Abstract
Multiple prion elements, which are transmitted as heritable protein conformations and often linked to distinct phenotypes, have been identified in the budding yeast, Saccharomyces cerevisiae. It has been shown that overproduction of a prion protein Swi1 can promote the de novo conversion of another yeast prion [PSI+] when Sup35 is co-overproduced. However, the mechanism underlying this Pin+ ([PSI+] inducible) activity is not clear. Moreover, how the Swi1 prion ([SWI+]) interacts with other yeast prions is unknown. Here, we demonstrate that the Pin+ activity associated with Swi1 overproduction is independent of Rnq1 expression or [PIN+] conversion. We also show that [SWI+] enhances the appearance of [PSI+] and [PIN+]. However, [SWI+] significantly compromises the Pin+ activity of [PIN+] when they coexist. We further demonstrate that a single yeast cell can harbor three prions, [PSI+], [PIN+], and [SWI+], simultaneously. However, under this condition, [SWI+] is significantly destabilized. While the propensity to aggregate underlies prionogenesis, Swi1 and Rnq1 aggregates resulting from overproduction are usually nonheritable. Conversely, prion protein aggregates formed in nonoverexpressing conditions or induced by preexisting prion(s) are more prionogenic. For [PSI+] and [PIN+] de novo formation, heterologous “facilitators,” such as preexisting [SWI+] aggregates, colocalize only with the newly formed ring-/rod-shaped Sup35 or Rnq1 aggregates, but not with the dot-shaped mature prion aggregates. Their colocalization frequency is coordinated with their prion inducibility, indicating that prion–prion interactions mainly occur at the early initiation stage. Our results provide supportive evidence for the cross-seeding model of prionogenesis and highlight a complex interaction network among prions in yeast.
Keywords: Saccharomyces cerevisiae, protein aggregation, prionogenesis, prion interactions, [SWI+]
PRIONS are host proteins with altered and infectious conformations. In mammals, all subtypes of prion diseases are associated with conformational changes of a single protein, prion protein (PrP) (Prusiner 1998). One unique property of PrP is its ability to exist as multiple stable conformations: a normal cellular conformation (PrPc) and abnormal pathogenic conformations (PrPSc) that manifest as a group of fatal neurodegenerative diseases known as transmissible spongiform encephalopathies or prion diseases (Prusiner 1998; Collinge and Clarke 2007; Weissmann 2009). Interestingly, a number of prions have also been discovered in Saccharomyces cerevisiae, among which are [PSI+] (Cox 1965), [URE3] (Lacroute 1971; Wickner 1994), [PIN+] (or [RNQ+]) (Derkatch et al. 1997; Sondheimer and Lindquist 2000), [SWI+] (Du et al. 2008), [OCT+] (Patel et al. 2009), [MOT3] (Alberti et al. 2009), [ISP+] (Rogoza et al. 2010), [MOD+] (Suzuki et al. 2012), and [NUP100+] (Halfmann et al. 2012), and their corresponding protein determinants are Sup35, Ure2, Rnq1, Swi1, Cyc8, Mot3, Sfp1, Mod5, and Nup100, respectively. Similar to PrPSc, these yeast prions are transmitted as altered protein conformations. Importantly, yeast prion proteins have diverse cellular functions and do not share significant sequence similarities with PrP (Crow and Li 2011; Li and Kowal 2012). Except for the newly identified Mod5, all of the above-mentioned yeast prion proteins contain a region that is highly rich in glutamine (Q) and/or asparagine (N) residues and is essential for prion formation and propagation (Alberti et al. 2009; Crow and Li 2011). These Q/N-rich regions are referred to as prion domains (PrD). Similar to PrPSc, yeast PrDs purified from an Escherichea coli recombinant source can form amyloid fibrils in vitro. Upon incubation with naïve cells, these test-tube assembled amyloid fibrils can be incorporated into yeast cells and serve as seeds for prion propagation (King and Diaz-Avalos 2004; Tanaka et al. 2004; Brachmann et al. 2005; Patel and Liebman 2007; Alberti et al. 2009; Du et al. 2010). Collectively, these data provide convincing evidence supporting the protein-only prion concept and demonstrating that amyloid architecture is the structural basis of prion-mediated infectivity.
The existence of multiple prion elements in yeast has made this unicellular organism an ideal system to study prion–prion interactions. The earliest work in this line of research is an investigation identifying the cellular factors required for [PSI+] de novo formation (Derkatch et al. 1997). The spontaneous conversion from [psi−] to [PSI+] is a rare event with an approximate frequency of ∼5.8 × 10−7 (Lund and Cox 1981; Chernoff et al. 1999; Allen et al. 2007; Lancaster et al. 2010). However, overproduction of Sup35 in the presence of [PIN+] dramatically increases [PSI+] de novo formation (Chernoff et al. 1993; Derkatch et al. 1996, 1997). Intriguingly, [PIN+] is required only for the de novo formation of [PSI+] but not for its propagation (Derkatch et al. 2000, 2001). Once [PSI+] is established, it can stably propagate in the absence of [PIN+]. In addition to [PIN+], presence of [URE3] or [NU+]—a prion form of the fusion protein New1PrD–Sup35MC—can also promote [PSI+] formation (Derkatch et al. 2001; Osherovich and Weissman 2001). In addition to prions, other events, for example, overproduction of one of several Q/N-rich yeast prion proteins, such as Ure2, Swi1, Cyc8, or New1, can also enhance [PSI+] de novo when Sup35 or its PrD is co-overproduced (Derkatch et al. 2001). Intriguingly, overproduction of Mod5, a non-Q/N-rich protein, can also substitute [PIN+] to facilitate [PSI+] conversion (Suzuki et al. 2012). In addition, such a [PSI+]-promoting phenotype (termed Pin+) can also be achieved by overproduction of some non-prion Q-rich proteins, such as the poly(Q)-containing domain of huntingtin (Q74 and Q103) (Derkatch et al. 2004). Although most reported prion–prion interactions are mutually promoting, antagonistic prion–prion interactions have also been reported. For example, the presence of [PSI+] can inhibit the de novo appearance of [URE3] (Bradley et al. 2002), and [URE3] can destabilize [PSI+] when they coexist (Schwimmer and Masison 2002). An antagonistic effect of [PIN+] on [PSI+] propagation has also been reported (Bradley and Liebman 2003). Although prion variants have been implied to be responsible for some of these contradictory observations, the underlying mechanisms of the complex prion interactions remain unclear.
We previously identified Swi1, a subunit of the evolutionarily conserved and ATP-dependent chromatin-remodeling complex—SWI/SNF, as a prion protein that can change conformation to become a prion termed [SWI+] (Du et al. 2008). Several noteworthy features of the [SWI+] prion have been reported: [SWI+] causes a knockdown phenotype of SWI/SNF; the propagation of [SWI+] is highly sensitive to the alteration of Hsp70 activities (Hines and Craig 2011; Hines et al. 2011); a Swi1 amino-terminal region with <40 amino acid residues that is Q free but N rich is sufficient to support [SWI+] propagation when fused to GFP (Crow et al. 2011); overproduction of Swi1 can function as a Pin+ factor to facilitate [PSI+] de novo formation (Derkatch et al. 2001; Du et al. 2008), and amyloid architecture is the structural basis of [SWI+] (Du et al. 2010). It is of importance to examine how [SWI+] interacts with other yeast prions, and we thus undertook this investigation. We have characterized the Pin+ features associated with Swi1 overproduction and [SWI+]. We also extended our study to examine how the non-prion and prion forms of Swi1 interact with Sup35 and Rnq1 during the de novo formation and maturation processes of [PSI+] and [PIN+].
Materials and Methods
Plasmids and primers
The plasmids used in this study are listed in Table 1. To construct the plasmid p304RNQ1CFP, the RNQ1 promoter (1.0-kb fragment upstream of the RNQ1 open reading frame) was amplified by PCR with a primer pair of Rnq1-up (CCGGGGTACCGTTCGAGCTCCAATTGTTGC) and Rnq1-dn (GCGCGGATCCACTAGTTTCAGATCTTTGCTATACG) from a 74D–694 strain, which was used to replace the GPD promoter of p304GPD–RNQ1CFP (Crow et al. 2011) through the SacI/SpeI sites. Plasmid p426GAL1–GFP was constructed by inserting the 0.75-kb GFP fragment from p426GPD–GFP (Du et al. 2008) to p426GAL1 (ATCC) through SpeI and XhoI sites. Plasmid pRS413CUP1–NM was generated by deleting the 0.6-kb GFP fragment of pRS413CUP1–NMGFP with SacI/SacII followed by blunt ending with T4 DNA polymerase and self-ligation. Constructs of pRS413CUP1–NMCFP and pRS316CUP1–NMCFP were created by replacing the NMGFP fragment of pRS413CUP1–NMGFP and pCUP1–NMGFP with the NMCFP fragment of pPD30.38–NMCFP (K. W. Park and L. Li, unpublished data) through SacII/SacI digestion. Similarly, pRS413CUP1–NMYFP and pRS316CUP1–NMYFP were constructed by replacing the NMGFP fragment of pRS413CUP1–NMGFP and pCUP1–NMGFP with the NMYFP fragment of pPD30.38–NMYFP (Nussbaum-Krammer et al. 2013) upon SacII/SacI digestion. Plasmid p426GAL1–SWI1 was created by cloning the SWI1 fragment from p416TEF–SWI1 (Du et al. 2008) into p426GAL1 (from ATCC) through SpeI/XhoI. Plasmid p426GAL1–SWI1YFP, p416GPD–SWI1YFP, and p426GPD–SWI1YFP were generated by subcloning the SWI1-YFP fragment from p416TEF–SWI1YFP into p426GAL1, p416GPD, and p426GPD (ATCC), respectively, through the SpeI/XhoI sites. Plasmid p413TEF–NQYFP was made by subcloning the SWI1-NQ-YFP fragment of the p416TEF–NQYFP into p413TEF (ATCC) through the sites of SpeI/XhoI.
Table 1. Plasmids used in this study.
| Plasmid | Auxotrophic marker | Origin | Promoter | Source |
|---|---|---|---|---|
| p304GPD-RNQ1CFP | TRP1 | N/A | GPD | Crow et al. (2011) |
| p304RNQ1CFP | TRP1 | N/A | RNQ1 | This study |
| p304TEF-RNQ1CFP | TRP1 | N/A | TEF1 | Crow et al. (2011) |
| p413TEF-NQYFP | HIS3 | CEN | TEF1 | This study |
| p416GPD-SWI1YFP | URA3 | CEN | GPD | This study |
| p416GPD-ure2NPDGFP | URA3 | CEN | GPD | Lindquist lab |
| p416TEF–GFP | URA3 | CEN | TEF1 | Du et al. (2010) |
| p416TEF–NQYFP | URA3 | CEN | TEF1 | Du et al. (2010) |
| p416TEF–SWI1 | URA3 | CEN | TEF1 | Du et al. (2008) |
| p416TEF–SWI1YFP | URA3 | CEN | TEF1 | This study |
| p426GAL1–SWI1YFP | URA3 | 2μ | GAL1 | This study |
| p426GAL1–SWI1 | URA3 | 2μ | GAL1 | This study |
| p426GAL1–GFP | URA3 | 2μ | GAL1 | This study |
| p426GPD–GFP | URA3 | 2μ | GPD | Du et al. (2008) |
| p426GPD–Q103GFP | URA3 | 2μ | GPD | Lindquist lab |
| p426GPD–SWI1 | URA3 | 2μ | GPD | Du et al. (2008) |
| p426GPD–SWI1YFP | URA3 | 2μ | GPD | This study |
| pCUP1–GFP | URA3 | CEN | CUP1 | Lindquist lab |
| pCUP1–NMGFP | URA3 | CEN | CUP1 | Park et al. (2006) |
| pCUP1–RNQ1GFP | URA3 | CEN | CUP1 | Sondheimer and Lindquist (2000) |
| pCUP1–YFP | URA3 | CEN | CUP1 | Lindquist lab |
| pRS316CUP1–NMCFP | URA3 | CEN | CUP1 | This study |
| pRS316CUP1–NMYFP | URA3 | CEN | CUP1 | This study |
| pRS316CUP1–RNQ1CFP | URA3 | CEN | CUP1 | Crow et al. (2011) |
| pRS413CUP1–GFP | HIS3 | CEN | CUP1 | Lindquist lab |
| pRS413CUP1–NM | HIS3 | CEN | CUP1 | This study |
| pRS413CUP1–NMCFP | HIS3 | CEN | CUP1 | This study |
| pRS413CUP1–NMGFP | HIS3 | CEN | CUP1 | Lindquist lab |
| pRS413CUP1–NMYFP | HIS3 | CEN | CUP1 | This study |
Yeast strains and media
Most yeast strains used in this study are derivatives of MATa 74D–694 (Chernoff et al. 1995) and have been previously described (Park et al. 2006; Fan et al. 2007; Du et al. 2008) except the RNQ1-CFP integrated strains. To generate RNQ1–CFP-integrated strains, plasmids of p304RNQ1CFP, p304TEF–RNQ1CFP, or p304GPD–RNQ1CFP were linearized by HindIII and introduced into the TRP1 locus of various 74D–694 strains by transformation. Successful integration was initially tested by the Trp+ phenotype and then confirmed by the presence of CFP signals in a fluorescence microscopic assay. The BY4741 SWI1–GFP-tagged strain is from ATCC. Standard YPD or synthetic complete (SC) media with different auxotrophic dropouts of amino acids were used based on the requirements of the strain. To activate the CUP1 promoter, 100 µM CuSO4 was supplemented in liquid media or SC agar plates unless specified. The raffinose media used for assaying the prion phenotype of [SWI+] was described previously (Du et al. 2008). For GAL1 promoter, log-phase cultures in liquid media using sucrose as the carbon source were supplemented with 2% galactose to induce the expression. Yeast strains were grown at 30° unless specified. If a growth time was longer than 24 hr, yeast cultures were refreshed with fresh media every 24 hr.
General fluorescence microscopic assays
GFP, YFP, and CFP fusions of Swi1, Sup35NM, and Rnq1 were expressed either ectopically or from a chromosomal locus. Fluorescence microscopic assays were carried out similarly to that described previously (Du et al. 2008). Mid-log-phase cultures were either directly used for fluorescence assay or spread to agar plates for colony purification. Individual colonies were directly used for fluorescence assay after suspended in water in some experiments or cultured in liquid media prior to fluorescence microscopic observations.
[PSI+] induction and spontaneous [PSI+] conversion experiments
[PSI+] induction experiments were conducted as described previously with minor modifications (Fan et al. 2007; Du et al. 2008). Briefly, SUP35NM–YFP, –GFP, or –CFP was overproduced ectopically from a cen-plasmid driven by the CUP1 promoter in a [psi−] 74D–694 strain upon supplementing the culture medium with CuSO4 when the culture reached the mid-log phase. The induction time was usually 48 hr. Following induction, equal number of cells of different treatments were spotted or spread onto SC–ade and SC plates after proper dilutions. For spotting assays, pictures were usually taken after 5–10 days of incubation on SC–ade plates but 3 days of growth on SC and YPD plates. Ade+ isolates were usually examined by up to three times of streaking or replica plating onto 5 mM guanidine hydrochloride (GdnHCl)-containing plates and subsequent growth on YPD and/or SC–ade plates to judge their curability. For spontaneous [PSI+] conversion assay, cells were typically grown in YPD medium and spread on SC and SC–ade plates after counting cell density and proper dilutions. Only the isolates whose colony color changes from white to red on YPD plate and accompanied by loss of growth in SC–ade upon 5 mM GdnCl treatment were scored as [PSI+].
To create isogenic strains of [SWI+][PIN+] and [swi−][PIN+] that carry no TEF1–RNQ1-CFP integration, a [SWI+][pin−] strain harboring the plasmid pCUP1–RNQ1GFP was streaked onto a SC–ura plate and incubated at 4° for 2 weeks. Colonies were then combined and spread onto SC–ura plates supplemented with 50 µM Cu2+. Individual colonies were then examined for their Rnq1-GFP fluorescence patterns and those with Rnq1-GFP aggregation were further spread onto SC–ura plates containing 50 µM Cu2+ and their Rnq1-GFP aggregation status was reexamined. Such traces for Rnq1-GFP aggregation were continued up to three times and those isolates containing stably inherited Rnq1-GFP aggregates were scored as [PIN+]. To create isogenic pairs of [swi−][PIN+] and [SWI+][PIN+] strains, several newly obtained [SWI+][PIN+] isolates from a single isolate of [SWI+][pin−] described above were screened for spontaneous loss of [SWI+] (Raf− → Raf+). The plasmid pCUP1–RNQ1GFP was then eliminated by 5-FOA counter selection. The plasmid p416TEF1–NQYFP was then reintroduced to verify their Swi1 prion status (Du et al. 2010). Those obtained isogenic pairs of [SWI+][PIN+] and [swi−][PIN+] strains carrying no TEF1–RNQ1-CFP integration were then compared for their Pin+ activities.
[PIN+] conversion and inheritability of Rnq1 aggregates
In this set of experiments, either the chromosomal TEF1–RNQ1-CFP or plasmid pCUP1-RNQ1GFP was used to report the Rnq1 aggregation status. With the chromosomal TEF1–RNQ1-CFP reporter, the Rnq1 aggregation status was examined using cells directly from plates or after cultured in liquid media. Copper was supplemented to induce RNQ1-GFP expression for cells harboring pCUP1RNQ1GFP. Only isolates that stably transmitted Rnq1-CFP/GFP aggregates were scored as [PIN+]. To investigate the effects of [PSI+] and [SWI+] on [PIN+] de novo formation, we first carried out [PSI+] induction experiments in TEF1–RNQ1-CFP integrated [pin−] strains upon Swi1 overproduction or in the presence of [SWI+]. The acquired Ade+ colonies were either directly investigated for Rnq1 aggregation state or done so after all plasmids were eliminated and the [PSI+] isolates were stabilized and verified. The stabilized [PSI+][pin−] isolates without plasmids were then selected to test if [PSI+] was able to induce [PIN+] appearance with a method described previously (Derkatch et al. 2001). Similarly, isogenic pairs of [SWI+] and [swi−] strains carrying an integrated copy of TEF1–RNQ1-CFP were compared for their capacities in promoting [PIN+] conversion. To verify that the [PIN+]-promoting activity of [SWI+] was independent of the chromosomal TEF1–RNQ1-CFP, the plasmid reporter pCUP1RNQ1GFP was also used to monitor the [PIN+] conversion in nonintegrated [SWI+] and [swi−] strains upon growth and incubation, using a method similar to that described previously (Derkatch et al. 2001). To test the inheritability of Rnq1CFP aggregates formed upon Swi1 overproduction, a non-prion strain with TEF1–RNQ1-CFP integration was transformed with p426GAL1–SWI1 or an empty vector. Rnq1-CFP aggregates were examined at indicated time points after galactose addition. The aggregate-carrying isolates were selected and spread onto YPD plates to form individual colonies after dilution. Their Rnq1-CFP fluorescence patterns were then examined and traced for inheritability. To investigate the inheritability of Rnq1-GFP aggregates formed upon different expression levels of Rnq1-GFP in the presence and absence of [PSI+], isogenic [psi−][pin−], [PSI+][pin−], and [psi−][PIN+] strains containing pCUP1–RNQ1GFP were spread to SC–ura plates that were supplemented with or without 100 μM CuSO4. After 3 days of growth at 30°, plates were transferred to 4° for up to 30 days. To check the Rnq1 status, at each indicated time point, at least 10 colonies were mixed, inoculated into liquid SC–ura medium, and cultured to log phase prior to adding CuSO4 to a final concentration of 50 µM. After 2–6 hr of induction, Rnq1–GFP aggregation frequency was examined. To investigate the inheritability of Rnq1–GFP aggregates, the mixed colonies upon 14 days of incubation at 4° were diluted and spread onto SC–ura plates containing 50 µM Cu2+ to give rise to single colonies. The resulting individual colonies were subsequently assayed for Rnq1–GFP aggregation. The Rnq1–GFP aggregate-carrying colonies were spread or streaked onto fresh SC–ura or SC–ura/ Cu2+ plates and the inheritability of the aggregates were examined. Only those successfully transmitted Rnq1 aggregates to progeny, which was comparable to a [PIN+] strain, were scored as heritable Rnq1GFP aggregates ([PIN+]).
Aggregation and inheritability of Swi1 aggregates
Plasmid-based TEF1–SWI1NQ-YFP constructs were used as reporters for [SWI+] in this study as described previously (Du et al. 2010). To express SWI1–YFP ectopically at different levels, cen–TEF1, cen–GPD, 2µ–GPD, and 2µ–GAL1 promoter were used. Swi1–YFP aggregation was assayed and the inheritability of Swi1–YFP was examined upon multiple times of passages on SC-selective plates with glucose (for 2µ–GPD) or raffinose/galactose (for 2µ–GAL1) as carbon sources/inducer. Liquid cultures were also used to verify some of the results. For fluorescence-activated cell sorting (FACS) analysis, a BY4741 SWI1–GFP-tagged strain was transformed with p426GAL1–SWI1 or p426GAL and the Swi1–GFP aggregation was examined after overnight (∼16 hr) induction. Prior to sorting, cells were collected, diluted into glucose-containing SC–ura medium to stop the SWI1 overexpression from the GAL1 promoter. The cell sorting was conducted in the Robert H. Lurie Comprehensive Cancer Center Flow Cytometry Core Facility at Northwestern University. The sorted cells harboring Swi1–GFP aggregates produced by endogenously tagged Swi1–GFP was verified by fluorescence microscopy and then spread onto SC–ura plates for further growth. Swi1–GFP aggregates were subsequently traced for their inheritability as described above upon passaging in the presence of glucose to turn off the GAL1 promoter. To get heritable Swi1–GFP aggregates ([SWI+] candidates), many rounds of colony purification in combination with fluorescence assay were conducted.
Centrifugation assay
Overnight cultures of cells grown in YPD were diluted into fresh YPD with a density of ∼106 cells/ml. After growing for additional 6 hr, cells were harvested. Spheroplasts were prepared, lysed, and centrifuged as described (Sondheimer and Lindquist 2000). Total lysate, soluble, and pellet fractions were analyzed by 10% SDS–PAGE followed by immunoblot analysis using a polyclonal anti-Rnq1 antibody, a gift from the Lindquist laboratory.
Results
SWI1 overexpression is a Pin+ factor independent of Rnq1
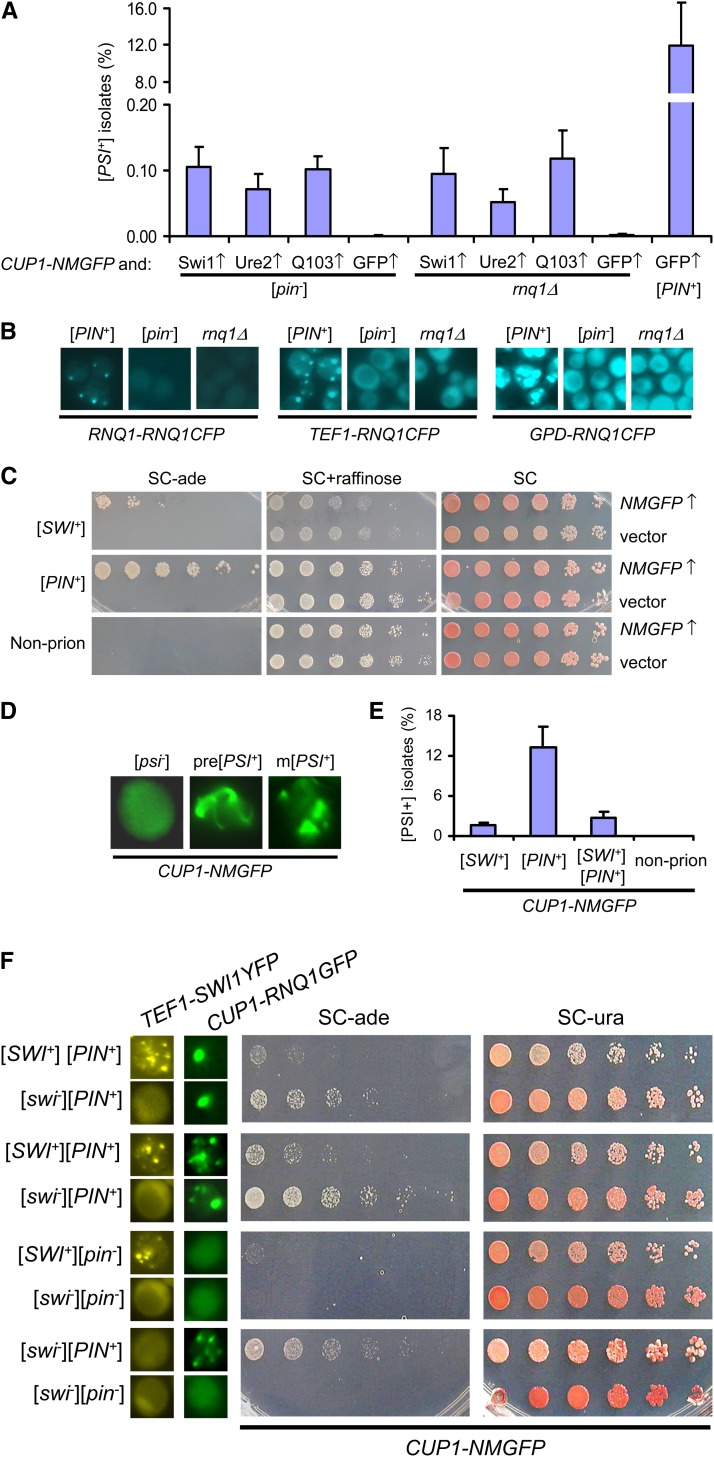
It was previously reported that overproduction of Swi1 could promote the de novo appearance of [PSI+] in [pin−] cells (Derkatch et al. 2001; Du et al. 2008). A few Q/N-rich proteins were recently shown to be able to promote [PSI+] conversion in rnq1Δ strains when overproduced (Chernova et al. 2011; Yang et al. 2013). To test if Swi1 overproduction can also promote [PSI+] conversion in the absence of Rnq1, we carried out [PSI+] induction experiments in isogenic [psi−][pin−] and [psi−] rnq1Δ strains by co-overproducing Sup35NM–GFP and Swi1. As demonstrated in Figure 1A and Supporting Information, Figure S1, overproduction of Swi1 was able to promote de novo [PSI+] formation in both strains with comparable efficiencies. Similar results were also obtained for Ure21–65–GFP, a GFP fusion of Ure2 prion domain, and Q103–GFP (Figure 1A and Figure S1). Quantitative spreading assay showed that the Pin+ activities conferred by overproductions of the three proteins were similarly low (0.05–0.15%) in both [pin−] and rnq1Δ cells, which was ∼100-fold lower than that conferred by [PIN+] under identical experimental conditions (Figure 1A). These results indicated that the overproduction of Swi1, Ure21–65–GFP, and Q103–GFP are weak Pin+ factors compared to [PIN+] and their Pin+ activities do not rely on Rnq1 protein or [PIN+] conversion.
Figure 1.
The effects of Swi1 overproduction and [SWI+] on de novo [PSI+] formation. (A) Sup35NM-GFP (from p413CUP1–NMGFP) and one of the indicated proteins were co-overproduced in a [pin−] or rnq1Δ strain to induce [PSI+]. Overproduction of Swi1 (Swi1↑), Ure21–65–GFP (Ure2↑), polyQ103–GFP (Q103↑), or GFP (GFP↑) was realized by yeast transformants containing plasmid p426GPD–SWI1, p416GPD–URE2NPDGFP, p426GPD–Q103GFP, or p426GPD–GFP. Log-phase cultures were induced for 48 hr upon addition of 100 μM CuSO4 (copper) before spreading onto SC–ade plates. Acquired ade+ isolates ([PSI+] candidates) were quantified for [PSI+] de novo formation frequencies after confirming their curability by 5 mM GdnHCl treatment. Shown are results from three independent experiments. (B) Rnq1-CFP signals of the indicated strains with a single copy of RNQ1-CFP integrated at the TRP1 locus, whose expression is driven by the indicated promoters. (C) [SWI+] can facilitate [PSI+] conversion. Sup35NM-GFP was overproduced with plasmid pCUP1–NMGFP (NMGFP↑). And “vector” represents an empty vector as control. [PSI+] induction was performed similarly to that described in A. Cultures were spotted to the indicated plates with a serial dilution and images were taken after 7 days of incubation for SC–ade plates and 3 days for other plates. (D) Sup35NM-GFP exhibits different aggregation patterns in [psi−], premature [PSI+] (pre[PSI+]), and mature [PSI+] (m[PSI+]) cells during [PSI+] prionogenesis facilitated by [SWI+]. (E) To quantify Pin+ activities, three independent spreading-based [PSI+] induction assays were conducted with a methodology similar to that described for experiments shown in A. (F) Nine isogenic pairs of [PIN+][SWI+] and [PIN+][swi−] strains and other control strains with the indicated prion backgrounds were compared for their Pin+ activities using a spotting assay similar to that used in B. Shown are results of two representative pairs of such strains and controls. Images were taken after 5 days incubation on plates. Left: prion statuses upon transformation of plasmid p416TEF–NQYFP (for Swi1) or pCUP1–RNQ1GFP (for Rnq1).
We further found that spontaneous [PSI+] conversion could also rarely occur in rnq1Δ cells even without Sup35 overproduction (Figure S1B, right). Similar to [PSI+] isolates derived from a wild-type strain, [PSI+] populations derived from an isogenic rnq1Δ strain included variants ranging from very strong to very weak, as judged by their distinct colors on YPD plates as well as different degrees of curing difficulty upon 5 mM GdnHCl treatment (Figure S1B). Earlier research has demonstrated that Sup35NM-GFP overproduction in [PIN+][psi−] cells can form aggregates that are ring and rod shaped (Zhou et al. 2001). These ring-/rod-shaped aggregates are usually formed in the initial phase of de novo [PSI+] formation and can subsequently be transformed into dot-like patterns that are indicatives of maturation or stabilization of [PSI+] (Zhou et al. 2001; Ganusova et al. 2006; Mathur et al. 2010; Tyedmers et al. 2010; Manogaran et al. 2011). We observed similar ring-/rod-shaped Sup35NM–GFP aggregates in both [psi−][pin−] and [psi−] rnq1Δ cells after 12 hr of induction of Sup35NM–GFP production (Figure S2, A and B). When [PSI+] isolates were stabilized upon passage after removal of the SWI1 expression plasmid, Sup35NM–GFP aggregates became mostly dot shaped (Figure S2C). Similar results were obtained for Ure21–65–GFP and Q103–GFP overproduction (Figure S2, B and C). These results indicated that the newly formed and stabilized [PSI+] aggregates have distinct patterns whose formation does not depend on the presence of Rnq1.
To further examine the Rnq1 independence of Pin+ activity associated with SWI1 overexpression, we needed a reporter system that could conveniently and faithfully record the aggregation status of Rnq1 during the [PSI+] de novo formation and maturation process. We constructed and compared three integrative RNQ1–CFP plasmids whose expressions were driven by GPD, TEF1, and RNQ1 promoters, respectively. When they were integrated in the TRP1 locus and expressed in [PIN+], [pin−], and rnq1Δ strains, TEF1–RNQ1-CFP produced the most optimal results for such a purpose in terms of faithfulness and robustness of the Rnq1–CFP signal (Figure 1B). Similar to a previous report (Vitrenko et al. 2007), we found that the Rnq1–CFP signals obtained under the native RNQ1 promoter were too weak to give a clear result and could be rapidly bleached during microscopic observation whereas the Rnq1–CFP signals driven by the GPD promoter were too strong to give a distinct outcome. With the commonly used cen-plasmid pCUP1–RNQ1-GFP, only ∼50–70% of [PIN+] cells showed Rnq1 aggregates. In comparison, TEF1–RNQ1-CFP strain showed an aggregation rate of ∼100% in [PIN+] cells and the Rnq1–CFP signals are robust and clear (Figure 1B); we thus used the TEF–RNQ1-CFP construct for the rest of our studies. The TEF1–RNQ1-CFP integration has no detectable effect on yeast fitness (data not shown). Under identical experimental conditions, the integrated TEF1–RNQ1-CFP strains of [PIN+], [pin−], and rnq1Δ gave similar Pin+ activities to that of nonintegrated strains upon co-overproduction of Sup35NM–GFP and Swi1 (data not shown). Most newly obtained [PSI+] isolates upon Sup35 and Swi1 co-overproduction showed diffuse Rnq1–CFP signals, indicating that they retained their [pin−] status (Figure S3A). Only ∼6% of the [PSI+] isolates harbored stably inherited Rnq1–CFP aggregates, indicative of [PIN+]. Taken together, these results indicate that Swi1 overproduction is a weak Pin+ factor whose activity can be independent of Rnq1 or [PIN+] conversion.
[SWI+] can facilitate [PSI+] conversion but weaken the Pin+ function of [PIN+]
It has been shown that like [PIN+], [URE3] can also function as a Pin+ factor to promote [PSI+] de novo formation in [pin−] cells (Derkatch et al. 2001). To examine if [SWI+] could also have a Pin+ function, we conducted [PSI+] induction experiments by overexpressing SUP35NM–GFP in a pair of isogenic strains of [SWI+][pin−][psi−] ([SWI+]) and [swi−][pin−][psi−] (non-prion). As shown in Figure 1C, [SWI+] significantly increased the frequency of [PSI+] appearance, indicating that [SWI+] can function as a Pin+ factor. We also observed that Sup35NM–GFP aggregation could be facilitated by [SWI+], which initially appeared as ring/rods and processed to become dots once [PSI+] was stabilized (Figure 1D).
Unexpectedly, when [PSI+] induction experiments were carried out in a [SWI+][PIN+][psi−] ([SWI+][PIN+]) strain, we saw that the presence of [SWI+] significantly reduced the Pin+ activity of [PIN+] compared to that seen in a [swi−][PIN+][psi−] ([PIN+]) strain (Figure 1E and data not shown), suggesting that [SWI+] antagonizes the Pin+ function of [PIN+] when they coexist. It is possible that this compromised Pin+ activity is not caused by [SWI+] but by a weaker [PIN+] variant in the tested [SWI+][PIN+][psi−] strain (the compared [SWI+][PIN+][psi−] and [swi−][PIN+][psi−] strains may harbor different [PIN+] variants). To rule out this possibility, we generated a set of spontaneously appearing [SWI+][PIN+][psi−] isolates from a single [SWI+][pin−][psi−] isolate. Each of these [SWI+][PIN+][psi−] isolates were subsequently screened for spontaneous loss of [SWI+] by their regained Raf+ phenotype and diffuse fluorescence pattern of Swi1NQ-YFP. The prion status of Rnq1 was also confirmed by fluorescence microscopy after transforming the pCUP1–RNQ1-GFP plasmid. Subsequently, nine pairs of isogenic [SWI+][PIN+][psi−] and [swi−][PIN+][psi−] isolates were acquired and compared for their Pin+ activities. As shown in Figure 1F (left), although the Rnq1–GFP aggregation patterns are distinct among the newly obtained [PIN+] isolates (single or multiple dots), indicative of the presence of different [PIN+] variants (Bradley and Liebman 2003), in all cases, [SWI+][PIN+] isolates had significantly lower Pin+ activities than that of their corresponding isogenic [swi−][PIN+] isolates (Figure 1F and data not shown). The presence of Rnq1 aggregation also clearly shows that the inhibition of [PIN+] activity by [SWI+] is not caused by loss of [PIN+]. Although it has been shown that different [PIN+] variants can have different [PSI+]-promoting (Bradley et al. 2002) or -destabilizing (Bradley and Liebman 2003) effects, the antagonizing effect of [SWI+] on [PIN+] that we have observed in this study is likely independent of [PIN+] variants.
[SWI+] facilitates de novo formation of [PIN+]
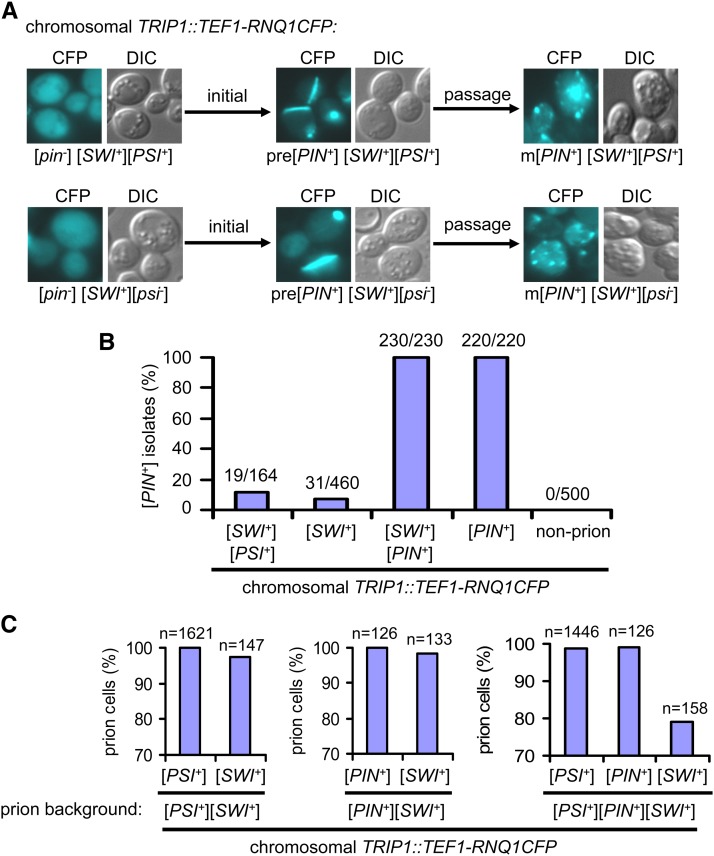
We next asked if [SWI+] was capable of promoting [PIN+] formation. We showed earlier that although the majority of [PSI+] isolates obtained from a [swi−][pin−][psi−] strain upon co-overexpression of SWI1 and SUP35NM-GFP remained [pin−], a small portion of them became [PIN+]. This indicates that [SWI+] might promote [PIN+] de novo appearance. The integrative TEF1–RNQ1-CFP provided a convenient tool with which the process of [PIN+] formation could be monitored by following the change of Rnq1-CFP aggregation patterns. Starting with a [SWI+][pin−][psi−] strain containing the integrative copy of TEF1–RNQ1-CFP, [PSI+] induction was conducted by overproduction of Sup35NM-YFP. We found that 40 of 150 examined [PSI+] isolates acquired [PIN+] as judged by the presence of inheritable Rnq1–CFP foci upon passage (Figure S3B). Rnq1-CFP aggregates could also arise occasionally in TEF1–RNQ1-CFP integrated [SWI+][pin−][PSI+] isolates containing no plasmids. In this case, most Rnq1-CFP aggregates initially appeared as a large single dot or rod-shaped fluorescence focus (premature [PIN+] aggregates) and later became multiple dots (mature [PIN+] aggregates) upon further growth and passage (Figure 2A, top). Under identical conditions, none of the examined [swi−][pin−][psi−] isolates showed heritable Rnq1-CFP aggregation and >95% of the [PSI+][PIN+] cells still retained Rnq1-CFP aggregation (Figure S3B and data not shown). A previous report showed that [PSI+] could induce [PIN+] appearance (Derkatch et al. 2001), and overproduction of Lsb2/Pin3 could also promote [PIN+] conversion (Chernova et al. 2011). Similarly, we also found that [PSI+] obtained by co-overproduction of Swi1 and Sup35NM-YFP from a [swi−][pin−][psi−] strain could promote de novo [PIN+] appearance as well (Figure S3, C and D). Our data suggest that like [PSI+], [SWI+] can also promote de novo [PIN+] conversion. For a further examination, a [SWI+][pin−][psi−] strain that contains an integrated copy of TEF1–RNQ1-CFP but carries no plasmids was grown and spread on YPD plates, individual colonies were examined for Rnq1-CFP fluorescence patterns. We observed that similar to what was observed in the [SWI+][pin−][PSI+] cells, the rod-shaped Rnq1-CFP aggregates appeared in a small fraction of [SWI+][pin−][psi−] cells, which subsequently became stable and transmissible dot-shaped aggregates (Figure 2A, bottom). To quantify, we assayed the [PIN+] conversion frequencies in a set of RNQ1-CFP integrated strains with different prion backgrounds in liquid medium. As shown in Figure 2B, after 48 hr of growth in YPD medium followed by spreading to YPD plates, ∼12% of [SWI+][pin−][PSI+] and 7% of [SWI+][pin−][psi−] colonies became [PIN+], as judged by the presence of heritable Rnq1-CFP aggregates.
Figure 2.
[SWI+] can significantly promote de novo formation of [PIN+]. Strains used in this figure all have an integrated TEF–RNQ1-CFP. (A) Top: a [pin−][SWI+][PSI+] isolate (a [PSI+] derivative of a [pin−][SWI+][psi−] strain upon overproduction of Sup35NM–YFP but no longer carrying any plasmids) that initially showed diffusible Rnq1-CFP signals gave rise to some rod-like Rnq1-CFP aggregates (pre[PIN+]) in a small portion of cells, which eventually became dot-shaped Rnq1-CFP foci in mature [PIN+] (m[PIN+]) isolates upon colony purification. Bottom: similar conformational changes of Rnq1-CFP and maturation process were observed in [pin−][SWI+][psi−] cells. Shown are representative images. (B) Quantification of spontaneous frequency of [PIN+] formation in the indicated strains, which were cultured in liquid YPD for 48 hr before spreading onto YPD plates. Individual colonies were examined for heritable Rnq1-CFP aggregation. Shown are combined results of percentages of examined colonies containing [PIN+] as judged by the presence of inheritable Rnq1GFP aggregates. The absolute numbers of [PIN+] colonies vs. the total examined are also shown. (C) Stability of [PSI+], [PIN+] and [SWI+] when two or three of them coexist. The indicated strains carrying different prions were grown in YPD medium for 48 hr and retested for their prion status after spreading onto YPD plates. The colony color on YPD plates, Rnq1-CFP signals, and raffinose utilization efficiency were used to score the status of [PSI+], [PIN+], and [SWI+], respectively. Combined results are shown (n, total number of colonies examined)
The above observations demonstrate that [SWI+] alone can induce the de novo formation of [PIN+] in the integrated strains, in which Rnq1-CFP was regulated by the TEF1 promoter and thus considerably overproduced compared to the endogenous Rnq1 expression (Figure 1B). To test whether the [PIN+]-promoting activity by [SWI+] could also occur in nonintegrated strains, [SWI+][pin−][psi−] and [swi−][pin−][psi−] strains were transformed with the plasmid pCUP1–RNQ1GFP, and the [PIN+] conversion was assayed both for fresh transformants and for cells after incubation on SC–ura plate without copper addition (see details in Materials and Methods). We found that 28 of 72 examined isolates (39%) of the [SWI+][pin−][psi−] strain turned to [SWI+][PIN+][psi−] (as indicated by heritable Rnq1-GFP aggregates) after 14 days of incubation at 4°. No inheritable Rnq1-GFP aggregates were observed among the 38 examined isolates of the [swi−][pin−][psi−] strain under identical conditions. Importantly, [PIN+] induction was also observed in a small number of cells of fresh transformants (data not shown). These results confirm that [PIN+]-promoting activity by [SWI+] can also occur in nonintegrated strains. Taken together, our data demonstrate that like [PSI+], [SWI+] can significantly promote de novo [PIN+] appearance under described experimental conditions.
Results in Figure 2, A and B, suggested that a single yeast cell was able to harbor three prions, [PSI+], [PIN+], and [SWI+]. To examine how their coexistence influences each other’s propagation, we next tested the stability of individual prions in three RNQ1-CFP integrated strains that harbor either two ([PSI+][SWI+] or [PIN+][SWI+]) or three prions ([SWI+][PSI+][PIN+]) (Figure 2C). After 24 or 48 hr of growth in YPD liquid medium, cultures were spread onto solid media to allow the formation of single colonies and examination of their prion status. Specifically, colony color change from white to red, acquired ability to use raffinose, and riddance of Rnq1-CFP aggregation were used as criteria for the loss of [PSI+], [SWI+], and [PIN+], respectively. About 97% of [SWI+][pin−][PSI+] ([SWI+][PSI+]) and [SWI+][PIN+][psi−] ([SWI+][PIN+]) isolates examined retained [SWI+], indicating that [SWI+] could stably coexist with [PSI+] or [PIN+]. For [PSI+][PIN+][SWI+] cells, >20% of them had lost [SWI+]; however, ∼98% of them retained their [PSI+] and [PIN+] prions (Figure 2C), suggesting that [SWI+] is unstable when coexisting with [PSI+] and [PIN+]. Our results demonstrated that yeast was capable of harboring three prions simultaneously ([PSI+], [PIN+], and [SWI+]), but the stability of [SWI+] significantly declined in this case.
Aggregates caused by overproduction of Swi1 are usually not heritable
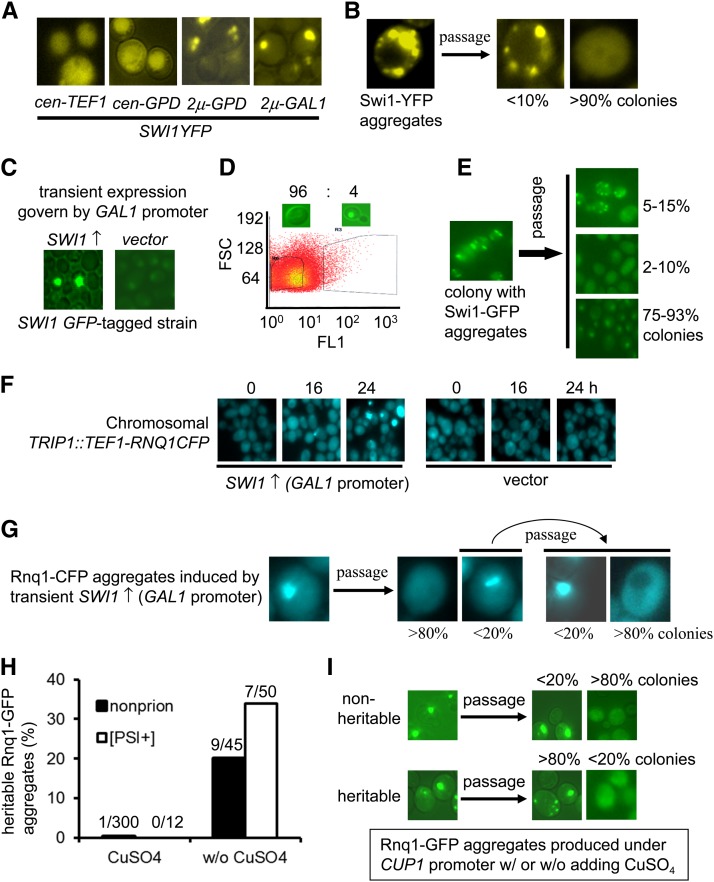
Aggregation is thought to underlie the prion propensity of amyloidogenic prion proteins. To understand the different Pin+ activities associated with SWI1 overexpression and [SWI+], we further investigated if the overexpression-caused aggregation of Swi1 is prionogenic. Swi1-YFP aggregation was observed only when SWI1-YFP was overexpressed from a 2µ-plasmid driven by GPD or GAL1 promoter, with an aggregation frequency of ∼15–40% (Figure 3A). Following colony purification, aggregate-containing cells were traced upon further growth and passaging. Such an attempt to obtain heritable Swi1-YFP aggregates ([SWI+] candidates) failed as for each round of restreaking, >90% of the newly formed colonies lost Swi1-YFP aggregates (Figure 3B). The failure to acquire [SWI+] might be, at least partially, due to the constant overproduction of Swi1-YFP because overproduction of a prion protein in the presence of its prion form might be toxic as known for other prions, e.g., [PSI+] (Dagkesamanskaya and Ter-Avanesyan 1991; Derkatch et al. 1996; Vishveshwara et al. 2009) and [PIN+] (Douglas et al. 2008; Stein and True 2011; Treusch and Lindquist 2012). We then tested if transient overproduction of Swi1 could lead to the aggregation of endogenous Swi1 and [SWI+] conversion. Using p426GAL1–SWI1, a 2µ-plasmid-carrying a GAL1–SWI1 expression cassette, transient overexpression of SWI1 in a SWI1GFP-tagged non-prion strain caused aggregation of endogenous Swi1-GFP (Figure 3C). When the SWI1 expression was turned off by transferring cells to a glucose-containing SC–ura medium and sorted by flow cytometry, ∼4% cells still contained Swi1-GFP aggregates (Figure 3D). These aggregate-carrying cells were collected and spread onto YPD plates, followed by examination of the inheritability of the Swi1-GFP aggregates as described in Materials and Methods. When individual colonies were examined, none contained heritable Swi1–GFP aggregates. Typically, only 5–15% of the resulting colonies retained aggregates, 2–10% colonies showed GFP signals diffusible in the entire cell (mislocalization), and the remaining population (75–93%) exhibited typical Swi1 nuclear localization (Figure 3E). When those aggregate-containing isolates were further selected and passaged, similar patterns were observed. Without selection, Swi1-GFP aggregates were rapidly lost upon growth. As a result, we failed to obtain any promising [SWI+] candidates in a period of ∼3 months after examining hundreds of cells harboring Swi1-GFP aggregate.
Figure 3.
Aggregates formed spontaneously or induced by a preexisting prion are more prionogenic compared to those formed upon overexpression. (A) Aggregation propensity of Swi1-YFP is depended on its expression levels. (B) Swi1-YFP aggregates acquired in the experiment shown in A were gradually lost upon passaging when Swi1-YFP was constitutively overproduced. (C) Transient overproduction of Swi1 led to aggregation of endogenous Swi1–GFP in a BY4741 SWI1–GFP tagged non-prion strain. (D) Cells harboring Swi1–GFP aggregate shown in C were sorted by flow cytometry. Forward scatter (FSC) height and green fluorescence emission (FL1) intensity are plotted as shown. (E) The nonheritable Swi1–GFP aggregates could not be stabilized upon passage. Hundreds of isolates were assayed and representative results are shown. (F) A TEF1–RNQ1–CFP-integrated [pin−] strain was transformed with p426GAL1–SWI1 or an empty vector. Rnq1-CFP aggregates were examined at the indicated time points after galactose addition. (G) Rnq1-CFP aggregates formed upon Swi1 overproduction shown in F could not be successfully transmitted to progeny upon passage. (H and I) Non-TEF1–RNQ1-CFP integrated [PSI+][pin−] ([PSI+]) and [psi−][pin−] (non-prion)) strains were transformed with plasmid pCUP1–RNQ1GFP. As described in Materials and Methods, Rnq1–GFP aggregates generated after incubation at 4° for 14 days on SC–ura with or without copper salt were assayed for inheritability. (H) Summary of percentages and absolute numbers of isolates harboring heritable Rnq1–GFP aggregate- vs. the total aggregate-containing isolates. (I) Representative result showing heritable and nonheritable aggregates upon passaging.
Intriguingly, we found that transient SWI1 overexpression can also promote Rnq1-CFP aggregation in TEF1–RNQ1-CFP integrated [pin−] cells, in which the Rnq1-CFP was relatively overproduced compared to the endogenous Rnq1 level (Figure 1B and Figure 3F). Unlike the rod-shaped prionogenic aggregates induced by [SWI+], these Rnq1-CFP aggregates are mostly dot shaped and not heritable upon further growth and passage as shown by colony purification and fluorescence microscopic assays (Figure 3G). This suggests that the Rnq1 aggregation caused by transient Swi1 overproduction is largely non-prionogenic. To further confirm our results, we compared the inheritability of Rnq1-GFP aggregates induced by different Rnq1-GFP production levels. Isogenic [swi−][pin−][psi−] (non-prion), [swi−][pin−][PSI+] ([PSI+]), and [swi−][PIN+][psi−] ([PIN+]) strains were transformed with pCUP1–RNQ1GFP followed by Rnq1-GFP aggregation assay of the obtained transformants in a 30-day period at 4° on SC–ura plates supplemented with (overproduction) or without (leaking expression) 100 µM CuSO4. As described in Materials and Methods, Rnq1-GFP aggregation was examined at each indicated time point. After 14 days of incubation at 4° in the presence of copper, an aggregation frequency of ∼93, 12.0, and 31.6% was observed for [PIN+], [PSI+], and non-prion cells, respectively. The frequency of Rnq1-GFP aggregation in the [PSI+] strain that was unexpectedly lower than that of the isogenic non-prion strain implies that overproduction of Rnq1-GFP in the presence of [PSI+] was toxic. Follow-up experiments indicated that ∼100% of the Rnq1 aggregates of the [PIN+] strain were stably inherited in the presence of copper. However, the Rnq1-GFP aggregates associated with copper addition were mostly not transmissible for [PSI+] and non-prion cells. Only ∼0 and 0.33% of their aggregates were heritable for [PSI+] and non-prion cells, respectively (Figure 3H). Typically, ∼80% of the progeny lost the Rnq1–GFP aggregates after each passage. We refer such Rnq1–GFP aggregates as nonheritable (Figure 3I, top). In comparison, significantly lower aggregation frequencies of Rnq1-GFP were observed for transformants after 14 days of incubation on plates without CuSO4 supplement: ∼64, 5.5, and ∼0.02% for the [PIN+], [PSI+], and non-prion strain, respectively. The higher aggregation frequency of the [PSI+] strain compared to that of the non-prion strain is consistent with previous findings that [PSI+] could facilitate [PIN+] formation (Derkatch et al. 2001) (also see Figure S3, C and D). We found that a significant portion of these aggregate-containing isolates (20% for non-prion; 34% for [PSI+]) were able to pass the aggregation to progeny to become stable [PIN+], although most of them were not (Figure 3H). Under this condition, ∼100% of the Rnq1-GFP aggregates were heritable for the [PIN+] strain. For these heritable Rnq1-GFP aggregates, as shown in Figure 3I (bottom), >80% progeny colonies retained Rnq1–GFP aggregates for each round of passage. Taken together, our results suggest that Swi1 and Rnq1 aggregates caused by overproduction are mostly nonheritable and distinct from those formed spontaneously or induced by a preexisting prion, which are more heritable. These findings are similar to the earlier reports that the aggregation induced by overproduction of Sup35 protein is mostly not heritable (Salnikova et al. 2005; Kushnirov et al. 2007).
Interaction of preexisting prion(s) with a newly generated heterologous prion during prionogenesis
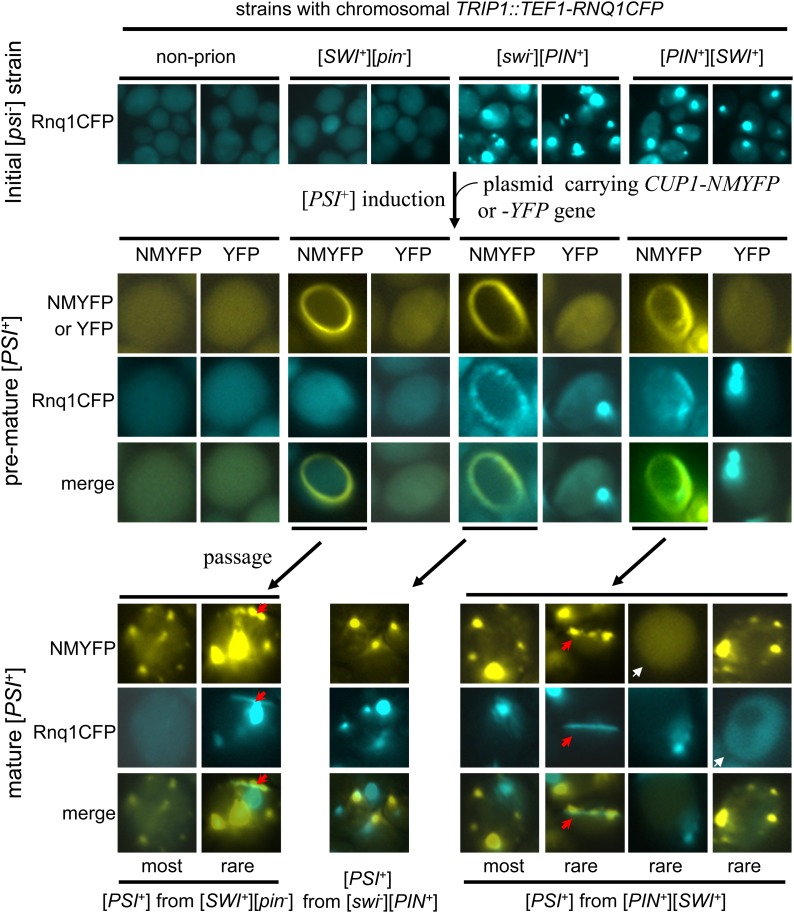
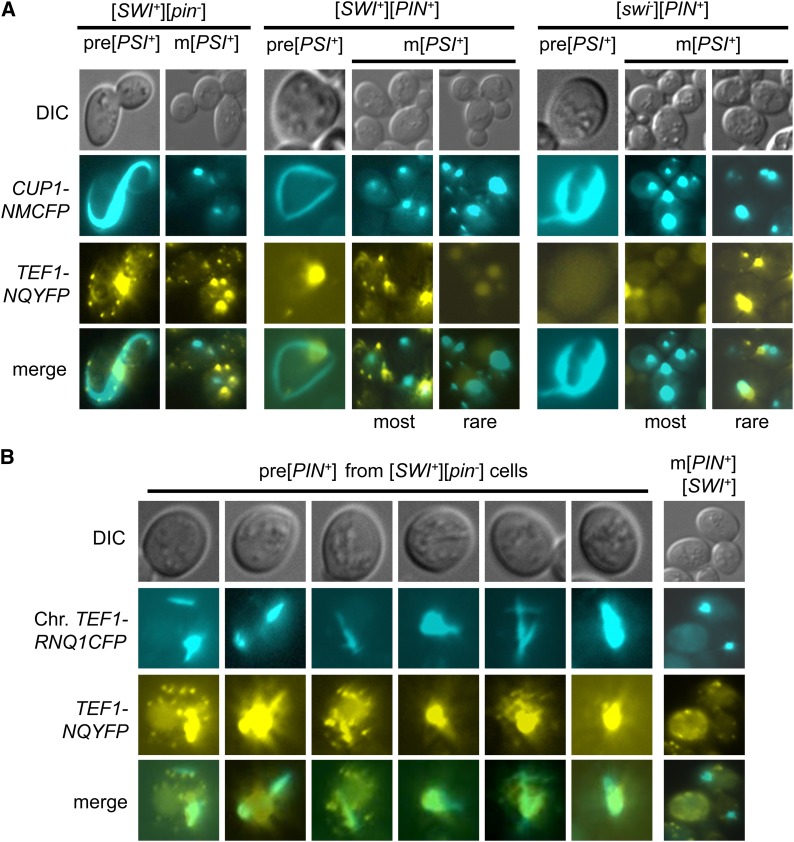
To examine how [SWI+] would influence the interactions of Sup35 and Rnq1 during the [PSI+] induction process in the presence and absence of [PIN+], Sup35NM-YFP was overproduced in four isogenic RNQ1-CFP-integrated strains, [psi−][pin−][swi−] (non-prion), [psi−][pin−][SWI+] ([SWI+]), [psi−][PIN+][swi−] ([PIN+]), and [psi−][PIN+][SWI+] ([PIN+][SWI+]). Sup35NM-YFP ring-/rod-shaped aggregates were observed only in strains containing [SWI+] or [PIN+] (Figure 4, middle, and Figure 5). Interestingly, the Rnq1-CFP aggregation pattern was dramatically altered from dot shaped to ring/rod shaped during [PSI+] induction, and the modulated Rnq1-CFP ring/rod aggregates were well colocalized with the newly formed Sup35NM-YFP ring/rods (Figure 4, middle, and Figure 5). In cells without Sup35NM-YFP ring/rods, Rnq1-CFP remained in diffusible ([pin−]) or dot-shaped ([PIN+]) patterns (Figure 4). In mature [PSI+] isolates, however, both Sup35NM-YFP and Rnq1-CFP aggregates were primarily observed as dots and were not colocalized (Figure 4, bottom). Moreover, the Rnq1-CFP aggregates and Sup35NM-YFP ring/rods showed decreased colocalization in [PIN+][SWI+] cells when compared to those in [PIN+] cells (Figure 4, middle, and Figure 5), which was consistent with the reduced Pin+ activity of [PIN+][SWI+] compared to that of [PIN+] (Figure 1F). Although most of the de novo formed [PSI+] cells derived from a [SWI+] strain remained [pin−], rod-shaped Rnq1-CFP aggregates occasionally appeared in the mature [PSI+][SWI+] cells, which often overlap with Sup35NM-YFP dots that are apparently morphologically changed (Figure 4, bottom left, red arrows). In cells containing three prions, [PSI+][PIN+][SWI+], Sup35NM-YFP, or Rnq1-CFP aggregations can be lost at a low frequency (Figure 4, white arrows). In rare cases, Rnq1–CFP rods could reappear in mature [PSI+] cells derived from the [PIN+][SWI+] strain, which largely overlapped the morphologically remodeled Sup35NM-YFP aggregates (Figure 4, right bottom, red arrows). These observations strongly suggested that architecture of the preexisting prion aggregates could be modified or decorated during the initiation process of another prion’s formation, likely a consequence of cross-seeding of the new prion using the protein determinant of the preexisting prion as a template. Apparently, the presence of [SWI+] can affect the interactions between Sup35 and Rnq1 during [PSI+] initiation and maturation process.
Figure 4.
Preexisting [PIN+] aggregates and their interaction with newly produced Sup35NMYFP aggregates during [PSI+] prionogenesis. Strains used in these experiments all carried an integrated copy of the TEF1–RNQ1-CFP. Shown are representative fluorescence patterns of YFP, Sup35-NMYFP (NMYFP), and Rnq1-CFP during [PSI+] initiation and maturation process for each indicated strain. Initial Rnq1-CFP aggregates in [PIN+] cells were dot shaped (top right). Upon NMYFP overproduction (from pRS316CUP1–NMYFP), NMYFP ring/rods appeared only in prion-containing strains (observed after 16 hr overproduction). Note that the ring-/rod-shaped NMYFP signals are significantly overlapped with the remodeled Rnq1-CFP aggregates in cells harboring [PIN+]. In mature [PSI+] cells, both of the NMYFP and Rnq1-CFP aggregates were mostly dot shaped and essentially not overlapping (bottom). In rare cases, Rnq1–CFP rod-like aggregates could appeared in mature [PSI+] cells that were derived from either [pin−] or [PIN+], and they overlap with Sup35NM–YFP aggregates (bottom, red arrows). Cells that occasionally lost [PIN+] or [PSI+] aggregates are indicated with white arrows.
Figure 5.
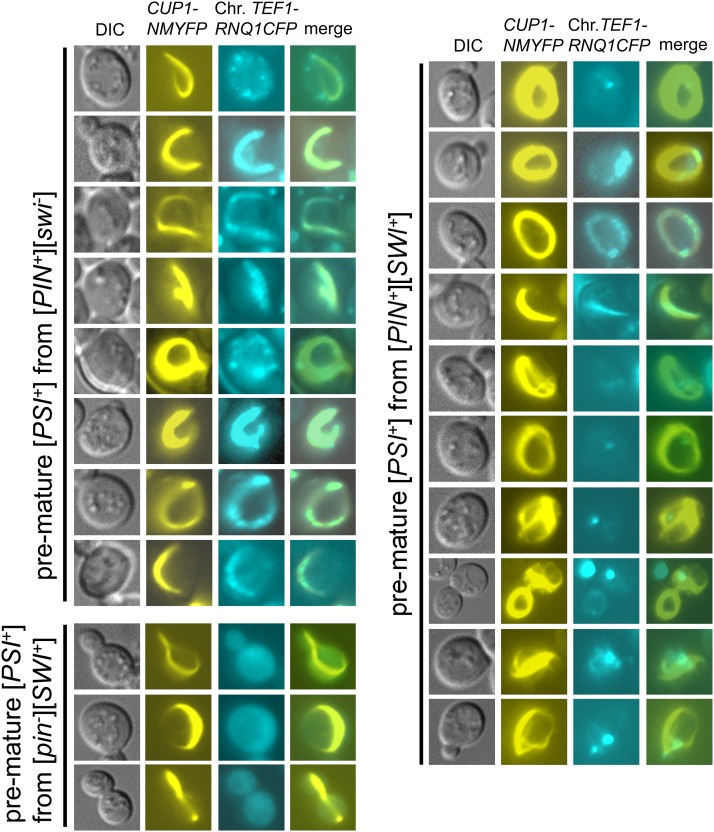
Interaction of newly generated premature [PSI+] aggregates with preexisting heterologous prion aggregates. The TEF1–RNQ1-CFP-integrated strains with indicated prion backgrounds were transformed with pCUP1–NMYFP and induced in liquid medium as described in the legend of Figure 4. The Sup35NM–YFP ring-/rod-like aggregates and Rnq1–CFP signals were pictured in premature [PSI+] cells and merged. Note that in the early stages of [PSI+] prionogenesis, there are remarkable overlapping signals between Sup35NM-YFP and Rnq1-CFP in [PIN+][swi−] strain. Such overlapping signals are significantly reduced in [PIN+][SWI+] cells and essential absent in [pin−][SWI+] cells.
We next investigated how [SWI+] prion aggregates interact with Sup35 and Rnq1 during the prionogenesis of [PSI+] and [PIN+]. We conducted [PSI+] induction experiments by overproducing Sup35NM-CFP in [SWI+][pin−][psi−] ([SWI+][pin−]), [swi−][PIN+][psi−] ([swi−][PIN+]), and [SWI+][PIN+][psi−] ([PIN+][SWI+]) cells containing p413TEF–NQYFP. Again, we saw that newly formed Sup35NM-CFP aggregates were ring/rod shaped in premature [PSI+] cells but became dot shaped in mature [PSI+] cells (Figure 6A and Figure S4). In [SWI+] cells, the morphology and distribution patterns of Swi1NQ-YFP aggregates became colocalized with the ring-/rod-shaped Sup35NM-CFP aggregates in premature [PSI+] cells (Figure 6A, left, and Figure S4). Noticeably, in agreement with a lower Pin+ activity of [SWI+], overlapping signals of Swi1NQ-YFP and Sup35NM-CFP in premature [PSI+] derivatives of [SWI+] cells were significantly less than that of Rnq1–CFP and Sup35NM-YFP in premature [PSI+] derivatives of [PIN+] cells, suggesting that [PIN+] can better seed Sup35 to induce [PSI+] than [SWI+]. Also, colocalization signals of Swi1NQ-YFP and Sup35NM-CFP were significantly reduced in [SWI+][PIN+] cells when compared to that in [SWI+] cells (Figure 6A, left and middle, and Figure S4). Although most Swi1NQ-YFP signals were diffused in mature [PSI+] cells derived from the [PIN+] strain, Swi1NQ-YFP aggregates were occasionally seen and to some extent, colocalized with Sup35NM-CFP aggregates (Figure 6A, right), suggesting that [SWI+] might be induced de novo in [PSI+][PIN+] cells. Moreover, Swi1NQ-YFP aggregates can be lost in a small portion of stabilized [PSI+] isolates derived from the [SWI+][PIN+] strain, an observation that is consistent with the decreased stability of [SWI+] when the three prions coexist (Figure 2C).
Figure 6.
Preexisting [SWI+] aggregates and their interaction with newly formed Sup35NM-CFP and Rnq1-CFP aggregates during [PSI+] and [PIN+] prionogenesis and maturation. (A) Yeast strains containing the indicated prion(s) were transformed with plasmids pRS316CUP1–NMCFP and p413TEF–NQYFP and induced for [PSI+] formation. Images were taken 16 hr after addition of CuSO4 when ring/rod-shaped Sup35NM-CFP (MNCFP) aggregates were formed in premature [PSI+] cells (pre[PSI+]) cells. The ring-/rod-shaped NMCFP signals are significantly overlapped with the remodeled Swi1NQ-YFP (NQYFP) aggregates in cells harboring [SWI+] (left). Mature [PSI+] (m[PSI+]) isolates were reexamined for NMCFP and NQYFP fluorescence patterns, which were mostly dot shaped and essentially not overlapping. Rare overlapping signals are indicated. See Figure S4 for more images. (B) An RNQ1-CFP-integrated [SWI+][pin−] strain was transformed with the plasmid p413TEF–NQYFP and grown in SC–his liquid medium for 2 days (∼40 hr) and screened for the rod-like premature Rnq1-CFP (pre[PIN+]) aggregates; NQYFP signals were also recorded. Upon colony purification, stably inherited aggregates of Rnq1-CFP and NQYFP were examined in mature [PIN+] (m[PIN+]) cells (right).
As [SWI+] was able to promote spontaneous [PIN+] appearance (Figure 2, A and B), we further investigated the interaction of Rnq1-CFP and Swi1NQ-YFP aggregates in premature and stabilized [PIN+] derived from [SWI+] cells. This was done by examining the appearance of Rnq1-CFP aggregation in a RNQ1-CFP-integrated [SWI+] strain ectopically expressing Swi1NQ-YFP. As shown in Figure 6B, in premature [PIN+] cells that harbor [SWI+], Rnq1-CFP rod-like aggregates were observed and colocalized with Swi1NQ-YFP foci. Morphology and distribution pattern of Swi1NQ-YFP aggregates were apparently modulated in the earlier stage of the Rnq1 prion generation. However, in stabilized [PIN+][SWI+] cells, the Rnq1-CFP and Swi1NQ-YFP aggregates were dot shaped and not colocalized (Figure 6B, right).
Discussion
The discovery of a number of amyloid-based prions in the budding yeast makes this organism a useful model to study heterologous prion interactions. This is of great interest as this line of research may generate valuable data to not only aid our understanding of prion formation and transmission events in yeast, but also shed light on the mechanisms underlying protein misfolding, aggregation, and pathogenesis in humans. Although prion aggregates usually template the conversion of identical protein isomers, different prion proteins and some non-prion Q/N-rich proteins can also interact and influence each other’s aggregation (Derkatch et al. 2001, 2004; Osherovich and Weissman 2001; Gonzalez Nelson and Ross 2011; Inoue et al. 2011). Two basic mechanisms have been proposed for the interactions of different prion proteins in yeast: the cross-seeding model and the titration model (Derkatch et al. 2001; Osherovich and Weissman 2001). In the cross-seeding model, direct protein–protein contacts are considered as a basis of interactions since almost all prionogenic proteins contain a transferrable region(s) similarly enriched in polar residues necessary for highly ordered amyloid formation (Ter-Avanesyan et al. 1994; Masison and Wickner 1995; Glover et al. 1997; King et al. 1997; Taylor et al. 1999; Derkatch et al. 2004; Alberti et al. 2009; Du et al. 2010). For example, direct cross-seeding was observed for Sup35 and Rnq1 in an in vitro fibril assembly assay (Derkatch et al. 2004; Sharma and Liebman 2013). As a consequence, preexisting prion aggregates or aggregates resulting from overproduction of a protein such as Q103 might serve as templates to allow de novo formation of a new prion (Derkatch et al. 2004). Alternatively, the titration model predicts that preexisting prion aggregates, or newly formed protein aggregates caused by overproduction, may compete for binding, or perhaps sequester anti-prion cellular factors, such as chaperones and proteases, and thereby increase the likelihood of a new prion conversion (Osherovich and Weissman 2001). Although both models have gained supportive evidence (Osherovich and Weissman 2001; Schwimmer and Masison 2002; Derkatch et al. 2004), our results support the cross-seeding model in general.
In this study, we show that although both [SWI+] and Swi1 overproduction act as Pin+ factors independent of Rnq1 or [PIN+], their Pin+ activities are significantly lower than that of [PIN+] (Figure 1, A and E). Although [PIN+] is an effective Pin+ factor (Derkatch et al. 1997), in an earlier study, several Q/N-rich proteins were found to be Pin+ when overproduced, which do not include Rnq1 (Derkatch et al. 2001), suggesting that overproduction of Rnq1 is not an effective Pin+ factor. In this study, we further showed that overproduction-mediated aggregation of Swi1 and Rnq1 is less heritable than that formed spontaneously or induced by a preexisting prion(s) (Figure 3), suggesting that aggregation resulting from overproduction is mostly amorphous aggregates that are ineffective in promoting prion conversions. This is in agreement with the fact that overproduction of Sup35 alone is ineffective in inducing [PSI+] de novo formation (Derkatch et al. 1997) and the aggregation induced by Sup35 is mostly not heritable (Salnikova et al. 2005; Kushnirov et al. 2007). A recent study suggested that [PIN+] variants differ mainly in their cross-seeding abilities but not in their seed numbers or other features in promoting [PSI+] conversion (Sharma and Liebman 2013). Likely, only a very small fraction of the aggregates resulting from overproduction can acquire prionogenic conformation(s). In addition, toxic effects associated with overproduction may also contribute to a lower prion propensity or seeding ability of a Q/N-rich protein in this study. Our finding that [PSI+] candidates obtained from [pin−] cells by co-overproduction of Sup35 and Swi1, Ure2–PrD–GFP or Q103–GFP appeared on –ade plates ∼2 to 3 days later than those obtained from [PIN+] by Sup35 overproduction seems to support this notion (Figure S1A). In addition, we found that when Rnq1-GFP was constitutively overproduced, a lower aggregation frequency was observed in [PSI+] cells compared to that in [psi−] cells. These results suggest that overproduction, especially constitutive overproduction of Swi1 and other Q/N-rich proteins in the presence of a preexisting prion, is stressful and toxic to the cells. This may explain some of our results in this study, for example, the noninheritability of Swi1-YFP and Rnq1-GFP aggregation under the condition of constant overproduction of Swi1-YFP and Rnq1-GFP, respectively (Figure 3, B and H). We also cannot exclude the possibility that suicidal [PSI+] (McGlinchey et al. 2011) may also contribute to the different Pin+ activities we observed in this study. The fact that most of the Swi1 and Rnq1 aggregates resulting from overproduction are not heritable does not necessarily mean that the heritable aggregation cannot be promoted by overproduction. The heritable aggregates may be buried in the larger amount of nonheritable aggregates concurrently induced and thereby are difficult to observe. Without a selection system, tracing aggregates generated by overproduction is therefore not an efficient way to identify prion candidates.
We showed that in addition to promoting the de novo formation of [PSI+] and [PIN+], [SWI+] can also antagonize [PIN+] to reduce its Pin+ activity when [PIN+] and [SWI+] coexist (Figure 1, E and F). Our observation that several [PIN+] isolates with distinct Rnq1 aggregation patterns all significantly reduced Pin+ function in the presence of [SWI+] (Figure 1F) indicates that this antagonizing effect is likely independent of [PIN+] variants. Although the underlying mechanism of this [SWI+]-mediated Pin+ reduction of [PIN+] remains to be elucidated, we speculate that [SWI+] and [PIN+] may compete each other for Sup35 to cross-seed and initiate [PSI+]. Recruiting of Sup35 by [SWI+] aggregates may have reduced the availability of free Sup35 to be cross-seeded by [PIN+], which is a stronger Pin+ factor than [SWI+] and thus may lead to less [PSI+] de novo appearance. Alternatively, interaction between [SWI+] and [PIN+] may reduce the quantity and/or quality of [PIN+] templates in cross-seeding Sup35.
We showed in this study that a single yeast cell can harbor three prions: [SWI+], [PSI+], and [PIN+] (Figure 2C). While [SWI+] can be stably maintained in >95% of cells that contain [PSI+] or [PIN+], we showed that when the three prions coexist, [SWI+] becomes significantly unstable with only <80% cells capable of maintaining [SWI+] (Figure 2C). The instability of maintaining three prion elements in a single yeast cell might also be a contributing factor to the reduced Pin+ activity of [PIN+] in [SWI+][PIN+] cells. Antagonizing effects have also been reported for cells containing two heterologous prions. For example, Bradley and Liebman (2003) found that some variants of [PIN+] can destabilize weak [PSI+]. Schwimmer and Masison (2002) have also shown that there are antagonistic interactions between [URE3] and [PSI+]. In this case, an elevated Hsp104 level was thought to be responsible for the partial loss of [PSI+] in [PSI+][URE3] cells (Schwimmer and Masison 2002).
[PSI+] prion aggregates formed at their earlier initiation stage have been shown to have unique ring-/rod-shaped structures, which are distinct from the dot-shaped aggregates observed at their later mature stage (Derkatch et al. 2001; Zhou et al. 2001). Upon overproduction, de novo formed Sup35 aggregates appeared as either ring/rod shaped or dot shaped, and usually the ring-/rod-shaped aggregates are prionogenic (Derkatch et al. 2001; Zhou et al. 2001; Alberti et al. 2009). The nonheritable dot-shaped Sup35 aggregates that appeared upon overproduction seem fundamentally different from the dot-shaped aggregates found in the mature [PSI+]. Similarly, we observed in this study that the initially formed aggregates of Swi1 or Rnq1 upon overproduction are mostly dot shaped and usually not heritable whereas the newly initiated prionogenic aggregates are usually ring/rod shaped (Figure 3, Figure 4, Figure 5, Figure 6, and Figure S2). Although we do not yet know if the initially formed Swi1 aggregates in pre-[SWI+] cells are also ring/rod like, current data seemingly suggest that only ring-/rod-shaped aggregates are prionogenic. Like the dot-shaped mature [PSI+] aggregates, the ring-/rod-shaped [PSI+] foci of Sup35 were shown to have bundled fibrillar amyloid structure (Kawai-Noma et al. 2010; Tyedmers et al. 2010). However, it is unclear whether the dot-shaped Sup35 aggregates promoted by overproduction contain amyloid-like fibrillar structures. It has been shown that the newly formed ring-/rod-shaped Sup35 aggregates were associated with actin-binding scaffold proteins as deletion mutants of cytoskeletonal components of SLA1 and SLA2 resulted in reduction of [PSI+] formation (Ganusova et al. 2006; Manogaran et al. 2011). A recent study shows that a mutation eliminating association of a stress-inducible QN-rich protein, Lsb2 with the actin cytoskeleton, inhibits its aggregation and prion-inducing ability (Chernova et al. 2011), confirming that the actin cytoskeleton likely serves as a platform for initial prion conversions. Interestingly, unlike the premature [PSI+] aggregates, which can be both ring and rod shaped, we saw that almost all newly formed [PIN+] aggregates appear rod shaped, and the ring-shaped aggregations are very rare in premature [PIN+] cells (Figure 2A and Figure 6B). These results suggest a structural difference between the premature aggregates of [PSI+] and [PIN+] and, perhaps, involvement of distinct cellular components in their de novo prionogenetic processes. Previous studies suggest that direct interactions of Rnq1 and Sup35 mainly happen at the early phase of [PSI+] induction, and after the de novo formed [PSI+] is stabilized, prion aggregates of [PSI+] and [PIN+] do not usually colocalize (Bagriantsev and Liebman 2004; Derkatch et al. 2004). Our data also suggest that prion–prion interactions mainly happen at the earlier prion induction stage (Figure 4, Figure 5, and Figure 6), a time when the colocalization of heterologous prion aggregates is noticeable. In cells containing established [PSI+] and [PIN+] prions, no significant colocalization of Sup35 and Rnq1 was observed (Figure 4). Intriguingly, the ability of one prion to enhance another prion’s de novo appearance is positively correlated with the colocalization frequency of the newly induced and the preexisting prion aggregates (Figure 1, Figure 2, Figure 5, Figure 6, and Figure S4), suggesting that direct protein–protein interactions (cross-seeding or decoration) are fundamental for heterologous prion induction. Even in the mature [PSI+][pin−][SWI+] cells, Rnq1-CFP aggregates can appear occasionally and when that happens, the Rnq1-CFP aggregates largely colocalize with the Sup35 aggregates (Figure 4). Similarly, in [PSI+][PIN+][swi+] cells, Swi1NQ-YFP aggregates are rarely formed, but if they do form, they also partially colocalize with the Sup35NM-CFP aggregates (Figure 6). Thus, our study provides evidence supporting the cross-seeding model, although the involvement of the titration mechanisms cannot be ruled out.
Acknowledgments
We thank Stephanie Valtierra and Anthony Kowal for critical comments. This work was supported by grants from the U.S. National Institutes of Health (R01NS056086) and U.S. National Science Foundation (MCB 1122135) to L.L.
Footnotes
Communicating editor: K. M. Arndt
Literature Cited
- Alberti S., Halfmann R., King O., Kapila A., Lindquist S., 2009. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137: 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen K. D., Chernova T. A., Tennant E. P., Wilkinson K. D., Chernoff Y. O., 2007. Effects of ubiquitin system alterations on the formation and loss of a yeast prion. J. Biol. Chem. 282: 3004–3013 [DOI] [PubMed] [Google Scholar]
- Bagriantsev S., Liebman S. W., 2004. Specificity of prion assembly in vivo. [PSI+] and [PIN+] form separate structures in yeast. J. Biol. Chem. 279: 51042–51048 [DOI] [PubMed] [Google Scholar]
- Brachmann A., Baxa U., Wickner R. B., 2005. Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J. 24: 3082–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley M. E., Liebman S. W., 2003. Destabilizing interactions among [PSI(+)] and [PIN(+)] yeast prion variants. Genetics 165: 1675–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley M. E., Edskes H. K., Hong J. Y., Wickner R. B., Liebman S. W., 2002. Interactions among prions and prion “strains” in yeast. Proc. Natl. Acad. Sci. USA 99(Suppl. 4): 16392–16399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff Y. O., Derkatch I. L., Inge-Vechtomov S. G., 1993. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr. Genet. 24: 268–270 [DOI] [PubMed] [Google Scholar]
- Chernoff Y. O., Lindquist S. L., Ono B., Inge-Vechtomov S. G., Liebman S. W., 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor. [PSI+] Science 268: 880–884 [DOI] [PubMed] [Google Scholar]
- Chernoff Y. O., Newnam G. P., Kumar J., Allen K., Zink A. D., 1999. Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone Ssb in formation, stability, and toxicity of the [PSI] prion. Mol. Cell. Biol. 19: 8103–8112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernova T. A., Romanyuk A. V., Karpova T. S., Shanks J. R., Ali M., et al. , 2011. Prion induction by the short-lived, stress-induced protein Lsb2 is regulated by ubiquitination and association with the actin cytoskeleton. Mol. Cell 43: 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge J., Clarke A. R., 2007. A general model of prion strains and their pathogenicity. Science 318: 930–936 [DOI] [PubMed] [Google Scholar]
- Cox B., 1965. [PSI], a cytoplasmic suppressor of super-suppression in yeast. Heredity 20: 505–521 [Google Scholar]
- Crow E. T., Li L., 2011. Newly identified prions in budding yeast, and their possible functions. Semin. Cell Dev. Biol. 22: 452–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow E. T., Du Z., Li L., 2011. A small, glutamine-free domain propagates the [SWI(+)] prion in budding yeast. Mol. Cell. Biol. 31: 3436–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagkesamanskaya A. R., Ter-Avanesyan M. D., 1991. Interaction of the yeast omnipotent suppressors SUP1(SUP45) and SUP2(SUP35) with non-Mendelian factors. Genetics 128: 513–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch I. L., Chernoff Y. O., Kushnirov V. V., Inge-Vechtomov S. G., Liebman S. W., 1996. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 144: 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch I. L., Bradley M. E., Zhou P., Chernoff Y. O., Liebman S. W., 1997. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 147: 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch I. L., Bradley M. E., Masse S. V., Zadorsky S. P., Polozkov G. V., et al. , 2000. Dependence and independence of [PSI+] and [PIN+]: a two-prion system in yeast? EMBO J. 19: 1942–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch I. L., Bradley M. E., Hong J. Y., Liebman S. W., 2001. Prions affect the appearance of other prions: the story of [PIN+] Cell 106: 171–182 [DOI] [PubMed] [Google Scholar]
- Derkatch I. L., Uptain S. M., Outeiro T. F., Krishnan R., Lindquist S. L., et al. , 2004. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc. Natl. Acad. Sci. USA 101: 12934–12939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas P. M., Treusch S., Ren H. Y., Halfmann R., Duennwald M. L., et al. , 2008. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc. Natl. Acad. Sci. USA 105: 7206–7211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., 2011. The complexity and implications of yeast prion domains. Prion 5: 311–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Park K. W., Yu H., Fan Q., Li L., 2008. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat. Genet. 40: 460–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Crow E. T., Kang H. S., Li L., 2010. Distinct subregions of Swi1 manifest striking differences in prion transmission and SWI/SNF function. Mol. Cell. Biol. 30: 4644–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q., Park K. W., Du Z., Morano K. A., Li L., 2007. The role of Sse1 in the de novo formation and variant determination of the [PSI+] prion. Genetics 177: 1583–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganusova E. E., Ozolins L. N., Bhagat S., Newnam G. P., Wegrzyn R. D., et al. , 2006. Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol. Cell. Biol. 26: 617–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J. R., Kowal A. S., Schirmer E. C., Patino M. M., Liu J. J., et al. , 1997. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 89: 811–819 [DOI] [PubMed] [Google Scholar]
- Gonzalez Nelson A. C., Ross E. D., 2011. Interactions between non-identical prion proteins. Semin. Cell Dev. Biol. 22: 437–443 [DOI] [PubMed] [Google Scholar]
- Halfmann R., Wright J. R., Alberti S., Lindquist S., Rexach M., 2012. Prion formation by a yeast GLFG nucleoporin. Prion 6: 391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines J. K., Craig E. A., 2011. The sensitive [SWI (+)] prion: new perspectives on yeast prion diversity. Prion 5: 164–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines J. K., Li X., Du Z., Higurashi T., Li L., et al. , 2011. [SWI], the prion formed by the chromatin remodeling factor Swi1, is highly sensitive to alterations in Hsp70 chaperone system activity. PLoS Genet. 7: e1001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Kawai-Noma S., Koike-Takeshita A., Taguchi H., Yoshida M., 2011. Yeast prion protein New1 can break Sup35 amyloid fibrils into fragments in an ATP-dependent manner. Genes Cells 16: 545–556 [DOI] [PubMed] [Google Scholar]
- Kawai-Noma S., Pack C. G., Kojidani T., Asakawa H., Hiraoka Y., et al. , 2010. In vivo evidence for the fibrillar structures of Sup35 prions in yeast cells. J. Cell Biol. 190: 223–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C. Y., Diaz-Avalos R., 2004. Protein-only transmission of three yeast prion strains. Nature 428: 319–323 [DOI] [PubMed] [Google Scholar]
- King C. Y., Tittmann P., Gross H., Gebert R., Aebi M., et al. , 1997. Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl. Acad. Sci. USA 94: 6618–6622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov V. V., Vishnevskaya A. B., Alexandrov I. M., Ter-Avanesyan M. D., 2007. Prion and nonprion amyloids: a comparison inspired by the yeast Sup35 protein. Prion 1: 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroute F., 1971. Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J. Bacteriol. 206: 519–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster A. K., Bardill J. P., True H. L., Masel J., 2010. The spontaneous appearance rate of the yeast prion [PSI+] and its implications for the evolution of the evolvability properties of the [PSI+] system. Genetics 184: 393–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Kowal A. S., 2012. Environmental regulation of prions in yeast. PLoS Pathog. 8: e1002973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund P. M., Cox B. S., 1981. Reversion analysis of [psi−] mutations in Saccharomyces cerevisiae. Genet. Res. 37: 173–182 [DOI] [PubMed] [Google Scholar]
- Manogaran A. L., Hong J. Y., Hufana J., Tyedmers J., Lindquist S., et al. , 2011. Prion formation and polyglutamine aggregation are controlled by two classes of genes. PLoS Genet. 7: e1001386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masison D. C., Wickner R. B., 1995. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 270: 93–95 [DOI] [PubMed] [Google Scholar]
- Mathur V., Taneja V., Sun Y., Liebman S. W., 2010. Analyzing the birth and propagation of two distinct prions, [PSI+] and [Het-s](y), in yeast. Mol. Biol. Cell 21: 1449–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlinchey R. P., Kryndushkin D., Wickner R. B., 2011. Suicidal [PSI+] is a lethal yeast prion. Proc. Natl. Acad. Sci. USA 108: 5337–5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum-Krammer C. I., Park K. W., Li L., Melki R., Morimoto R. I., 2013. Spreading of a prion domain from cell-to-cell by vesicular transport in Caenorhabditis elegans. PLoS Genet. 9: e1003351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherovich L. Z., Weissman J. S., 2001. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI+] prion. Cell 106: 183–194 [DOI] [PubMed] [Google Scholar]
- Park K. W., Hahn J. S., Fan Q., Thiele D. J., Li L., 2006. De novo appearance and “strain” formation of yeast prion [PSI+] are regulated by the heat-shock transcription factor. Genetics 173: 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel B. K., Liebman S. W., 2007. “Prion-proof” for [PIN+]: infection with in vitro-made amyloid aggregates of Rnq1p-(132–405) induces. [PIN+] J. Mol. Biol. 365: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel B. K., Gavin-Smyth J., Liebman S. W., 2009. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat. Cell Biol. 11: 344–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner S. B., 1998. Prions. Proc. Natl. Acad. Sci. USA 95: 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogoza T., Goginashvili A., Rodionova S., Ivanov M., Viktorovskaya O., et al. , 2010. Non-Mendelian determinant [ISP+] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1. Proc. Natl. Acad. Sci. USA 107: 10573–10577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikova A. B., Kryndushkin D. S., Smirnov V. N., Kushnirov V. V., Ter-Avanesyan M. D., 2005. Nonsense suppression in yeast cells overproducing Sup35 (eRF3) is caused by its non-heritable amyloids. J. Biol. Chem. 280: 8808–8812 [DOI] [PubMed] [Google Scholar]
- Schwimmer C., Masison D. C., 2002. Antagonistic interactions between yeast [PSI(+)] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell. Biol. 22: 3590–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma J., Liebman S. W., 2013. Exploring the basis of [PIN(+)] variant differences in [PSI(+)] induction. J. Mol. Biol. 425: 3046–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondheimer N., Lindquist S., 2000. Rnq1: an epigenetic modifier of protein function in yeast. Mol. Cell 5: 163–172 [DOI] [PubMed] [Google Scholar]
- Stein K. C., True H. L., 2011. The [RNQ+] prion: a model of both functional and pathological amyloid. Prion 5: 291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki G., Shimazu N., Tanaka M., 2012. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science 336: 355–359 [DOI] [PubMed] [Google Scholar]
- Tanaka M., Chien P., Naber N., Cooke R., Weissman J. S., 2004. Conformational variations in an infectious protein determine prion strain differences. Nature 428: 323–328 [DOI] [PubMed] [Google Scholar]
- Taylor K. L., Cheng N., Williams R. W., Steven A. C., Wickner R. B., 1999. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science 283: 1339–1343 [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan M. D., Dagkesamanskaya A. R., Kushnirov V. V., Smirnov V. N., 1994. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [PSI+] in the yeast Saccharomyces cerevisiae. Genetics 137: 671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treusch S., Lindquist S., 2012. An intrinsically disordered yeast prion arrests the cell cycle by sequestering a spindle pole body component. J. Cell Biol. 197: 369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J., Treusch S., Dong J., McCaffery J. M., Bevis B., et al. , 2010. Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. Proc. Natl. Acad. Sci. USA 107: 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishveshwara N., Bradley M. E., Liebman S. W., 2009. Sequestration of essential proteins causes prion associated toxicity in yeast. Mol. Microbiol. 73: 1101–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitrenko Y. A., Pavon M. E., Stone S. I., Liebman S. W., 2007. Propagation of the [PIN+] prion by fragments of Rnq1 fused to GFP. Curr. Genet. 51: 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann C., 2009. Thoughts on mammalian prion strains. Folia Neuropathol. 47: 104–113 [PubMed] [Google Scholar]
- Wickner R. B., 1994. [URE3] as an altered Ure2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264: 566–569 [DOI] [PubMed] [Google Scholar]
- Yang Z., Hong J. Y., Derkatch I. L., Liebman S. W., 2013. Heterologous gln/asn-rich proteins impede the propagation of yeast prions by altering chaperone availability. PLoS Genet. 9: e1003236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Derkatch I. L., Liebman S. W., 2001. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI+] and [PIN+]. Mol. Microbiol. 39: 37–46 [DOI] [PubMed] [Google Scholar]