Abstract
Systems biology is an approach to dissection of complex traits that explicitly recognizes the impact of genetic, physiological, and environmental interactions in the generation of phenotypic variation. We describe comprehensive transcriptional and metabolic profiling in Drosophila melanogaster across four diets, finding little overlap in modular architecture. Genotype and genotype-by-diet interactions are a major component of transcriptional variation (24 and 5.3% of the total variation, respectively) while there were no main effects of diet (<1%). Genotype was also a major contributor to metabolomic variation (16%), but in contrast to the transcriptome, diet had a large effect (9%) and the interaction effect was minor (2%) for the metabolome. Yet specific principal components of these molecular phenotypes measured in larvae are strongly correlated with particular metabolic syndrome-like phenotypes such as pupal weight, larval sugar content and triglyceride content, development time, and cardiac arrhythmia in adults. The second principal component of the metabolomic profile is especially informative across these traits with glycine identified as a key loading variable. To further relate this physiological variability to genotypic polymorphism, we performed evolve-and-resequence experiments, finding rapid and replicated changes in gene frequency across hundreds of loci that are specific to each diet. Adaptation to diet is thus highly polygenic. However, loci differentially transcribed across diet or previously identified by RNAi knockdown or expression QTL analysis were not the loci responding to dietary selection. Therefore, loci that respond to the selective pressures of diet cannot be readily predicted a priori from functional analyses.
Keywords: metabolic syndrome, metabolomics, evolve-and-resequence, genotype-by-environment, adaptation
TO what extent does genetic variation flow linearly through the transcriptome, proteome, and metabolome to generate phenotypes? Under the simplest model, additive genetic variation for transcript abundance and protein activity should correlate directly with variation in protein abundance and metabolite abundance, and in turn with organismal phenotypes (Lehner 2013; Civelek and Lusis 2013). We would generally expect an increase in variance at each successive physiological level such that the strength of genetic association decreases from transcript to metabolite to phenotype. This model has been used to support the notion that modules of gene activity, for example, may often associate with complex traits (Ayroles et al. 2009; Harbison et al. 2009). However, evidence that contradicts this model, and even suggests that modular reorganization may occur at successive levels of molecular function, is beginning to appear, considerably complicating the mapping of genotype to phenotype. For example, protein abundance often diverges between primate species to a lesser degree than transcript abundance (Khan et al. 2013), highly modular gene expression in Drosophila wing imaginal discs does not associate with wing shape (Dworkin et al. 2009), and eQTL map poorly onto protein and metabolite levels in Arabidopsis thaliana (Atwell et al. 2010), where it was argued that phenotypic buffering muddles the association with molecular measures.
Furthermore, environmental variation may also modify genotypic effects. An area of particular concern for human health is the increasingly prevalent metabolic syndrome (MetS), a collection of symptoms including obesity, insulin resistance, and heart disease (Eckel et al. 2005; Alberti et al. 2006). The dramatic increase in MetS and its related diseases in westernized nations can be largely blamed on environmental (lifestyle) effects rather than genetic effects alone (Schulz et al. 2006). However, individuals with a high genetic risk score for BMI accumulate weight more rapidly when they consume sugary beverages than their low-risk counterparts, indicating a genotype-by-diet interaction effect (Qi et al. 2012). We have also found that for MetS-like phenotypes in Drosophila, there is a large genotype-by-diet interaction effect contributing to variation in these traits (Reed et al. 2010). Many genes have been identified as being functionally linked to obesity, diabetes, and heart disease in flies, and those effects can be exacerbated by diet (Diop and Bodmer 2012). The questions thus arise as to whether these dietary influences on wild-type flies manifest themselves at the transcriptional or metabolic levels, whether modularity at these levels is comparable, and whether it can explain phenotypic variation for metabolism in flies.
One of the obstacles to be overcome in understanding these complex disease phenotypes is the variable correlation between genotype and phenotype over life history due to interaction effects between each genome and the individual’s environment. These can be complicated by behavioral choice, such as the ability of flies to modify their nutritional geometry, namely the ratio of protein to carbohydrate intake, apparently consistent with optimizing lifetime fecundity (Lee et al. 2008). Concordantly, the relationship between mRNA, protein, and metabolite abundance varies across genotypes and environments. In the realm of medicine, inclusion of clinical and functional genomic data may enhance risk classification beyond what can be done with genomic data alone (Patel et al. 2013), particularly where the relationship between the static genome and dynamic gene function changes over the lifetime of an organism.
These considerations led us to ask how genetic and environmental effects influence patterns of transcriptional, metabolic, and phenotypic variation and whether those patterns bear any relationship to the response of a natural population to artificial selection on different diets, using the well-characterized model organism Drosophila melanogaster. Here we report two complementary experiments. First we asked how dietary perturbation in a genetically variable population affects transcriptional, metabolic, and phenotypic profiles. Second, using an evolve-and-resequence approach (Burke et al. 2010), we quantified genomic responses to artificial dietary selection. We then contrast the core lab-adaptation and diet-specific genotypic changes with functional genomic data from other studies.
Materials and Methods
Systems biology
Experimental analysis was performed on 20 lines representing the diversity of dietary reaction norms for pupal weight and larval lipid storage phenotypes identified from an initial screen of 146 inbred lines sampled from North Carolina and Maine populations (Reed et al. 2010). Lines were raised on four dietary treatments used in the analysis following a rationale described in detail in Reed et al. (2010). All diets were cornmeal based but varied in their sugar and fat content. The base for all diets is by weight 0.7% agar, 6.5% cornmeal, and 1.3% inactive yeast into water. In the normal diet (maintenance diet for fly stocks in many labs in addition to our own) the major source of calories is 4% (0.117 M) sucrose and is made from the standard cornmeal-based lab food with the addition of 58 ml of molasses. Sang (1956) reports that the maximum rate of development in Drosophila larvae is achieved at a sugar concentration of 0.75% by weight, while a 4% sugar diet produced a decreased developmental rate. We found that the type of sugar (e.g., sucrose vs. glucose), in addition to the sugar concentration, affects the metabolic health of the larvae (Reed et al. 2010); maximum weight was achieved at a concentration of <1% of glucose or sucrose, and survival decreased dramatically at higher (>8%) glucose. To both reflect the past research on dietary variation in flies and specifically target the insulin pathway through glucose metabolism we added glucose to the standard base at a concentration of 0.75% glucose by weight (0.042 M) to make our low-sugar diet and a 4% glucose by weight (0.222 M) diet to make the high-sugar diet. Note that the total calories are approximately the same between the normal and high-sugar diets but differ in which sugar is providing those calories. The fat content of the diet base is <0.2% fat, so we supplemented the low-sugar diet with 3% coconut oil by weight to make the high-fat diet; coconut oil is nearly 100% fat (85% saturated, 15% mono- and polyunsaturated). The specific names of the diets are not intended to have an absolute significance in comparison to other studies and signify only the relative sugar and fat concentrations used within this study.
Fifty first-instar larvae were seeded into each food vial and six gross phenotypes were measured for each line on each of the four diets. Samples of third-instar larvae were pooled across a minimum of three food vials for each treatment to be measured for their triglyceride and trehalose content and preserved for expression and metabolomics analysis. Homogenates of six randomly selected larvae were characterized using a 96-well spectrophotometer to determine total triglyceride content using the Sigma triglyceride determination kit and to determine trehalose content (the primary circulating sugar) by treating with trehalase to produce glucose was determined by Sigma glucose determination kit (Clark and Keith 1988; Rulifson et al. 2002; Deluca et al. 2005; Reed et al. 2010). Up to 15 male pupae from each of two food vials per treatment were weighed individually to determine the weight phenotype. Larval and pupal survival and the time to pupation (development time) were also scored. Samples were generated in randomized blocks of four synchronized lines per week on all four diet treatments; each genetic line was replicated at three time points.
Whole-genome expression profiles for high-quality RNA samples were determined using Nimblegen 12-plex microarrays using the manufacturers protocols and software (Nuwaysir et al. 2002). The gene expression data from this publication are deposited in the GEO database under accession no. GSE50745 (http://www.ncbi.nlm.nih.gov/geo/). Metabolomic profiling was performed by gas chromatography–mass spectrometry (GC–MS) on samples of exactly six larvae. Samples were homogenized in 60:40 methanol:water, dried down, and then TMS derivitized in acetonitrile. Samples were processed in daily randomized blocks of 15–22, along with pooled standards. Samples were run in daily sets in a randomized order into a Thermo Scientific DSQ II Series single quadrupole GC-MS with an electron impact source and an Agilent DB-5 column run in splitless mode with a 30-min temperature ramp. Over the course of the experiment, a minimum of five distinct biological samples were analyzed for each genotype and diet. The Kovats retention index (RI) was calculated for each chromatographic peak, and chromatograms were then aligned to a consensus list of internally determined candidate target profiles. Chromatographic peaks were initially cataloged using AnalyzerPro (http://www.spectralworks.com/analyzerpro.html) and were then hand curated to develop a list of potential analytes. Relative concentrations for each chromatographic peak were determined from the standard curve produced by a pooled standard. After quality control filtering, 187 putative metabolites were proposed as being reliably detectable. Chemical category of those 187 putative metabolites was determined by the searching profiles against the publically available National Institute of Standards and Technology (NIST) database, identifying candidate compounds, and then running candidate compounds on the GC–MS system to confirm profile matches. Using this approach, we were able to identify with confidence the exact molecule for 60 of the putative metabolites; another 124 were matched with confidence to chemical class (e.g., amino acid or monosaccharide), while the remaining 3 are presently unknown.
All statistical analyses were performed using JMP Genomics (SAS Institute, Cary NC). Individual metabolites were first normalized to the mean value for the metabolite in one pooled standard for the day the sample was run. They were then log2 transformed and the median centered standardized residuals of a day-of-run ANOVA model on the individual metabolites composed the final data set. Of a potential 15595 genes, 11650 were expressed at detectable levels in this data set. The log2 transformed expressed gene values were median centered, and then the standardized residuals of the hybridization block ANOVA model were themselves median centered and used for all subsequent gene expression analysis. In this analysis, means for each line and diet combination (n = 80) were determined and those values used for all subsequent analyses. Principal components analysis on the correlation matrix among metabolites and expression profiles was performed using the “correlation and principal components” function in JMP Genomics and the first 10 principal components were estimated. Our sensitivity to detection of rare metabolites was lower than to detection of rare expressed gene products but we have no a priori reason to anticipate the variance structure in the rare metabolites to be fundamentally different from the detectable metabolites. In addition, the numerous metabolite and gene expression variables greatly outnumber the principal components calculated; thus the principal components should be robust estimates of the variance in both data sets. Very similar results were obtained after normalization using the supervised normalization of microarrays algorithm (http://www.bioconductor.org/packages/2.12/bioc/html/snm.html). The mean values for all phenotypes, including gene expression and metabolites, calculated for each genotype-by-diet combination are available at the authors’ website as data sets S1–S3 (http://www.gibsongroup.biology.gatech.edu/supplemental-data-reed-et-al).
Laboratory adaptation
Two hundred gravid isofemale flies were trapped on the campus of Georgia Tech over a 10-day period in June of 2010. They were placed in individual vials with a male on standard laboratory food, and as progeny emerged, males were examined for D. melanogaster genital arch morphology, and two sibling virgin females and males were transferred to bottles. In addition, we set up a fresh cross of one virgin female to a standard D. melanogaster male to provide further confirmation that the wild flies were predominantly D. melanogaster. We estimated a frequency of ∼15% D. simulans, and contamination of the baseline population is likely <1%.
Once male and female progeny had been collected from 150 isofemales, 12 bottles were seeded with 12 flies of each sex and allowed to lay up to 100 eggs. The next generation, 36 virgin flies of each sex were collected and mixed with the opposite sex flies from a different bottle, and at this point adaptation to the different diets was commenced. Two dozen progeny of each bottle were collected, and in the third generation they were split into two parallel experiments, each with 12 bottles for each of three diets (low sugar, high sugar, and high fat as described above), and an average of 12–15 flies of both sexes in each bottle. A bottle rotation scheme was established to maintain outbreeding, ensuring that the effective population size of each of the six parallel evolving populations (that is, three diets in duplicate) was ∼300. Selection of 12 virgins from each bottle in each generation was presumably a random sample of the 100–200 emerging flies. At generations 5 and 6, fecundity of the high-fat diet dropped, indicating strong purifying selection and necessitating a reduction in the fat content from coconut oil from 3 to 1.5%, at which point viability and fecundity returned. No attempt was made to estimate the intensity of selection, which could have occurred at any stage of the life cycle, noting that the flies are constantly reared on the specific diet. The stocks were maintained at room temperature, ∼22°, with 15:9 hr light:dark cycle.
Sequence analysis
To determine the allele frequencies of the SNPs that are polymorphic in our Georgia Tech population, we mapped paired end reads from whole-genome sequencing to the D. melanogaster reference genome (build 5.33) using Bowtie2, and subsequently Samtools mpileup was used to generate the pileup files. A minimum mapping quality score of 30 (Phred scale) and base quality score of 15 were set as lower thresholds for variant calling. Calls were subsequently performed using the default parameters in Varscan downloaded from http://varscan.sourceforge.net/. This is BioProject (http://www.ncbi.nlm.nih.gov/bioproject/) accession no. PRJNA194129 and the raw resequencing data are available under accession no. SRA143721 at the Sequencing Read Archive (http://www.ncbi.nlm.nih.gov/sra) and the variant calls used in this analysis are available as data set S4 at the authors’ website (http://www.gibsongroup.biology.gatech.edu/supplemental-data-reed-et-al).
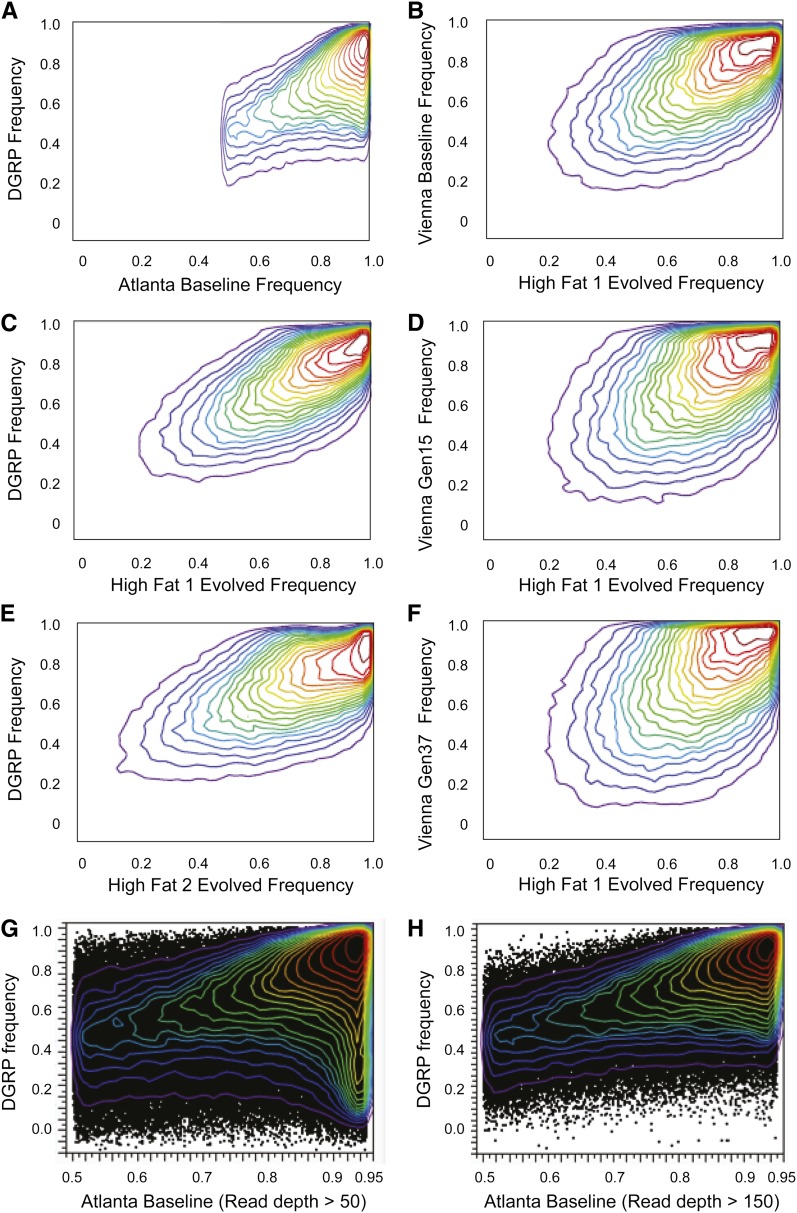
Single nucleotide variant calls were then imported into JMP Genomics and further filtered for quality control. Only biallelic loci were considered in subsequent analyses, and these represented >99% of all SNV calls. Read depth ranged from 150 to 463 (with the exception of two outliers corresponding to the chorion complexes) and had a standard deviation of 47. Given our high-mapping-quality threshold (MAPQ ≥ 30), it is unlikely that many SNV positions in the genome would have high depth because of being in repetitive regions. We also excluded all variants with minor allele frequency (MAF) < 0.05 in baseline; this was done to remove likely sequencing errors, but likely also removed many low-MAF variants since our focus is on selection acting on common variants in the natural population. Since SNVs with both low estimated allele frequency (<0.1) and low read depth in the baseline tended to change the most in the evolving populations, low-MAF SNPs with low coverage seem to have poor allele frequency estimates, as also seen in Figure 1 contrasting the Georgia Tech and Raleigh Drosophila Genetic Reference Panel (DGRP) estimates. This is theoretically supported by the notion that the variance of the estimate of allele frequency of a given SNP is linearly dependent on the number of the alleles sampled. Consequently, positions with depth <150 in the baseline population were excluded in the final analyses reported in the manuscript.
Figure 1.
Comparison of laboratory adaptation between studies in Atlanta, Raleigh, and Vienna. Density plots contrast the allele frequency for all SNPs with q50 read depth >150 in the Atlanta Baseline population that are also documented in the Raleigh and Vienna studies, considering only the major allele in the Atlanta Baseline also with minor allele frequency >0.05. (A) Raleigh DGRP against Atlanta Baseline showing predominant density along the expect equivalence of major allele frequencies between 0.5 and 0.95. (C and E) Raleigh DGRP against high-fat evolved populations 1 and 2, showing convergence of frequencies across the full range from 0.0 to 1.0, suggesting parallel laboratory adaptation. (B, D, and F) Schlotterer lab Vienna allele frequencies against Atlanta high fat 1 evolved frequencies, at Baseline, and after 15 and 37 generations of lab adaptation, respectively. The Vienna Baseline is actually derived from a Portuguese population that was maintained as isofemale lines for 5 generations before initiation of lab adaptation under an elevated fluctuating temperature regime from 18° to 28°. Clearly the lab-adapted allele frequencies again converge, but ongoing laboratory adaptation causes some alleles to diverge with respect to dietary selection. (G and H) Impact of read depth on estimation of baseline allele frequency. Including all SNPs with a q50 read depth of at least 50 identifies a large density of less common variants (major allele frequency >0.9, vertical density at right of A that appear to have very different frequencies than those observed in the DGRP reference population. However, when the read depth is increased to 150 this set of polymorphisms largely disappears (B), suggesting that many of the less common variants called at low depth are sequencing or alignment errors.
Since one replicate of each of the evolved populations was sequenced using a different technology, we confirmed that there was no platform effect by performing a paired two-sample t-test of all frequencies between replicates of the evolved populations. Each SNP is represented by six measures, namely the one replicate of each diet measured on the GAiix platform and the second replicate on the HiSeq2000 platform. The values for each diet were paired and a t-test for the difference between platforms was assessed. The results shown in the Q–Q plot in Supporting Information, Figure S1 confirm that there is no enrichment of variants that tend to show a higher or lower frequency between the GAiix and HiSeq platforms.
After quality-control steps, we identified the SNPs that likely evolved in frequency under laboratory adaptation, by finding alleles in the evolved populations that differed more than expected under the assumption of drift compared to the baseline population. We did so by deriving the effective population size from the expectation that the variance in each allele frequency is proportional to the baseline frequency and the number of generations at a given effective population size
| (1) |
which upon rearrangement, recognizing that it is measured from the sum of squared deviations between each observed yi after evolution and its expected frequency
| (2) |
yields
| (3) |
where n is the number of SNPs (∼16,000 per bin), and t is the number of generations of laboratory adaptation (14 for replicate 1, 17 for replicate 2). The expected value under the null hypothesis of no sequence evolution, is just the baseline frequency. The effective population size of each evolved population was initially estimated assuming that most SNPs in our data set were not evolving.
The value is calculated independently for every SNP because each SNP has a different starting allele frequency. The average estimated effective population size was calculated for bins of varying initial allele frequencies (Figure S2) and across all bins is ∼60 flies for all six populations. The estimates for effective population size both support the consistency in the fly cultivation techniques employed in the course of the experiments and corroborate the effective population size estimated from the experimental procedures.
Since it is likely that at least some alleles are under selective pressure in our evolving populations, this procedure certainly underestimates Ne. We examined the effect of removing the alleles that changed the most in frequency on our estimate of Ne. Removing 20% of the most extreme SNPs increased our estimate of effective population size to between 80 and 100 flies per generation. Another source of variance inflation is the contribution of technical variance. Technical variance is the component of the total variance, which is introduced by the fact that each allele-frequency estimate is derived from high-throughput sequencing read depths and has a defined variance. Since we had technical replicates for the baseline population (alternate lanes of the GAiix), we estimated the technical variance introduced by random sampling of alleles. We then calculated a more accurate estimate for biological variance due to drift, which increased our estimates of Ne to consistently ∼100–120 flies. Consequently, we note that while our estimates of Ne may be >100, we use this value in our subsequent analysis as a conservative estimated parameter.
The SNPs on the X chromosome suggest a higher effective population size than for those of the autosomes. This effect may partially be explained by the fact that we used a scaling factor of 3/2 instead of 2 in the estimation of Ne for the X chromosome because there are only 1.5 chromosomes per effective individual of the population. However, scaling by 2 for the X chromosome led to an underestimate of the Ne relative to the autosomes.
After estimating Ne for each evolved population, we then used this value to calculate the variance expected under drift using Equation 1. The expected variance is also dependent upon the initial allele frequency. Since our analysis suggested that the accuracy of the estimates of allele frequency decreased with decreasing minor allele frequency, we set a minimum value of p × q of 0.85 × 0.15 = 0.1275, which would prevent alleles with low MAF from benefiting from both a poor estimate of p and a decreased level of expected variance due to drift. We then divided the allele frequency change by the standard deviation (the square root of the variance), which yielded a z-score that was subsequently used to calculate a P-value. To correct for multiple testing, we used a Bonferroni correction of six tests for 143,975 SNPs at P < 0.05, which yields a genome-wide significant threshold of P < 4.00 × 10−8, which we reiterate is a conservative threshold.
Since the median baseline major allele frequency is 0.85, the response to lab adaptation is highly asymmetric as many sites go to fixation under drift. Our test for selection requires changes of frequency at least 0.3, so effectively only analyzes reduction in the major allele frequency. Table 1 illustrates, however, that similar (but more variable, as expected) results are observed for alleles increasing in frequency.
Table 1. Number of SNPs evolving in frequency under lab adaptation.
| NLP 5a | NLP 1.3a | > 0.3b | <−0.3c | Fixedd | |
|---|---|---|---|---|---|
| High fat 1 | 3,292 | 6,653 | 16,805 | 996 | 7,628 |
| High fat 2 | 3,501 | 6,634 | 22,036 | 1,889 | 14,289 |
| High sugar 1 | 3,725 | 6,859 | 18,906 | 1,073 | 7,003 |
| High sugar 2 | 3,395 | 6,385 | 22,328 | 1,491 | 11,099 |
| Low sugar 1 | 2,705 | 6,090 | 14,413 | 830 | 5,821 |
| Low sugar 2 | 3,259 | 6,345 | 20,919 | 1,807 | 14,719 |
| High fat (both) | 1,662 | 5,676 | 8,502 | 295 | 3,613 |
| High sugar (both) | 1,862 | 5,592 | 9,217 | 298 | 3,025 |
| Low sugar (both) | 1,245 | 4,958 | 6,697 | 235 | 2,619 |
| High calorie | 609 | 4,318 | 3,568 | 47 | 852 |
| Sugar diet | 451 | 3,652 | 2,549 | 27 | 635 |
| All six populations | 232 | 3,032 | 1,614 | 7 | 302 |
Number of SNPs of the 8343 significant at NLP 5 in any replicate, significant at NLP 1.3 (P < 0.05) for the indicated contrast with baseline.
Major alleles at baseline that decreased in frequency by >0.3.
Major alleles at baseline that increased by at least 0.3 (compare with footnote d).
Major alleles at baseline that increased to fixation.
Results
Genotype-by-diet effects on physiological phenotypes
Both theoretical and classical work on Drosophila point to genotype-by-environment interactions as an important aspect of the genetic architecture of complex traits (Takano et al. 1987; Barnes et al. 1989; Zhou et al. 2012). In our earlier study of multiple phenotypes related to growth and development in 146 highly inbred wild-type lines of D. melanogaster (Reed et al. 2010), diet effects across all lines were negligible, whereas genotype-by-diet (G × D) interaction effects accounted for a substantial amount of the variation. To test whether this pattern derives from interaction effects at the level of gene activity, we here performed metabolite and whole transcriptome profiling on third-instar larvae from 20 of the highly inbred lines chosen to represent the range of phenotypic diversity in our larger panel (Reed et al. 2010). The larvae were grown on each of four diets: normal cornmeal–molasses (N, 4% sucrose by weight), high fat (F, 0.75% glucose and 3% coconut oil by weight), high sugar (4% glucose by weight), and low sugar (C, 0.75% glucose by weight). We expected one of two possible outcomes: (i) that some component(s) of transcriptional and/or metabolite variation would be closely associated with metabolic phenotypes and predictable across diets or (ii) that such associations would be diet specific, reflecting the observed genotype dependence of dietary response.
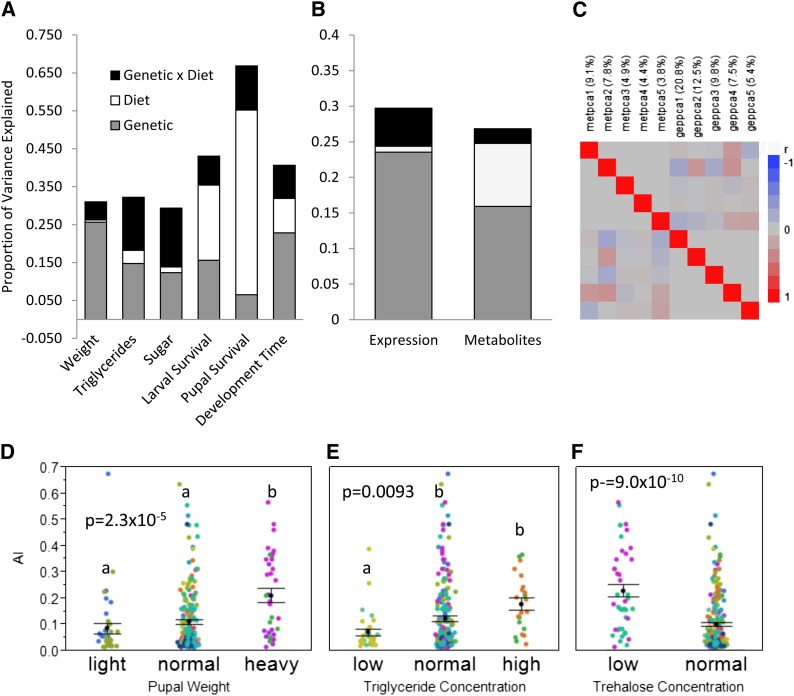
The pattern of variation for genetic, dietary, and G × D interaction effects for all of the measured organismal phenotypes (e.g., weight and development time) were consistent with the previous results observed in the larger panel of lines (Figure 2A and Table S1). Gene expression was measured on Nimblegen Drosophila transcriptome arrays (Nuwaysir et al. 2002) using total RNA from between three and six replicates of 30 larvae pooled from different vials. The metabolome of larvae reared on the same food vials was measured by GC–MS, resulting in 187 high confidence metabolites, many of which were assigned to specific chemical structures by comparison to chemical standards. The transcriptome and GC–MS-derived metabolomes have very different variance components (Figure 2B). Whereas the transcriptome shows highly significant genotype (24% of the variance) and G × D interaction effects (5.3%), but no main effect of diet (<1%), the metabolome shows highly significant genotype (16%) and diet (9%) effects but only limited interaction (2%). In other words, the transcriptomic response to dietary shift is highly genotype dependent, whereas the metabolome has a much stronger dietary component.
Figure 2.
Architecture of phenotypic, gene expression, and metabolome profiles. (A) Variance partition for the phenotypes measured on 20 genetic lines across four diets. Weight, triglyceride, and sugar phenotypes show low levels of variance due to diet while survival and development time traits show large effects of diet. (B) Variance partition on the first 10 principal components for gene expression and metabolite profiles. Expression profiles show a small dietary component while metabolite profiles show a large dietary component. (C) Correlations of the first five principal components of gene expression (gep) and metabolite (met) profiles. Only small correlations are observed and none are significant at a Bonferroni corrected P-value (the smallest raw P-value observed was 0.003 between metpc2 and geppc4). (D–F) Phenotypic classification of metabolic phenotypes explains significant variation in heart arrhythmia index (AI). Data points are colored by genetic line, error bars indicate the mean ±1 SE, phenotypic groups are defined as being light/low <1 SD below, heavy/high >1 SD above, and normal within 1 SD of the mean of the lines across all diets. AI data points are measures of individual flies. (D) Lines grouped by pupal weight category vs. AI measures. Heavy genetic lines had significantly higher AI: AI means are low 0.09, normal 0.11, high 0.21. (E) Lines grouped by third-instar triglyceride content. Genetic lines with low triglyceride levels had significantly lower AI: AI means are low 0.07, normal 0.12, high 0.18. (F) Lines grouped by third-instar trehalose content genetic lines with low trehalose levels had significantly higher AI (mean 0.23 vs. 0.10).
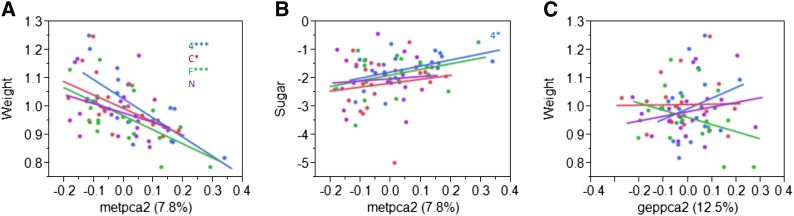
The first five principal components of the metabolite and gene expression profiles are only mildly correlated with one another (Figure 2C), suggesting that there is only weak predictive value when comparing one functional physiological level to the next. However, the second metabolite principal component (metpc2) is mildly correlated with each of the first four expression principal components (0.003 < P < 0.04; Table S2), which suggests that the metabolites with heavy loadings for metpc2 are the most likely to be tied mechanistically to changes in gene expression (e.g., potential cofactors or upstream regulators of gene transcription). The metabolites most strongly correlated with the metpc2 include six amino acids, two monosaccharides, and the components of dopamine metabolism, l-dopa and n-arachidonoyl dopamine (Table S3).
There are a number of significant associations between the principal components and individual metabolic traits as documented in Table 2. This is particularly evident for metpc2, which is also significantly correlated with the four traits of development time, triglycerides, sugar, and body weight. Six of the first 10 principal components for both gene expression and for metabolites are significantly correlated with one or more phenotypic traits. For five of the six traits, the total variance explained by the first 10 principal components is greater for the metabolite profiles than for gene expression (Table S4). The metabolite most strongly associated with metpc2 is glycine (P = 1.9 × 10−9, R2 = 0.64), which in turn is associated with weight (P = 4.6 × 10−5, R2 = 0.27), triglycerides (P = 0.0005, R2 = 0.14), and nominally with sugar (P = 0.0327, R2 = 0.057). Specifying heavy weight as a function of glycine produces an area under the ROC curve of 0.86, with peak sensitivity of 92% achieved with 78% specificity.
Table 2. Correlation of gene expression and metabolite PC with phenotypes at P < 0.05.
| Phenotype | Principal componenta | Correlation | P-value |
|---|---|---|---|
| Development time | geppc1 (20.8%) | −0.224 | 0.046 |
| Development time | geppc4 (7.5%) | 0.305 | 0.006 |
| Sugar | geppc4 (7.5%) | 0.236 | 0.035 |
| Weight | geppc4 (7.5%) | −0.316 | 0.004 |
| Larval survival | geppc5 (5.4%) | −0.311 | 0.005 |
| Sugar | geppc5 (5.4%) | −0.276 | 0.013 |
| Weight | geppc7 (3.1%) | 0.289 | 0.009 |
| Arrhythmia index | geppc8 (2.7%) | −0.523 | 1x10−4 |
| Development time | geppc8 (2.7%) | −0.222 | 0.048 |
| Arrhythmia index | geppc10 (1.9%) | −0.274 | 0.041 |
| Sugar | geppc10 (1.9%) | 0.225 | 0.045 |
| Larval survival | metpc1 (9.1%) | 0.314 | 0.005 |
| Pupal survival | metpc1 (9.1%) | 0.278 | 0.013 |
| Arrhythmia index | metpc2 (7.8%) | −0.333 | 0.012 |
| Development time | metpc2 (7.8%) | 0.437 | 0.011 |
| Triglycerides | metpc2 (7.8%) | −0.321 | 0.004 |
| Sugar | metpc2 (7.8%) | 0.286 | 0.010 |
| Weight | metpc2 (7.8%) | −0.559 | 0.009 |
| Arrhythmia index | metpc4 (4.4%) | −0.397 | 0.002 |
| Triglycerides | metpc4 (4.4%) | −0.281 | 0.012 |
| Sugar | metpc4 (4.4%) | 0.255 | 0.022 |
| Development time | metpc5 (3.8%) | 0.358 | 0.001 |
| Pupal survival | metpc5 (3.8%) | −0.302 | 0.007 |
| Arrhythmia index | metpc6 (3.5%) | −0.362 | 0.006 |
| Larval survival | metpc6 (3.5%) | −0.496 | 0.002 |
| Pupal survival | metpc6 (3.5%) | −0.507 | 3x10−4 |
| Weight | metpc6 (3.5%) | −0.239 | 0.033 |
| Development time | metpc8 (2.9%) | −0.250 | 0.025 |
| Triglycerides | metpc8 (2.9%) | −0.227 | 0.043 |
geppc refers to gene expression principal component (describing the indicated percentage of variation), while metpc refers to principal components of the metabolome.
Despite the above examples, in general the relationship between gene expression or metabolomic variation and phenotype is confounded by dietary interactions, so not predictive across all diets, as illustrated in Figure 3. Nevertheless, we next tested whether the variation in phenotypes, metabolite profiles, or transcript profiles can predict a disease outcome. Metabolic imbalance in Drosophila due to extreme diets or genetic manipulation can cause an increase in cardiac arrhythmias (Birse et al. 2010; Lim et al. 2011; Na et al. 2013), which we quantified as the arrhythmia index (AI) from high-speed movies of semi-intact heart preparations (Ocorr et al. 2007). The AI was measured on 15 of the 20 lines used in the G × D analysis on individual 1-week-old adult females raised on a normal diet. ANOVA of AI according to phenotype categories found that lines that had high weight and triglyceride concentrations as larvae had more than twice the average adult AI (P = 2.3 × 10−5 and P = 0.0093, respectively; Figure 2, D and E) than the lines in the lowest categories. In addition, lines with low larval sugar (total trehalose) levels had a mean AI two-and-a-half times greater than the lines with high sugar levels (P = 9.0 × 10−10; Figure 2F). This result shows that line (hence genotype) trait measurements in larvae can be predictive of disease state in a different developmental stage, in this case heart dysfunction in adult flies.
Figure 3.
Associations between metabolites and phenotypes. Each plot shows the correlation between phenotype on the y-axis and metabolic or transcriptional measure, colored by the diet (F, high fat, green; 4, high sugar, blue; N, normal cornmeal–molasses, purple; C, low sugar, red) on which the 20 lines were grown. Letters to the side indicate significance of the correlation on each diet (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. (A) Weight (mg) is significantly correlated with metabolite PC2 on three diets. (B) Sugar (log transformed) is negatively correlated with metabolite PC2 on 1 diet. (C) Weight is not significantly correlated with expression PC2 on any diet.
To relate AI to the metabolite and transcriptome variance, we performed linear regression on the principal components of variation. Third-instar gene expression principal component 8 (geppc8) was highly significantly correlated with adult AI (P = 1.3 × 10−4), explaining >27% of the variance (Table 2). AI was also correlated with three metabolite principal components, including metpc2 (P = 0.012), the principal component that is also strongly correlated with weight, triglycerides, sugar, and development time. Collectively the first 10 principal components for gene expression and metabolite profiles explain comparable amounts of the variation in AI (45 and 38%, respectively), so unlike some of the other traits, both gene expression and metabolite profiles are useful in predicting disease risk at a later developmental stage. In addition, third-instar glycine levels are significantly negatively correlated with adult heart AI (R2 = 0.148, P = 0.0048).
Adaptive response to diet
We collected 200 gravid female D. melanogaster on the campus of Georgia Tech in summer 2010 and sequenced the population to an average depth of 300× to estimate genotype frequencies at millions of polymorphic sites genome wide. The second-generation progeny founded six parallel populations each with an expected effective population size of 180 virgin females and 180 males, which were adapted in duplicate to three diets with a deliberate schema to prevent inbreeding. The high-fat diet resulted in high morbidity and loss of fecundity, necessitating a reduction in coconut oil concentration at generation five, but no gross loss of viability was observed for the high-sugar and low-sugar diets. After 14–17 generations, we resequenced both replicates of each population to ∼ 70× depth, facilitating estimation of allele frequency changes in response to adaptation to each diet.
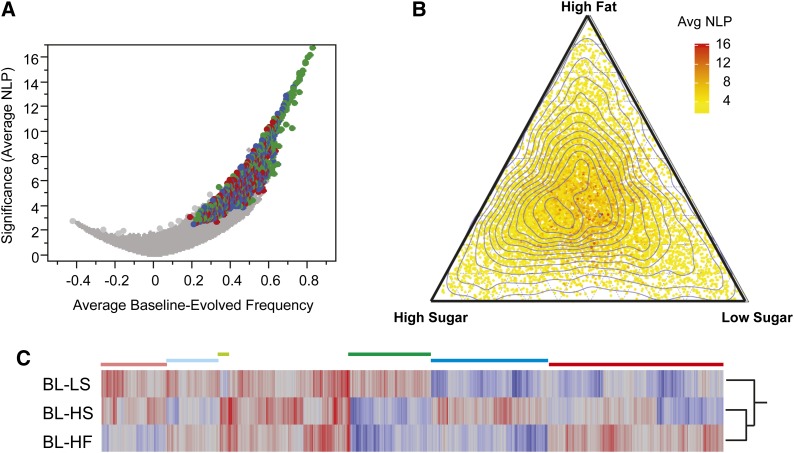
We identified 1,474,945 biallelic SNPs and here report data for a filtered set of 143,975 high-confidence common polymorphisms where the minor allele frequency at baseline was at least 0.05 (estimated from 150 or more reads), and all short-read alignments had a sequence quality score corresponding to an error rate of 10−3 or better. To detect selection, we devised a method by which to calculate the actual effective population size and subsequently the expected allele frequency under drift. We then used the magnitude of the change in allele frequency from baseline to each of the evolved populations to ascertain which SNPs were evolving more than expected by drift alone (Figure S3). A total of 8343 SNPs had significantly different allele frequencies between the baseline and at least one of the evolved populations at P < 4 × 10−8; of these, 36% were in common to all six evolved populations at P < 0.05 (Figure 4A and Table 1).
Figure 4.
General and diet-specific laboratory adaptation. (A) Volcano plot of the average significance (NLP) against average difference between baseline and evolved allele frequency across all six laboratory adaptation experiments. Only the major allele at baseline is shown, which results in the asymmetric distribution since there is a greater range over which SNPs can change in frequency. Colored dots represent 2734 SNPs that were significant at NLP > 6 in both high fat (red), both HIGH Sugar (blue), or both LOW Sugar (green) replicates. (B) Ternary plot of average significance of adaptation in both replicates of each diet, with 27,459 SNPs significant at NLP > 4 in at least one experiment, colored with respect to significance (yellow, low; red, high). Contour lines show the density of SNPs, showing a greater number of sites significant in both high-calorie diets and relatively independent evolution on low sugar. (C) Heat map of average difference between baseline and evolved allele frequency on the three diets, with red representing higher frequency of the major allele at baseline and blue representing higher frequency in the evolved population. Clusters of sites showing largely diet-specific changes are indicated by horizontal bars. Plot includes all SNPs with average NLP > 6 for both replicates of at least one diet.
We also observed significant enrichment for diet-specific responses, with 692 SNPs showing significant deviation in allele frequencies between both replicates of one diet and baseline (at P < 10−5) but not in the other diets. A total of 571 genes are located in the immediate vicinity of SNPs that are significant for high fat, 661 for high sugar, and 484 for low sugar. In addition, 48% of all SNPs found to change frequency significantly on one diet also showed significant adaptation on the second dietary replicate (Figure S2), an overwhelming proportion given the independent population dynamics in each replicate. The allele frequencies among the replicates were most highly correlated between the within-diet replicates (Figure S4). Thus, parallel adaptation to a specific food source occurs in conjunction with the stronger overall adaptation to laboratory culture. The dietary response is genome wide, resulting in correlated changes in allele frequency comparing all three diets to baseline or to one another. For the pairwise comparisons of each diet, across all SNPs for the allele frequency difference between diet and baseline, correlations are 0.5 or greater.
In addition, the two high-calorie diets (high fat and high sugar) resulted in much more similar allele frequency profiles than either did to the low-calorie diet (Figure 4B). Figure 4C highlights distinct clusters of genes that show diet-specific responses, particularly those that changed in the opposite directions on the high-fat, high-sugar, and low-sugar diets. Responses were distributed across all chromosomes (Figure 5A and Figure S5). The mean size of selected blocks was between 100 and 200 kb (Figure 5B), which is expected given the very-low linkage disequilibrium observed in outbred Drosophila (Haddrill et al. 2005) coupled with the measured effective population size between 100 and 150 female flies per generation. There was no evidence for selective sweeps on large blocks surrounding a rare variant.
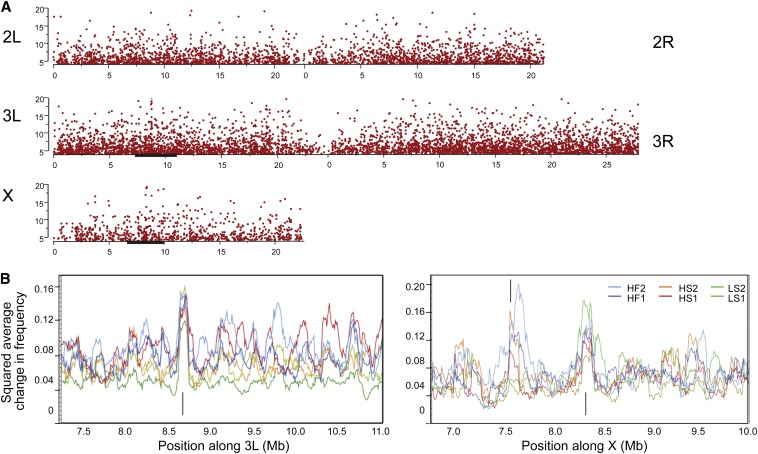
Figure 5.
Change in allele frequency by chromosomal position. (A) Manhattan plots of first replicate of the high-fat lab adaptation showing significance of the change in frequency against chromosomal position, for the indicated chromosome arms. (B) Sliding window plots of average change in frequency (expressed as the squared change difference with baseline), in two less than 5-Mb intervals on 3L and X indicated by solid black bars in A. Each of the six replicates, two per diet, is shown as a different color. Upward arrows show location of the two chorion complexes, indicating parallel evolution on all diets, while the adjacent downward arrow on the X shows a region that only changes in frequency on the high-calorie (not the low-sugar) diets. Most sliding window peaks are between 100 and 150 kb in width, indicating that this is the extent of chromosome selected.
We conclude that there is pervasive standing genetic variation available for dietary response, with selection differentials ranging from 10% or more for sites that change as much as 50% in frequency, to very small differentials responsible for the overall genomic response. Figure 5B highlights two representative regions showing adaptation to both the laboratory and diet-specific selection regimes. Both chorion complexes on X and 3L showed consistent adaptation across all diets, likely because substrate differences between wild (rotting fruit) and laboratory (solid agar media) settings would dramatically alter the fitness of egg chorion structural alleles because of differences in both water tension and oxygen availability. The chorion complex undergoes early embryonic amplification that could introduce a technical bias (Orozco-Terwengel et al. 2012), but this unusual feature of the loci also underscores their critical role in organismal physiology. For comparison, a third representative region adjacent to the chorion complex on the X chromosome (Figure 5B) showed a selective response only on the high-calorie diets, implying that genes in that region are especially relevant in a nutrient-rich environment.
Adaptation and gene expression
Although selection presumably acts on individual sites, the resolution of our experiment was to no less than 100-kb intervals that often include multiple genes. Consequently, we sought experimental support for the function of a subset of variants. We first confirmed genotypic divergence for >80% of a sample of 48 polymorphisms in regulatory regions by genotyping 96 flies from one of each of the evolved populations on three diets using Fluidigm arrays (Spurgeon et al. 2008). Next, we measured gene expression in whole adults from eight inbred lines derived from each of the three populations and observed significant divergence in Ct counts from the Fluidigm nanoscale qRT–PCR assays for 23 of the 41 transcripts (Table S5), indicating that dietary adaptation has modified gene expression at these target loci, consistent with selection on the regulatory polymorphisms. For example, BG4 shows a fourfold downregulation after adaptation to the high-fat diet relative to the low-sugar diet (Figure S6).
As a second approach to linking the genotypic adaptive responses to the transcriptomic analyses, we asked whether there is significant enrichment of shared genes (i) in the vicinity of selected loci, (ii) in the list of differentially expressed transcripts across diets, (iii) among the 505 genes found to regulate larval lipid levels in an RNAi screen (Pospisilik et al. 2010), or (iv) among a published list of 486 regulatory eQTL detected in adults of both sexes (Massouras et al. 2012). Surprisingly, for all of these pairwise comparisons, the overlap of genes was no greater than expected by chance (Table S6).
Discussion
We find that the transcriptome as a whole is more susceptible to genotype-by-environment (diet) interactions than is the metabolome, but certain transcripts and metabolites can nevertheless predict phenotype: for example, heart arrhythmia susceptibility in adults is correlated with the metabolite glycine (as well as triglycerides, weight, and blood sugar) in larvae. In parallel, evolve-and-resequence contrasts of populations of recently trapped wild flies demonstrated pervasive genome-wide variation for adaptation to three different diets that is superimposed on rapid adaptation to the lab environment. There was, however, little overlap with loci inferred to influence metabolic traits from gene expression profiles or from RNAi knockdown. Thus, environmental influences on the genotype to phenotype map involve somewhat independent organization of transcriptional and metabolic profiles. Similarly, eQTL associate poorly with expected metabolite abundances in Arabidopsis (Atwell et al. 2010), and metabolic flux is not well predicted by metabolic enzyme transcript levels in bacteria (Chubukov et al. 2013).
It is noteworthy that glycine was correlated with MetS-like phenotypes and heart arrhythmia in our study, since it is a gluconeogenic amino acid, low levels of which in serum have been associated with type 2 diabetes risk in humans (Floegel et al. 2013). NMR spectroscopy of cardiac tissue from patients with persistent atrial fibrillation also revealed elevated levels of ketogenic amino acids including glycine (Mayr et al. 2008). In addition, a recent systems genomics analysis of cancer cell lines pinpointed glycine as a key regulator of cancer-cell proliferation by acting as a rate-limiting mediator of DNA synthesis (Jain et al. 2012).
Given the strong signal of G × D interaction effects across a panel of wild-type Drosophila lines, we wondered whether it was possible to identify genetic loci responsible for the interaction effect through artificial dietary selection on a wild population of flies and thus predict the genetic evolutionary trajectory of a population after a shift in diet. Specifically, we hypothesized that it would be possible to identify SNPs or genomic regions that evolve similarly in response to similar diets. We found that rather than selection being limited to a handful of genetic variants, adaptation to the artificial diets was highly polygenic, but a large portion of those regions that did respond to selection did so in parallel across dietary replicates. Published evolve and resequence experiments mostly utilizing previously lab-adapted Drosophila have demonstrated substantial genome-wide genetic variation for artificial selection on courtship song (Turner and Miller 2012), late-reproductive capacity (Burke et al. 2010), and body size (Turner et al. 2011), as well as adaptation to the lab environment in general (Orozco-Terwengel et al. 2012).
It appears that the genomic adaptation to the laboratory may occur consistently as soon as flies are trapped, since allele frequencies of our evolved lines are similar to those reported in two other laboratory adapted samples. In comparing our baseline allele frequencies to those found in the DGRP isofemale lines (Mackay et al. 2012), we found a strong positive correlation, indicating that the standing genetic variation present in the Raleigh Farmers Market (Raleigh, NC) and on the Georgia Tech campus (Atlanta, GA) are very similar despite being 650 km apart (Figure 1A). This correlation was even stronger following lab adaptation (Figure 1, C and E), implying caution about the use of isogenic lines for population genetic inference in natural populations. A similar though weaker correlation between a Portuguese baseline population (Orozco-Terwengel et al. 2012) and our evolved samples was also observed, but this became weaker as their flies experienced the effects of strong lab-specific selection (Figure 1, B, D, and F). It should also be noted that an excess of high-frequency variants was observed in the Atlanta population relative to the DGRP (Mackay et al. 2012) when all polymorphisms with a 50× or greater read depth were considered, but that excess disappeared when read depth was constrained to be 150× or greater (Figure 1, G and H). Thus, even high-depth resequencing is prone to significant error and read depths exceeding 150× may be needed to accurately genotype a population.
While we did find that a substantial portion of the tested loci that showed a response to selection at cis-regulatory regions showed a corresponding change in gene expression, we did not observe a significant overlap between loci showing significant variation for genotype-by-diet expression effects and loci responding to selection in our study nor between those loci and genes independently functionally characterized as effecting obesity traits. At a gross level, this implies that the genes that can be artificially manipulated to cause an obesity phenotype under standard laboratory conditions are not necessarily the ones that have altered expression as the diet changes or have regulatory variants that respond to adaptation to different diets. (See File S1, File S2, File S3, and File S4.)
Collectively, then, in natural populations of Drosophila, there are extensive genotype-by-diet interaction effects at the gene expression, metabolomic, and phenotypic levels. However, there is not always direct or clear linkage between the source of the genetic variation (the genome) and the (endo)phenotypes (transcription, metabolites, traits). Different physiological levels are buffered against dietary perturbation to differing degrees, and every genetic line tested has a different reaction norm to dietary variation. Direct ties between physiological markers and phenotypic outcomes are most robust when they are measured in the same environment, but some general patterns can be linked across environments such as predicting the risk of disease (heart arrhythmia) from a genotype’s metabolic or expression profile. Others have also documented genotypic differences in the magnitude of environmental or genetic perturbations on the transcriptome that complicate the mapping of genotype onto phenotype (Dworkin et al. 2009; Jumbo-Lucioni et al. 2010; Doroszuk et al. 2012; Zhou et al. 2012; Civelek and Lusis 2013).
In summary, we tell both a cautionary and a hopeful tale. We show the limited power of particular genetic loci to predict phenotypes or evolutionary response and the tremendous variance that is introduced when a genome is challenged by different environments. The presence of such complexity in a disease phenotype, such as MetS, suggests that we have much work to do before a complete picture of the mechanisms of the disease can be resolved. However, we also share the optimistic perspective that, despite the complexity of interacting biological factors, by using a systems biology approach it is still possible to find many useful signals. We know that for most complex traits any given genetic locus can explain only a small percentage of the total genetic variation, and thus most traits of pressing biomedical or evolutionary interest have a highly polygenic basis. Genome-wide association studies are very powerful for finding the largest-effect loci, but provide only a static picture of a small component of standing variation that has limited capacity to predict organismal responses in evolutionary or ecological settings. We do not believe that previous findings about the mechanisms underlying of MetS should be disregarded; what has already been determined about the relationship between dietary influences and metabolic health is extremely useful, and important public health recommendations are being made as a result. Instead, we mean to address the causes of and possible strategies to closing the remaining gaps in our understanding. Systems biology provides an orthogonal and more holistic approach to association mapping for identification of predictive factors of phenotypic outcomes.
Acknowledgments
The authors thank T. Reid, G. Doho, M. Bouzyk, Z. Johnson, and N. Patel at Emory University for the Illumina DNA sequencing, C. Ward for assistance with establishing the natural population, J. Anderson, S. Williams, K. Dew-Budd, and K. Davis for assistance in generating the metabolomics and gene expression samples, and I. Dworkin and C. Gunter for constructive criticism of the manuscript. This project was funded by National Institutes of Health grants R01-HL08548 (to G.G. and R.B.) and R01-GM61600 (to G.G.). L.K.R., R.B., and G.G. designed the experiments and wrote the manuscript, L.R. and A.P. performed the selection experiment, L.K.R., N.D., and N.G. performed the metabolomics, L.K.R. performed and G.G. assisted in analysis of the gene expression profiling, K.L. and B.H. analyzed the DNA sequence data, and Z.Z. and R.B. measured and analyzed the heart rate variability. The authors declare no conflicts of interest.
Footnotes
Communicating editor: E. Petretto
Literature Cited
- Alberti K., Zimmet P., Shaw J., 2006. Metabolic syndrome: a new world-wide definition: a consensus statement from the International Diabetes Federation. Diabet. Med. 23: 469–480 [DOI] [PubMed] [Google Scholar]
- Atwell S., Huang Y. S., B. J. Vilhja ´lmsson, G. Willems, M. Horton et al, 2010. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroles J. F., Carbone M. A., Stone E. A., Jordan K. W., Lyman R. F., et al. , 2009. Systems genetics of complex traits in Drosophila melanogaster. Nat. Genet. 41: 299–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes P. T., Holland B., Courreges V., 1989. Genotype-by-environment and epistatic interactions in Drosophila melanogaster: the effects of Gpdh allozymes, genetic background and rearing temperature on larval developmental time and viability. Genetics 122: 859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse R. T., Choi J., Reardon K., Rodriguez J., Graham S., et al. , 2010. High-fat-diet-induced obesity and heart dysfunction are regulated by the TOR pathway in Drosophila. Cell Metab. 12: 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke M. K., Dunham J. P., Shahrestani P., Thornton K. R., Rose M. R., et al. , 2010. Genome-wide analysis of a long-term evolution experiment with Drosophila. Nature 467: 587–590 [DOI] [PubMed] [Google Scholar]
- Chubukov V., Uhr M., Le Chat L., Kleijn R. J., Jules M., et al. , 2013. Transcriptional regulation is insufficient to explain substrate-induced flux changes in Bacillus subtilis. Mol. Syst. Biol. 9: 709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelek M., Lusis A. J., 2013. Systems genetics approaches to understand complex traits. Natl. Rev. 15: 34–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. G., Keith L. E., 1988. Variation among extracted lines of Drosophila melanogaster in triacylglycerol and carbohydrate storage. Genetics 119: 595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M., Yi N., Allison D. B., Leips J., Ruden D. M., 2005. Mapping quantitative trait loci affecting variation in Drosophila triacylglycerol storage. Obes. Res. 13: 1596–1605 [DOI] [PubMed] [Google Scholar]
- Diop S. B., Bodmer R., 2012. Drosophila as a model to study the genetic mechanisms of obesity-associated heart dysfunction. J. Cell. Mol. Med. 16: 966–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroszuk A., Jonker M. J., Pul N., Breit T. M., Zwaan B. J., 2012. Transcriptome analysis of a long-lived natural Drosophila variant: a prominent role of stress- and reproduction- genes in lifespan extension. BMC Genomics 13: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin I. M., Kennerly E., Tack D., Hutchinson J., Brown J., et al. , 2009. Genomic consequences of background effects on scalloped mutant expressivity in the wing of Drosophila melanogaster. Genetics 181: 1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel R. H., Grundy S. M., Zimmet P. Z., 2005. The metabolic syndrome. Lancet 365: 16–22 [DOI] [PubMed] [Google Scholar]
- Floegel A., Stefan N., Yu Z., Mühlenbruch K., Drogan D. et al, 2013. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes 62: 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill P. R., Thornton K. R., Charlesworth B., Andolfatto P., 2005. Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res. 15: 790–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison S. T., Carbone M. A., Ayroles J. F., Stone E. A., Lyman R. F. et al, 2009. Co-regulated transcriptional networks contribute to natural genetic variation in Drosophila sleep. Nat. Genet. 41: 371–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M. M., Nilsson R., Sharma S., Madhusudhan N., Kitami T., et al. , 2012. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 336: 1040–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumbo-Lucioni P., Ayroles J. F., Chambers M. M., Jordan K. W., Leips J., et al. , 2010. Systems genetics analysis of body weight and energy metabolism traits in Drosophila melanogaster. BMC Genomics 11: 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan Z., Ford M. J., Cusanovich D. A., Mitrano A., Pritchard J. K., et al. , 2013. Primate transcript and protein expression levels evolve under compensatory selection pressures. Science 342: 1100–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. P., Simpson S. J., Clissold F. J., Brooks R., Ballard J. W. O. et al, 2008. Lifespan and reproduction in Drosophila: new insights from nutritional geometry. Proc. Natl. Acad. Sci. USA 105: 2498–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B., 2013. Genotype to phenotype: lessons from model organisms for human genetics. Nat. Rev. Genet. 14: 168–178 [DOI] [PubMed] [Google Scholar]
- Lim H.-Y., Wang W., Wessells R. J., Ocorr K., Bodmer R., 2011. Phospholipid homeostasis regulates lipid metabolism and cardiac function through SREBP signaling in Drosophila. Genes Dev. 25: 189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster Genetic Reference Panel. Nature 482: 173–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massouras A., Waszak S. M., Albarca-Aguilera M., Hens K., Holcombe W. et al, 2012. Genomic variation and its impact on gene expression in Drosophila melanogaster. PLoS Genet. 8: e1003055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr M., Yusuf S., Weir G., Chung Y. L., Mayr U., et al. , 2008. Combined metabolomic and proteomic analysis of human atrial fibrillation. J. Am. Coll. Cardiol. 51: 585–594 [DOI] [PubMed] [Google Scholar]
- Na J., Musselman L. P., Pendse J., Baranski T. J., Bodmer R., et al. , 2013. A Drosophila model of high sugar diet-induced cardiomyopathy. PLoS Genet. 9: e1003175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuwaysir E. F., Huang W., Albert T. J., Singh J., Nuwaysir K., et al. , 2002. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res. 12: 1749–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocorr K., Reeves N. L., Wessells R. J., Fink M., Chen H. S. et al, 2007. KCNQ potassium channel mutations cause cardiac arrhythmias in Drosophila that mimic the effects of aging. Proc. Natl. Acad. Sci. USA 104: 3943–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-terWengel P., Kapun M., Nolte V., Kofler R., Flatt T., et al. , 2012. Adaptation of Drosophila to a novel laboratory environment reveals temporally heterogeneous trajectories of selected alleles. Mol. Ecol. 21: 4931–4941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel C. J., Sivadas A., Tabassum R., Preeprem T., Zhao J., et al. , 2013. Whole genome sequencing in support of wellness and health maintenance. Genome Med. 5: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospisilik J.A., Schramek D., Schnidar H., Cronin S. J., Nehme N. T. et al, 2010. Drosophila genome-wide obesity screen reveals hedgehog as a determinant of brown vs. white adipose cell fate. Cell 140: 148–160 [DOI] [PubMed] [Google Scholar]
- Qi Q., A. Y. Chu, J. H. Kang, M. K. Jensen, G. C. Curhan et al, 2012. Sugar-sweetened beverages and genetic risk of obesity. N. Engl. J. Med. 367: 1387–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L. K., Williams S., Springston M., Brown J., Freeman K., et al. , 2010. Genotype-by-diet interactions drive metabolic phenotype variation in Drosophila melanogaster. Genetics 185: 1009–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson E. J., Kim S. K., Nusse R., 2002. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science 296: 1118–1120 [DOI] [PubMed] [Google Scholar]
- Sang J. H., 1956. The quantitative nutritional requirements of Drosophila melanogaster. J. Exp. Biol. 33: 45–72 [Google Scholar]
- Schulz L., Bennett P.H., Ravussin E., Kidd J. R., Kidd K.K. et al, 2006. Effects of traditional and Western environments on prevalence of type 2 diabetes in Pima Indians in Mexico and the U.S. Diabetes Care 29: 1866–1871. [DOI] [PubMed] [Google Scholar]
- Spurgeon S. L., Jones R. C., Ramakrishnan R., 2008. High throughput gene expression measurement with real time PCR in a microfluidic dynamic array. PLoS ONE 3: e1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T., Kusakabe S., Mukai T., 1987. The genetic structure of natural populations of Drosophila melanogaster. XX. Comparison of genotype-environment interaction in viability between a northern and a southern population. Genetics 117: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Miller P. M., P.M., 2012. Investigating natural variation in Drosophila courtship song by the evolve and resequence approach. Genetics 191: 633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Stewart A. D., Fields A. T., Rice W. R., A. M. Tarone, 2011. Population-based resequencing of experimentally evolved populations reveals the genetic basis of body size variation in Drosophila melanogaster. PLoS Genet. 7: e1001336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Campbell T. G., Stone E. A., Mackay T. F. C., Anholt R. R. H., 2012. Phenotypic plasticity of the Drosophila transcriptome. PLoS Genet. 8: e1002593. [DOI] [PMC free article] [PubMed] [Google Scholar]