series of deletion mutants that lack the apospory locus (called LOSS OF APOMEIOSIS or LOA) and an additional locus responsible for fertilization-independent seed development (called LOSS OF PARTHENOGENESIS or LOP) were developed. Deletion of either LOA or LOP alone sees the return of the sexual pathway for that component. For example, plants with LOA deleted no longer produce diploid embryo sacs via apospory as they lack apomeiosis function. Instead, the megaspore mother cell undergoes meiosis and as aposporous initial cells are not formed, a functional haploid embryo sac develops, and the egg and central cell form an embryo and endosperm, respectively, in the absence of fertilization. Deletion of both the LOA and LOP loci results in complete reversion to sexual development (Catanach et al. 2006; Koltunow et al. 2011). Apomixis in aposporous Hieracium therefore seems to be superimposed on the sexual pathway, suggesting that apomixis may redirect the fate of cells with gametic potential, rather than being a detached and completely independent pathway (Koltunow et al. 2013). This theory is consistent with the known coexistence of apomixis and sex, suggesting that if the apomixis pathway is not initiated in an ovule, the sexual pathway remains functional. One interpretation is that apomixis may be thought of as an alteration to the spatial and temporal expression of sexual reproductive processes, where the sexual pathway is initiated at a different time and/or in a different cell. For diplospory, the megaspore mother cell appears to be specified with gametic potential; however, the sexual pathway is altered so that the events of meiosis do not initiate, or meiosis is altered so that it is not successfully completed. For apospory, an altered sexual pathway appears to be expressed in additional cells within the ovule, which results in diploid somatic cells mitotically forming an embryo sac, which is usually the role of the haploid functional megaspore in sexual reproduction. During the development of the diploid diplosporous and aposporous embryo sacs, the egg cell and central cell differentiate with a capability to directly transition into an embryo and endosperm, respectively, if seed formation is completely fertilization independent. Studies using various reproductive markers support these hypotheses (Tucker et al. 2003; Rodrigues et al. 2010).
Most apomicts are polyploid, and apomixis has previously been proposed to be a consequence of hybridization and/or genome doubling, i.e., the events of polyploidization (Carman 1997). However, polyploidization alone is not sufficient to induce apomixis, as not all polyploids are apomicts. Apomixis has previously been considered as an evolutionary dead end, as it is associated with loss of the ability to generate genetic variation through recombination, which renders apomictic populations incapable of adapting to environmental change. However, the incomplete penetrance of apomixis and maintenance of a normal sexual pathway in most apomicts provides opportunity for genetic diversification and evolution via sexual recombination (Chapman et al. 2003; Hörandl and Hojsgaard 2012).
Characteristics of Apomixis-Related Loci
Attempts to identify apomixis loci through map-based cloning approaches have proven challenging in many apomictic species, as recombination is often suppressed around apomixis loci. This distortion of recombination frequency at apomixis loci suggests strong divergence of alleles at the apomixis-related genomic regions. In the most extreme cases, the divergence has been associated with hemizygosity of the associated loci e.g., Hieracium (Okada et al. 2011), Pennisetum (Akiyama et al. 2005), and Paspalum (Calderini et al. 2006). Some apomixis loci have also been associated with heterochromatin and/or substantial repetitive sequences. In apomictic Pennisetum squamulatum, the apospory-specific genomic region (ASGR) is located at a heterochromatic and nonsynaptic telomeric region of a single chromosome (Akiyama et al. 2004). Chromosomes carrying the ASGR in Pennisetum and the LOA locus in Hieracium are associated with extensive repetitive sequence and transposon-rich regions (Akiyama et al. 2004; Okada et al. 2011). Similarly, the hemizygous apomictic controlling locus (ACL) of Paspalum, which shows strong suppression of recombination, has undergone large-scale rearrangements due to transposable elements, when compared to a syntenic region in rice (Calderini et al. 2006). These commonalities of repetitive, heterochromatic regions associated with apomixis loci have led to suggestions that such a repetitive chromosomal structure may have a functional role in apomixis. For example, one hypothesis is that the repetitive sequences may act as a sink to sequester factors involved in the sexual reproductive pathway, thereby altering the expression of sexual reproductive processes, and possibly causing apomixis (Koltunow and Grossniklaus 2003). However, recent results suggest that the abundant repetitive sequences on a chromosome carrying the Hieracium apospory locus (LOA) are not required for apospory to successfully occur. In this study, progeny arising from a cross between sexual H. pilosella and apomictic H. praealtum that are recombinant for LOA-linked markers are missing the extensive repetitive sequence structure associated with the locus, yet still remain apomictic (Kotani et al. 2013). If the repetitive chromosomal structure associated with some apomixis loci is not required for function of the locus, it is possible that these structural features and allele divergence are a consequence of asexual reproduction and suppressed recombination, which may have evolved to maintain genic elements required for apomixis. The chromosomal structure and degree of recombination may reflect the evolutionary age of apomixis in a given species. For instance, those species for which the apomixis components can be separated by recombination are likely to be evolutionarily recent apomicts [e.g., Poa (Albertini et al. 2001), Taraxacum (Vijverberg et al. 2010), Hypericum (Schallau et al. 2010), Erigeron (Noyes and Rieseberg 2000), and Panicum (Kaushal et al. 2008)]. By contrast, evolutionarily older apomicts are likely to be those characterized by apomixis associated loci found in heterochromatic regions that display suppressed recombination [e.g., Pennisetum (Ozias-Akins et al. 1998), Brachiaria (Pessino et al. 1998), Paspalum (Labombarda et al. 2002), and Tripsacum (Grimanelli et al. 1998)].
Genes Linked with Apomixis Loci
Despite the suppressed recombination observed in some apomictic species, loci genetically linked to components of apomixis have been identified in various species, and sequencing of these loci has revealed a number of genes with the potential to have critical roles in apomixis.
The ASGR of Pennisetum was originally identified following random amplified polymorphic DNA (RAPD)-based analysis of a selection of apomictic and sexual plants, which revealed a set of apomixis-specific markers that define the ASGR (Ozias-Akins et al. 1998). Shotgun sequencing of Pennisetum and Cenchrus bacterial artificial chromosome (BAC) clones from the ASGR identified 40 putative protein coding regions, two of which had sequence similarity to the rice BABY BOOM (BBM) gene (Conner et al. 2008). BBM was originally identified in Brassica napus as an AP2-domain transcription factor, and overexpression in Arabidopsis results in embryo development from vegetative tissue (Boutilier et al. 2002). The ASGR–BBM-like genes, which are present in the ASGR of both Pennisetum and Cenchrus, are therefore candidate genes with strong potential to have a role in the induction and/or maintenance of apomixis events (Conner et al. 2008).
In Hypericum, the HYPERICUM APOSPORY (HAPPY) locus was identified following an amplified fragment length polymorphism (AFLP)-based screen of a selection of apomictic and sexual plants. When an AFLP, which perfectly cosegregated with apospory, was used to screen a Hypercium BAC library, a single clone that contained, among other genes, a ubiquitin-mediated E3 ligase was identified (Schallau et al. 2010). This ARIADNE 7-like E3 ligase (HpARI) has been postulated as a strong candidate for the HAPPY locus, as one of the four HpARI alleles within the tetraploid apomict is truncated compared to sexual plants. The truncated HpARI gene may act in a dominant-negative fashion, interacting with the three remaining intact alleles. E3 ligases are involved in ubiquitin-mediated protein degradation and are known to have a role in embryo sac development. For instance, the MATH-BTB protein MAB interacts with an E3 ubiquitin ligase component (Cullin 3a) and is required for mitotic spindle function and nuclear fate in developing maize (Z. mays) embryo sacs (Juranić et al. 2012). Alteration in expression or function of the HpARI E3 ligase might therefore affect embryo sac development in Hypericum, leading to apomixis events.
Differentially Expressed Genes Associated with Apomixis
Next-generation sequencing has fueled gene expression-based approaches in a range of apomictic species, the reasoning being that gene transcripts increased or decreased in abundance in apomicts relative to sexual individuals will provide clues to the differential transcriptional pathways involved in apomixis. If apomixis is a consequence of spatial and temporal changes in expression of sexual pathway-related genes, comparative gene expression provides a method with which to identify such genes. This approach is complicated by the nature of asexual reproduction that prevents the generation of isogenic lines that differ solely with regard to reproductive mode. Nevertheless, transcriptome comparisons between apomictic and closely related sexual individuals from reproductive tissues at different developmental stages have uncovered pathways potentially central to apomixis including protein degradation, transcription, stress response, and cell-to-cell signaling (Albertini et al. 2005; Laspina et al. 2008; Sharbel et al. 2010; Silveira et al. 2012; Okada et al. 2013).
It is hoped that comparative gene expression studies in aposporous plants may reveal genes and pathways involved in processes such as cell-fate specification of the aposporous initial cell, reprogramming of a somatic cell to a germ cell fate, and degradation of sexual products (e.g., the megaspores). In the aposporous grass Brachiaria brizantha, low-depth expressed sequence tag (EST) libraries made from ovaries of sexual and apomictic individuals revealed ESTs that show similarity to genes with known involvement in female embryo sac development (Silveira et al. 2012). Further characterization of some of these ESTs identified genes that are expressed in the region where aposporous initial cells appear. These genes include a helicase (BbrizHelic), a MADS-box transcription factor (BbrizAGL6), and a stress-induced protein (BbrizSti1), suggesting that stress-related genes may play a role in the induction of aposporous initial cells (Silveira et al. 2012; Guimarães et al. 2013). In aposporous Hieracium praealtum, stress-like genes are also expressed in individual aposporous initial cells and early stage embryo sacs that contain two to four nuclei (Okada et al. 2013). In situ hybridization also reveals that at least three stress-associated genes are upregulated in the aposporous initial cell, but are undetectable in sexual ovules. It is plausible that stress-associated pathways may be involved in cell-to-cell signaling during the appearance of aposporous initial cells, or possibly in the degeneration of cells derived from the sexual pathway. Components related to the ubiquitin-mediated proteasome pathway were also identified as enriched in aposporous initial cells and the early stage aposporous embryo sac when compared to other somatic ovule cells (Okada et al. 2013). This finding, combined with the identification of the HpARI E3 ubiquitin ligase within the Hypericum apospory locus, makes the ubiquitin-mediated pathway a candidate for involvement in aposporous initial cell initiation and growth.
Other gene expression studies in aposporous species have used cDNA–AFLPs to identify differentially expressed transcripts. In Poa pratensis, this led to the identification and characterization of two candidate genes: SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE (PpSERK) and APOSTART (Albertini et al. 2004; Albertini et al. 2005). Both PpSERK and APOSTART were isolated in two copies, although further copies or alleles are present within the P. pratensis genome. PpSERK is a tyrosine kinase proposed as the switch that allows the aposporous initial cell to form and develop into an embryo sac. In apomictic P. pratensis, PpSERK is expressed in cells neighboring the megaspore mother cell, which is the region where aposporous initial cells appear (Albertini et al. 2005). This suggests PpSERK may play a role in specifying an embryo sac fate to somatic cells. APOSTART contains a lipid-binding START domain and is believed to have a role in meiosis. APOSTART may also be related to programmed cell death and the degeneration of the nonfunctional megaspores, with one of the two isolated copies (APOSTART1) overexpressed in sexual lines relative to apomicts. The Arabidopsis APOSTART1 ortholog is expressed in mature female embryo sacs and developing embryos, and the phenotype of APOSTART1/APOSTART2 double mutants suggests that this gene has a role in embryo and seed development (Barcaccia and Albertini 2013).
Within diplosporous species, comparative gene expression studies performed during embryo sac development may identify genes that allow the megaspore mother cell to avoid or modify meiosis. Few transcriptome studies comparing diplosporous and related sexual taxa have been reported, although Selva et al. (2012) identified genes differentially expressed between diplosporous and sexual Eragrostis curvula, which include retrotransposon proteins, but no obvious candidates known to affect meiosis. Sharbel et al. (2010) compared gene expression between apomictic and sexual Boechera species across four stages of development to investigate whether a heterochronic change in gene expression is the mechanism by which a diploid embryo sac can develop from the megaspore mother cell in a sexual background. This study reported a global decrease in gene expression within the apomict compared to the sexual species during the early stages of development. While not identifying any individual candidate genes, the authors hypothesize that apomeiosis is correlated with a global downregulation of gene expression during megaspore mother cell formation.
Knowledge Gained from Sexual Species
If apomixis represents an alteration in the spatial and temporal expression of the sexual pathway and recruits genes involved in sex, it follows that conversion of the sexual pathway to an apomictic one in nonapomictic species should be possible. This assumption is supported by the characterization of sexual mutants that mimic different aspects of apomixis, thus providing candidate genes that may be involved in apomixis. Recent reviews have described many of these genes and provide comprehensive lists of genes and mutants that show apomictic phenotypes (Barcaccia and Albertini 2013; Koltunow et al. 2013).
Mutants that develop embryo sacs without meiosis (apomeiosis phenotype)
Mutants that display a phenotype reminiscent of diplospory involve genes that affect meiosis. For example, the maize elongate1 mutant produces functional diploid embryo sacs by skipping meiosis II (Barrell and Grossniklaus 2005). Similarly, the dyad mutant of Arabidopsis can produce diploid embryo sacs following disrupted meiosis, where only a single equational division occurs followed by an arrest in progression (Ravi et al. 2008). Three recessive Arabidopsis mutants that have altered meiotic processes have been identified and, when combined in a triple mutant, are able to replace meiosis with mitosis, mimicking the apomeiosis phenotype. This triple mutant, called MiMe (Mitosis instead of Meiosis) combines mutants in the omission of second division 1 (Osd1), Atspo11-1, and Atrec8 genes and produces viable diploid gametes that are genetically identical to their mother (D’Erfurth et al. 2009). The MiMe and dyad mutants have both been used in an attempt to engineer apomixis in Arabidopsis as a proof of concept (Marimuthu et al. 2011). Instead of replicating parthenogenesis, MiMe and dyad mutants were crossed with an Arabidopsis line that contains a manipulated centromere-specific histone (CENH3), which has previously been demonstrated to cause genome elimination (Ravi and Chan 2010). These combinations resulted in diploid progeny that were genetically identical to the maternal parent, as the paternal genome had been eliminated. While the outcome of this synthesis is the same as in apomixis, crossing is still required. Nevertheless, this elegant system clearly demonstrates the feasibility of engineering clonal propagation by seeds into sexual species.
Specification of the functional megaspore is known to be affected by both cytokinin and auxin signaling in Arabidopsis, suggesting that both these signaling pathways may be important in aposporous initial cell initiation. Arabidopsis cytokinin receptor triple mutants show disrupted embryo sac development. The absence of a functional megaspore-specific marker in mutant ovules suggests that disruption of cytokinin signaling blocks functional megaspore specification (Cheng et al. 2013). Similarly, the auxin efflux carrier gene PIN-FORMED 1 (PIN1) is necessary for embryo sac development in Arabidopsis (Ceccato et al. 2013). Moreover, inhibition of polar auxin transport in Hieracium also affects the timing, position, and frequency of aposporous initial cell formation (Tucker et al. 2012). Alterations in cytokinin and/or auxin signaling may therefore be important in specifying an embryo sac identity to aposporous initial cells of aposporous plants.
Genes with a known role in epigenetic regulation have been implicated in the regulation of ovule cell-fate specification and embryo sac development. Mutation of ARGONAUTE genes within sexual plants can lead to changes in the number of cells that have the capacity to initiate embryo sac development (Olmedo-Monfil et al. 2010; Singh et al. 2011). ARGONAUTE (AGO) proteins carry small RNAs (sRNAs) forming complexes with other proteins that cleave mRNA or lead to RNA dependent DNA methylation (Meister 2013). In Arabidopsis, ago9 mutants appear to form multiple cells with the potential to mitotically form embryo sacs within a single ovule, a phenotype reminiscent of apospory (Olmedo-Monfil et al. 2010). Similar phenotypes are also observed in other sRNA pathway mutants including rna-dependent rna polymerase 6 (rdr6) and suppressor of gene silencing 3 (sgs3) (Olmedo-Monfil et al. 2010). Inactivation of DNA methyltransferases DMT102 and DMT103 in maize also results in apomictic-like phenotypes, including the production of diploid gametes and multiple embryo sacs within a single ovule, instead of the usual single embryo sac (Garcia-Aguilar et al. 2010). These results suggest that DNA methyltransferases may have a role in the cell-fate specification of embryo sac precursor cells. These apomixis-like phenotypes further suggest that differentiation between apomictic and sexual reproduction may be epigenetically regulated via alterations in chromatin state.
In maize, mutation of AGO104 similarly results in an apomixis-like phenotype. However, the ago104 phenotype mimics diplospory, with a single megaspore mother cell per ovule undergoing mitosis rather than meiosis, resulting in diploid gametes (Singh et al. 2011). AGO104 protein accumulates in somatic cells surrounding the megaspore mother cell, implying that a mobile signal may be responsible for repressing a somatic fate in germ cells. AGO genes have also been implicated in germ-cell identity in rice. For example, MEIOSIS ARRESTED AT LEPTOTENE1 (MEL1) is an ARGONAUTE gene expressed specifically in gamete precursor cells within ovules of rice and is essential for normal sporogenesis and meiosis in these cells (Nonomura et al. 2007).
Following the characterization of these mutants and the subsequent hypothesis that apomixis may be epigenetically regulated, approaches that compare expression of sRNAs in natural apomicts to relative sexual plants have begun, although have not yet yielded causal links between sRNAs and apomixis (Amiteye et al. 2011; Galla et al. 2013). Given the accumulation of evidence supporting the potential epigenetic regulation of apomixis obtained from sexual mutants, studies exploring differential large RNA expression combined with changes in small RNA are likely to provide important insights into the genetic regulation of apomixis.
It is possible that apomixis represents reversible, epigenetic silencing of the sexual pathway. Epigenetic regulation of apomixis is an attractive hypothesis as it potentially accounts for the facultative nature of apomixis and the ability of apomicts to revert to sexuality. In an evolutionary sense, it may be that such epigenetic changes occur following polyploidization and subsequently activate the expression of apomixis and suppression of sexual reproduction. In this scenario, all angiosperms may be considered as having “potential” for apomixis (Hörandl and Hojsgaard 2012).
Mutants capable of fertilization-independent embryo and endosperm development
Genes that may be involved in either fertilization-independent embryo or endosperm development have also been identified through sexual mutants. Interestingly, many of these genes are associated with the Polycomb-group (PcG) chromatin modeling complex. In particular, the Polycomb repressive complex 2 (PRC2) is known to have a role in suppressing seed development in the absence of fertilization. The PRC2 is conserved between plants and animals and is responsible for repressing gene expression via trimethylation of histone H3 at lysine 27 (H3K27me3). When core PRC2 genes are mutated in Arabidopsis, phenotypes of fertilization-independent seed development have been observed. For example, the fertilization-independent seed (FIS) PRC2 complex (FIS-PRC2) consists of the genes MEDEA (MEA), FIS2, FERTILIZATION-INDEPENDENT ENDOSPERM (FIE), and MULTICOPY SUPPRESSOR OF IRA1 (MSI1). Loss of function of any of these genes results in fertilization-independent endosperm development and, in the case of MSI1 mutants, parthenogenetic embryo initiation; however, viable seeds are not formed (Chaudhury et al. 1997; Ohad et al. 1999; Guitton and Berger 2005; Schmidt et al. 2013). The role of some of the FIS–PRC2 genes has been investigated in Hieracium, one of the few apomicts that develop endosperm without fertilization. Downregulation of Hieracium FIE (HFIE) does not result in fertilization-independent seed development in sexual plants, although it is required for fertilization-independent embryo and endosperm development in apomicts (Rodrigues et al. 2008). A Hieracium homolog of another FIS–PRC2 gene, HMSI1, is expressed in apomictic and sexual ovaries within Hieracium, but is unlikely to be linked to the LOSS OF PARTHENOGENESIS (LOP) locus. The authors subsequently propose that HMSI1 may be involved in fertilization-independent seed development downstream of LOP activity (Rodrigues et al. 2010). These results demonstrate that although sexual developmental mutants provide promising gene candidates that initiate apomixis-like events, the natural apomictic pathway may be somewhat more complex with additional or alternative regulators that promote maturation of viable embryos and endosperm.
Conclusions
The genetic and molecular control of apomixis is intriguing from both developmental and evolutionary perspectives, and it attracts significant interest because of its potential value to agriculture if it can be harnessed for plant breeding. The coexistence of both apomixis and sexual reproduction within individual plants suggests that apomixis is reversibly superimposed upon the sexual pathway. Studies involving natural apomicts and sexual mutants displaying apomixis-like phenotypes have led to the identification of apomixis loci and candidate genes that may be responsible for superimposing apomixis on the sexual pathway. However, causal genes have not yet been identified from apomictic species. The possibility of apomixis being epigenetically regulated is an attractive hypothesis with growing support from studies in sexual plants where mutations in epigenetic pathways lead to apomixis-like phenotypes. Epigenetic regulation will continue to be a key focus of future studies in natural apomicts. The synthesis of apomixis in crops could also be attempted using developed tools to switch the sexual pathway to an apomictic-like route. Genes identified from natural apomicts may augment this synthetic route and perhaps ultimately allow flexibility to switch apomixis on and off for plant breeding purposes.
Acknowledgments
We thank Matthew Tucker, Jennifer Taylor, Susan Johnson, Steven Henderson, and David Rabiger for critical reading and comments on the review. Research in the Anna M. G. Koltunow laboratory is currently supported by a grant from the Science and Industry Endowment Fund.
Footnotes
Communicating editor: J. Rine
Literature Cited
- Akiyama Y., Conner J. A., Goel S., Morishige D. T., Mullet J. E., et al. , 2004. High-resolution physical mapping in Pennisetum squamulatum reveals extensive chromosomal heteromorphism of the genomic region associated with apomixis. Plant Physiol. 134: 1733–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama Y., Hanna W. W., Ozias-Akins P., 2005. High-resolution physical mapping reveals that the apospory-specific genomic region (ASGR) in Cenchrus ciliaris is located on a heterochromatic and hemizygous region of a single chromosome. Theor. Appl. Genet. 111: 1042–1051 [DOI] [PubMed] [Google Scholar]
- Albertini E., Porceddu A., Ferranti F., Reale L., Barcaccia G., et al. , 2001. Apospory and parthenogenesis may be uncoupled in Poa pratensis: a cytological investigation. Sex. Plant Reprod. 14: 213–217 [DOI] [PubMed] [Google Scholar]
- Albertini E., Marconi G., Barcaccia G., Raggi L., Falcinelli M., 2004. Isolation of candidate genes for apomixis in Poa pratensis L. Plant Mol. Biol. 56: 879–894 [DOI] [PubMed] [Google Scholar]
- Albertini E., Marconi G., Reale L., Barcaccia G., Porceddu A., et al. , 2005. SERK and APOSTART: candidate genes for apomixis in Poa pratensis. Plant Physiol. 138: 2185–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiteye S., Corral J. M., Vogel H., Sharbel T. F., 2011. Analysis of conserved microRNAs in floral tissues of sexual and apomictic Boechera species. BMC Genomics 12: 500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo A. C. G., Mukhambetzhanov S., Pozzobon M. T., Santana E. F., Carneiro V. T. C., 2000. Female gametophyte development in apomictic and sexual Braciaria brizantha (Poaceae). Rev. Cytol. Biol. Veg. Botan. 23: 13–28 [Google Scholar]
- Barcaccia G., Albertini E., 2013. Apomixis in plant reproduction: a novel perspective on an old dilemma. Plant Reprod 26: 159–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell P. J., Grossniklaus U., 2005. Confocal microscopy of whole ovules for analysis of reproductive development: the elongate1 mutant affects meiosis II. Plant J. 43: 309–320 [DOI] [PubMed] [Google Scholar]
- Berger F., Hamamura Y., Ingouff M., Higashiyama T., 2008. Double fertilization: caught in the act. Trends Plant Sci. 13: 437–443 [DOI] [PubMed] [Google Scholar]
- Boutilier K., Offringa R., Sharma V. K., Kieft H., Ouellet T., et al. , 2002. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14: 1737–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderini O., Chang S. B., De Jong H., Busti A., Paolocci F., et al. , 2006. Molecular cytogenetics and DNA sequence analysis of an apomixis-linked BAC in Paspalum simplex reveal a non pericentromere location and partial microcolinearity with rice. Theor. Appl. Genet. 112: 1179–1191 [DOI] [PubMed] [Google Scholar]
- Carman J. G., 1997. Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol. J. Linn. Soc. 61: 51–94 [Google Scholar]
- Catanach A. S., Erasmuson S. K., Podivinsky E., Jordan B. R., Bicknell R., 2006. Deletion mapping of genetic regions associated with apomixis in Hieracium. Proc. Natl. Acad. Sci. USA 103: 18650–18655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccato L., Masiero S., Sinha Roy D., Bencivenga S., Roig-Villanova I., et al. , 2013. Maternal control of PIN1 is required for female gametophyte development in Arabidopsis. PLoS ONE 8: e66148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman H., Houliston G. J., Robson B., Iline I., 2003. A case of reversal: the evolution and maintenance of sexuals from parthenogenetic clones in Hieracium pilosella. Int. J. Plant Sci. 164: 719–728 [Google Scholar]
- Chaudhury A. M., Ming L., Miller C., Craig S., Dennis E. S., et al. , 1997. Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94: 4223–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.-Y., Mathews D. E., Eric Schaller G., Kieber J. J., 2013. Cytokinin-dependent specification of the functional megaspore in the Arabidopsis female gametophyte. Plant J. 73: 929–940 [DOI] [PubMed] [Google Scholar]
- Conner J. A., Goel S., Gunawan G., Cordonnier-Pratt M. M., Johnson V. E., et al. , 2008. Sequence analysis of bacterial artificial chromosome clones from the apospory-specific genomic region of Pennisetum and Cenchrus. Plant Physiol. 147: 1396–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner J. A., Gunawan G., Ozias-Akins P., 2013. Recombination within the apospory specific genomic region leads to the uncoupling of apomixis components in Cenchrus ciliaris. Planta 238: 51–63 [DOI] [PubMed] [Google Scholar]
- Curtis M. D., Grossniklaus U., 2008. Molecular control of autonomous embryo and endosperm development. Sex. Plant Reprod. 21: 79–88 [Google Scholar]
- D’Erfurth I., Jolivet S., Froger N., Catrice O., Novatchkova M., et al. , 2009. Turning meiosis into mitosis. PLoS Biol. 7: e1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews G., Koltunow A. M., 2011. The female gametophyte: the Arabidopsis book 9: e0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galla G., Volpato M., Sharbel T., Barcaccia G., 2013. Computational identification of conserved microRNAs and their putative targets in the Hypericum perforatum L. flower transcriptome. Plant Reprod. 26: 209–229 [DOI] [PubMed] [Google Scholar]
- García R., Asíns M. J., Forner J., Carbonell E. A., 1999. Genetic analysis of apomixis in Citrus and Poncirus by molecular markers. Theor. Appl. Genet. 99: 511–518 [DOI] [PubMed] [Google Scholar]
- Garcia-Aguilar M., Michaud C., Leblanc O., Grimanelli D., 2010. Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. Plant Cell 22: 3249–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimanelli D., Leblanc O., Espinosa E., Perotti E., González De León D., et al. , 1998. Mapping diplosporous apomixis in tetraploid Tripsacum: one gene or several genes? Heredity 80: 33–39 [DOI] [PubMed] [Google Scholar]
- Guimarães L., Dusi D. A., Masiero S., Resentini F., Gomes A. M., et al. , 2013. BbrizAGL6 is differentially expressed during embryo sac formation of apomictic and sexual Brachiaria brizantha plants. Plant Mol. Biol. Rep. DOI: . 10.1007/s11105-013-0618-8 [DOI] [Google Scholar]
- Guitton A. E., Berger F., 2005. Loss of function of MULTICOPY SUPPRESSOR of IRA 1 produces nonviable parthenogenetic embryos in Arabidopsis. Curr. Biol. 15: 750–754 [DOI] [PubMed] [Google Scholar]
- Hörandl E., Hojsgaard D., 2012. The evolution of apomixis in angiosperms: a reappraisal. Plant Biosyst. 146: 681–693 [Google Scholar]
- Juranić M., Srilunchang K.-o., Krohn N. G., Leljak-Levanić D., Sprunck S., et al. , 2012. Germline-specific MATH-BTB substrate adaptor MAB1 regulates spindle length and nuclei identity in maize. Plant Cell 24: 4974–4991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal P., Malaviya D. R., Roy A. K., Pathak S., Agrawal A., et al. , 2008. Reproductive pathways of seed development in apomictic guinea grass (Panicum maximum Jacq.) reveal uncoupling of apomixis components. Euphytica 164: 81–92 [Google Scholar]
- Koltunow A. M., 1993. Apomixis: embryo sacs and embryos formed without meiosis or fertilization in ovules. Plant Cell 5: 1425–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow A. M., Grossniklaus U., 2003. Apomixis: a developmental perspective. Annu. Rev. Plant Biol. 54: 547–574 [DOI] [PubMed] [Google Scholar]
- Koltunow A. M., Soltys K., Nito N., McClure S., 1995. Anther, ovule, seed, and nucellar embryo development in Citrus sinensis cv. Valencia. Can. J. Bot. 73: 1567–1582 [Google Scholar]
- Koltunow A. M., Hidaka T., Robinson S. P., 1996. Polyembryony in Citrus: accumulation of seed storage proteins in seeds and in embryos cultured in vitro. Plant Physiol. 110: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow A. M. G., Johnson S. D., Rodrigues J. C. M., Okada T., Hu Y., et al. , 2011. Sexual reproduction is the default mode in apomictic Hieracium subgenus Pilosella, in which two dominant loci function to enable apomixis. Plant J. 66: 890–902 [DOI] [PubMed] [Google Scholar]
- Koltunow A. M., Ozias-Akins P., Siddiqi I., 2013. Apomixis, pp. 83–110 in Seed Genomics, edited by Becraft P. W. Wiley, New York [Google Scholar]
- Kotani Y., Henderson S. T., Suzuki G., Johnson S. D., Okada T., et al. , 2013. The LOSS OF APOMEIOSIS (LOA) locus in Hieracium praealtum can function independently of the associated large-scale repetitive chromosomal structure. New Phytol. DOI: . 10.1111/nph.12574 [DOI] [PubMed] [Google Scholar]
- Labombarda P., Busti A., Caceres M. E., Pupilli F., Arcioni S., 2002. An AFLP marker tightly linked to apomixis reveals hemizygosity in a portion of the apomixis-controlling locus in Paspalum simplex. Genome 45: 513–519 [DOI] [PubMed] [Google Scholar]
- Laspina N. V., Vega T., Seijo J. G., González A. M., Martelotto L. G., et al. , 2008. Gene expression analysis at the onset of aposporous apomixis in Paspalum notatum. Plant Mol. Biol. 67: 615–628 [DOI] [PubMed] [Google Scholar]
- Marimuthu M. P. A., Jolivet S., Ravi M., Pereira L., Davda J. N., et al. , 2011. Synthetic clonal reproduction through seeds. Science 331: 876. [DOI] [PubMed] [Google Scholar]
- Meister G., 2013. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 14: 447–459 [DOI] [PubMed] [Google Scholar]
- Nogler G., 1984. Gametophytic apomixis, pp. 475–518 in Embryology of Angiosperms, edited byJohri B. M. Springer, Berlin [Google Scholar]
- Nonomura K. I., Morohoshi A., Nakano M., Eiguchi M., Miyao A., et al. , 2007. A germ cell-specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 19: 2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes R. D., Rieseberg L. H., 2000. Two independent loci control agamospermy (apomixis) in the triploid flowering plant Erigeron annuus. Genetics 155: 379–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa D., Johnson S. D., Henderson S. T., Koltunow A. M., 2013. Genetic separation of autonomous endosperm formation (AutE) from two other components of apomixis in Hieracium. Plant Reprod. 26: 113–123 [DOI] [PubMed] [Google Scholar]
- Ohad N., Yadegari R., Margossian L., Hannon M., Michaeli D., et al. , 1999. Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11: 407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T., Ito K., Johnson S. D., Oelkers K., Suzuki G., et al. , 2011. Chromosomes carrying meiotic avoidance loci in three apomictic eudicot Hieracium subgenus Pilosella species share structural features with two monocot apomicts. Plant Physiol. 157: 1327–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T., Hu Y., Tucker M. R., Taylor J. M., Johnson S. D., et al. , 2013. Enlarging cells initiating apomixis in Hieracium praealtum transition to an embryo sac program prior to entering mitosis. Plant Physiol. 163: 216–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo-Monfil V., Durán-Figueroa N., Arteaga-Vázquez M., Demesa-Arévalo E., Autran D., et al. , 2010. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464: 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozias-Akins P., Roche D., Hanna W. W., 1998. Tight clustering and hemizygosity of apomixis-linked molecular markers in Pennisetum squamulatum implies genetic control of apospory by a divergent locus that may have no allelic form in sexual genotypes. Proc. Natl. Acad. Sci. USA 95: 5127–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel M. D., Carman J. G., Leblanc O., 1997. Megasporocyte callose in apomictic buffelgrass, Kentucky bluegrass, Pennisetum squamulatum Fresen, Tripsacum L., and weeping lovegrass. Crop Sci. 37: 724–732 [Google Scholar]
- Pessino S. C., Evans C., Ortiz J. P. A., Armstead I., Do Valle C. B., et al. , 1998. A genetic map of the apospory-region in Brachiaria hybrids: identification of two markers closely associated with the trait. Hereditas 128: 153–158 [Google Scholar]
- Ravi M., Chan S. W. L., 2010. Haploid plants produced by centromere-mediated genome elimination. Nature 464: 615–618 [DOI] [PubMed] [Google Scholar]
- Ravi M., Marimuthu M. P. A., Siddiqi I., 2008. Gamete formation without meiosis in Arabidopsis. Nature 451: 1121–1124 [DOI] [PubMed] [Google Scholar]
- Rodrigues J. C. M., Tucker M. R., Johnson S. D., Hrmova M., Koltunow A. M. G., 2008. Sexual and apomictic seed formation in Hieracium requires the plant polycomb-group gene FERTILIZATION INDEPENDENT ENDOSPERM. Plant Cell 20: 2372–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues J. C. M., Okada T., Johnson S. D., Koltunow A. M., 2010. A MULTICOPY SUPPRESSOR OF IRA1 (MSI1) homologue is not associated with the switch to autonomous seed development in apomictic (asexual) Hieracium plants. Plant Sci. 179: 590–597 [Google Scholar]
- Savidan Y., 2000. Apomixis: Genetics and Breeding, pp. 13–86 in Plant Breeding Reviews, edited by Janick J. Wiley, New York [Google Scholar]
- Schallau A., Arzenton F., Johnston A. J., Hähnel U., Koszegi D., et al. , 2010. Identification and genetic analysis of the APOSPORY locus in Hypericum perforatum L. Plant J. 62: 773–784 [DOI] [PubMed] [Google Scholar]
- Schmidt A., Wöhrmann H. J. P., Raissig M. T., Arand J., Gheyselinck J., et al. , 2013. The Polycomb group protein MEDEA and the DNA methyltransferase MET1 interact to repress autonomous endosperm development in Arabidopsis. Plant J. 73: 776–787 [DOI] [PubMed] [Google Scholar]
- Selva J. P., Pessino S., Meier M., Echenique V., 2012. Identification of candidate genes related to polyploidy and/or apomixis in Eragrostis curvula. Am. J. Plant Sci. 3: 403–416 [Google Scholar]
- Sharbel T. F., Voigt M. L., Corral J. M., Galla G., Kumlehn J., et al. , 2010. Apomictic and sexual ovules of Boechera display heterochronic global gene expression patterns. Plant Cell 22: 655–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira E. D., Guimarães L. A., de Dusi D. M. A., da Silva F. R., Martins N. F., et al. , 2012. Expressed sequence-tag analysis of ovaries of Brachiaria brizantha reveals genes associated with the early steps of embryo sac differentiation of apomictic plants. Plant Cell Rep. 31: 403–416 [DOI] [PubMed] [Google Scholar]
- Singh M., Goel S., Meeley R. B., Dantec C., Parrinello H., et al. , 2011. Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 23: 443–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M. R., Araujo A. C. G., Paech N. A., Hecht V., Schmidt E. D. L., et al. , 2003. Sexual and apomictic reproduction in Hieracium subgenus Pilosella are closely interrelated developmental pathways. Plant Cell 15: 1524–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M. R., Okada T., Johnson S. D., Takaiwa F., Koltunow A. M. G., 2012. Sporophytic ovule tissues modulate the initiation and progression of apomixis in Hieracium. J. Exp. Bot. 63: 3229–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk P., Vijverberg K., 2005. The significance of apomixis in the evolution of the angiosperms: a reappraisal, pp. 101–116 in Plant Species-Level Systematics: New Perspectives on Pattern and Process, edited byBakker F., Chatrou L., Gravendeel B., Pelser P. Gantner Verlag, Ruggell, Lichtenstein [Google Scholar]
- Van Dijk P. J., Tas I. C. Q., Falque M., Bakx-Schotman T., 1999. Crosses between sexual and apomictic dandelions (Taraxacum). II. The breakdown of apomixis. Heredity 83: 715–721 [DOI] [PubMed] [Google Scholar]
- Vijverberg K., Milanovic-Ivanovic S., Bakx-Schotman T., Van Dijk P. J., 2010. Genetic fine-mapping of DIPLOSPOROUS in Taraxacum (dandelion; Asteraceae) indicates a duplicated DIP-gene. BMC Plant Biol. 10: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]