Abstract
Successful sequencing experiments require judicious sample selection. However, this selection must often be performed on the basis of limited preliminary data. Predicting the statistical properties of the final sample based on preliminary data can be challenging, because numerous uncertain model assumptions may be involved. Here, we ask whether we can predict “omics” variation across many samples by sequencing only a fraction of them. In the infinite-genome limit, we find that a pilot study sequencing 5% of a population is sufficient to predict the number of genetic variants in the entire population within 6% of the correct value, using an estimator agnostic to demography, selection, or population structure. To reach similar accuracy in a finite genome with millions of polymorphisms, the pilot study would require ∼15% of the population. We present computationally efficient jackknife and linear programming methods that exhibit substantially less bias than the state of the art when applied to simulated data and subsampled 1000 Genomes Project data. Extrapolating based on the National Heart, Lung, and Blood Institute Exome Sequencing Project data, we predict that 7.2% of sites in the capture region would be variable in a sample of 50,000 African Americans and 8.8% in a European sample of equal size. Finally, we show how the linear programming method can also predict discovery rates of various genomic features, such as the number of transcription factor binding sites across different cell types.
Keywords: rare variants, capture–recapture, population genetics, linear programming, sequencing
PREDICTING the genetic makeup of a large population sample based on a small subsample serves two distinct purposes. First, it can facilitate study design by providing the expected number of samples needed to achieve a given discovery goal, be it enough markers for a custom array design or enough rare variants to perform a well-powered burden test. Second, such predictions serve as a useful test for our statistical and evolutionary hypotheses about the population. Because evolutionary experiments for long-lived organisms are extremely difficult, predictions about evolution are hard to falsify. By contrast, predictions about the outcome of sequencing experiments can be easily tested, due to the rapid advances in sequencing technology. This opportunity to test our models should be taken advantage of. Here, we show that such predictions can be easily generated to high accuracy and in a way that is robust to many model assumptions such as mating patterns, selection, and population structure.
We are interested in predicting the number of sites that are variable for some “omic” feature across samples. Features may be of different types (SNPs, indels, binding sites, epigenetic markers, etc.), and samples may be cells, cell types, whole organisms, or even entire populations or species. For definiteness, we focus primarily on predicting the discovery rate of genetic variants (SNPs or indels) in a population. Because variant discovery is central to many large-scale sequencing efforts, many methods have been proposed to predict the number of variants discovered as a function of sample size in a given population. Some methods require explicit modeling of complex evolutionary scenarios, fitting parameters to existing data (Eberle and Kruglyak 2000; Durrett and Limic 2001; Gutenkunst et al. 2009; Gravel et al. 2011; Lukić et al. 2011). These approaches enable model testing, but they are complex and computationally intensive. The interpretation of model parameters can also be challenging (Myers et al. 2008). Ionita-Laza et al. (2009) pointed out a similarity between the variant discovery problem and a well-studied species counting problem in ecology (Pollock et al. 1990), and this led to the development of tractable heuristic approaches that rely on simple assumptions about underlying distributions of allele frequencies (Ionita-Laza et al. 2009; Ionita-Laza and Laird 2010; Gravel et al. 2011). These methods are easy to use and often accurate, but the validity of the heuristic assumptions is uncertain, and departures from these models can lead to uncontrolled errors (see Link 2003 and the debate in Holzmann et al. 2006).
In this article, we build on results of previous studies and propose improved estimators, quantifying their uncertainties and biases (Ionita-Laza et al. 2009; Ionita-Laza and Laird 2010; Gravel et al. 2011). Even though fully nonparametric estimators were deemed impossible in the ecology problem (see Link 2003 and Discussion), we obtain a nonparametric estimator based on linear programming (LP) that is asymptotically optimal in the infinite-genome limit, in the sense that the estimated confidence intervals contain precisely the values that are consistent with the data. These LP estimators are similar to estimators developed in the slightly different context of vocabulary size estimation (see Efron and Thisted 1976 and Discussion). Whereas parametric approaches were needed to get meaningful predictions beyond 10-fold extrapolation in the vocabulary problem, the nonparametric LP approach provides estimates of the number of genetic variants within 6% of the correct value under 20-fold sample increases in a realistic genetic model in the infinite-genome limit and within 35% when 107 polymorphisms are present in the population. We also present a jackknife estimator and provide strategies to estimate both the sampling uncertainty (via bootstrap) and bounds to the bias of the estimator. By applying the estimators to data generated by the 1000 Genomes Project (1000G) and the National Heart, Lung, and Blood Institute Exome Sequencing Project (ESP), we find that both estimators compare favorably with the state of the art for computational efficiency, accuracy, and robustness to biases.
We provide examples of how these estimators can be used after preliminary data have been obtained to decide on the sample size required to achieve a given discovery goal, to estimate the impact of sample composition on projected study outcomes, and to predict the proportion of synonymous to nonsynonymous sites as a function of sample size. Experimental design decisions require weighing many different factors, some of which must be estimated from incomplete information. Simple and robust estimates of the composition of the final sample should provide a useful tool for scientists seeking to obtain a clearer picture of the expected outcomes of different experimental strategies.
Finally, because nonparametric approaches do not depend on a specific evolutionary or biochemical model, they can be applied to a variety of genomic features. As an illustration, we apply the LP approach to predict the number of DNaseI footprints to be identified as a function of the number of cell types studied. Thus, the number of occupied transcription factor binding sites across all cell types in an organism can be estimated directly (and accurately) from a randomly selected sample of cell types. In addition to being a tool for study design, the discovery rate can answer fundamental biological questions, such as the total proportion of DNA that is bound by any or all transcription factors in any cell type.
Software is available through the author’s webpage.
Methods
Capture–recapture experiments use statistical inference to estimate population sizes without observing every individual. They use the overlap among random subsamples to estimate redundancy and therefore how much new information is to be found in unobserved samples. For example, the size of a rabbit population may be estimated by tagging R1 randomly selected rabbits and counting the proportion p of tagged rabbits in a subsequent random sample from the population. If rabbits and samplings are uniform and independent, the total population can be estimated as R1/p. In practice, a number of complications may arise: sampling conditions may vary across field trips, rabbits can join or leave the population, and they can become afraid or fond of the capture device. As a result, an extensive literature on statistical methods accounts for these complications (Pollock et al. 1990). A particularly challenging situation occurs when rabbits vary in their probability of capture. In this case, no amount of data can rule out the existence of a large number of very uncatchable rabbits. Based on this intuition, it has been argued that an unbiased estimator for this problem required prior knowledge of the distribution of capture probability (Holzmann et al. 2006).
Ionita-Laza et al. (2009) pointed out that predicting the number of genetic variants that are present in a population is closely related to this rabbit-counting problem. In the analogy between the genetic and ecological cases, displayed in Table 1, rabbits to be counted are replaced by genetic variants; the capture of a rabbit is replaced by the variant identification in a sequenced (haploid) genome; and the probability of capturing a given rabbit on a given field trip is replaced by the population frequency of the variant.
Table 1. Some analogies between rabbit and genetic variant counting.
| Polymorphic loci | Rabbits |
|---|---|
| Sequenced chromosome | Sampling expedition |
| Nonreference genotype | Capture |
| Allele frequency | Rabbit catchability |
| Rare variant | Rascally rabbit |
Whereas the ecological problem requires us to take into account the distribution of catchabilities among rabbits, the genetics problem requires us to consider the distribution of allele frequencies among genomic loci. This distribution, Φ(f), depends on past mutation rates, demographic history, and selection and thus provides a natural testing ground for evolutionary models (see, e.g., Gutenkunst et al. 2009 and Lukić et al. 2011 and references therein). The variant discovery rate therefore depends on many evolutionary parameters, but it is also limited by basic sampling statistics: in all models, the discovery rate per sample is expected to decrease as the sample size is increased. The goal of this article is to formalize this intuition and develop quantitative prediction methods.
We consider a general model of “omic” diversity that we describe in terms of genotype diversity. A haploid genome has L independent loci. Each locus i has a genotype gi that is a “reference” with probability 1 − fi and a “nonreference” with probability fi. This nonreference allele frequency fi is drawn from an underlying frequency distribution Φ(f). To generate n samples in this model, we first draw each {fi}i=1,… ,L from Φ(f). Then, for each locus i, we generate n independent genotypes. We consider a variant “discovered” if the nonreference allele has been discovered. An alternate definition, where a variant is discovered if both alleles have been observed in the sample, requires only minor modifications to what follows.
We do not know Φ(f) and wish to learn about it from the data. What we do observe is the sample site-frequency spectrum (SFS), the histogram {ϕn(j)}j=1,…,n counting loci where exactly j of n chromosomes have the nonreference allele in our sample. In the limit of infinite L,
| (1) |
The SFS is a sufficient statistic for the unknown distribution Φ(f). We are now interested in predicting V(N), the total number of variants discovered in a large sample of finite size N. Consider the number of undiscovered variants:
| (2) |
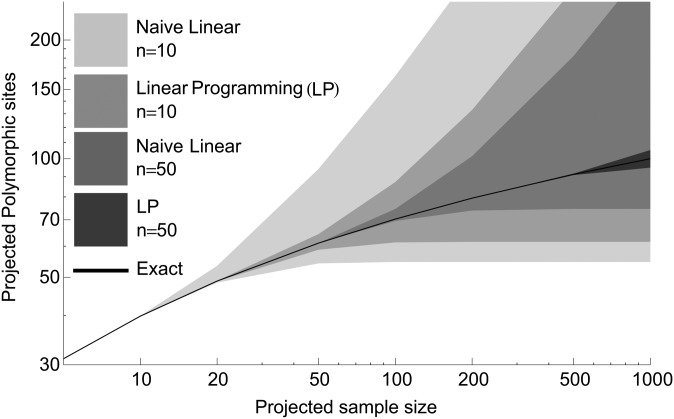
This is bounded below by 0, since the number of discovered variants must be positive. Because the rate of variant discovery per sample is expected to decrease with sample size, this quantity can also be bounded above. In Supporting Information, File S1 we provide simple bounds that are based on generalizations of this argument and expressed as linear combinations of the ϕn(j); we refer to those as naive linear bounds. Even though they are mathematically interesting, we see in Figure 1 that naive linear bounds do not provide the best practical bounds.
Figure 1.
Bounds on the number of polymorphic sites to be discovered, based on discoveries in samples of size n = 10 and n = 50 from model (4), using second-order naive linear and linear programming (LP) bounds. The shaded areas correspond to strict upper and lower bounds in the infinite-sites model and do not rely on any assumption about distribution (4). Note that LP provides dramatically tighter confidence intervals for both sample sizes and that order-of-magnitude extrapolations can be performed for LP and n = 50 with high accuracy.
Linear programming
Rather than think of our sample of size n as drawn from an infinite population, imagine that it is drawn from a larger sample of size N > n, with allele frequency distribution ΦN(i). In the limit of an infinite genome, the problem of finding values of that are consistent with the observed ϕn(j) can be formulated as the linear program displayed in Table 2. This infinite-genome linear program always has a solution if the subsample was indeed generated by hypergeometric sampling from a distribution ΦN(i). Since we have shown that V(N) is bounded, the solution to the linear program is precisely the finite interval of values that are consistent with the data. The existence of such an interval settles the question of whether estimates can be obtained without assumptions about the underlying frequency distribution (Holzmann et al. 2006): point estimators require assumptions about ΦN(i), but interval estimators can be obtained using LP. If N = ∞, the intervals are semi-infinite. In practice, we can efficiently calculate tight bounds on V(N) for N in the thousands through the revised simplex method (see, e.g., Kasana and Kumar 2004; here we use a version implemented in Mathematica).
Table 2. The linear program formulation.
| Variables | ΦN(i), i ∈ {0 … , N} |
|---|---|
| Objective function | |
| Observational constraints | |
| Nonnegativity constraints | ΦN(i) ≥ 0. |
An LP formulation of the capture–recapture problem was also used in a related problem of vocabulary estimation, where the sampling process is Poisson rather than hypergeometric (Efron and Thisted 1976). By contrast to the Poisson case, where the unknown distribution of frequencies Φ is arbitrary, the underlying function ΦN(i) in the genetics problem is usually drawn from a larger population of size M, and this imposes additional constraints on ΦN(i) that can be incorporated into the linear program to improve accuracy. We now have
| (3) |
We wish to find an upper and a lower bound to the total number of variants. We must therefore solve two linear programs with the same constraints but opposite objective functions: ±cN.AM,NΦM, where cN = {0, 1, 1, … , 1}. The resulting interval is the best possible estimator in the infinite-sites model for extrapolating from n to N in a population of size M, without using assumptions on the underlying model. In cases where we require a point estimator, we simply use the average of the upper and the lower bound. This is not entirely arbitrary—given the current constraints, the solutions at the constraint boundary have a frequency spectrum that reaches zero for some frequency (Figure S6). We expect the correct value to lie in the interior of the interval.
Linear estimators
The LP bounds are the best we can do without assumptions about ΦN. However, these may be computationally intensive for very large N. Given the general success of the Burnham–Overton (BO) jackknife estimators (Burnham and Overton 1979), it is worth asking whether similar estimators could be successful here. However, the BO assumptions that fail even for a panmictic, neutrally evolving constant-size population [i.e., the standard neutral model, where V(n) ≃ log(n)].
In Gravel et al. (2011), we proposed an expansion of the form with the (n − 1)st harmonic number. A simpler and more principled expansion is We show in File S1 that both expansions yield the same jackknife estimates, but the latter is more tractable. Even though more general expansions could be considered, this particular expansion is practical because (a) it provides exact results at linear order in the standard neutral model, (b) it allows the modeling of a diversity of functions that increase slowly but do not converge, and (c) it performs well in simulations (Figure S3). We refer to the resulting estimate as the harmonic jackknife.
Finite genome
Two complications can arise as a consequence of the finiteness of the genome. First, the infinite-genome approximation underlying the standard neutral model expression V(n) ∼ log(n) that serves as a starting point for the jackknife expansion may not hold: for a large enough sample size, we will run out of sites. The BO estimator might eventually become a better choice. The LP approach would not be sensitive to this problem, as it does not rely on the standard neutral model.
The second complication introduced by a finite genome is that the observed site-frequency spectrum is now a random variable, as there are a finite number of observations per frequency bin. For the jackknife estimator, this may result in large, uncontrolled inaccuracies, especially if high-order estimators are used. The infinite-sites LP problem, by contrast, is likely to be infeasible in the presence of noise. Under the random Poisson field approximation, one may attempt to maximize the likelihood
under the constraint Φ ≥ 0, where P(μ, x) is the Poisson distribution with mean μ. The maximizing Φ may or may not be unique (so that we may have either a point or an interval estimator). Unfortunately, because the optimizing problem is now nonlinear, the general optimization problem is intractable in its exact form.
To take advantage of the LP formalism, we may wish to relax some of the constraints imposed as equalities in the infinite-L limit, in such a way that realizable vectors exist and the LP problem can be solved. One approach is to turn equality constraints into range constraints (Efron and Thisted 1976), with width informed by the expected fluctuation sizes in each bin. However, a more efficient option is to coarsen the least informative bins. Since most of the unobserved variants are rare, we do not care for the precise frequency of the common variants. We use a bin-merging strategy, collapsing bins containing common variants into a smaller set of coarser bins. This has the added benefit of reducing the number of constraints, making the problem numerically more tractable. We use a simple scheme in which we keep the p lowest-frequency bins intact and then merge the next two bins, then the following four bins, and so on, increasing the bin size exponentially until all bins have been taken into account. We then choose p as high as possible without making the LP problem infeasible. Fortunately, Figure S2 shows that it is not necessary to use a large number of bins to obtain tight bounds.
This procedure will result in a predicted range for the number of discovered polymorphisms. This range accounts for uncertainties about the underlying distribution, but not for sampling uncertainty. To account for sampling uncertainty, we can bootstrap the data, each bootstrap iteration providing a confidence interval. We can then define confidence intervals by using 95% confidence intervals on both the upper and lower bounds. Such confidence intervals on bounds are expected to be more conservative than confidence intervals on point estimates.
Multiple populations
The strategies described above do not require random mating assumptions. They can therefore predict the number of variants in samples drawn from multiple populations if subsamples from the subpopulations are available. The LP approach can be generalized to bound any linear function of the joint SFS, including the number of variants private or shared across samples. However, the number of variables grows rapidly with the number of populations, and such a linear program would require careful optimization. We use a simpler strategy and form a subsample with the appropriate ancestry proportions and extrapolate using the single-population strategies outlined above. In Figure S5, we show extrapolations based on 100 African and 100 European haplotypes, using 1000 Genomes Yoruba (YRI) and European-American (CEU) populations, as well as results based on equal-sized samples using a known simulated demographic history. As expected, we find that discovery rates are higher in mixed populations, and the mixing proportion that maximizes discovery depends on the total sample size.
Alternate approaches
We compare the results of the methods presented above to three different strategies: (a) the parametric model of Ionita-Laza et al. (2009), which supposes that the allele frequency distribution can be modeled as a beta-distribution with parameters fitted to the observed distribution of allele frequencies; (b) the standard Burnham–Overton estimator of order 3, which supposes that the proportion of missed variants at sample size N can be expanded as a third-order polynomial in and (c) a fully model-based approach, using ∂a∂i (Gutenkunst et al. 2009) to fit a three-parameter, one-population demographic model to the observed SFS. The model involved two periods of constant population size, N1 and N2, and instantaneous change between the two values at time t.
Results
Simulations
To study the predictive power of different methods in the infinite-sites limit, we generated expected frequency spectra in a population of M = 1000 individuals, with
| (4) |
and for subsamples of size n ∈ {10, 20, 50}. Extrapolations were attempted to N ∈ {20, 50, 100, 200, 500, 1000}. Figure 1 presents extrapolations based on samples of size 10 and 50, using naive linear and LP bounds, and Table 3 shows confidence intervals for extrapolations to N = 200, using two different naive linear bounds: LP and LP using the M > N strategy. To facilitate comparison, the predicted number of polymorphisms is expressed as a percentage of the variants in the population of 1000 individuals. Because these simulations closely follow the harmonic jackknife assumptions, harmonic jackknife estimates are essentially perfect, but this is not representative. Harmonic and Burnham–Overton jackknife estimates with different underlying distributions are presented in Figure S3 and in the 1000 Genomes example below.
Table 3. Confidence intervals for extrapolating the number of polymorphic sites discovered in 200 chromosomes, based on samples of size n = 10, 20, and 50 and four different approaches, in the infinite-sites limit.
| N = 200 | Sample size (n) | ||
|---|---|---|---|
| Method | 10 | 20 | 50 |
| Naive linear, d = 2 | 69–375 | 79–215 | 93–128 |
| Naive linear, d = 2, 3 | 69–203 | 79–139 | 93–106 |
| LP, M = N | 76–173 | 92–109 | 99.98–100.01 |
| LP, M = 1000 | 78–167 | 96–106 | 99.9999–100.0001 |
The intervals represent 100% confidence intervals expressed as percentages of the correct value from the model given in Equation 4.
LP approaches provide significantly tighter bounds than second-order naive linear bounds and, surprisingly, allow for accurate extrapolations over more than an order of magnitude in sample size. However, these simulations assume a nearly infinite genome, and the convergence to this limit may be slow. Figure S7 shows the slow increase in prediction accuracy with sample size. In a sample with 10 million polymorphisms, the 20-fold extrapolations are not very precise, but 8-fold extrapolations provide conservative lower bounds 4% below the correct value and upper bounds 16% above it.
Subsampling 1000 Genomes data
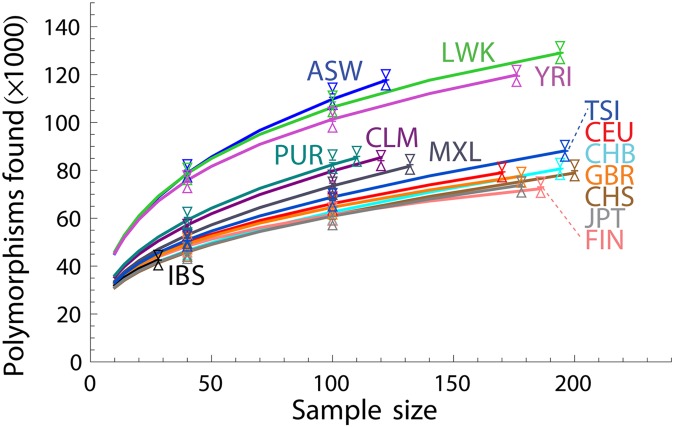
The 1000 Genomes Project (2012) has released exome-capture data for 1092 individuals from 14 populations: some from predominantly European [European-American (CEU), Tuscan (TSI), British (GBR), Finnish (FIN), and Iberian (IBS)], African [Yoruba (YRI) and Luhya (LWK)], and East Asian [Han Chinese (CHB, CHS) and Japanese (JPT)] ancestry and others of mixed continental ancestry [African-American (ASW), Mexican (MXL), Colombian (CLM), and Puerto Rican (PUR)]. Figure 2 shows the number of nonreference variants discovered as a function of sample size in each population.
Figure 2.
Number of nonreference variants discovered for each of the 1000 Genomes Project populations (solid lines). Linear programming (LP) predictions (shown as vertical intervals) are based on random subsamples of diploid individuals corresponding to 20, 40, and 100 haploid genomes. The triangle tips indicate the 95% confidence maxima for the LP upper bounds and 95% confidence minima for the LP lower bounds from 50 bootstrap runs. The short horizontal lines between triangles represent the width of the confidence interval for a single LP run; it is thinner than the line width in most instances. Each displayed interval uses the maximum subsample size available.
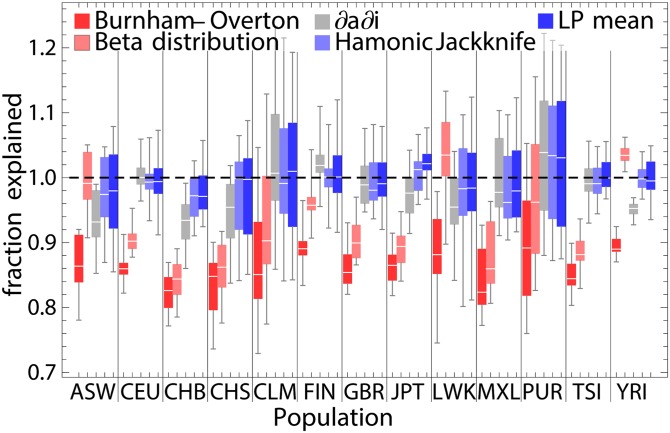
To estimate the accuracy of the capture–recapture strategies, we randomly drew subsamples of 10, 20, and 50 diploid individuals and extrapolated the number of discoveries from each subsample size to the next larger subsample size or to the full population size. We find that the LP approach and the harmonic jackknife provide accurate estimates to within a few percent of the true values (Figure 2 and Figure 3), whereas the BO and beta-distribution estimators underestimate the number of variants for most populations (Figure 3). The demographic model approach is only slightly more biased than LP and harmonic jackknife, but it is also more intensive computationally and technically.
Figure 3.
Predicted number of new variants discovered in N = 100 haplotypes based on multiple subsamples of 20 diploid individuals from 1000 Genomes populations, expressed as a proportion of the correct value. We display the existing Burnham–Overton (Burnham and Overton 1979) and beta-distribution (Ionita-Laza et al. 2009; Ionita-Laza and Laird 2010) predictors, a prediction based on a three-parameter demographic model fitted using ∂a∂i, and the harmonic jackknife and linear programming (LP) approaches presented here.
Even though the harmonic jackknife and LP approaches appear unbiased for all populations, the variance of the estimate depends on the population, with recently admixed populations (ASW, CLM, MXL, and PUR) showing the most variance, followed by populations with known cryptic relatedness (LWK and CHS). This variance indicates that the relatively small subsamples have “personality” in these populations—if a sample contains an individual with a particularly high European ancestry proportion or a pair of closely related individuals, it may sway the estimate in a way that would not occur in a more uniform sample. If we consider confidence intervals based on Poisson random field (PRF) parametric bootstrap, which assumes a perfectly homogeneous sample, 95% confidence intervals contain the observed data in 76% of cases, whereas the harmonic jackknife confidence intervals contain the true value 68% of the time (see also Figure S1). If we exclude populations with admixture and relatedness, the proportion of confidence intervals containing the correct value increases to 92% for LP and 86% for the jackknife. Inhomogeneity effects are expected to decrease with sample size.
Importantly, both the harmonic jackknife and LP estimators appear to remain unbiased and accurate even for small inhomogeneous samples. This is in stark contrast to the BO jackknife and the parametric beta-distribution approach of Ionita-Laza et al. (2009) and Ionita-Laza and Laird (2010), which exhibit substantial bias for most populations (Figure 3).
Extrapolations using 1000 Genomes data
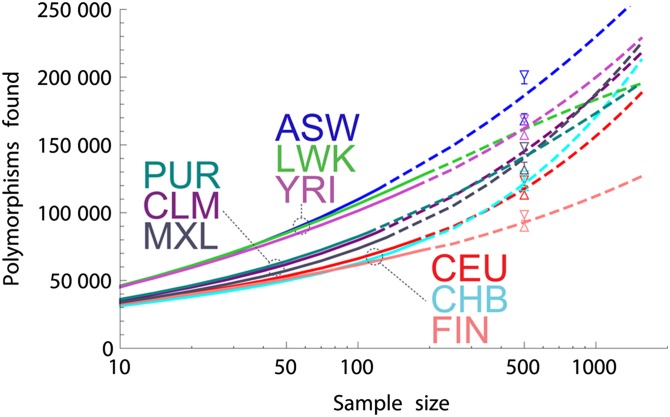
Extrapolations from the 1000G data are shown in Figure 4. The harmonic jackknife and LP estimates are in good agreement. As in Nelson et al. (2012), we find that the African-American population (ASW), with predominantly West African and European continental ancestries, has the highest predicted discovery rate. This is a joint effect of the high diversity of the African source population and of the contribution of two continental populations. By contrast, the FIN population shows the least amount of diversity, consistent with a smaller recent effective population size. Whereas the populations tend to cluster by continental ancestry at low sample sizes, reflecting shared histories, continental ancestry becomes less informative as sample sizes are increased, revealing consequences of the more recent histories of the sampled populations.
Figure 4.
Predictions of nonreference exomic SNPs to be discovered in a selection of the 1000G populations as a function of the number of chromosomes sequenced, using the harmonic jackknife (dashed lines) and linear programming (LP) (vertical intervals, shown for a subset of populations) on the full data set. The triangle tips indicate the 95% confidence maxima for the LP upper bounds and the 95% confidence minima for the LP lower bounds from 50 bootstrap runs. The short horizontal lines between triangle tips represent the width of the confidence interval for the nonbootstrapped sample.
The Exome Sequencing Project example
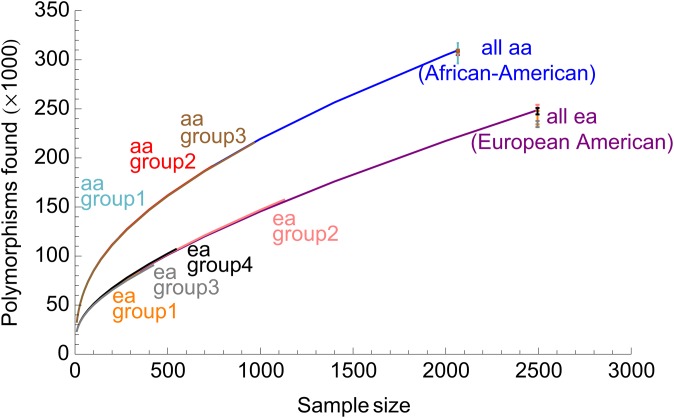
To test whether the approach is applicable to cross-cohort prediction, we applied the method to data from the first 2500 sequenced individuals of the Exome Sequencing Project (Tennessen et al. 2012), which combined data across different cohorts and sequencing centers. Figure 5 shows the total number of variants based on variants observed by four different sequencing groups (focusing on 1-LDL, 2-EOMI, 3-BMI and EOS, 4-Lung diseases; see Tennessen et al. 2012 for cohort and project descriptions). We find excellent agreement for predictions based on these subsets. The largest departure is from the European-American sample for group 3, which is also the smallest subset.
Figure 5.
Extrapolations based on different African-American (aa) and European-American (ea) subcohorts of the ESP metacohort on the full sample size. The different subcohorts correspond to data obtained from different projects and sequencing centers, as explained in the text.
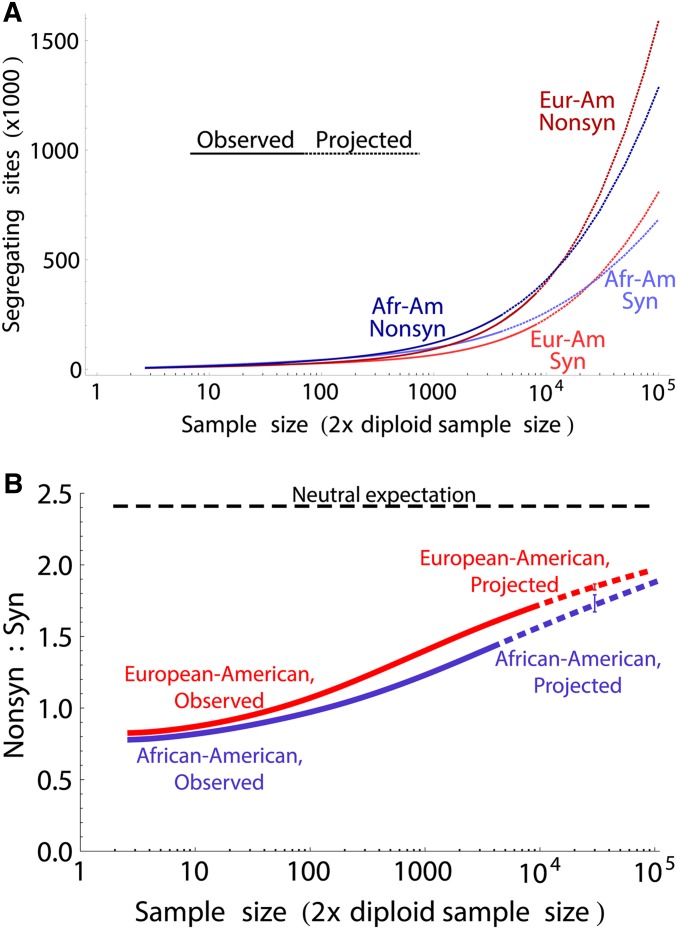
Finally, to obtain the prediction for the largest possible sample, we considered the most recent data released by the ESP project, including >6500 individuals of European-American and African-American descent, and generated predictions based on samples of 2000 African Americans and 4000 Europeans, for sites with mean coverage >40. Even though African-American populations have the most variable sites in present-day samples, we predict that this will no longer be the case in samples of 50,000 diploid individuals, with 8.7% of target sites predicted to be variable in European Americans, compared to 7.2% in African Americans. The crossover is predicted to occur between 7500 and 10,000 individuals. The predicted number of variants is higher in European Americans for both synonymous and nonsynonymous variants (Figure 6A), but the proportion of nonsynonymous variants is likely to remain higher in Europeans than in African Americans (Figure 6B). The nonsynonymous:synonymous ratio will remain considerably lower than the neutral expectation under a Hwang–Green mutational model (Hwang and Green 2004) until samples in the millions are considered.
Figure 6.
(A) Projections for the number of synonymous and nonsynonymous sites in African Americans and European Americans based on the ESP sample. (B) Observed and projected ratio of nonsynonymous to synonymous variants in the two populations.
DNaseI footprinting
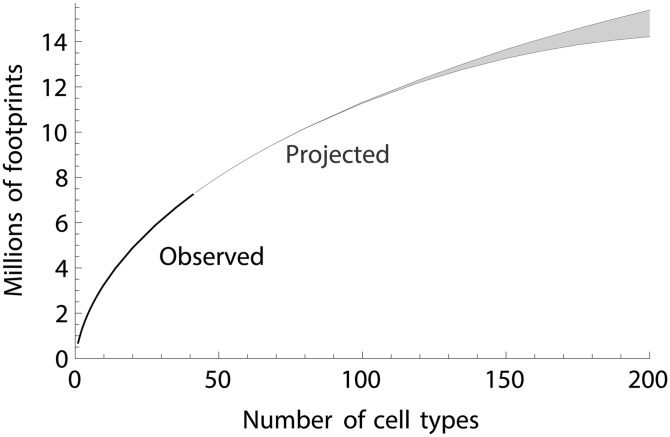
Because the LP approach is nonparametric, it can be applied to any genomic feature that is present genome-wide and across samples. To illustrate this, we consider DNaseI footprints, which indicate sites where transcription factors bind to DNA and protect against cleavage by DNaseI. Encode produced a genome-wide map of such features across 41 different cell types (Thurman et al. 2012). Using the same software as above, we are able to predict the number of transcription factor binding sites that will be identified as the number of cell types is increased. We define “sites” as contiguous genomic regions where at least one cell type has a footprint. The LP bounds are particularly tight in this example (Figure 7), and the main source of uncertainty in this problem is the degree to which the choice of cell types in the Encode study is representative of the remaining cell types with respect to transcription factor binding.
Figure 7.
Observed and projected number of DNaseI footprints, marking putative transcription factor binding sites, as a function of the number of cell types studied. Projections use the LP approach, and the confidence interval represents the difference between the LP upper and lower bounds.
Discussion
Theoretical and statistical considerations
Jackknife and LP approaches for finite and infinite extrapolation for the species-counting problem have been discussed before (Efron and Thisted 1976). The sampling processes, binomial for the rabbit-counting problem, Poisson for the species-counting problem, and hypergeometric in the genetics context, lead to fundamental differences. For example, in the Poisson case, an infinite number of data points are available because each species can be observed an arbitrary number of times. This allows for a (possibly divergent) formal expansion of the number of unobserved variants in terms of the {φ(i)}i=0,…,∞ (Efron and Thisted 1976). In the binomial and hypergeometric cases, we have only a finite number of observations {φ(i)}i=0,…,n, making it clear that the series expansion cannot provide an exact result. In addition, the size M of the population from which our sample was drawn determines how accurately we can perform extrapolations to sizes N < M, a situation that does not have a direct analog in the Poisson case.
A difference between the genetics problem and both the species- and rabbit-counting problems is the target extrapolation size: in many ecological problems, the number of field trips itself is not a variable of interest, and the ultimate goal is to extrapolate to infinite sample sizes. In such a case, the resulting confidence interval would be semi-infinite. Intuitively, we can never exclude the possibility that a very large number of very uncatchable rabbits have eluded detection. As a result, all point estimates require implicit or explicit assumptions about the existence of such sneaky rabbits. This led to the correct statement (Link 2003) that nonparametric point estimates are impossible in the rabbit-counting problem. Nonparametric point estimates are still impossible in the finite extrapolation context studied here: there is a finite interval of values equally consistent with the data, and any choice implies parametric assumptions. However, if this finite interval is narrow enough, we may not need point estimates: in many cases, the predicted consistency interval is narrower than other uncertainty sources. Nonparametric point estimates do not exist, but this may not be important: LP provides a practical, nonparametric interval estimator.
Some of the strategies that we have proposed may translate back to the ecology problems. One example is the coarsening strategy used in the finite-genome problem, in which we merge bins of less-informative common variants to improve computational performance and accuracy. We have found that extrapolations can be accurate beyond 20-fold increases in sample size, a finding surprising in the light of previous work. The accuracy of projections as a function of sampling scheme, sample size, and model assumptions remains a largely open question of considerable theoretical interest.
We have discussed five different extrapolation strategies in this article and found that two of these (the harmonic jackknife and LP) outperformed the others (beta-distribution, demographic modeling, and BO jackknife). The beta-distribution and demographic modeling suffer from their attempt to model the entire allele frequency distribution via a few-parameter family of models. With larger data sets, departures from these model families become more significant and lead to the observed biases. By contrast, the jackknife approaches fit a similar number of parameters but model only the rare end of the frequency spectrum, which contains most of the information about future discovery rates. In that sense, they make better use of fitting parameters, but the assumptions of the BO jackknife differ too much from realistic genetic scenarios. The assumptions of the harmonic jackknife, by contrast, include realistic genetic scenarios, and as a result the extrapolations are quite accurate. Finally, linear programming does not require any assumptions about allele frequency distribution and as a result is much more broadly applicable than the other methods. Furthermore, in the infinite-genome limit, it uses all the information available in the data, and we have found it to be surprisingly accurate. Thus, the nonparametric and less parametric methods fare very well in this comparison. This is because the large data set is very informative about the underlying distribution, making parametric assumptions both less useful and more risky.
Practical aspects
Where computationally tractable, the linear programming approach has important advantages, the main one being the easy transferability to different types of problems. However, from a practical standpoint, jackknife estimators are not to be discounted. They are extremely fast and, even though the underlying assumptions may be difficult to interpret in terms of the fundamental processes involved, they tend to produce accurate estimators in a wide range of scenarios. Comparison of the exact and jackknife weights (Figure S4 and File S1) provides good intuition for this relative robustness. Finally, even though the LP bounds are asymptotically optimal among nonparametric estimators, a visual inspection of the underlying distributions (Figure S6) suggests that even fairly conservative biological assumptions can produce narrower bounds. For example, requiring that the large population be drawn from an even larger population resulted in improved intervals (Table 3). Some other assumptions, such as smoothness or monotonicity over a range of frequencies, can easily be accommodated in a linear program and would be worth exploring. In cases where LP and jackknife are applicable, we suggest using both methods; if the jackknife falls outside the LP bounds, we know that its assumptions were not met, and the LP estimator should be used. Otherwise, the jackknife estimator is probably the most principled guess among the values allowed by LP.
The most crucial assumption underlying the extrapolation methods presented here is random sampling—we must be able to consider the existing sample as a random subset of the larger population. By contrast, we found that recent admixture, population structure, and cryptic relatedness do not seem to cause substantial biases, and the LP approach should be applicable to data sets whose evolution is fundamentally different from that of SNPs. We found that some of these factors change the variance of LP estimates: populations with more sample inhomogeneity and cryptic relatedness lead to more variable estimates. We expect these effects to decrease when the sample size is increased. We do not expect linkage disequilibrium (LD) to bias our estimates, because LD does not affect the expected frequency spectrum that is the starting point of our estimates. Furthermore, we are mostly concerned with rare variants, which typically are not in high LD with each other. Thus, both the expectation and variance of genome-wide estimates should be little affected by LD. There may be applications where variances are more affected by correlations: in the transcription factor binding example, we may imagine that cell-type-specific transcription factor binding sites cluster, in which case the Poisson random field that we used to estimate confidence intervals may become a poor approximation. In such cases, leave-one-out experiments should be performed to assess confidence intervals.
The random subsampling assumption remains a demanding one—in practice, subtle differences in sampling make it likely that results extrapolated from one sample will not apply to another one. Witness the 1000 Genomes data (Figure 4), which sampled largely distinct populations. In this case, very different discovery rate estimates reflect the different recent histories of the populations. On the other hand, we also find that results in the large medical cohorts from the ESP are exquisitely reproducible across cohorts, even though these are definitely not subsamples of each other. By contrast with the 1000 Genomes data, the ESP metacohort was assembled using comparable (even sometimes overlapping) cohorts (Tennessen et al. 2012). This emphasizes how the methods presented here are applicable to make predictions across panels that are similar but not identical.
Large sequencing efforts such as the 1000 Genomes Project often start with a pilot project aimed at testing the technology, identifying possible issues and providing funding bodies and stakeholders a preview of the full project. The methods presented here provide a straightforward and well-calibrated approach to estimating a key deliverable in the final project. As the project is completed, the results can be compared to the initial predictions, assessing the impact of methodological and sampling changes between the pilot and the main phase. Of course, the final results can be extrapolated to serve as a baseline prediction for the next set of experiments.
Predicting the number of variants to be discovered in a new sample is one of the few areas where population geneticists studying long-lived organisms can make experimental predictions and as such is an important tool for population genetics hypothesis validation. The success of the nonparametric methods presented here shows that this can be performed to high accuracy. However, the success of nonparametric methods and their robustness to linkage, demography, population structure, and selection suggest that accurate model-based predictions of future discovery rates do not necessarily provide additional evidence that these effects are correctly taken into account. Overfitted models that are consistent with the data should provide predictions within the LP confidence intervals. Model-based predictions should therefore improve upon the LP predictions to validate the model. By contrast, the LP prediction provides a strong test of whether the initial sample can be considered a random subsample of the full population, a commonly used assumption in population genetics models. This work therefore demonstrates that nontrivial falsifiable predictions can easily be generated and tested against future genomics experiments. I hope that it will encourage more genomicists to put their heads on the prediction block.
Acknowledgments
The authors acknowledge S. Baharian, R. Gutenkunst, J. Kelley, O. Cornejo, S. Shringarpure, and M. Carpenter for useful comments; E. E. Kenny and C. D. Bustamante for discussions and help with data access; the support of the National Heart, Lung, and Blood Institute (NHLBI); and the contributions of the research institutions, study investigators, field staff, and study participants in creating this resource for biomedical research. Funding for the Gene Ontology Exome Sequencing Project was provided by NHLBI grants RC2 HL-103010 (HeartGO), RC2 HL-102923 (LungGO), and RC2 HL-102924 (WHISP). The exome sequencing was performed through NHLBI grants RC2 HL-102925 (BroadGO) and RC2 HL-102926 (SeattleGO). This research was undertaken, in part, thanks to funding from the Canada Research Chairs program.
Footnotes
Communicating editor: N. A. Rosenberg
Literature Cited
- Burnham K., Overton W., 1979. Robust estimation of population size when capture probabilities vary among animals. Ecology 60(5): 927–936. [Google Scholar]
- Durrett R., Limic V., 2001. On the quantity and quality of single nucleotide polymorphisms in the human genome. Stoch. Proc. Appl. 93(1): 1–24. [Google Scholar]
- Eberle M. A., Kruglyak L., 2000. An analysis of strategies for discovery of single nucleotide polymorphisms. Genet. Epidemiol. 19(S1): S29–S35. [DOI] [PubMed] [Google Scholar]
- Efron B., Thisted R., 1976. Estimating the number of unseen species: How many words did Shakespeare know? Biometrika 63(3): 435–447. [Google Scholar]
- Gravel S., Henn B. M., Gutenkunst R. N., Indap A. R., Marth G. T., et al. , 2011. Demographic history and rare allele sharing among human populations. Proc. Natl. Acad. Sci. USA 108(29): 11983–11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutenkunst R. N., Hernandez R. D., Williamson S. H., Bustamante C. D., 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5(10): e1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmann H., Munk A., Zucchini W., 2006. On identifiability in capture-recapture models. Biometrics 62(3): 934–936, discussion 936–939. [DOI] [PubMed] [Google Scholar]
- Hwang D. G., Green P., 2004. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc. Natl. Acad. Sci. USA 101(39): 13994–14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita-Laza, I. and N. M. Laird, 2010 On the optimal design of genetic variant discovery studies. Stat. Appl. Genet. Mol. Biol. 9(1): Article 33. [DOI] [PMC free article] [PubMed]
- Ionita-Laza I., Lange C., Laird N. M., 2009. Estimating the number of unseen variants in the human genome. Proc. Natl. Acad. Sci. USA 106(13): 5008–5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasana, H. S., and K. D. Kumar (Editors), 2004 Introductory Operations Research: Theory and Applications. Springer-Verlag, Heidelberg, Germany. [Google Scholar]
- Link W. A., 2003. Nonidentifiability of population size from capture-recapture data with heterogeneous detection probabilities. Biometrics 59(4): 1123–1130. [DOI] [PubMed] [Google Scholar]
- Lukić S., Hey J., Chen K., 2011. Non-equilibrium allele frequency spectra via spectral methods. Theor. Popul. Biol. 79(4): 203–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S., Fefferman C., Patterson N., 2008. Can one learn history from the allelic spectrum? Theor. Popul. Biol. 73(3): 342–348. [DOI] [PubMed] [Google Scholar]
- Nelson M. R., Wegmann D., Ehm M. G., Kessner D., St Jean P., et al. , 2012. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science 337(6090): 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock, K., J. Nichols, C. Brownie, and J. E. Hines, 1990 Statistical Inference for Capture-Recapture Experiments Wildlife Monographs, 107.
- Tennessen J. A., Bigham A. W., O’Connor T. D., Fu W., Kenny E. E., et al. , 2012. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337(6090): 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium, 2012 An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman R. E., Rynes E., Humbert R., Vierstra J., Maurano M. T., et al. , 2012. The accessible chromatin landscape of the human genome. Nature 489(7414): 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]