Abstract
The extent of the innate immune response is regulated by many positively and negatively acting signaling proteins. This allows for proper activation of innate immunity to fight infection while ensuring that the response is limited to prevent unwanted complications. Thus mutations in innate immune regulators can lead to immune dysfunction or to inflammatory diseases such as arthritis or atherosclerosis. To identify novel innate immune regulators that could affect infectious or inflammatory disease, we have taken a comparative genomics RNAi screening approach in which we inhibit orthologous genes in the nematode Caenorhabditis elegans and murine macrophages, expecting that genes with evolutionarily conserved function also will regulate innate immunity in humans. Here we report the results of an RNAi screen of approximately half of the C. elegans genome, which led to the identification of many candidate genes that regulate innate immunity in C. elegans and mouse macrophages. One of these novel conserved regulators of innate immunity is the mRNA splicing regulator Eftud2, which we show controls the alternate splicing of the MyD88 innate immunity signaling adaptor to modulate the extent of the innate immune response.
Keywords: innate immunity, C. elegans, macrophage, LPS, MyD88, complex genetics, tolerance, complex immunity, infection, resistance
PROPER regulation of the innate immune response is critical to prevent disease. Activation of the innate immune response can involve the recruitment and activation of phagocytic cells that produce antimicrobial compounds as well as cytokines and chemokines that recruit and stimulate other immune cells (Kaufmann 2004). A strong innate immune response also is required for a robust adaptive immune response (Lee and Iwasaki 2007; Iwasaki and Medzhitov 2010). Thus, defects in innate immune signaling can lead to enhanced susceptibility to bacterial or fungal pathogens in human patients (Picard et al. 2011). On the other hand chronic innate immune activation can affect a myriad of diseases with an inflammatory component including atherosclerosis, asthma, Crohn’s disease, and cancer (Cook et al. 2004; Arcaroli et al. 2005; Karin and Greten 2005; Mullick et al. 2005; Goh and Midwood 2012). It is therefore critical that innate immunity be tightly regulated for proper human health, active when needed and inactive when not. Thus, identifying novel regulators of innate immunity could aid in the development of novel therapeutic and diagnostic options for a range of infectious and inflammatory diseases (Cook et al. 2004; Connolly and O’Neill 2012).
Several organisms including the nematode Caenorhabditis elegans have been used as models to investigate innate immunity and identify novel innate immunity regulators. While C. elegans lacks an adaptive immune response, it has a robust innate immune response, and has become an established model for innate immunity studies (Engelmann and Pujol 2010; Irazoqui and Ausubel 2010; Pukkila-Worley and Ausubel 2012). In response to infection, C. elegans produces numerous antimicrobial proteins including lysozymes, saposin-domain-containing proteins, defensin-like molecules, and many others (Ewbank 2006; Ewbank and Zugasti 2011). Production of these antimicrobials occurs in tissues exposed to pathogens and is regulated by conserved innate immune signaling pathways, including a p38 MAPK cascade, an insulin signaling pathway, a TGFβ cascade, and others (Mochii et al. 1999; Mallo et al. 2002; Murphy et al. 2003; Couillault et al. 2004; Huffman et al. 2004; O’Rourke et al. 2006; Shapira et al. 2006; Troemel et al. 2006; Alper et al. 2007). Inhibition or mutation of genes in these conserved signaling pathways diminishes production of antimicrobial genes and enhances susceptibility to infection.
We previously used an RNAi screen to target all of the genes on C. elegans chromosome I to identify genes that regulate expression of a clec-85::gfp fusion in transgenic nematodes (Alper et al. 2008). clec-85 encodes a C-type lectin whose expression is induced by the bacterial pathogen Serratia marcescens (Mallo et al. 2002). clec-85 expression is regulated by several known innate immunity signaling pathways in C. elegans including the p38 MAPK, insulin signaling, and TGFβ signaling pathways in the presence of either pathogenic Pseudomonas aeruginosa or nonpathogenic Escherichia coli (Alper et al. 2007; Alper et al. 2008). We identified several genes on chromosome I that were required for robust clec-85 expression in the presence of the nonpathogenic bacterium E. coli. Follow-up studies using RNAi in mouse macrophages, and knockouts in C. elegans and mice validated many of these genes as novel regulators of innate immunity (Alper et al. 2008; De Arras et al. 2012, 2013). This demonstrated the utility of this comparative genomics approach for innate immune regulator gene discovery.
Here we extend this approach to C. elegans chromosomes II–IV, representing 10,862 genes and find 32 additional genes that, when inhibited by RNAi, affect expression of clec-85::gfp. In mouse macrophages, we find that 8 of 20 mammalian orthologs of these genes affect lipopolysaccharide (LPS)-induced cytokine production in mammalian cells. Overexpression studies confirm that two of these genes, Eftud2 and Atp6v1c1, regulate the response to LPS in murine macrophages. RNAi studies in C. elegans confirm that Eftud2 affects host defense in vivo. We further show that Eftud2, a component of the U5 small nuclear ribonucleoprotein (snRNP), which works with the rest of the spliceosome to regulate mRNA splicing (Kramer 1996; Fabrizio et al. 1997; Bartels et al. 2002, 2003; Brenner and Guthrie 2006; Small et al. 2006; Sperling et al. 2008; Wahl et al. 2009), regulates the innate immune response in mouse macrophages, at least in part, by controlling the alternative mRNA splicing of MyD88, a critical signaling adaptor in multiple Toll-like receptor (TLR) signaling pathways (Kawai and Akira 2010; Takeuchi and Akira 2010).
Materials and Methods
C. elegans chromosome II–IV RNAi screen to identify regulators of clec-85::gfp production
RNAi was performed in liquid culture in 96-well format as described previously (Alper et al. 2007, 2008) using the Ahringer laboratory E. coli RNAi feeding library (Kamath and Ahringer 2003; Kamath et al. 2003). In brief, dsRNA-producing bacteria were inoculated from frozen stocks into LB, grown overnight at 37°, dsRNA synthesis was induced using IPTG, the bacteria were centrifuged and resuspended in nematode growth medium, nematode eggs isolated by bleaching (Wood 1988) were added to the wells, and the nematodes were allowed to grow at 25° in the presence of the dsRNA producing bacteria for 3 days. After this incubation, each nematode was assayed using the COPAS Biosort (Union Biometrica), which assays and reports time of flight (TOF) (nematode length, a measure of nematode age) and green fluorescence (due to the clec-85::gfp fusion) in each animal. Because nematodes can sometimes get “trapped” in the Biosort and leak between wells, we performed our screen in 48 wells of each 96-well plate with a wash well between screening wells. The data were analyzed in R using slight modifications of our previously published scripts (Alper et al. 2007) (full scripts available upon request). These scripts first remove obvious outliers and noise in the data that are not due to nematodes; then the scripts generate scatterplots and boxplots, which allow us to compare nematode length and fluorescence between RNAi treatments, and report mean fluorescence and the number of worms in each well. For the genomic RNAi screen of chromosomes II–IV, scatterplots were manually screened for RNAi treatments that diminished nematode fluorescence. RNAi treatments that obviously inhibited nematode development were not considered as potential positives; these were recognized as treatments that hindered both fluorescence and nematode growth (as reflected in a decrease in time of flight). Because we presumed that any individual RNAi treatment was unlikely to have an effect, each test RNAi on the plate was compared to the other 47 RNAi treatments on that plate to look for differences.
Of the 10,862 RNAi treatments tested in the initial genomic II–IV screen, 430 genes were retested in follow-up assays. These 430 genes were reassayed by RNAi a minimum of two additional times. For the retests, bacteria and nematodes were prepared as described above, although in this case negative control dsRNA-producing bacteria (Dillin et al. 2002) were included on the assay plates. Only those genes that, when inhibited, altered clec-85::gfp fluorescence in both retests were considered positive hits. Table 1 reports the 32 positive genes with mean fluorescence compared to control. Table 1 also displays the mean fluorescence divided by the mean time of flight, which should control for variations in nematode populations. Data for the 430 genes retested, including the 32 positive hits, is presented in Supporting Information, Table S1, with the data broken down by each individual test well.
Table 1. Genes on C. elegans chromosomes II–IV that regulate clec-85::gfp expression.
| Clone | Worm gene | GFP (%) | GFP/TOF (%)b | n | Mouse ortholog | Mouse ortholog description |
|---|---|---|---|---|---|---|
| 89H12 | Y82E9BR.13 | 43 | 46 | 278 | Als2cr12 | Amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 12 |
| 83A3 | cbp-1 | 48 | 53 | 155 | Ep300 | E1A binding protein p300 |
| 92F5 | rps-24 | 49 | 63 | 82 | Rps24 | Ribosomal protein S24 |
| 121H1 | vha-11 | 49 | 70 | 33 | Atp6v1c1 | ATPase, H+ transporting, lysosomal V1 subunit C1 |
| 76G3 | rpn-6.1 | 50 | 63 | 29 | Psmd11 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| 74G4 | eftu-2 | 51 | 61 | 53 | Eftud2 | Elongation factor Tu GTP binding domain containing 2 |
| 31D5 | T25D3.3 | 52 | 64 | 139 | — | |
| 81E9 | far-1 | 53 | 74 | 149 | — | |
| 80H9 | zfp-1 | 54 | 63 | 119 | Mllt10 | Myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| 87B6 | rpt-6 | 55 | 71 | 64 | Psmc5 | Protease (prosome, macropain) 26S subunit, ATPase 5 |
| 70D5 | R74.2 | 55 | 61 | 137 | — | |
| 70E11 | mcm-5 | 57 | 69 | 57 | Mcm5 | Minichromosome maintenance deficient 5, cell division cycle 46 (S. cerevisiae) |
| 80B7 | C30C11.4 | 58 | 68 | 74 | — | |
| 75H11 | rpt-3 | 59 | 68 | 61 | Psmc4 | Proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
| 69H1 | rpn-1a | 60 | 66 | 88 | Psmd2 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| 107B12 | rpn-7 | 63 | 80 | 101 | Psmd6 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| 92G7 | W03G1.5 | 63 | 74 | 61 | Catsper1 | Cation channel, sperm associated 1 |
| 92C9 | dpy-9 | 64 | 72 | 190 | — | |
| 51E1 | hel-1 | 65 | 69 | 126 | Ddx39a | DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 |
| 71H8 | F26A1.3 | 67 | 72 | 374 | Ttbk1 | Tau tubulin kinase 1 |
| 67G1 | fbxa-30 | 68 | 70 | 185 | — | |
| 84H7 | W09D10.1 | 69 | 78 | 151 | Smap1 | Stromal membrane-associated protein 1 |
| 31D11 | T01D1.5 | 69 | 74 | 253 | — | |
| 76D7 | lim-8 | 71 | 74 | 92 | Lmo7 | LIM domain only 7 |
| 31D7 | T01D1.1 | 72 | 80 | 182 | — | |
| 107F12 | spp-18a | 74 | 82 | 334 | — | |
| 86D2 | C24H11.1 | 76 | 88 | 71 | Ppp1cb | Protein phosphatase 1, catalytic subunit, beta isoform |
| 84B9 | T07A5.7a | 76 | 88 | 108 | — | Fukutin |
| 82D4 | clu-1 | 78 | 86 | 169 | 1300001I01Rik | — |
| 107C12 | T20D3.6 | 80 | 82 | 325 | Higd1b | HIG1 domain family, member 1B |
| 56B4 | C07E3.3 | 81 | 84 | 214 | — | |
| 116C12 | tag-273a | 84 | 84 | 379 | — |
Sequencing of the RNAi clone identified a different gene than the predicted gene; the correct (sequenced) gene is listed here.
GFP/TOF is the mean GFP level normalized to nematode size.
The identity of all E. coli RNAi clones that produced a phenotype was verified by DNA sequencing; clones that differed from the reported sequence are indicated in Table 1 and Table S1. We list the identity of the sequence-verified clone in these tables.
Use of RNAi in mouse macrophages to test candidate genes
Orthologs of C. elegans genes were identified using either the Homologene database at the National Center for Biotechnology Information or the Panther database from within WormBase (Mi et al. 2010; Sayers et al. 2012; Yook et al. 2012). RNAi was performed largely as described previously (Alper et al. 2008). Briefly, either pools of siRNA duplexes or individual siRNA duplexes (Dharmacon) were transfected into the mouse macrophage cell line RAW264.7 using the Amaxa 96-well nucleofector shuttle. Cells were then plated at either 100,000 cells/well in 96-well format (for ELISAs) or 200,000 cells/well in 24-well format (for qPCR). Negative control siRNAs included Dharmacon’s nontargeting siRNA pool no. 1 and nontargeting siRNA no. 1. Twenty-four hours after transfection, cells were incubated in the presence of 20 ng/ml LPS (List Biological Labs), and supernatants were collected for ELISA (R&D Systems). In the primary screen, we monitored the production of IL-6 as we have found this to be a more responsive readout than TNFα or other cytokines in response to most RNAi treatments (Alper et al. 2008; De Arras et al. 2013). The cells were then either subjected to viability analysis using fluorescien diacetate as described (Fernandez-Botran and Větvička 2001) or lysed in RLT buffer (Qiagen) for RNA preparation for qPCR and RT–PCR. RNA was purified using the RNeasy kit (Qiagen), and qPCR was performed using the Quantitect SYBR Green RT–PCR kit (Qiagen) on an ABI 7900 thermocycler. Conditions for qPCR and RT-PCR to monitor MyD88L and MyD88S were as described previously (De Arras and Alper 2013). Expression levels were normalized using β-actin as a control; all primer sequences are listed in Table S2.
In one set of experiments, both Eftud2- and MyD88S-specific siRNA were delivered simultaneously. In the controls for this experiment when only a single siRNA was used, an equivalent volume of nontargeting control siRNA was included as well.
Overexpression studies in mouse macrophages
Plasmids containing full-length cDNAs cloned downstream of the CMV promoter were obtained for Eftud2 (Origene); Rps24, Atp6v1c1, and Mcm5 (Open Biosystems); and negative control chloramphenicol acetyltransferase (CAT, Invitrogen). Transient transfections were performed using 3.75 μl Fugene HD (Roche) with 100,000 cells in 24-well format and contained 300 ng IL-6-luciferase, 100 ng SV40-rluc (Promega), and 600 ng of overexpression plasmid. Twenty-four hours after transfection, cells were stimulated with LPS for 6 hr and then luciferase activity was monitored using the Dual luciferase assay kit (Promega) using SV40-rluc as a normalization control for transfection efficiency. Stable Eftud2-overexpressing lines were generated by transfecting the CMV-Eftud2 plasmid (200 ng into 200,000 cells, 12-well format) and selecting with G418.
C. elegans survival assays
Survival assays in the presence of either P. aeruginosa strain PA14 (Rahme et al. 1995; Mahajan-Miklos et al. 1999; Tan et al. 1999) or E. coli strain OP50 (Wood 1988) were performed largely as described previously (Alper et al. 2010) with the following modifications. The RNAi hypersensitive strain GR1373eri-1(mg366) IV (Kennedy et al. 2004) was synchronized by collecting hatchlings at 15° (Wood 1988), the hatchlings were moved to E. coli-expressing dsRNA and incubated at 25° for 24 hr until they were in the late L4 stage, and then the nematodes were exposed to either PA14 or OP50 at 25° in the presence of 5 μg/ml of the sterilizing agent 5-fluoro-2-deoxyuridine (Mitchell et al. 1979) on modified NGM plates (Powell and Ausubel 2008). In the heat-killed bacteria experiment, a liquid OP50 culture was concentrated fivefold, incubated at 80° for 3 hr, and plated on modified NGM supplemented with kanamycin. Inhibition of eftu-2 or vha-11 in the eri-1 mutant background caused the nematodes to develop smaller and thinner than control-treated animals.
Statistical analyses
The analyses of the nematode RNAi data are described above. C. elegans survival assays were performed in triplicate and were analyzed using GraphPad Prism 5. All macrophage experiments were performed with a minimum of three independent biological replicates, and in the case of the follow-up assays, were performed numerous additional times. Statistical analyses were performed using unpaired t-tests in GraphPad Prism 5; significance was considered P < 0.05.
Results
Genomic RNAi screen of C. elegans chromosomes II–IV
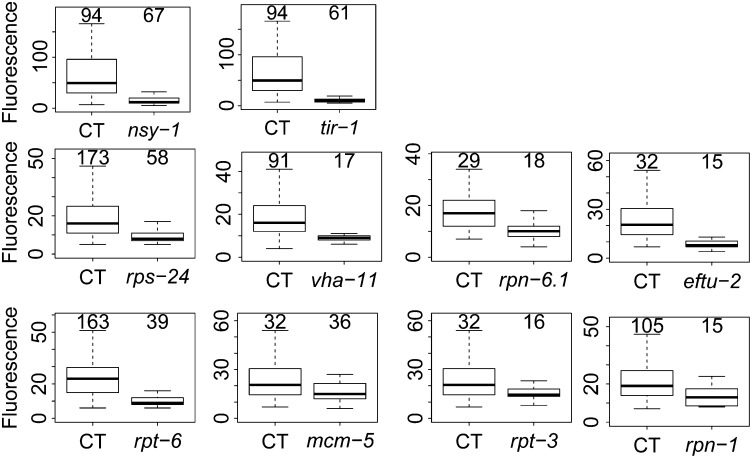
We used an E. coli library that expresses dsRNA corresponding to most genes in the C. elegans genome (Kamath and Ahringer 2003; Kamath et al. 2003) to systematically inhibit each gene on chromosomes II–IV, and then monitored the effect on fluorescence in clec-85::gfp transgenic nematodes using the COPAS Biosort (essentially a nematode flow cytometer). As a control, we found that clec-85::gfp expression is strongly diminished when either nsy-1 or tir-1 are inhibited (Figure 1, top). nsy-1 is the MAPKKK that acts upstream of the p38 MAP kinase in C. elegans; tir-1 is the sole C. elegans MyD88 family member. Both nsy-1 and tir-1 are required for expression of numerous antimicrobial genes and resistance to Gram negative and positive bacteria and fungi (Kim et al. 2002; Couillault et al. 2004; Liberati et al. 2004; Alper et al. 2007; Bolz et al. 2010; Muhammed et al. 2012).
Figure 1.
Identification of genes on C. elegans chromosomes II–IV that regulate clec-85::gfp expression. clec-85::gfp transgenic nematodes were subjected to either the indicated test RNAi treatment or control (CT) RNAi treatment and fluorescence in these nematodes was assayed using the COPAS Biosort, which reports GFP-mediated fluorescence and time of flight (TOF, which measures nematode length, a measure of nematode age) for each animal. The boxplots display fluorescence data for TOF ranging from 150 to 350 with the black bar representing the median and the box extending from the 25th to the 75th percentiles. Individual n values are displayed above each boxplot.
We screened through ∼10,862 genes by RNAi and monitored fluorescence in these nematodes. Of these 10,862 genes tested, we identified 430 genes whose inhibition led to diminished clec-85::gfp fluorescence without significantly diminishing overall nematode size. RNAi treatments that affect development or overall fitness are predicted to diminish both fluorescence as well as nematode size (a marker of growth and development) and were not considered further. We retested these 430 gene candidates, performing the assays a minimum of two more times. Of these 430 retested candidates, RNAi-mediated inhibition of 32 genes led to decreased clec-85::gfp fluorescence in both retests as well as in the original genomic screen [list of 32 hits in Table 1, boxplots for genes that also affect macrophage immunity (see below) in Figure 1 rows 2 and 3, compilation of all retest data in Table S1]. Thus, we identified 32 genes on C. elegans chromosomes II–IV that affected production of the candidate antimicrobial gene clec-85 and that are therefore potentially novel innate immune regulators. Many of these candidates function in various catabolic pathways (Table S3, analysis performed using the Gather Utility) (Chang and Nevins 2006), suggesting that catabolism could modulate innate immunity in C. elegans.
RNAi in mouse macrophages to monitor innate immune regulatory function of novel candidate genes
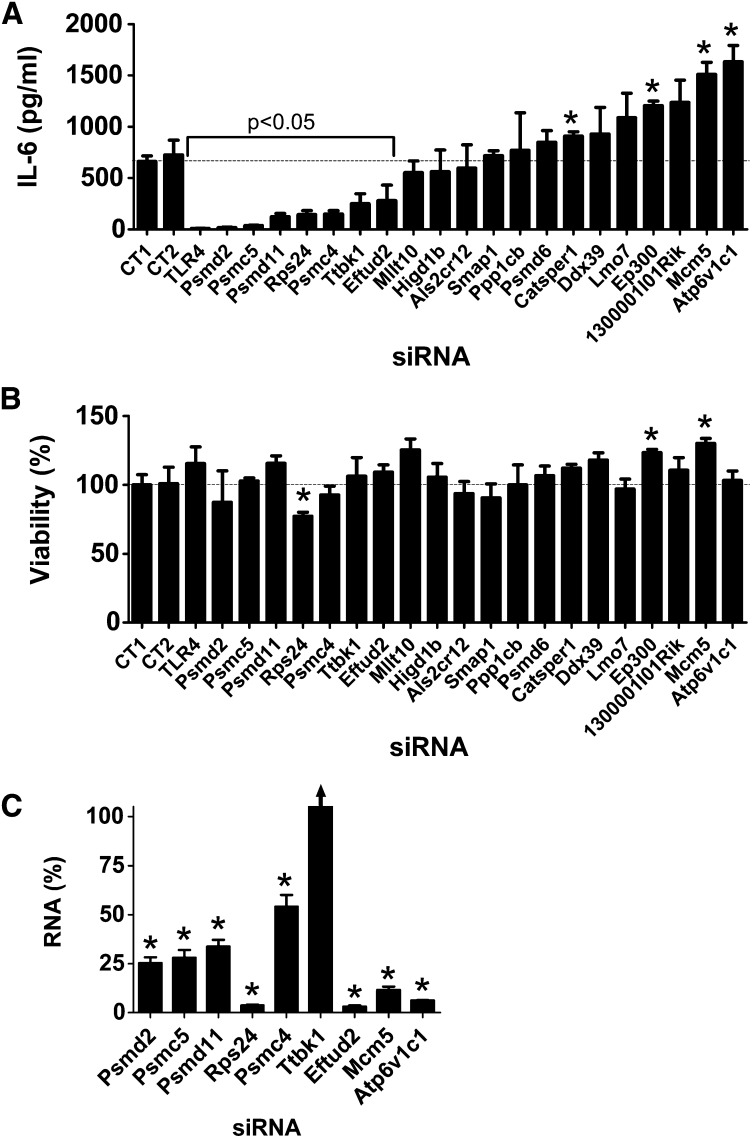
Twenty of these 32 novel candidates identified in C. elegans have an ortholog in mice (Table 1). To examine the potential innate immune regulatory function of these mammalian genes, we inhibited these 20 genes using RNAi in the mouse macrophage cell line RAW264.7, stimulated the cells with LPS, and monitored production of the proinflammatory cytokine IL-6. As a positive control, we found that treatment of these cells with siRNA that targeted TLR4, the LPS receptor, strongly inhibited LPS-induced IL-6 production (Figure 2A). Inhibition of 7 of our 20 novel candidates using pools of siRNA duplexes targeting each gene led to a statistically significant decrease in IL-6 production (Figure 2A); in contrast, inhibition of several other genes significantly increased IL-6 production (Figure 2A). As a control, we found that these siRNA treatments, with the exception of Rps24 siRNA, did not significantly diminish cell viability (Figure 2B).
Figure 2.
Use of siRNA pools to identify genes that regulate LPS-induced IL-6 production in murine macrophages. Pools of siRNA duplexes targeting the indicated genes (or nontargeting control pools CT1 or CT2) were transfected into the mouse macrophage cell line RAW264.7; the cells were stimulated for 6 hr with 20 ng/ml LPS; and IL-6 protein production (A), cell viability (B), or siRNA-mediated gene knockdown (C) was monitored. All plots display mean ± SEM. Asterisks in this figure and subsequent figures indicate values that were significantly different from control (P < 0.05). The arrow for Ttbk1 in C indicates an RNA value that is off scale (156%).
We took several steps to confirm these RNAi data and to confirm that the effects were due to specific inhibition of the corresponding endogenous gene and not an off-target siRNA effect. We focused on the seven genes whose inhibition led to decreased IL-6 production and the two genes whose inhibition had the strongest effect on increased IL-6 production. First, we used qPCR to demonstrate that the siRNA treatments were inhibiting production of the corresponding endogenous gene; we found that gene-specific knockdown was statistically significant for eight of these nine siRNAs, but could not validate knockdown for one gene, Ttbk1 (Figure 2C). To further verify that the effects were due to inhibition of the corresponding endogenous gene, we also demonstrated that at least two individual siRNA duplexes targeting each gene induced a similar phenotype. We were able to validate the data for eight of the nine gene targets, with the exception of Ttbk1, where none of the individual siRNA duplexes diminished LPS-induced IL-6 production (Figure S1). Multiple siRNA duplexes targeting the same gene induced the same phenotype, which suggested that the altered LPS-induced cytokine production was due to inhibition of that gene. Thus, we conclude that with the exception of Ttbk1, eight of these nine novel candidate genes do regulate the extent of LPS-induced IL-6 production. Four of the genes that affected the LPS response were components of the 26S proteasome (Psmd2, Psmc5, Psmd11, Psmc4); however, siRNA-mediated inhibition of another 26S proteasome subunit (Psmd6) did not affect the LPS response (Figure 2A), despite relatively strong Psmd6 knockdown as assayed with either of two sets of qPCR primers (Figure S2).
Overexpression studies in mouse macrophages to monitor innate immune regulatory function of novel candidate genes
Our RNAi studies identified eight potential candidate innate immunity regulators, including six genes whose inhibition diminished LPS-induced IL-6 production (four proteasome subunits Psmd2, Psmc5, Psmd11, and Psmc4; the ribosomal protein Rps24; and the mRNA splicing regulator Eftud2) and two genes whose inhibition enhanced LPS-induced IL-6 production (Mcm5 and Atp6v1c1). To further investigate the potential innate immune regulatory function of these genes, we chose to overexpress some of them and monitor the effect on IL-6 transcription using an IL-6-luciferase reporter construct (Baccam et al. 2003). If overexpression induced a phenotype opposite to that caused by RNAi, that would confirm the importance of that gene in innate immunity regulation. In contrast, lack of a phenotype is not necessarily informative.
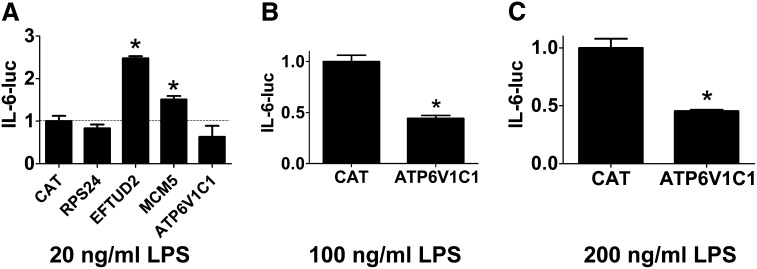
The 26S proteasome is known to regulate the LPS response (Shen et al. 2006), so we chose to focus on the other four potentially novel candidates. We transiently transfected IL-6-luciferase, SV40-rluc for normalization, and plasmids that overexpress each of Rps24, Eftud2, Mcm5, and Atp6v1c1 using the strong CMV promoter into the mouse macrophage cell line RAW264.7 and monitored IL-6-luc production in the presence of 20 ng/ml LPS. As a negative control, we also transfected a plasmid that overexpressed CAT, which should not affect innate immunity. Rps24 overexpression did not have a significant effect on IL-6-luc production (Figure 3A), and Mcm5 overexpression induced a moderate increase in IL-6-luc production (Figure 3A), which is similar to the effect observed with RNAi, so we cannot draw any further conclusions about the role of Rps24 or Mcm5 in innate immunity based on these overexpression data. In contrast, overexpression of Eftud2 strongly enhanced IL-6-luc production (Figure 3A), a phenotype opposite to that of Eftud2 siRNA. Similarly, Atp6v1c1 overexpression decreased IL-6-luc expression, a phenotype that also is opposite to the RNAi-induced phenotype, although this decrease was not statistically significant (Figure 3A). To explore the function of Atp6v1c1 further, we overexpressed Atp6v1c1 in the presence of higher doses of LPS and found that Atp6v1c1 did significantly inhibit IL-6-luc expression under those conditions (Figure 3, B and C). Thus, the RNAi and overexpression data indicate that wild-type Eftud2 enhances LPS-induced IL-6 expression while wild-type Atp6v1c1 diminishes LPS-induced IL-6 expression.
Figure 3.
Overexpression studies demonstrate that Eftud2 and Atp6v1c1 regulate IL-6 expression. Plasmids overexpressing the indicated genes were cotransfected along with plasmids expressing IL-6-luciferase and SV40-rluc into RAW264.7 cells, the cells were stimulated with the indicated quantities of LPS for 6 hr, and IL-6-luc activity (normalized to SV40-rluc) was monitored.
eftu-2/Eftud2 regulates host defense in C. elegans
To test the potential innate immune regulatory function of Eftud2 and Atp6v1c1 in vivo, we inhibited these two genes by RNAi in C. elegans, exposed these nematodes to the nematode and human pathogen P. aeruginosa strain PA14, and then monitored survival of the animals. We inhibited either eftu-2 or vha-11 (the C. elegans orthologs of Eftud2 and Atp6v1c1, respectively) in nematodes harboring a mutation in eri-1, which enhances nematode RNAi sensitivity. Inhibition of either gene diminished survival of nematodes exposed to P. aeruginosa compared to animals exposed to control RNAi bacteria (Figure 4A). However, inhibition of vha-11 also diminished survival in the presence of the nonpathogenic E. coli strain (Figure 4B). Because live E. coli can be slightly pathogenic to C. elegans (Garsin et al. 2001; Tenor and Aballay 2007), we also monitored survival of vha-11(RNAi) animals in the presence of heat-killed E. coli. Under these conditions, inhibition of vha-11 still diminished nematode survival (Figure 4C). Thus, we cannot differentiate the effects of vha-11 in C. elegans between an effect on fitness or an effect on both fitness and host defense. In contrast, nematodes exposed to eftu-2 RNAi lived as long as or slightly longer than control RNAi-treated nematodes in the presence of nonpathogenic E. coli (Figure 4B). Thus, eftu-2/Eftud2 is specifically affecting host defense and not fitness in C. elegans.
Figure 4.
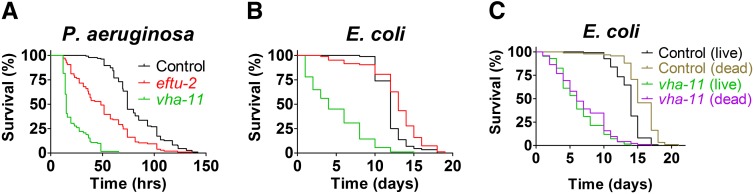
eftu-2/Eftud2 regulates host defense in C. elegans. Depicted are survival plots for eri-1 mutant nematodes treated with either control RNAi, eftu-2 RNAi, or vha-11 RNAi (as indicated) that were subsequently exposed to either P. aeruginosa strain PA14 (A), live E. coli strain OP50 (B and C), or heat-killed OP50 (C). Further statistical data (medians, n, and P-values) are presented in Table S4.
Eftud2 is a novel innate immunity regulator in mouse macrophages
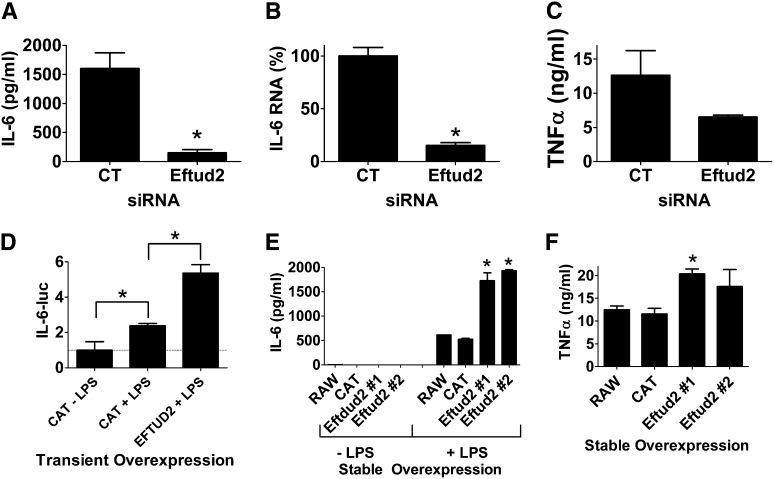
Based on the siRNA and overexpression data in macrophages and the in vivo survival data in C. elegans, we chose to investigate the function of Eftud2 further. First, we performed additional siRNA experiments in macrophages using a slightly higher dose of Eftud2 siRNA to see if this could enhance the phenotype; we found that this led to a greater inhibition in LPS-induced IL-6 protein production (Figure 5A). Inhibition of Eftud2 also diminished IL-6 mRNA production (Figure 5B). The effect of Eftud2 was not limited to IL-6 as production of the proinflammatory cytokine TNFα also was diminished when Eftud2 was inhibited by siRNA (Figure 5C). As further confirmation that transient Eftud2 overexpression enhanced IL-6-luciferase expression, we overexpressed Eftud2 in the presence of a second higher LPS dose of 50 ng/ml (Figure 5D) and found that Eftud2 enhanced IL-6 transcription at this higher LPS dose as well. To further test the function of Eftud2, we generated stable RAW264.7 lines overexpressing Eftud2, and as expected, found that stable Eftud2 overexpression enhanced LPS-induced production of IL-6 and TNFα (Figure 5, E and F). All these data indicate that wild-type Eftud2 enhances LPS-induced IL-6 production by enhancing IL-6 expression.
Figure 5.
Eftud2 regulates the innate immune response in mouse macrophages. (A–C) Cells were treated with either Eftud2 siRNA or control (CT) siRNA; were exposed to 20 ng/ml LPS for 6 hr; and IL-6 protein (A), IL-6 mRNA (B), or TNFα protein (C) was monitored. (D) Cells were transiently transfected with plasmids expressing IL-6-luciferase, SV40-rluc, and either Eftud2 or CAT; were exposed to 50 ng/ml LPS for 6 hr where indicated; and IL-6-luc production (normalized relative to SV40-rluc) was monitored. (E and F) Cell lines stably overexpressing Eftud2, CAT, or the parent RAW264.7 cell line were stimulated with 20 ng/ml LPS for 6 hr and cytokine production was monitored.
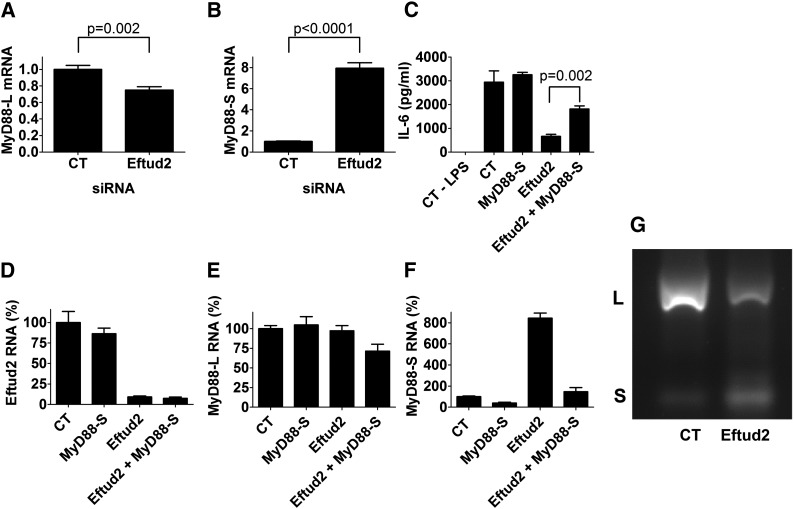
Eftud2 regulates alternative splicing of MyD88 to control the innate immune response in macrophages
Eftud2 is a component of the U5 snRNP, which functions with the rest of the spliceosome to control mRNA splicing (Kramer 1996; Fabrizio et al. 1997; Bartels et al. 2002, 2003; Brenner and Guthrie 2006; Small et al. 2006; Sperling et al. 2008; Wahl et al. 2009). We previously found that SF3a and SF3b, which function with the U2 snRNP to control mRNA splicing (Kramer et al. 2005; Sperling et al. 2008; Collins et al. 2009; Rino and Carmo-Fonseca 2009; Wahl et al. 2009), also regulate the extent of the innate immune response (De Arras and Alper 2013). SF3a and SF3b affect innate immunity in mouse macrophages, at least in part, by regulating alternative splicing of MyD88 (De Arras and Alper 2013). MyD88 is a signaling adaptor that functions downstream of most TLRs (Kawai and Akira 2010; Takeuchi and Akira 2010). Full-length MyD88 is encoded by a five-exon mRNA (long form or MyD88L). However, a shorter MyD88 mRNA (MyD88S) that lacks exon 2 encodes a negative regulator of TLR signaling (Janssens et al. 2002; Burns et al. 2003; Janssens et al. 2003; Adib-Conquy et al. 2006; Mendoza-Barbera et al. 2009). Inhibition of SF3a or SF3b using siRNA or a pharmacological agent diminishes LPS-induced IL-6 and TNFα production, at least in part, by enhancing production of the negatively acting MyD88S splice form (De Arras and Alper 2013). We therefore chose to monitor if the Eftud2 mRNA splicing regulator likewise affects alternative splicing of MyD88.
To monitor MyD88L and MyD88S levels when Eftud2 was inhibited, we used a qPCR assay and primer sets that can distinguish between the two mRNA splice forms (De Arras and Alper 2013). We inhibited Eftud2 using siRNA, stimulated the cells with 20 ng/ml LPS for 6 hr, and then monitored the two MyD88 mRNA splice forms by qPCR. Eftud2 inhibition led to a moderate decrease in MyD88L levels and a substantial increase in MyD88S levels (Figure 6, A and B). In contrast, Eftud2 inhibition did not affect all mRNA splicing events as intron 3 of actin was excised normally (data not shown).
Figure 6.
Eftud2 regulates innate immunity by controlling the alternative mRNA splicing of MyD88. (A and B) Cells were treated with either Eftud2 siRNA or control (CT) nontargeting siRNA, were stimulated with 20 ng/ml LPS for 6 hr, and MyD88L and MyD88S mRNA levels were monitored by qPCR. (C–F) Cells were treated with the indicated siRNAs, were stimulated with 20 ng/ml LPS for 6 hr, and either IL-6 was monitored by ELISA (C) or Eftud2, MyD88L, and MyD88S mRNA levels were monitored by qPCR (D–F). mRNA levels were normalized such that 100% indicates mRNA in the presence of control RNAi in the absence of LPS. (G) Cells were exposed to either Eftud2 siRNA or control nontargeting siRNA (CT), were then treated with 20 ng/ml LPS for 6 hr, RNA was extracted, RT–PCR was performed to amplify the indicated mRNAs, and the amplified products were analyzed by agarose gel electrophoresis. Representative image is shown of PCR products using primers that bracket MyD88 exon 2 and that therefore simultaneously generate a 280-bp product corresponding to MyD88L and a 145-bp product corresponding to MyD88S.
We also used semiquantitative RT–PCR and subsequent agarose gel electrophoresis to monitor the two MyD88 mRNA splice forms. RT–PCR using primers specific to the MyD88S mRNA splice form demonstrated that inhibition of Eftud2 by RNAi led to increased MyD88S production (Figure S3, A and B), confirming the qPCR data. In contrast, production of β-actin was not significantly affected (Figure S3, A and C). Using primers that bracketed exon 2, we also were able to amplify products corresponding to both MyD88L and MyD88S simultaneously. As observed previously (De Arras and Alper 2013), there is substantially more MyD88L than MyD88S in unstimulated cells (Figure 6G). Inhibition of Eftud2 by RNAi led to a significant decrease in MyD88L and a concomitant increase in the smaller MyD88S form (Figure 6G and Figure S3, D and E).
The increased amount of the MyD88S inhibitory splice form could explain in whole or in part the weakened LPS response when Eftud2 is inhibited. To directly test if Eftud2’s effect on innate immunity is mediated by changes in MyD88S levels, we chose to inhibit the MyD88S mRNA splice form when Eftud2 was inhibited in an attempt to restore LPS-induced IL-6 production. To decrease MyD88S mRNA levels, we used a siRNA that targets the unique exon 1–exon 3 splice junction that is present in MyD88S but absent in MyD88L (De Arras and Alper 2013). Treatment of cells with this MyD88S siRNA leads to a moderate increase in LPS-induced IL-6 production (Figure 6C; in other experiments not shown, this increase was more significant). We found that treatment of cells with Eftud2 siRNA alone strongly diminished LPS-induced IL-6 production while treatment of cells with both Eftud2 siRNA and MyD88S siRNA simultaneously strongly rescued LPS-induced IL-6 production Figure 6C). As a control, we verified that the siRNAs were inducing similar knockdown under the various conditions (Figure 6, D–F). This suggests that much of the effect of Eftud2 on the LPS response is mediated by altered MyD88 mRNA splicing.
Discussion
Identification of novel conserved regulators of innate immunity using comparative genomics
Our hypothesis is that genes with evolutionarily conserved innate immune regulatory function in C. elegans and murine macrophages will likely also affect humans and are therefore candidate regulators of human infectious and inflammatory disease. Our comparative genomics RNAi screening approach in C. elegans and mouse macrophages has identified many such candidate innate immune regulators (this study and Alper et al. 2008; De Arras et al. 2013). One advantage of this approach is that it may overcome the high false-positive rate present in mammalian RNAi screens due to off-target effects (Editorial 2003). Thus far, we have validated many of these dual RNAi hits (nematodes and macrophages) using knockout mutations in C. elegans and have validated two dual RNAi hits using mouse knockouts (Alper et al. 2008; De Arras et al. 2012, 2013).
The C. elegans screening approach is an efficient one for identifying innate immunity regulators, as we see a 30–40% hit rate translating genes from nematodes to macrophages (Alper et al. 2008; De Arras et al. 2013; and the current study, which is far higher than other approaches that we have taken (for example, we had a 2% hit rate monitoring a set of 100 genes identified as candidate innate immunity regulators based on a computational analysis of their expression levels in the Gene Expression Omnibus database, data not shown). One unanswered question is: Are these genes regulating innate immunity in a similar fashion in both species? This is particularly relevant as many of the candidates identified in the current study are components of the 26S proteasome, which regulates innate immunity in Drosophila and mammals by degrading IκB, thereby allowing NFκB activation (Coux et al. 1996; Foley and O’Farrell 2004). However, while C. elegans has an IκB homolog, as yet no direct NFκB homolog has been identified (Pujol et al. 2001). It could be that another gene serves a similar function in C. elegans, or it could be that the 26S proteasome regulates innate immunity in C. elegans by a different mechanism. Nevertheless, this comparative genomics approach seems to be an efficient way to find novel genes that matter.
There is precedent in C. elegans for innate immunity genes in C. elegans acting in both conserved and novel fashion. The p38 MAPK pathway acts similarly in both C. elegans and mammals (Kim et al. 2002). In contrast, while the Xbp1 transcription factor enhances host defense in both mammals and C. elegans, it does so by different mechanisms: by acting as a TLR-signaling dependent transcriptional activator that functions with NFκB in mice (Martinon et al. 2010) and by playing a detoxification role in the immune response in C. elegans (Richardson et al. 2010). Thus, while we see a high degree of “conservation of function” of innate immunity regulators, it is as yet unclear if there is “conservation of mechanism,” although given the reported differences in C. elegans innate immunity, we speculate that many of these genes may use different mechanisms in the different species to effect host defense.
Many of our candidate genes encoded components of the 26S proteasome, which is already the target of potential therapeutic pharmacological options (Elliott et al. 2003; Wang and Maldonado 2006; Calzado et al. 2007). We note that while siRNA-mediated inhibition of several 26S proteasome subunits strongly diminished LPS-induced cytokine production, the inhibition of Psmd6 did not. This raises the possibility of either differential function or differential sensitivity of different 26S proteasome subunits to inhibition, suggesting that targeting individual subunits of the 26S proteasome using siRNA could be another approach to treat inflammatory diseases.
The role of Atp6v1c1 in innate immunity regulation
Our RNAi and overexpression data indicate that Atp6v1c1 limits the innate immune response by diminishing IL-6 transcription. Atp6v1c1 is the VMA5 subunit of the vacuolar H+-ATPase (V-ATPase) (Smith et al. 2003). The V-ATPase is a large multiprotein complex that mediates acidification of intracellular organelles and can therefore affect numerous biological processes (Stevens and Forgac 1997; Forgac 1999; Nishi and Forgac 2002). Atp6v1c1 helps mediate the assembly of the catalytic and membrane portions of the V-ATPase (Ho et al. 1993; Drory et al. 2004). Inhibition of the V-ATPase in macrophages using the pharmacological agent bafilomycin A1 enhances LPS-induced NFκB activation and cytokine production (Bidani and Heming 1995; Conboy et al. 1999), which is consistent with our RNAi and overexpression data. This suggests that activating individual V-ATPase subunits could also be a potential therapeutic option for inflammatory diseases.
The Eftud2 mRNA splicing regulator controls innate immunity by altering MyD88 mRNA splicing
Another novel innate immunity regulator identified by our comparative genomics approach is Eftud2, a component of the U5 snRNP in the spliceosome that mediates pre-mRNA splicing (Kramer 1996; Fabrizio et al. 1997; Bartels et al. 2002, 2003; Brenner and Guthrie 2006; Small et al. 2006; Sperling et al. 2008; Wahl et al. 2009). Consistent with its role as a general mRNA splicing regulator, C. elegans eftu-2/Eftud2 has been implicated in a variety of developmental processes, including the organization of myofilaments and P granule development (Meissner et al. 2009; Updike and Strome 2009). In humans, mutations in Eftud2 cause craniofacial conditions including mandibulofacial dysostosis and esophageal atresia (Gordon et al. 2012; Lines et al. 2012; Luquetti et al. 2013; Voigt et al. 2013).
In the present study, we found that inhibition of Eftud2 diminished LPS-induced IL-6 and TNFα production while overexpression of Eftud2 enhanced cytokine production. At least part of this effect occurs at the IL-6 transcriptional level. Importantly, inhibition of Eftud2 did not affect overall cell viability, suggesting that at this level of gene inhibition, the innate immune response is far more sensitive to Eftud2 levels than are other general cell functions.
These data are reminiscent of the effects of the SF3a and SF3b complexes that interact with the U2 snRNP to facilitate mRNA splicing (Kramer et al. 2005; Sperling et al. 2008; Collins et al. 2009; Rino and Carmo-Fonseca 2009; Wahl et al. 2009). We previously reported that inhibition of SF3a or SF3b by RNAi or a pharmacological agent diminished LPS-induced cytokine production in mouse macrophages (De Arras and Alper 2013). SF3a and SF3b regulate innate immunity in part by modulating alternative splicing of MyD88 (De Arras and Alper 2013). The five exon full-length long form MyD88L encodes a critical signaling adaptor in multiple TLR response pathways. In contrast, the shorter splice form MyD88S that lacks the 135-bp exon 2 encodes an in-frame protein that is a negative regulator of TLR signaling, which prevents activation of the downstream signaling IRAK proteins (Janssens et al. 2002; Burns et al. 2003; Mendoza-Barbera et al. 2009). Because of the similarity in the effects of SF3a/b and Eftud2 inhibition, we monitored MyD88 mRNA splicing when Eftud2 was inhibited and found that the inhibitory splice form MyD88S was increased dramatically, which is consistent with the resulting diminished cytokine response. Moreover, we were able to partially rescue the defect caused by Eftud2 inhibition by normalizing MyD88S levels with a second MyD88S-specific siRNA. Thus, SF3a, SF3b, and Eftud2 all are required for retention of exon 2 in MyD88, as this exon is lost and MyD88S is produced when any of these spliceosome components are inhibited. This result is consistent with other reports that show that inhibition of specific spliceosome subunits leads to alternative splicing and exon skipping (Corrionero et al. 2011; Fan et al. 2011; An and Henion 2012; Visconte et al. 2012). Moreover, the current data show that regulators of the U2 snRNP (SF3a and SF3b), which affect 3′ splice site choice, and the U5 snRNP (Eftud2), which joins the spliceosome at a later stage of assembly (Newman 1997; Turner et al. 2004; Wahl et al. 2009), both have similar effects on MyD88 mRNA splicing.
The C. elegans and mouse orthologs of Eftud2, SF3a1, and SF3b1 exhibit 70, 45, and 67% identity, respectively. While Eftud2 regulates innate immunity in both species, it is unclear how much mechanistic conservation there is. While the sole C. elegans MyD88 family member tir-1 also exhibits alternate splicing, no alternate functions have been reported for these splice forms in innate immunity, and the relevant intron–exon junctions in MyD88 do not appear to be conserved in tir-1 (data not shown).
MyD88S levels are increased ∼10-fold in monocytes from septic patients (Adib-Conquy et al. 2006), suggesting that a better understanding of the regulation of MyD88 mRNA splicing could contribute to our understanding and ability to treat inflammatory disease. The identification of Eftud2 as a novel regulator of MyD88 mRNA splicing and innate immunity should further this approach.
Acknowledgments
We thank Gail Bishop for the IL-6-luciferase reporter construct. This work was supported by R21ES019256 from the National Institute of Environmental Health Sciences, the Intramural Research Programs of the National Heart Lung and Blood Institute, the National Institute of Environmental Health Sciences (Z01ES102045), and a Butcher Seed grant.
Footnotes
Communicating editor: B. Lazzaro
Literature Cited
- Adib-Conquy M., Adrie C., Fitting C., Gattolliat O., Beyaert R., et al. , 2006. Up-regulation of MyD88s and SIGIRR, molecules inhibiting Toll-like receptor signaling, in monocytes from septic patients. Crit. Care Med. 34: 2377–2385 [DOI] [PubMed] [Google Scholar]
- Alper S., McBride S. J., Lackford B., Freedman J. H., Schwartz D. A., 2007. Specificity and complexity of the C. elegans innate immune response. Mol. Cell. Biol. 27: 5544–5553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper S., Laws R., Lackford B., Boyd W. A., Dunlap P., et al. , 2008. Identification of innate immunity genes and pathways using a comparative genomics approach. Proc. Natl. Acad. Sci. USA 105: 7016–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper S., McElwee M., Apfeld J., Lackford B., Freedman J., et al. , 2010. Germline proliferation regulates distinct signaling pathways in C. elegans to control lifespan and innate immunity. J. Biol. Chem. 285: 1822–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An M., Henion P. D., 2012. The zebrafish sf3b1b460 mutant reveals differential requirements for the sf3b1 pre-mRNA processing gene during neural crest development. Int. J. Dev. Biol. 56: 223–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcaroli J., Fessler M. B., Abraham E., 2005. Genetic polymorphisms and sepsis. Shock 24: 300–312 [DOI] [PubMed] [Google Scholar]
- Baccam M., Woo S. Y., Vinson C., Bishop G. A., 2003. CD40-mediated transcriptional regulation of the IL-6 gene in B lymphocytes: involvement of NF-kappa B, AP-1, and C/EBP. J. Immunol. 170: 3099–3108 [DOI] [PubMed] [Google Scholar]
- Bartels C., Klatt C., Luhrmann R., Fabrizio P., 2002. The ribosomal translocase homologue Snu114p is involved in unwinding U4/U6 RNA during activation of the spliceosome. EMBO Rep. 3: 875–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels C., Urlaub H., Luhrmann R., Fabrizio P., 2003. Mutagenesis suggests several roles of Snu114p in pre-mRNA splicing. J. Biol. Chem. 278: 28324–28334 [DOI] [PubMed] [Google Scholar]
- Bidani A., Heming T. A., 1995. Effects of bafilomycin A1 on functional capabilities of LPS-activated alveolar macrophages. J. Leukoc. Biol. 57: 275–281 [DOI] [PubMed] [Google Scholar]
- Bolz D. D., Tenor J. L., Aballay A., 2010. A conserved PMK-1/p38 MAPK is required in C. elegans tissue-specific immune response to Y. pestis infection. J. Biol. Chem. 285: 10832–10840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner T. J., Guthrie C., 2006. Assembly of Snu114 into U5 snRNP requires Prp8 and a functional GTPase domain. RNA 12: 862–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns K., Janssens S., Brissoni B., Olivos N., Beyaert R., et al. , 2003. Inhibition of interleukin 1 receptor/Toll-like receptor signaling through the alternatively spliced, short form of MyD88 is due to its failure to recruit IRAK-4. J. Exp. Med. 197: 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzado M. A., Bacher S., Schmitz M. L., 2007. NF-kappaB inhibitors for the treatment of inflammatory diseases and cancer. Curr. Med. Chem. 14: 367–376 [DOI] [PubMed] [Google Scholar]
- Chang J. T., Nevins J. R., 2006. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics 22: 2926–2933 [DOI] [PubMed] [Google Scholar]
- Collins L. J., Kurland C. G., Biggs P., Penny D., 2009. The modern RNP world of eukaryotes. J. Hered. 100: 597–604 [DOI] [PubMed] [Google Scholar]
- Conboy I. M., Manoli D., Mhaiskar V., Jones P. P., 1999. Calcineurin and vacuolar-type H+-ATPase modulate macrophage effector functions. Proc. Natl. Acad. Sci. USA 96: 6324–6329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly D. J., O’Neill L. A., 2012. New developments in Toll-like receptor targeted therapeutics. Curr. Opin. Pharmacol. 12: 510–518 [DOI] [PubMed] [Google Scholar]
- Cook D. N., Pisetsky D. S., Schwartz D. A., 2004. Toll-like receptors in the pathogenesis of human disease. Nat. Immunol. 5: 975–979 [DOI] [PubMed] [Google Scholar]
- Corrionero A., Minana B., Valcarcel J., 2011. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev. 25: 445–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couillault C., Pujol N., Reboul J., Sabatier L., Guichou J. F., et al. , 2004. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 5: 488–494 [DOI] [PubMed] [Google Scholar]
- Coux O., Tanaka K., Goldberg A. L., 1996. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65: 801–847 [DOI] [PubMed] [Google Scholar]
- De Arras L., Alper S., 2013. The Sf3a mRNA splicing complex mediates a MyD88-dependent negative feedback loop that limits the innate immune response. PLoS Genet. 9: e1003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Arras L., Yang I. V., Lackford B., Riches D. W., Prekeris R., et al. , 2012. Spatiotemporal Inhibition of Innate Immunity Signaling by the Tbc1d23 RAB-GAP. J. Immunol. 188: 2905–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Arras L., Seng A., Lackford B., Keikhaee M., Bowerman B., et al. , 2013. An evolutionarily conserved innate immunity protein interaction network. J. Biol. Chem. 288: 1967–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A., Crawford D. K., Kenyon C., 2002. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science 298: 830–834 [DOI] [PubMed] [Google Scholar]
- Drory O., Frolow F., Nelson N., 2004. Crystal structure of yeast V-ATPase subunit C reveals its stator function. EMBO Rep. 5: 1148–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott P. J., Zollner T. M., Boehncke W. H., 2003. Proteasome inhibition: a new anti-inflammatory strategy. J. Mol. Med. 81: 235–245 [DOI] [PubMed] [Google Scholar]
- Engelmann I., Pujol N., 2010. Innate immunity in C. elegans. Adv. Exp. Med. Biol. 708: 105–121 [DOI] [PubMed] [Google Scholar]
- Ewbank, J. J., 2006 Signaling in the immune response (January 23, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.83.1, http://www.wormbook.org [DOI] [PMC free article] [PubMed]
- Ewbank J. J., Zugasti O., 2011. C. elegans: model host and tool for antimicrobial drug discovery. Dis. Model. Mech. 4: 300–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P., Laggerbauer B., Lauber J., Lane W. S., Luhrmann R., 1997. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 16: 4092–4106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L., Lagisetti C., Edwards C. C., Webb T. R., Potter P. M., 2011. Sudemycins, novel small molecule analogues of FR901464, induce alternative gene splicing. ACS Chem. Biol. 6: 582–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Botran R., Větvička V., 2001. Methods in Cellular Immunology, CRC Press, Boca Raton [Google Scholar]
- Foley E., O’Farrell P. H., 2004. Functional dissection of an innate immune response by a genome-wide RNAi screen. PLoS Biol. 2: E203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgac M., 1999. Structure and properties of the vacuolar (H+)-ATPases. J. Biol. Chem. 274: 12951–12954 [DOI] [PubMed] [Google Scholar]
- Garsin D. A., Sifri C. D., Mylonakis E., Qin X., Singh K. V., et al. , 2001. A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. USA 98: 10892–10897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh F. G., Midwood K. S., 2012. Intrinsic danger: activation of Toll-like receptors in rheumatoid arthritis. Rheumatology (Oxford) 51: 7–23 [DOI] [PubMed] [Google Scholar]
- Gordon C. T., Petit F., Oufadem M., Decaestecker C., Jourdain A. S., et al. , 2012. EFTUD2 haploinsufficiency leads to syndromic oesophageal atresia. J. Med. Genet. 49: 737–746 [DOI] [PubMed] [Google Scholar]
- Ho M. N., Hill K. J., Lindorfer M. A., Stevens T. H., 1993. Isolation of vacuolar membrane H(+)-ATPase-deficient yeast mutants; the VMA5 and VMA4 genes are essential for assembly and activity of the vacuolar H(+)-ATPase. J. Biol. Chem. 268: 221–227 [PubMed] [Google Scholar]
- Huffman D. L., Abrami L., Sasik R., Corbeil J., van der Goot F. G., et al. , 2004. Mitogen-activated protein kinase pathways defend against bacterial pore-forming toxins. Proc. Natl. Acad. Sci. USA 101: 10995–11000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui J., Ausubel F., 2010. 99th Dahlem Conference on infection, inflammation, and chronic inflammatory disorders: Caenorhabditis elegans as a model to study tissues involved in host immunity and microbial pathogenesis. Clin. Exp. Immunol. 160: 48–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., Medzhitov R., 2010. Regulation of adaptive immunity by the innate immune system. Science 327: 291–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens S., Burns K., Tschopp J., Beyaert R., 2002. Regulation of interleukin-1- and lipopolysaccharide-induced NF-kappaB activation by alternative splicing of MyD88. Curr. Biol. 12: 467–471 [DOI] [PubMed] [Google Scholar]
- Janssens S., Burns K., Vercammen E., Tschopp J., Beyaert R., 2003. MyD88S, a splice variant of MyD88, differentially modulates NF-kappaB- and AP-1-dependent gene expression. FEBS Lett. 548: 103–107 [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Ahringer J., 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321 [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., et al. , 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237 [DOI] [PubMed] [Google Scholar]
- Karin M., Greten F. R., 2005. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 5: 749–759 [DOI] [PubMed] [Google Scholar]
- Kaufmann, S. H. E., R. Medzhitov, S. Gordon, 2004 The Innate Immune Response to Infection ASM Press, Washington D.C. [Google Scholar]
- Kawai T., Akira S., 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11: 373–384 [DOI] [PubMed] [Google Scholar]
- Kennedy S., Wang D., Ruvkun G., 2004. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427: 645–649 [DOI] [PubMed] [Google Scholar]
- Kim D. H., Feinbaum R., Alloing G., Emerson F. E., Garsin D. A., et al. , 2002. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297: 623–626 [DOI] [PubMed] [Google Scholar]
- Kramer A., 1996. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65: 367–409 [DOI] [PubMed] [Google Scholar]
- Kramer A., Ferfoglia F., Huang C. J., Mulhaupt F., Nesic D., et al. , 2005. Structure-function analysis of the U2 snRNP-associated splicing factor SF3a. Biochem. Soc. Trans. 33: 439–442 [DOI] [PubMed] [Google Scholar]
- Lee H. K., Iwasaki A., 2007. Innate control of adaptive immunity: dendritic cells and beyond. Semin. Immunol. 19: 48–55 [DOI] [PubMed] [Google Scholar]
- Liberati N. T., Fitzgerald K. A., Kim D. H., Feinbaum R., Golenbock D. T., et al. , 2004. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc. Natl. Acad. Sci. USA 101: 6593–6598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lines M. A., Huang L., Schwartzentruber J., Douglas S. L., Lynch D. C., et al. , 2012. Haploinsufficiency of a spliceosomal GTPase encoded by EFTUD2 causes mandibulofacial dysostosis with microcephaly. Am. J. Hum. Genet. 90: 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luquetti D. V., Hing A. V., Rieder M. J., Nickerson D. A., Turner E. H., et al. , 2013. “Mandibulofacial dysostosis with microcephaly” caused by EFTUD2 mutations: expanding the phenotype. Am. J. Med. Genet. A. 161A: 108–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan-Miklos S., Tan M. W., Rahme L. G., Ausubel F. M., 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96: 47–56 [DOI] [PubMed] [Google Scholar]
- Mallo G. V., Kurz C. L., Couillault C., Pujol N., Granjeaud S., et al. , 2002. Inducible antibacterial defense system in C. elegans. Curr. Biol. 12: 1209–1214 [DOI] [PubMed] [Google Scholar]
- Martinon F., Chen X., Lee A. H., Glimcher L. H., 2010. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat. Immunol. 11: 411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner B., Warner A., Wong K., Dube N., Lorch A., et al. , 2009. An integrated strategy to study muscle development and myofilament structure in Caenorhabditis elegans. PLoS Genet. 5: e1000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Barbera E., Corral-Rodriguez M. A., Soares-Schanoski A., Velarde M., Macieira S., et al. , 2009. Contribution of globular death domains and unstructured linkers to MyD88.IRAK-4 heterodimer formation: an explanation for the antagonistic activity of MyD88s. Biochem. Biophys. Res. Commun. 380: 183–187 [DOI] [PubMed] [Google Scholar]
- Mi H., Dong Q., Muruganujan A., Gaudet P., Lewis S., et al. , 2010. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 38: D204–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D. H., Stiles J. W., Santelli J., Sanadi D. R., 1979. Synchronous growth and aging of Caenorhabditis elegans in the presence of fluorodeoxyuridine. J. Gerontol. 34: 28–36 [DOI] [PubMed] [Google Scholar]
- Mochii M., Yoshida S., Morita K., Kohara Y., Ueno N., 1999. Identification of transforming growth factor-beta- regulated genes in Caenorhabditis elegans by differential hybridization of arrayed cDNAs. Proc. Natl. Acad. Sci. USA 96: 15020–15025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammed M., Fuchs B. B., Wu M. P., Breger J., Coleman J. J., et al. , 2012. The role of mycelium production and a MAPK-mediated immune response in the C. elegans-Fusarium model system. Med. Mycol. 50: 488–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullick A. E., Tobias P. S., Curtiss L. K., 2005. Modulation of atherosclerosis in mice by Toll-like receptor 2. J. Clin. Invest. 115: 3149–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C. T., McCarroll S. A., Bargmann C. I., Fraser A., Kamath R. S., et al. , 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424: 277–283 [DOI] [PubMed] [Google Scholar]
- Newman A. J., 1997. The role of U5 snRNP in pre-mRNA splicing. EMBO J. 16: 5797–5800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi T., Forgac M., 2002. The vacuolar (H+)-ATPases–nature’s most versatile proton pumps. Nat. Rev. Mol. Cell Biol. 3: 94–103 [DOI] [PubMed] [Google Scholar]
- O’Rourke D., Baban D., Demidova M., Mott R., Hodgkin J., 2006. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 16: 1005–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard C., Casanova J. L., Puel A., 2011. Infectious diseases in patients with IRAK-4, MyD88, NEMO, or IkappaBalpha deficiency. Clin. Microbiol. Rev. 24: 490–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J. R., Ausubel F. M., 2008. Models of Caenorhabditis elegans infection by bacterial and fungal pathogens. Methods Mol. Biol. 415: 403–427 [DOI] [PubMed] [Google Scholar]
- Pujol N., Link E. M., Liu L. X., Kurz C. L., Alloing G., et al. , 2001. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr. Biol. 11: 809–821 [DOI] [PubMed] [Google Scholar]
- Pukkila-Worley R., Ausubel F. M., 2012. Immune defense mechanisms in the Caenorhabditis elegans intestinal epithelium. Curr. Opin. Immunol. 24: 3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulverer B., 2003. Whither RNAi? Nat Cell Biol 5: 489–90 [DOI] [PubMed] [Google Scholar]
- Rahme L. G., Stevens E. J., Wolfort S. F., Shao J., Tompkins R. G., et al. , 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268: 1899–1902 [DOI] [PubMed] [Google Scholar]
- Richardson C. E., Kooistra T., Kim D. H., 2010. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature 463: 1092–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rino J., Carmo-Fonseca M., 2009. The spliceosome: A self-organized macromolecular machine in the nucleus? Trends Cell Biol. 19: 375–384 [DOI] [PubMed] [Google Scholar]
- Sayers E. W., Barrett T., Benson D. A., Bolton E., Bryant S. H., et al. , 2012. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 40: D13–D25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira M., Hamlin B. J., Rong J., Chen K., Ronen M., et al. , 2006. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc. Natl. Acad. Sci. USA 103: 14086–14091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Reis J., Morrison D. C., Papasian C., Raghavakaimal S., et al. , 2006. Key inflammatory signaling pathways are regulated by the proteasome. Shock 25: 472–484 [DOI] [PubMed] [Google Scholar]
- Small E. C., Leggett S. R., Winans A. A., Staley J. P., 2006. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol. Cell 23: 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. N., Lovering R. C., Futai M., Takeda J., Brown D., et al. , 2003. Revised nomenclature for mammalian vacuolar-type H+ -ATPase subunit genes. Mol. Cell 12: 801–803 [DOI] [PubMed] [Google Scholar]
- Sperling J., Azubel M., Sperling R., 2008. Structure and function of the Pre-mRNA splicing machine. Structure 16: 1605–1615 [DOI] [PubMed] [Google Scholar]
- Stevens T. H., Forgac M., 1997. Structure, function and regulation of the vacuolar (H+)-ATPase. Annu. Rev. Cell Dev. Biol. 13: 779–808 [DOI] [PubMed] [Google Scholar]
- Takeuchi O., Akira S., 2010. Pattern recognition receptors and inflammation. Cell 140: 805–820 [DOI] [PubMed] [Google Scholar]
- Tan M. W., Mahajan-Miklos S., Ausubel F. M., 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. USA 96: 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenor J. L., Aballay A., 2007. A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO Rep. 9: 103–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel E. R., Chu S. W., Reinke V., Lee S. S., Ausubel F. M., et al. , 2006. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2: e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner I. A., Norman C. M., Churcher M. J., Newman A. J., 2004. Roles of the U5 snRNP in spliceosome dynamics and catalysis. Biochem. Soc. Trans. 32: 928–931 [DOI] [PubMed] [Google Scholar]
- Updike D. L., Strome S., 2009. A genomewide RNAi screen for genes that affect the stability, distribution and function of P granules in Caenorhabditis elegans. Genetics 183: 1397–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visconte V., Rogers H. J., Singh J., Barnard J., Bupathi M., et al. , 2012. SF3B1 haploinsufficiency leads to formation of ring sideroblasts in myelodysplastic syndromes. Blood 120: 3173–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt C., Megarbane A., Neveling K., Czeschik J. C., Albrecht B., et al. , 2013. Oto-facial syndrome and esophageal atresia, intellectual disability and zygomatic anomalies: expanding the phenotypes associated with EFTUD2 mutations. Orphanet J. Rare Dis. 8: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl M. C., Will C. L., Luhrmann R., 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wang J., Maldonado M. A., 2006. The ubiquitin-proteasome system and its role in inflammatory and autoimmune diseases. Cell. Mol. Immunol. 3: 255–261 [PubMed] [Google Scholar]
- Wood, W. B., 1988 The Nematode Caenorhabditis elegans, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Yook K., Harris T. W., Bieri T., Cabunoc A., Chan J., et al. , 2012. WormBase 2012: more genomes, more data, new website. Nucleic Acids Res. 40: D735–D741 [DOI] [PMC free article] [PubMed] [Google Scholar]