Abstract
Chromosomal sex determination is phylogenetically widespread, having arisen independently in many lineages. Decades of theoretical work provide predictions about sex chromosome differentiation that are well supported by observations in both XY and ZW systems. However, the phylogenetic scope of previous work gives us a limited understanding of the pace of sex chromosome gain and loss and why Y or W chromosomes are more often lost in some lineages than others, creating XO or ZO systems. To gain phylogenetic breadth we therefore assembled a database of 4724 beetle species’ karyotypes and found substantial variation in sex chromosome systems. We used the data to estimate rates of Y chromosome gain and loss across a phylogeny of 1126 taxa estimated from seven genes. Contrary to our initial expectations, we find that highly degenerated Y chromosomes of many members of the suborder Polyphaga are rarely lost, and that cases of Y chromosome loss are strongly associated with chiasmatic segregation during male meiosis. We propose the “fragile Y” hypothesis, that recurrent selection to reduce recombination between the X and Y chromosome leads to the evolution of a small pseudoautosomal region (PAR), which, in taxa that require XY chiasmata for proper segregation during meiosis, increases the probability of aneuploid gamete production, with Y chromosome loss. This hypothesis predicts that taxa that evolve achiasmatic segregation during male meiosis will rarely lose the Y chromosome. We discuss data from mammals, which are consistent with our prediction.
Keywords: Coleoptera, Karyotype, comparative methods, sex chromosome, genetics of sex
CHROMOSOMAL sex determination has evolved independently in many lineages (Bull 1983). In addition to their role in gender determination, sex chromosomes are fascinating because the homologs often differ in gene content and morphology (Vallender and Lahn 2004; Graves 2006; Arunkumar et al. 2009; Betrán et al. 2012). Their unequal distribution between sexes also means that sex-linked genes experience and respond to evolutionary forces in different ways compared with autosomes (Charlesworth et al. 1987; Rice 1987; Charlesworth 1991; Rice 1994). The sex chromosomes can encompass the extremes of evolutionary rate. For example, the average divergence between human and chimpanzee X and Y chromosomes is lower and higher, respectively, than average autosomal divergence (Mikkelsen et al. 2005). Sex chromosomes can also play a special role in the origin of species, where the hemizygous sex often suffers the consequences of hybridization disproportionately (Haldane’s rule) (Haldane 1922; Watson and Demuth 2012) and X-linked introgressions have larger effects on hybrid fitness than autosomal introgressions (large X-effect) (Presgraves 2008; Phillips and Edmands 2012). Finally, sex chromosomes that are confined to the heterogametic sex (Y or W in male or female heterogametic species, respectively) are also particularly interesting for their apparent dispensability in some taxa but not others.
The canonical view of sex chromosome evolution assumes that a sex-determining region evolves that leads to a pair of ancestral autosomes evolving into proto sex chromosomes (Westergaard 1958). Most models suggest that the resulting proto Y(W) will degenerate as a consequence of reduced effective population size (as these chromosomes are only found in one sex) and lack of recombination near the sex-determining locus. The nonrecombining region can expand to adjacent portions of the chromosome. The selective force for this is thought to be selection to maintain linkage between sexually antagonistic loci (those polymorphic for alleles that benefit one sex at the expense of the other) and the sex determination locus. Recombination suppression may involve chromosomal rearrangements (e.g., inversions) that include the sex determining locus (Charlesworth et al. 2005). Once recombination is suppressed, the Y(W) chromosome is subject to evolutionary forces that are expected to lead to loss of the chromosome’s genes (Charlesworth and Charlesworth 2000). The phylogenetically widespread observation of XO (ZO) species (Makino 1951) indicates that degeneration of the Y(W) may ultimately result in its complete loss; yet despite considerable work on the molecular evolution of particular Y chromosomes (Lahn et al. 2001; Bachtrog et al. 2008; Hughes et al. 2012) we still have a relatively poor understanding of the factors that govern the rates of Y(W) chromosome gain and loss.
Forces Promoting Y(W)-Chromosome Degeneration
In principle, the forces responsible for decay of these chromosomes include: Muller’s ratchet, background selection, Hill–Robertson effect, and genetic hitchhiking (Bachtrog 2013). The predicted inevitable decay of Y and W chromosomes has led to the idea that they are “born to be destroyed” (Steinemann and Steinemann 2005) and indeed these chromosomes are often dispensable [e.g., Lepidoptera (Traut et al. 2008), nematodes (Bull 1983), Orthoptera (Castillo et al. 2010), and Odonata (Kiauta 1969)]. Some groups, such as Coleoptera and Diptera, exhibit multiple independent losses of the Y chromosome (White 1977). In Drosophila, the tenuous persistence of the Y chromosome is evident in that the ancestral Y was likely lost long ago in an ancestor of Drosophila melanogaster while the current Y is likely a secondarily captured B chromosome (Carvalho and Clark 2005). In fact, recent analysis indicates that the ancestral Y may have been lost as part of a sex chromosome reversal where a formerly autosomal pair of chromosomes became the determinants of sex allowing the ancestral X to be fixed in the Drosophila lineage as an autosome (the dot chromosome) (Vicoso and Bachtrog 2013). In D. pseudoobscura the existing Y is homologous with an ancestral autosome, suggesting that the sex chromosomes fused with an autosome and the ancestral Y region was subsequently lost (Carvalho and Clark 2005). Even among taxa with generally persistent XY chromosome systems such as those in mammals, there is precedent for Y dispensability; both mole voles (Just et al. 1995) and spiny rats (Arakawa et al. 2002) have lost the ancestral Y chromosome.
Forces Promoting Y(W)-Chromosome Retention
Several lines of evidence, however, suggest that the evolution of Y(W) chromosomes is more complex than just inevitable decay. For instance, frequent turnover in the sex-determining chromosome (i.e., changes in the linkage group responsible for sex determination, so that a chromosome is not involved long enough for gene loss to occur) and/or intermittent recombination between sex chromosomes may play a role in persistence of the homomorphic sex chromosomes observed among most amphibians and fish (Stein et al. 2002; Woram et al. 2003; Van Doorn and Kirkpatrick 2007; Perrin 2009; Blaser et al. 2012; Guerrero et al. 2012). Sex-specific gene regulation may ameliorate situations with sexually antagonistic polymorphisms (Prince et al. 2010) and may further contribute to retention of old homomorphic sex chromosomes, as recently suggested for the emu (Vicoso et al. 2013).
In systems that retain the Y(W) chromosome despite considerable degeneration, selection may prevent complete gene loss and/or promote recruitment of genes from elsewhere in the genome. For example, degeneration of the human Y chromosome occurred in five waves over 200–300 million years of mammalian evolution (Hughes et al. 2012). Linear extrapolation, using the average rate of gene loss, predicts that the human Y would be lost within 10 million years (Aitken and Graves 2002); however, as the number of sites decline, so should the rate at which genes are lost (Bachtrog 2008). Recent analyses show that a few genes have been conserved due to purifying selection (Hughes et al. 2012) and that new genes that are important for male fertility have been transferred to the Y (Lahn et al. 2001). Retention of these “essential” male genes is aided by their frequent occurrence in palindromes where intrachromosomal gene conversion decreases the chance of loss and may also foster fixation of new genes by adaptive evolution (Betrán et al. 2012).
The strength of selection to retain Y(W)-linked genes should also be affected by the evolution of dosage compensation. If X(Z)-linked genes are expressed at low levels in males, this may lower males’ fitness, and purifying selection will then act against loss of Y(W) homologs unless dosage compensation evolves (Ohno 1967). While chromosome-wide (global) dosage compensation is the norm in most mammals and Drosophila, considerable data now show that it is incomplete in a broad range of animals, including trematodes (Vicoso and Bachtrog 2011), lepidopterans (Harrison et al. 2012), birds (Itoh et al. 2007), fish (Leder et al. 2010), and monotremes (Deakin et al. 2008). In these groups, the Y or W chromosome should decay more slowly since loss-of-function mutations will not be masked by increased expression of the X or Z copy.
Sex Chromosome Evolution in Coleoptera
The model systems for studying Y chromosome evolution, Drosophila and mammals, are ill suited to explore hypotheses about the tempo and mode of Y chromosome turnover because there are few transitions among sex chromosome states. Here we use comparative methods in the order Coleoptera to explore the evolution of Y chromosomes and generate hypotheses. Beetles are the most speciose order of eukaryotes and we have compiled karyotype data for thousands of species (available at www.uta.edu/karyodb).
To analyze sex chromosome changes in a comparative framework, we use DNA sequences for >1000 species in our karyotype database to estimate the phylogeny of Coleoptera. There have been few explicitly phylogenetic analyses of karyotype data (Flores et al. 2008; Leache and Sites 2009; Henning et al. 2011; Maddison and Leduc-Robert 2013), and to our knowledge, our analysis provides the first estimates of transition rates for sex chromosome turnover, Y chromosome decay, and Y chromosome loss, over such a large number of species. We find distinctly different patterns and rates of sex chromosome transitions between the two main suborders of beetles (Adephaga and Polyphaga). We propose that the much lower rate of Y chromosome loss in Polyphaga can be explained by the evolution of distance-pairing sex chromosomes that ensure proper meiotic segregation, even when no recombination occurs between the sex chromosomes.
Methods
Data collection
Karyotypes:
We performed a thorough literature search and compiled a comprehensive record of published Coleoptera karyotypes. To the extent possible, we reconciled historical karyotype data with currently accepted Coleopteran taxonomy (North American species: Arnett and Thomas 2000; Arnett et al. 2002; outside of North America: Beutel and Leschen 2005; Leschen et al. 2010].
Coleoptera are male heterogametic, and in most beetles the Y chromosome is smaller than the X (Smith and Virkki 1978). The most common sex chromosome systems in the literature are XY, XO, and Xy+ (see below). Here we denote sex chromosomes that undergo synapsis during meiosis as XY. In the vast majority of XY taxa, the synaptic chromosomes also form chiasmata (i.e., contain at least one region that can recombine) (Smith and Virkki 1978). However, achiasmatic male meiosis—where all chromosomes in males form synapses but do not recombine—has evolved four times in the suborder Adephaga. Two instances of achiasmatic male meiosis appear to involve only one or a few species and are probably of recent origin (Serrano 1981; Yadav and Burra 1987), while the other two instances are probably old and appear to be synapomorphies for the clades Trechitae and Cicindelini + Colyrinae (Galian et al. 2002; Maddison and Ober 2011).
Sex chromosome systems that form distance-pairing sex bivalents are denoted as Xy+. Such X and Y chromosomes are entirely nonrecombining. In these species the autosomes undergo normal synapsis and crossing over in both sexes and the X chromosomes do so in females. However, in males the X and Y chromosome pair at a distance with no synapsis and no opportunity for crossing over. In Xy+ species the Y is usually very small, often being described as “punctiform.” We denote species that have completely lost the Y chromosome as XO.
Sequence data:
Sequences for two mitochondrial genes (16s and COI) and five nuclear genes (18s, 28s, elongation factor 1, arginine kinase, and wingless) from 1140 operational taxonomic units (OTUs) representing members of 47 of the 59 families with karyotype data were downloaded from GenBank table of sequences available from Dryad Digital Repository at http://doi.org/10.5061/dryad.g8010. The karyotype database and the sequences available from GenBank contain overlapping, but nonidentical sets of species, so the sequence data were treated as follows. When the karyotype database and GenBank had a match at a level higher than species, we created “chimeric OTUs.” In these cases a composite branch was created by assigning all sequences and sex chromosome states found in that clade to the single higher level group. In this way, we created 280 genus level OTUs and 14 family level OTUs. To increase overlap in the matrix of gene sequences, we also created 18 anchoring OTUs. In these cases, relationships among several members of a monophyletic group in the karyotype database had sequences of a single gene, but another member of the taxon (not present in the karyotype database) had sequences for additional target genes. In these cases, an arbitrarily chosen member of the monophyletic group that was sampled for the single gene and was present in the karyotype database was assigned sequences for all of the otherwise unrepresented target genes. This effectively “anchors” the monophyletic group within the larger Coleoptera tree without impacting resolution within the group. For example, in the genus Curculio, we have karyotype data for three species, but those species only have sequence data available for COI. There is an additional species (Curculio niveopictus) with sequence data available for the 18S and 28S genes, but it does not have karyotype data. To increase overlap in our matrix, one of the species with COI sequence, C. nucum, was arbitrarily chosen to act as the anchor by having the 18s and 28s sequences from C. niveopictus assigned to it. This anchors the genus Curculio within the larger Coleopteran phylogeny, while preserving resolution within the genus. Both chimeric and anchoring OTUs are indicated as such in supplemental data files.
All sequences were aligned in MAFFT (Katoh et al. 2009). RNA genes (16s, 18s, and 28s) were then filtered with the program Gblocks to remove ambiguously aligned sites (Talavera and Castresana 2007). This resulted in alignments for 16s, 18s, and 28s of 544 bp, 1964 bp, and 404 bp in length, respectively. We used MEGA to manually adjust the alignments of protein coding genes (COI, wingless, elongation factor 1, and arginine kinase) to insure that the reading frame was maintained (Tamura et al. 2011); these alignments were 1567 bp, 585 bp, 1189 bp, and 810 bp in length, respectively. Finally all alignments were checked for poorly aligned taxa using GUIDANCE (Penn et al. 2010); 14 taxa were found to have few unambiguously aligned sites and were removed from our dataset. The alignments for our seven target genes were concatenated into a sparse supermatrix that contained 1126 OTUs and was 7063 bp in length. Most taxa do not have sequences for all genes, and the mean number of alignment sites with information was 1870.
Phylogenetic inference:
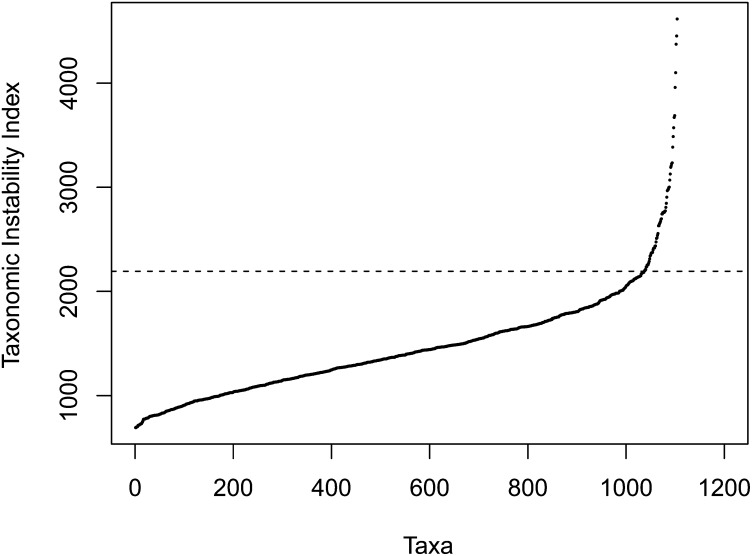
Inconsistency in the placement of a small subset of taxa among trees with equally probable topologies (rogue taxa) is a common problem in phylogenetic inference from sparse supermatrices (Thomson and Shaffer 2010). The problem is magnified by the computational burden of optimizing over the large number of OTUs in our dataset (e.g., 14,000 CPU hours on CIPRES (Miller et al. 2010) servers to complete the first phase below). Therefore, we divided our phylogenetic inference into two phases. The first phase used maximum likelihood inferences to build a collection of trees that we used to assess taxon instability. We computed 500 maximum likelihood trees using RAxML v 7.2.8 (Stamatakis 2009). Based on the resulting collection of trees, the instability index for all taxa was calculated (Aberer et al. 2013). High index values indicate that a taxon’s placement is variable among trees. The distribution of instability indices shown in Figure 1 indicates that 92% of taxa have indices <2194 but that above this, indices increase quickly. The 84 taxa with scores above this cutoff were removed from subsequent analyses, resulting in a dataset containing 744 Polyphaga taxa, 296 Adephaga taxa, and two outgroup species.
Figure 1.
Taxonomic instability indices based on 500 maximum likelihood tree inferences. The dashed line shows the chosen cutoff, an index of 2194. Most taxa, 93%, fall below this while above this value instability increases quickly.
The second phase of our phylogenetic inference employed Bayesian methods to produce a posterior sampling of ultrametric trees. The best maximum likelihood phylogram from the first phase was converted to an ultrametric tree using nonparametric rate smoothing in the R package APE (Paradis 2011). The resulting tree was subsequently used as input for two independent inferences in BEAST (v1.7.5) (Drummond and Rambaut 2007; Suchard and Rambaut 2009). We assumed a log-normal relaxed clock and used normal distributions to place priors on the age of seven nodes. The seven nodes represent the age of the order (Coleoptera = 285 my), both major suborders (Adephaga = 237.2 and Polyphaga = 270.5 my) and four arbitrarily chosen clades (Hydradephaga = 219.8 my, Elatridae = 139.9 my, Brentidae = 137.5 my, and Passalidae = 121.4 my). The standard deviations of the priors were set to reflect the 95% confidence interval of previous estimates (McKenna and Farrell 2009).
The two independent MCMC analyses required ∼70 million generations to converge on a parameter space with equal likelihood; to insure that they had reached stationarity, they were allowed to run for an additional 40 million generations. The phylogeny inferred from our sparse supermatrix is largely consistent with the previously most comprehensive family level analysis for Coleoptera (Hunt et al. 2007). Because of the computational demands of analyzing evolutionary rates over such large trees, parameter estimates for the evolutionary models below were marginalized over 100 randomly selected trees from the stationary phase of our two chains (henceforth referred to as “sampled trees”). The sampled trees had high resolution; a maximum clade credibility tree exhibited posterior probabilities >90% at 76% of the nodes. The 100 sampled trees were subsequently used to model sex chromosome evolution in Polyphaga and Adephaga and are publicly available from Dryad Digital Repository at http://doi.org/10.5061/dryad.g8010.
Trait Evolution:
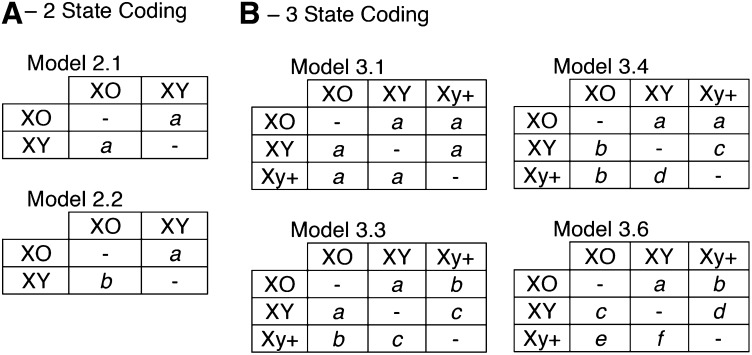
Sex chromosome systems are reported as discrete states (e.g., XY, Xy+, and XO) despite representing a fundamentally continuous, though probably not linear, process of differentiation. In modeling their evolution across a phylogeny, we must determine which states to include in the matrix of transition probabilities. For instance, if Xy+ is biologically equivalent to XY, then including the additional transition probabilities for that state will only add noise to the inferred rate of Y chromosome gain and loss. On the other hand, if Xy+ is a distinct state with different rates of transition to and from the XO state, then allowing for independent transition probabilities should provide a significantly better fit to the observed data. To assess how many states and rates best describe Y chromosome evolution in Coleoptera, we estimated transition probabilities under both two-state (XY/XO) and three-state (XY/Xy+/XO) models (Figure 2). For the two-state models, all XY and Xy+ taxa were both coded as XY. Species where literature reports note “NeoXY” and other complex sex chromosome systems (i.e., those with multiple X and/or Y chromosomes) were also included in the analysis based on whether the sex chromosomes form a synapse during male meiosis (XY) or not (Xy+). Of the 1042 OTUs in our tree, 88 taxa do not possess sex chromosome data and were coded as missing data, and do not affect our estimated rates of Y chromosome changes. Included in these are 23 parthenogenetic taxa as well as one haplodiploid taxon. The remaining 64 taxa have only the chromosome number available, and while homomorphic chromosomes are not reported in Coleoptera, some of these species may have sex chromosomes of this type. However, in most cases the investigators describe the chromosome squashes as inadequate to resolve the sex chromosomes; therefore, it would appear that sex chromosomes in beetles are rarely in a homomorphic state, and this should not bias our results.
Figure 2.
Models of sex chromosome system transitions. (A) Two-state coding model with taxa partitioned between XO and XY. Using this coding we fit models with one rate (model 2.1) and two rates (model 2.2). (B) Three-state coding model with taxa partitioned between XO, XY, and Xy+. Using this coding we fit models with 1, 2, 4, and 6 rate parameters. Model 3.4 is a constrained model allowing for comparison between two-state and three-state coding in Polyphaga. If XY and Xy+ are equivalent states, models 3.4 and 3.6 should perform equally well.
We estimated transition rates using BayesTraits, which allowed us to marginalize over uncertainty in phylogenetic inference and uncertainty in tip states (Pagel et al. 2004). For the tw-state coding we estimated rates assuming that all rates are equal (2.1) and also assuming that all rates differ (2.2). For the three-state coding, we again estimated rates assuming that all rates are equal (3.1), plus a time reversible model (3.3), a four-rate model (3.4), and a model with all six rates different (3.6). Finally model 3.4 is a nested version of model 3.6 in which we force both states XY and Xy+ to have a single rate of transition to XO and a single rate of transition back to XY from XO; this is equivalent to using two-state coding for the data. Comparing models 3.4 and 3.6 tests whether XY and Xy+ have significantly different transition rates to and from XO.
To improve computational feasibility of the rate estimates, we first used BayesTraits to perform a preliminary maximum likelihood analysis of the sex chromosome transition rates across all sampled trees for the two major suborders, Polyphaga and Adephaga. Since estimated transition rates were always <0.05, we conservatively set uniform priors between 0 and 0.1 on all transition rates for subsequent Bayesian analyses. None of our estimates were bounded by these priors. We adjusted the RateDev parameter for each run to insure that the acceptance rate of moves was between 20 and 40%. The marginal likelihood of each model was computed as the harmonic mean of the post burn-in likelihoods across all sampled trees. To compare models we used the marginal likelihoods (LS) to calculate the log Bayes Factor (LBF): LBF = 2(LS1−LS0), where LS1 is the more complex model and LS0 is the less complex model. We interpret LBF = 2–6 as positive support for the more complex model, 6–10 as strong support, and >10 as very strong support (Kass and Raftery 1995).
To further assess adequacy of our chosen models, we performed posterior predictive simulations (PPSs) (Rubin 1984). PPS datasets were created in the “R” environment (R Development Core Team 2013) using a custom function available in package evobiR. Briefly, we extracted rate matrices and associated trees from 1500 random points during the post burn-in phase of the BayesTraits MCMC runs for the best two-state and three-state models; each extraction was used to create a simulated dataset by evolving sex chromosomes over the extracted tree with the extracted rate estimates. The root state for each simulation was set so that the distribution of roots across each set of 1500 simulations matched the distribution of root states inferred during original parameterization of the model. We used these simulated datasets to compare the frequency of each tip state with the observed data. This same process was repeated on two subtrees within the suborder Adephaga to test whether specific subgroups had significantly different patterns and rates of transitions compared to what is expected based on rates estimated for the full suborder. Transition rates are reported as the mean probability of a transition per 100 million years ± the standard error.
Results
Coleopteran karyotypes
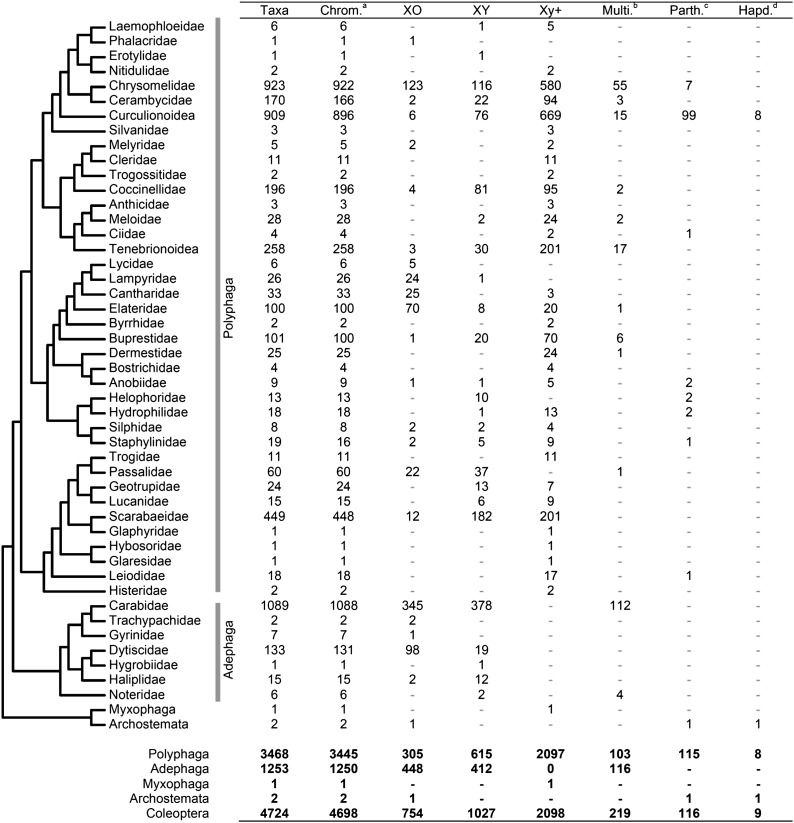
Our comprehensive database of Coleoptera karyotypes includes 4724 records based on 208 literature sources (Figure 3). The database is available online at www.uta.edu/karyodb and can be interrogated for any combination of: suborder, family, genus, sex chromosome system, presence of B chromosomes, and/or reproductive mode. Tables of selected data can be downloaded from the website. Karyotypes follow the format of Smith and Virkki (1978), the previously most comprehensive compilation of Coleopteran karyotypes. In the Coleoptera cytogenetic literature, distance-pairing sex bivalents are usually denoted with a lower case letter that describes how they are oriented during meiosis (e.g., “p” stands for parachute and indicates a large X chromosome with a small Y chromosome that appears suspended from it; “r” stands for rod and indicates that the X and Y are oriented end to end.) This format is maintained in our database, but for clarity and consistency with the broader literature, all distance-pairing sex bivalents are denoted by Xy+ in the present study. Whenever possible the meioformula is given in the database. For example 9+Xy+ means a haploid autosome count of 9 and distance-pairing sex chromosomes. The meioformula is not available for 470 species in the database where only the diploid number is reported in the literature, or for 9 haplodiploid records, and 116 parthenogenetic records. In total, data on sex chromosomes were available for 4223 species. Since records were available for only three species in the relatively small Coleopteran suborders Archostemata and Myxophaga, we analyzed only the major suborders Adephaga and Polyphaga.
Figure 3.
Cladogram illustrating the available cytogenetic data and distribution of sex chromosome systems in Coleoptera. Number of species in the karyotype database in each sex chromosome (or sex determination) state. Data and references are available at www.uta.edu/karyodb. The footnote symbols represent: athe number of species with chromosome number available; bsex chromosome systems with multiple X and or Y chromosomes; cspecies with parthenogenetic reproduction; and dspecies with haplodiploidy sex determination.
There is a striking difference between Adephaga and Polyphaga in the number of taxa with distance-pairing sex bivalents (Xy+). None of the 1253 Adephaga taxa in our dataset have Xy+ systems (Figure 3). Xy+ has been reported seven times in Adephaga, but subsequent investigations failed to replicate the observations (Serrano and Yadav 1984; Hughes and Angus 1999; Aradottir and Angus 2004). In contrast to Adephaga, 60% of Polyphaga species (2097/3468) exhibit Xy+.
Models of sex chromosome evolution
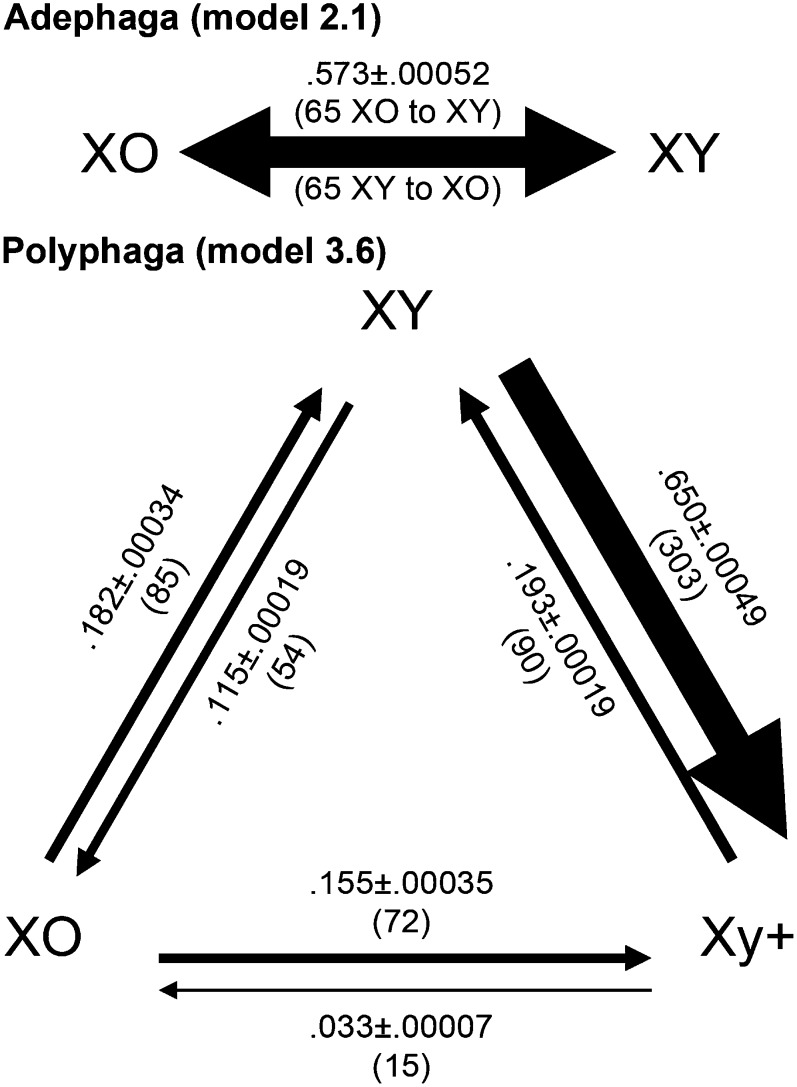
The difference in frequency of Xy+ systems between Adephaga and Polyphaga suggests that different biological mechanisms may act in each suborder. Therefore, we inferred parameters for models of sex chromosome evolution independently for each suborder. For Adephaga, we find that Y chromosomes are gained and lost at a rate of 0.573 ± 0.00052 gains and losses per 100 million years (Figure 4). Since only XY and XO states are observed in Adephaga, the model comparisons reduce to the difference between models with a single transition rate (model 2.1) and two rates (model 2.2; Figure 2). Comparison of marginal likelihoods (Table 1) reveals that both models fit the data equally well in Adephaga (LBF = 1.9; Table 1). Indeed, the two-rate model estimates nearly identical rates for XY to XO and the reverse (0.574 and 0.572, respectively). To verify that the best model is able to recapitulate the distribution of empirical data in Adephaga, we conducted PPSs under model 2.1. The simulated datasets are centered on the observed distribution of sex chromosomes (Figure 5A), indicating that the estimated parameterization of model 2.1 can produce outcomes similar to the observed data.
Figure 4.
Sex chromosome system transition rate estimates. Rates are reported as the probability of transition per 100 million years ± the standard error. Parentheses indicate the mean number of transitions inferred. In Adephaga the mean number of transitions is the sum of transitions between both states.
Table 1. Marginal likelihoods and model comparisons for models of sex chromosome evolution.
| Suborder | Model | Marginal likelihood | Comparison | Log Bayes factor |
|---|---|---|---|---|
| Adephaga | 2.1 | −103.5 | 2.1 vs. 2.2 | 1.9 |
| 2.2 | −102.6 | |||
| Polyphaga | 2.1 | −131.2 | 2.1 vs. 2.2 | 11.8a |
| 2.2 | −125.3 | 3.1 vs. 3.3 | 30.5a | |
| 3.1 | −393.8 | 3.3 vs. 3.6 | 21.5a | |
| 3.3 | −378.6 | 3.4 vs. 3.6 | 10.7a | |
| 3.4 | −367.8 | |||
| 3.6 | −362.4 |
Indicates very strong support for the more complex model.
Figure 5.
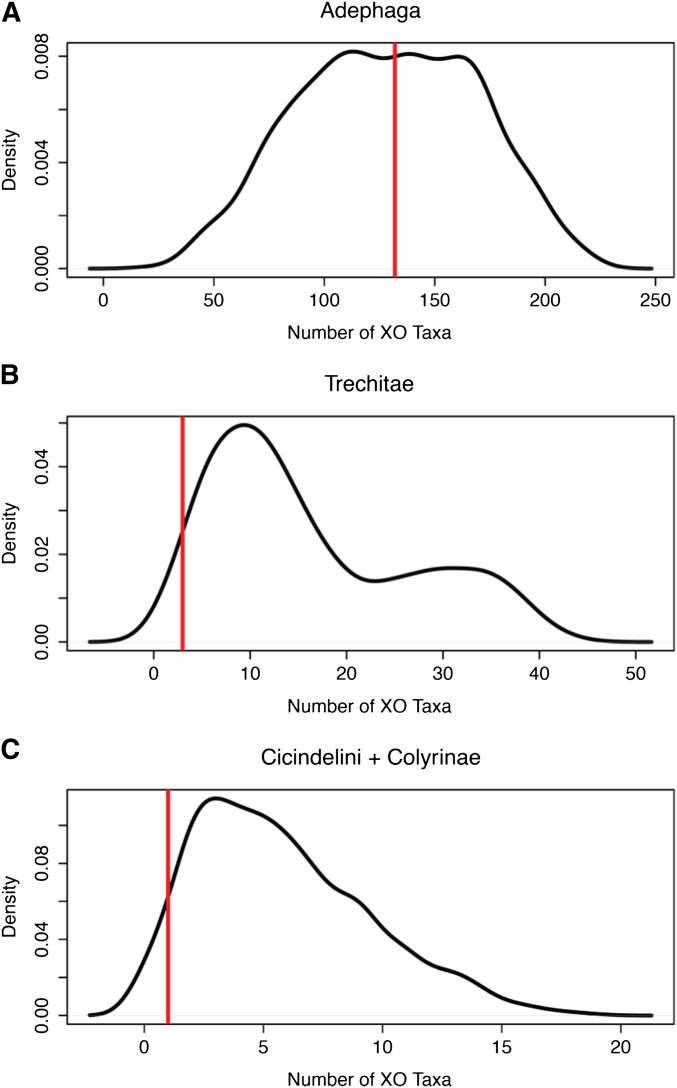
Distribution of PPS datasets in the suborder Adephaga. The black lines indicate the density of simulated datasets; the vertical red lines indicate the number of taxa observed in the XY state. (A) Adequate performance of model 2.1 in Adephaga is evident by the concentration of datasets similar to the observed data. The poor performance of model 2.1 in the subtrees composed of the clades Trechitae (B) and Cicindelini + Colyrinae (C) is evidence that these clades have higher retention rates of the Y chromosome than is expected for groups in the suborder Adephaga.
For the Polyphaga, the more complex of the two-state models (2.2) is preferred in comparison with model 2.1 (LBF = 11.8; Table 1). Likewise, the most complex three-state model (3.6; LBF = 10.7–30.5; Table 1) is preferred in comparison to all other three-state models. Comparison of models 3.4 and 3.6 indicates that the three-state coding is more appropriate than two-state coding (LBF = 10.7; Table 1). Therefore, Xy+ and XY states differ biologically in terms of the rates of Y chromosome changes they undergo. Changes from XY to Xy+ have the highest estimated rate among all transitions in Coleoptera (0.65 ± 0.00049), while the rates of transitions from Xy+ to any other state are the lowest (to XO = 0.033 ± 0.00007; to XY = 0.19 ± 0.00019; Figure 4), i.e., Xy+ distance-pairing sex chromosomes in Polyphaga are the most evolutionarily stable sex chromosome state in Coleoptera in our analyses. PPSs under model 3.6 show that our phylogenies and model parameterization can produce outcomes similar to the observed data (Figure 6).
Figure 6.
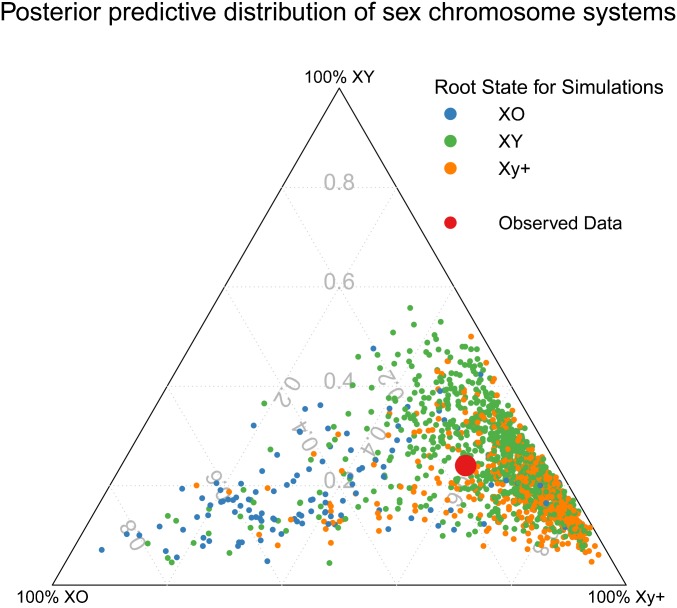
Distribution of PPS datasets in the suborder Polyphaga. Each circle represents a simulation based on the parameter estimates from model 3.6 and are colored to reflect the root state chosen for the simulation. The larger red circle indicates the observed data. Axes represent the percent of terminal taxa in each of the three sex chromosome states. The empirical observation being near the most dense part of the distribution of simulation results indicates that model 3.6 adequately predicts sex chromosome evolution in Polyphaga. The tail of simulations with a high proportion of XO taxa arises in large part from runs where XO was assigned as the root state, which is unlikely to be the true ancestral state in Polyphaga.
Discussion
The rates and patterns of Y chromosome turnover are distinctly different between the two largest Coleoptera suborders, Polyphaga and Adephaga. We suggest that important differences in the meiotic machinery, and particularly sex chromosome pairing, between these lineages indicates that previously unappreciated evolutionary forces may influence Y chromosome evolution.
In the suborder Adephaga, a Y chromosome has a 57% chance of being lost or gained per 100 million years. The 65 Y chromosome losses (Figure 4) in Adephaga, are not difficult to explain. Presumably, these losses reflect the standard population genetic forces promoting Y decay noted earlier, and that gene losses ultimately reach the point where the Y chromosome becomes dispensable (Steinemann and Steinemann 2005). Our estimate of an equal number of Y chromosome gains in Adephaga is more interesting. While some sex chromosomes are famous for avoiding decay and remaining homomorphic [e.g., ratite birds (Adolfsson and Ellegren 2013; Vicoso et al. 2013) and anurans (Stock et al. 2011; Stock et al. 2013)], the situation in Adephaga is different. In this group, new Y chromosomes are evolving at the same rate they are lost.
There are two mechanisms for gaining a Y chromosome: (1) fusion of all or part of an autosome to the X (White 1977; Charlesworth and Charlesworth 1980; Watson et al. 1991; Veltsos et al. 2008) or (2) capture of a supernumerary (B) chromosome (Carvalho 2002). To determine whether transitions from XO to XY are the result of fusions between the X chromosome and a whole autosome we used stochastic character mapping (Huelsenbeck et al. 2003; Revell 2012) of chromosome number (downloaded from uta.edu/karyodb/) and sex chromosome system, to calculate the proportion of branches where a Y chromosome gain co-occurs with a reduction in the number of autosomes. This method suggests that at least 49% of the Y chromosome gains in Adephaga are the result of fusions between the X chromosome and a whole autosome. This is a far more frequent co-occurrence than the 2.7% expected if gaining a new Y chromosome was independent of decreasing chromosome number (We infer that Y chromosomes are gained on 22.7% of branches in the Adephaga tree, and we infer chromosome losses on 11.9% of branches; the probability of independent co-occurrence is then 0.227*0.119 = 0.027). This may indicate that sexually antagonistic loci are sufficiently common in Adephaga genomes that fusions of an autosome to the X are often favored. Without data for Y chromosome homology, we cannot say whether the remaining 51% of branches are primarily due to translocations (i.e., fusion of partial chromosomes) or B-chromosome captures.
In contrast to Adephaga, we estimate that Y chromosomes in the suborder Polyphaga are more than twice as readily gained as lost (34% probability of gain per 100 my, 157 total gains; 15% probability of loss per 100 my, 69 total losses; Figure 4). To investigate the source of Y chromosome gains, we mapped transitions from the XO state and transitions in chromosome number as we did for Adephaga above. We find that only 27% of the Y gains in Polyphaga coincide with reductions in chromosome number, but that this is still far more frequent than the 0.5% expected if the events were independent (We infer Y chromosomes gains on 6.8% of branches in the Polyphaga tree, and chromosome losses on 7.6% of branches; the probability of independent co-occurrence is then 0.068*0.076 = 0.005). While fusions clearly coincide with Y chromosome gains in both suborders, in comparison with Adephaga, a much larger proportion of Polyphaga Y chromosome gains appear on branches where no reduction in chromosome number is inferred. This suggests that relative to Adephaga, a larger proportion of Y chromosome gains in Polyphaga result from either B-chromosome capture or the fusion of only a portion of an autosome to the X chromosome. The presence of an existing mechanism for segregation of unpaired sex chromosomes in Polyphaga may facilitate the capture of B chromosomes and contribute to the difference in Y chromosome origins between beetle suborders.
Rates of Y chromosome loss are also interesting in Polyphaga because they are so low. The nonrecombining Xy+ sex chromosomes in Polyphaga species do not contain a PAR and thus the entire Y chromosome is subject to the population genetic forces promoting Y decay. If decay followed by loss is the dominant source of XO species, as often suggested, it is surprising that Xy+ systems lose their Y 3.5 times less frequently than XY systems with a PAR (Figure 4). Consequently, we suggest that some evolutionary force(s) promoting retention must be acting in Polyphagan Xy+ systems. As noted in the introduction, frequent turnover in the sex-determining chromosome and or intermittent recombination can promote retention of homomorphic sex chromosomes, but these mechanisms do not apply in species with highly degenerate Xy+ sex chromosomes. Other hypotheses for retention that could apply to the situation in Polyphaga involve purifying selection either due to “essential” male genes or haploinsufficiency (Li et al. 2013). Although little is known about the genes present on the Y chromosomes of Coleoptera, it seems unlikely that genes required for male viability are widespread on the Y chromosome, since XO species occur in 24 of 59 Coleoptera families studied, and our estimates indicate that the Y chromosome has been independently lost ∼69 and 65 times in Polyphaga and Adephaga, respectively (Figure 4).
The argument for retention of the Y chromosome due to haploinsufficiency of X-linked genes in males depends on whether dosage compensation occurs and to what extent. However, it seems unlikely to explain our results. Dosage compensation has been studied in only a single species of Coleopteran, the red flour beetle Tribolium castaneum, a polyphagan beetle. In this species, chromosome wide dosage compensation of the X occurs in males, such that, on average, expression from one X equals that from two autosomes (Prince et al. 2010). This type of chromosome-wide upregulation should provide haplosufficiency for all the genes on the X chromosome, reducing purifying selection to maintain Y homologs. We lack information for most beetle taxa, but the fact that the Y in Xy+ species is typically punctiform also indicates that most X chromosome genes must be haplosufficient so that haploinsufficiency is unlikely to be a general explanation for the exceedingly rare loss of Y chromosomes from Xy+ species.
What then can explain the relative stability of Y chromosomes in Polyphaga? We propose the fragile Y hypothesis: when proper segregation of the sex chromosomes depends on chiasmata, recurring selection to reduce recombination between loci in the PAR (e.g., loci with sexually antagonistic polymorphisms) and the sex-determining locus (1) reduces the size of the PAR and consequently opportunities for chiasma formation, and (2) this leads to an increased probability of producing aneuploid gametes (Raudsepp et al. 2012) creating increased opportunities for Y loss (hence, fragile Y).
Our hypothesis makes two predictions: (1) As the PAR shrinks, selection should favor segregation mechanisms that do not rely on chiasmata. (2) Taxa that evolve achiasmatic pairing should have lower rates of Y chromosome loss. We tested within the suborder Adephaga for evidence of this pattern. While no adephagans have distance-pairing sex chromosomes of the type found in Polyphaga, complete achiasmatic meiosis has arisen at least four times independently in Adephaga. Two of these origins involve only one or a handful of species in the genera Egadroma and Calasoma, suggesting that they arose relatively recently (Serrano 1981). However two of the origins involve the larger clades of Trechitae (Maddison and Ober 2011) and Cicindelini + Colyrinae (Galian et al. 2007), and must be older. Both these clades lose the Y more rarely than expected, consistent with our predictions. In our dataset a total of 45 Trechitae species are represented. Within these, at most three changes from XY to XO have occurred; for the clade including Cicindelini + Colyrinae we have 21 species and only a single such change. PPS analyses for these clades suggest that both groups have fewer XO species (i.e., fewer Y chromosome losses) than expected, based on the overall transition rates for Adephaga. In Trechitae 95% of simulations predict more XO species than we observe (mean expected by simulation = 16.5, vs. 3 observed; Figure 5B). In Cicindelini + Colyrini 92% of the simulations predict more XO species than the empirical observation (mean expected by simulation = 5.8, vs. 1 observed; Figure 5C).
Mammals:
An additional opportunity to test the fragile Y hypothesis is available in mammals. The infraclasses Eutheria (placental mammals) and Metatheria (marsupials) offer a parallel example to Adephaga and Polyphaga in beetles. The sex chromosomes of metatherian mammals segregate in males without the presence of a PAR or chiasmata (Page et al. 2006) and no cases of Y loss are reported. In contrast, the Eutherians generally require a PAR region that forms chiasmata to faithfully segregate the sex chromosomes, and in taxa with small PARs, the Y is occasionally lost (Fernández-Donoso et al. 2010). Among eutherian mammals, the rodents have the smallest documented PAR (Raudsepp and Chowdhary 2008), and it is within the rodents that we see multiple independent responses to the forces we ascribe to a fragile Y. First, within the family Cricetidae, the genus Microtus exhibits at least three origins of achiasmatic sex chromosomes, and as we expect, there are no reported Y chromosome losses (Table 2) (Borodin et al. 2012). In contrast, the closely related mole vole genus Ellobius has not evolved achiasmatic meiosis but shows at least one, and possibly two instances of Y chromosome loss (Just et al. 1995). Second, within the largest family of mammals, Muridae, we find additional origins of achiasmatic male meiosis and Y chromosome loss. The subfamily Gerbillinae has evolved achiasmatic sex chromosomes and Y losses are not reported, whereas in a related subfamily Murinae, which has not evolved achiasmatic meiosis, the spiny rat genus Tokudaia has three species, two of which have lost the Y chromosome (Arakawa et al. 2002), and a third whose Y chromosome is fused with an autosome, rejuvenating the PAR and escaping potential difficulty in segregation during male meiosis (Murata et al. 2012). While this is a small sample, it is worth noting that in mammals the Y often carries genes essential for male viability and, all else being equal, is thus likely to be under stronger selection to be retained than in Coleoptera. The repeated evolution of either achiasmatic meiosis, or Y chromosome loss, in eutherians with the smallest PAR size is precisely what the fragile Y hypothesis predicts.
Table 2. Summary of mammals with achiasmatic X–Y segregation or Y chromosome losses.
| Clade | X–Y segregation mechanism in males | Taxa | Independent Y losses | Citation | |
|---|---|---|---|---|---|
| Cricetidae | Microtus | Predominately achiasmatica | 29 | 0 | Borodin et al. (2012) |
| Ellobius | Chiasmatic | 3 | 1–2 | Just et al. (1995) | |
| Muridae | Gerbillinae | Achiasmatic | 9b | 0 | Ratomponirina et al. (1986, 1989) |
| Tokudaia | Chiasmatic | 3 | 1 | (Arakawa et al. (2002) | |
Seventeen species are achiasmatic; 12 are chiasmatic with at least three independent origins of achiasmatic segregation.
Four species of Gerbillinae have experienced autosome sex chromosome fusions, the autosomal portion of which undergoes crossover.
Conclusion
Our analysis suggests that meiotic mechanisms play an important, previously unappreciated role in the tempo of Y chromosome gain and loss. Additionally, given the relatively widespread loss of Y chromosomes among Coleopterans, sex determination seems likely to often involve an X counting system such as in D. melanogaster (Bridges 1921) where the Y plays little role in sex determination (otherwise it would not be dispensable) (Bachtrog 2013). Finally, despite being the largest analysis of its kind, our analysis of sex chromosome evolution based on available karyotype data is relatively coarse. A more nuanced understanding of sex chromosome evolution, one that tests the predictions of this study, would benefit from genomic data that allow for assignment of chromosomal homologies. Given the large number of novel Y chromosomes arising in Coleoptera (225; Figure 4), many of which are fusions or potential B-chromosome captures, it will be interesting to investigate whether some genes (or chromosomes) are recurrently recruited to Y chromosomes.
Acknowledgments
We thank D. Charlesworth, E. Betran, A. Williford, and two anonymous reviewers for helpful comments on the manuscript. Michael Landis provided valuable advice on model comparisons. This work was supported by National Institutes of Health grant R01GM065414.
Footnotes
Communicating editor: D. Charlesworth
Literature Cited
- Aberer A. J., Krompass D., Stamatakis A., 2013. Pruning rogue taxa improves phylogenetic accuracy: an efficient algorithm and webservice. Syst. Biol. 62: 162–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolfsson S., Ellegren H., 2013. Lack of dosage compensation accompanies the arrested stage of sex chromosome evolution in ostriches. Mol. Biol. Evol. 30: 806–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken R. J., Graves J. A. M., 2002. Human spermatozoa: the future of sex. Nature 415: 963. [DOI] [PubMed] [Google Scholar]
- Aradottir G., Angus R., 2004. A chromosomal analysis of some water beetle species recently transferred from Agabus Leach to Ilybius Erichson, with particular reference to the variation in chromosome number shown by I. montanus Stephens (Coleoptera: Dytiscidae). Hereditas 140: 185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa Y., Nishida-Umehara C., Matsuda Y., Sutou S., Suzuki H., 2002. X-chromosomal localization of mammalian Y-linked genes in two XO species of the Ryukyu spiny rat. Cytogenet. Genome Res. 99: 303–309 [DOI] [PubMed] [Google Scholar]
- Arnett R., Thomas M., 2000. American Beetles; Archostemata, Myxophaga, Adephaga, Polyphaga: Staphyliniformia. CRC Press, Boca Raton, FL [Google Scholar]
- Arnett R., Thomas M., Skelley P., Frank J., 2002. American Beetles; Polyphaga: Scarabaeoidea Through Curculionoidea. CRC Press, Boca Raton, FL [Google Scholar]
- Arunkumar K., Mita K., Nagaraju J., 2009. The silkworm Z chromosome is enriched in testis-specific genes. Genetics 182: 493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D., 2008. The temporal dynamics of processes underlying Y chromosome degeneration. Genetics 179: 1513–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D., 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 14: 113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D., Hom E., Wong K. M., Maside X., de Jong P., 2008. Genomic degradation of a young Y chromosome in Drosophila miranda. Genome Biol. 9: R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betrán E., Demuth J. P., Williford A., 2012. Why chromosome palindromes? Int. J. Evol. Biol., 207958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutel, R., and R. Leschen, 2005 Handbook of Zoology, Band IV, Part 38, Coleoptera, Beetles. Vol. 1: Morphology and Systematics (Archostemata, Adephaga, Myxophaga, Polyphaga partim) Walter de Gruyter, Berlin. [Google Scholar]
- Blaser O., Grossen C., Neuenschwander S., Perrin N., 2012. Sex-chromosome turnovers induced by deleterious mutation load. Evolution 67: 635–645 [DOI] [PubMed] [Google Scholar]
- Borodin P. M., Basheva E. A., Torgasheva A. A., Dashkevich O. A., Golenishchev F. N., et al. , 2012. Multiple independent evolutionary losses of XY pairing at meiosis in the grey voles. Chromosome Res. 20: 259–268 [DOI] [PubMed] [Google Scholar]
- Bridges C., 1921. Triploid intersexes in Drosophila. Science 54: 252–254 [DOI] [PubMed] [Google Scholar]
- Bull J. J., 1983. Evolution of Sex Determining Mechanisms. Benjamin/Cummings Publishing, Menlo Park, CA. [Google Scholar]
- Carvalho A. B., 2002. Origin and evolution of the Drosophila Y chromosome. Curr. Opin. Genet. Dev. 12: 664–668 [DOI] [PubMed] [Google Scholar]
- Carvalho A. B., Clark A. G., 2005. Y chromosome of D. pseudoobscura is not homologous to the ancestral Drosophila Y. Science 307: 108–110 [DOI] [PubMed] [Google Scholar]
- Castillo E. R., Marti D. A., Bidau C. J., 2010. Sex and neo-sex chromosomes in Orthoptera: a review*. J. Orthoptera Res. 19: 213–231 [Google Scholar]
- Charlesworth B., 1991. The evolution of sex chromosomes. Science 251: 1030. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 2000. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Coyne J., Barton N., 1987. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 130: 113–146 [Google Scholar]
- Charlesworth D., Charlesworth B., 1980. Sex differences in fitness and selection for centric fusions between sex-chromosomes and autosomes. Genet. Res. 35: 205–214 [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., Marais G., 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95: 118–128 [DOI] [PubMed] [Google Scholar]
- Deakin J. E., Hore T. A., Koina E., Graves J. A. M., 2008. The status of dosage compensation in the multiple X chromosomes of the platypus. PLoS Genet. 4: e1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A., 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Donoso, R., S. Berríos, J. S. Rufas and J. Page, 2010 Marsupial sex chromosome behaviour during male meiosis, pp. 187–206 in Marsupial Genetics and Genomics Springer, New York.
- Flores S. V., Evans A. L., McAllister B. F., 2008. Independent origins of new sex-linked chromosomes in the melanica and robusta species groups of Drosophila. BMC Evol. Biol. 8: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galian J., Hogan J. E., Vogler A. P., 2002. The origin of multiple sex chromosomes in tiger beetles. Mol. Biol. Evol. 19: 1792–1796 [DOI] [PubMed] [Google Scholar]
- Galian J., Proenca S. J., Vogler A. P., 2007. Evolutionary dynamics of autosomal-heterosomal rearrangements in a multiple-X chromosome system of tiger beetles (Cicindelidae). BMC Evol. Biol. 7: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves J. A. M., 2006. Sex chromosome specialization and degeneration in mammals. Cell 124: 901–914 [DOI] [PubMed] [Google Scholar]
- Guerrero R., Kirkpatrick M., Perrin N., 2012. Cryptic recombination in the ever-young sex chromosomes of Hylid frogs. J. Evol. Biol. 25: 1947–1954 [DOI] [PubMed] [Google Scholar]
- Haldane J. B., 1922. Sex ratio and unisexual sterility in hybrid animals. J. Genet. 12: 101–109 [Google Scholar]
- Harrison P. W., Mank J. E., Wedell N., 2012. Incomplete sex chromosome dosage compensation in the Indian meal moth, Plodia interpunctella, based on de novo transcriptome assembly. Genome Biol. Evol. 4: 1118–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning F., Moyses C. B., Calcagnotto D., Meyer A., de Almeida-Toledo L. F., 2011. Independent fusions and recent origins of sex chromosomes in the evolution and diversification of glass knife fishes (Eigenmannia). Heredity (Edinb) 106: 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J. P., Nielsen R., Bollback J. P., 2003. Stochastic mapping of morphological characters. Syst. Biol. 52: 131–158 [DOI] [PubMed] [Google Scholar]
- Hughes C., Angus R., 1999. The karyotype of the squeak beetle, Hygrobia hermanni (F.)(Coleoptera: Hygrobiidiae). Koleopterol. Rundsch. 69: 41–45 [Google Scholar]
- Hughes J. F., Skaletsky H., Brown L. G., Pyntikova T., Graves T., et al. , 2012. Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature 483: 82–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt T., Bergsten J., Levkanicova Z., Papadopoulou A., John O. S., et al. , 2007. A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science 318: 1913–1916 [DOI] [PubMed] [Google Scholar]
- Itoh Y., Melamed E., Yang X., Kampf K., Wang S., et al. , 2007. Dosage compensation is less effective in birds than in mammals. J. Biol. 6: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Just W., Rau W., Vogel W., Akhverdian M., Fredga K., et al. , 1995. Absence of Sry in species of the vole Ellobius. Nat. Genet. 11: 117. [DOI] [PubMed] [Google Scholar]
- Kass R. E., Raftery A. E., 1995. Bayes factors. J. Am. Stat. Assoc. 90: 773–795 [Google Scholar]
- Katoh K., Asimenos G., Toh H., 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 537: 39–64 [DOI] [PubMed] [Google Scholar]
- Kiauta B., 1969. Sex chromosomes and sex determining mechanisms in Odonata, with a review of the cytological conditions in the family Gomphidae, and references to the karyotypic evolution in the order. Genetica 40: 127–157 [DOI] [PubMed] [Google Scholar]
- Lahn B. T., Pearson N. M., Jegalian K., 2001. The human Y chromosome, in the light of evolution. Nat. Rev. Genet. 2: 207–216 [DOI] [PubMed] [Google Scholar]
- Leache A. D., Sites J. W., Jr, 2009. Chromosome evolution and diversification in North American spiny lizards (genus Sceloporus). Cytogenet. Genome Res. 127: 166–181 [DOI] [PubMed] [Google Scholar]
- Leder E. H., Cano J. M., Leinonen T., O’Hara R. B., Nikinmaa M., et al. , 2010. Female-biased expression on the X chromosome as a key step in sex chromosome evolution in threespine sticklebacks. Mol. Biol. Evol. 27: 1495–1503 [DOI] [PubMed] [Google Scholar]
- Leschen, R. A. B., R. Beutel, J. F. Lawrence, and S. A. Ślipiński, 2010 Handbook of Zoology, Band IV, Part 39, Coleoptera: Morphology and Systematics, Elateroidea, Bostrichiformia, Cucujifornia Partim De Gruyter, Berlin. [Google Scholar]
- Li G., Davis B., Raudsepp T., Wilkerson A. P., Mason V., et al. , 2013. Comparative analysis of mammalian Y chromosomes illuminates ancestral structure and lineage-specific evolution. Genome Res. 23: 1486–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison, D. R., and K. A. Ober, 2011 Phylogeny of minute carabid beetles and their relatives based upon DNA sequence data (Coleoptera, Carabidae, Trechitae). Zookeys: 229–260. [DOI] [PMC free article] [PubMed]
- Maddison W. P., Leduc-Robert G., 2013. Multiple origins of sex chromosome fusions correlated with chiasma localization in Habronattus jumping spiders (Araneae: Salticidae). Evolution 67: 2258–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., 1951. An Atlas of the Chromosome Numbers in Animals. Iowa State College Press, Ames, Iowa [Google Scholar]
- McKenna, D. D., and B. D. Farrell, 2009 Beetles (Coleoptera), pp. 278 in The Timetree of Life Oxford University Press, New York. [Google Scholar]
- Mikkelsen T. S., Hillier L. W., Eichler E. E., Zody M. C., Jaffe D. B., et al. , 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437: 69–87 [DOI] [PubMed] [Google Scholar]
- Miller, M. A., W. Pfeiffer, and T. Schwartz, 2010 Creating the CIPRES Science Gateway for inference of large phylogenetic trees, pp. 1–8 in Gateway Computing Environments Workshop (GCE) IEEE, New York. [Google Scholar]
- Murata C., Yamada F., Kawauchi N., Matsuda Y., Kuroiwa A., 2012. The Y chromosome of the Okinawa spiny rat, Tokudaia muenninki, was rescued through fusion with an autosome. Chromosome Res. 20: 111–125 [DOI] [PubMed] [Google Scholar]
- Ohno S., 1967. Sex Chromosomes and Sex-Linked Genes, Springer, New York [Google Scholar]
- Page J., Viera A., Parra M. T., de la Fuente R., Suja J. A., et al. , 2006. Involvement of synaptonemal complex proteins in sex chromosome segregation during marsupial male meiosis. PLoS Genet. 2: e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M., Meade A., Barker D., 2004. Bayesian estimation of ancestral character states on phylogenies. Syst. Biol. 53: 673–684 [DOI] [PubMed] [Google Scholar]
- Paradis E., 2011. Analysis of Phylogenetics and Evolution with R, Springer, New York [Google Scholar]
- Penn O., Privman E., Ashkenazy H., Landan G., Graur D., et al. , 2010. GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Res. 38: W23–W28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin N., 2009. Sex reversal: A fountain of youth for sex chromosomes? Evolution 63: 3043–3049 [DOI] [PubMed] [Google Scholar]
- Phillips B. C., Edmands S., 2012. Does the speciation clock tick more slowly in the absence of heteromorphic sex chromosomes? Bioessays 34: 166–169 [DOI] [PubMed] [Google Scholar]
- Presgraves D. C., 2008. Sex chromosomes and speciation in Drosophila. Trends Genet. 24: 336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince E. G., Kirkland D., Demuth J. P., 2010. Hyperexpression of the X chromosome in both sexes results in extensive female bias of X-linked genes in the flour beetle. Genome Biol. Evol. 2: 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, 2013 R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna (http://www.R-project.org). [Google Scholar]
- Ratomponirina C., Viegas-Pequignot E., Dutrillaux B., Petter F., Rumpler Y., 1986. Synaptonemal complexes in Gerbillidae: probable role of intercalated heterochromatin in gonosome-autosome translocations. Cytogenet. Cell Genet. 43: 161–167 [DOI] [PubMed] [Google Scholar]
- Ratomponirina C., Viegaspequignot E., Petter F., Dutrillaux B., Rumpler Y., 1989. Synaptonemal complex study in some species of Gerbillidae without heterochromatin interposition. Cytogenet. Cell Genet. 52: 23–27 [DOI] [PubMed] [Google Scholar]
- Raudsepp T., Chowdhary B. P., 2008. The horse pseudoautosomal region (PAR): characterization and comparison with the human, chimp and mouse PARs. Cytogenet. Genome Res. 121: 102–109 [DOI] [PubMed] [Google Scholar]
- Raudsepp T., Das P., Avila F., Chowdhary B., 2012. The pseudoautosomal region and sex chromosome aneuploidies in domestic species. Sex Dev. 6: 72–83 [DOI] [PubMed] [Google Scholar]
- Revell L. J., 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3: 217–223 [Google Scholar]
- Rice W. R., 1987. Genetic hitchhiking and the evolution of reduced genetic activity of the Y sex chromosome. Genetics 116: 161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice W. R., 1994. Degeneration of a nonrecombining chromosome. Science 263: 230–232 [DOI] [PubMed] [Google Scholar]
- Rubin D. B., 1984. Bayesianly justifiable and relevant frequency calculations for the applies statistician. Ann. Stat. 12: 1151–1172 [Google Scholar]
- Serrano J., 1981. Male achiasmatic meiosis in Caraboidea (Coleoptera, Adephaga). Genetica 57: 131–137 [Google Scholar]
- Serrano J., Yadav J., 1984. Chromosome numbers and sex-determining mechanisms in adephagan Coleoptera. Coleopt. Bull. 38: 335–357 [Google Scholar]
- Smith, S. G., and N. Virkki, 1978 Animal Cytogenetics: Vol. 3. Insecta. Coleoptera Borntraeger, Berlin. [Google Scholar]
- Stamatakis A., 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Stein J., Reed K. M., Wilson C. C., Phillips R. B., 2002. A sex-linked microsatellite locus isolated from the Y chromosome of lake charr, Salvelinus namaycush. Environ. Biol. Fishes 64: 211–216 [Google Scholar]
- Steinemann S., Steinemann M., 2005. Y chromosomes: born to be destroyed. Bioessays 27: 1076–1083 [DOI] [PubMed] [Google Scholar]
- Stock M., Horn A., Grossen C., Lindtke D., Sermier R., et al. , 2011. Ever-young sex chromosomes in European tree frogs. PLoS Biol. 9: e1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock M., Savary R., Betto-Colliard C., Biollay S., Jourdan-Pineau H., et al. , 2013. Low rates of X-Y recombination, not turnovers, account for homomorphic sex chromosomes in several diploid species of Palearctic green toads (Bufo viridis subgroup). J. Evol. Biol. 26: 674–682 [DOI] [PubMed] [Google Scholar]
- Suchard M. A., Rambaut A., 2009. Many-core algorithms for statistical phylogenetics. Bioinformatics 25: 1370–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G., Castresana J., 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56: 564–577 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., et al. , 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson R. C., Shaffer H. B., 2010. Sparse supermatrices for phylogenetic inference: taxonomy, alignment, rogue taxa, and the phylogeny of living turtles. Syst. Biol. 59: 42–58 [DOI] [PubMed] [Google Scholar]
- Traut W., Sahara K., Marec F., 2008. Sex chromosomes and sex determination in Lepidoptera. Sex Dev. 1: 332–346 [DOI] [PubMed] [Google Scholar]
- Vallender E. J., Lahn B. T., 2004. How mammalian sex chromosomes acquired their peculiar gene content. Bioessays 26: 159–169 [DOI] [PubMed] [Google Scholar]
- Van Doorn G., Kirkpatrick M., 2007. Turnover of sex chromosomes induced by sexual conflict. Nature 449: 909–912 [DOI] [PubMed] [Google Scholar]
- Veltsos P., Keller I., Nichols R. A., 2008. The inexorable spread of a newly arisen neo-Y chromosome. PLoS Genet. 4: e1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B., Bachtrog D., 2011. Lack of global dosage compensation in Schistosoma mansoni, a female-heterogametic parasite. Genome Biol. Evol. 3: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B., Bachtrog D., 2013. Reversal of an ancient sex chromosome to an autosome in Drosophila. Nature 499: 332–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B., Kaiser V. B., Bachtrog D., 2013. Sex-biased gene expression at homomorphic sex chromosomes in emus and its implication for sex chromosome evolution. Proc. Natl. Acad. Sci. USA 110: 6453–6458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E. T., Demuth J. P., 2012. Haldane’s rule in marsupials: What happens when both sexes are functionally hemizygous? J. Hered. 103: 453–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J. M., Spencer J. A., Riggs A. D., Graves J. A. M., 1991. Sex chromosome evolution: platypus gene mapping suggests that part of the human X chromosome was originally autosomal. Proc. Natl. Acad. Sci. USA 88: 11256–11260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergaard M., 1958. The mechanism of sex determination in dioecious flowering plants. Adv. Genet. 9: 217–281 [DOI] [PubMed] [Google Scholar]
- White, M. J. D., 1977 Animal Cytology and Evolution, Cambridge University Press, Cambridge, UK. [Google Scholar]
- Woram R. A., Gharbi K., Sakamoto T., Hoyheim B., Holm L.-E., et al. , 2003. Comparative genome analysis of the primary sex-determining locus in salmonid fishes. Genome Res. 13: 272–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav J. S., Burra M. R., 1987. Chromosomes of Calosoma weber (caraboidea: Coleoptera) with comments on achiasmate meiosis. Genet. Iber. 39: 9–16 [Google Scholar]