Abstract
Pancreatitis is an inflammatory condition of the pancreas which, in its chronic form, involves tissue destruction, exocrine and endocrine insufficiency, increased risk of pancreatic cancer, and an extensive fibrotic pathology which is due to unrelenting collagen deposition by pancreatic stellate cells (PSC). In response to noxious agents such as alcohol—excessive consumption of which is a major cause of pancreatitis in the West—normally quiescent PSC undergo a phenotypic and functional transition to activated myofibroblasts which produce and deposit collagen at high levels. This process is regulated by connective tissue growth factor (CCN2), expression of which is highly up-regulated in activated PSC. We show that CCN2 production by activated PSC is associated with enhanced expression of microRNA-21 (miR-21) which was detected at high levels in activated PSC in a murine model of alcoholic chronic pancreatitis. A positive feedback loop between CCN2 and miR-21 was identified that resulted in enhancement of their respective expression as well as that of collagen α1(I). Both miR-21 and CCN2 mRNA were present in PSC-derived exosomes, which were characterized as 50–150 nm CD9-positive nano-vesicles. Exosomes from CCN2-GFP- or miR-21-GFP-transfected PSC were taken up by other PSC cultures, as shown by direct fluorescence or qRT-PCR for GFP. Collectively these studies establish miR-21 and CCN2 as participants in a positive feedback loop during PSC activation and as components of the molecular payload in PSC-derived exosomes that can be delivered to other PSC. Thus interactions between cellular or exosomal miR-21 and CCN2 represent novel aspects of fibrogenic regulation in PSC. Summary Chronic injury in the pancreas is associated with fibrotic pathology which is driven in large part by CCN2-dependent collagen production in pancreatic stellate cells. This study shows that CCN2 up-regulation in PSC is associated with increased expression of miR-21 which, in turn, is able to stimulate CCN2 expression further via a positive feedback loop. Additionally miR-21 and CCN2 were identified in PSC-derived exosomes which effected their delivery to other PSC. The cellular and exosomal miR-21-CCN2 axis is a novel component in PSC fibrogenic signaling.
Keywords: Alcohol, Connective tissue growth factor, Fibrosis, miR, Microvesicle, Pancreas
Introduction
Chronic injury induces fibrotic responses in virtually every organ system and is frequently associated with transforming growth factor beta-1 (TGF-β1)-dependent connective tissue growth factor (CCN2) pro-fibrotic signaling in mesenchymal cells. About 80,000 people in the USA are diagnosed each year with chronic pancreatitis (CP), a disorder characterized by progressive destruction of pancreatic tissue, deposition of fibrous scar material, and increased risk of developing pancreatic cancer (Charrier and Brigstock 2013). CCN2 mRNA is enhanced in pancreata of human CP patients (di Mola et al. 1999) and in mouse experimental CP (Charrier and Brigstock 2010). Pancreatic stellate cells (PSC) are the principal pro-fibrogenic cell type in the pancreas (Apte et al. 1998, 1999) and CCN2 plays an important role in driving fibrogenic pathways in these cells after injury (Gao and Brigstock 2005, 2006). CCN2 acts via autocrine and paracrine pathways to stimulate PSC migration, proliferation, mitogenesis and collagen production (Gao and Brigstock 2005). An important feature of CCN2 production by PSC is that it is restricted to times of PSC activation, the process by which these normally quiescent fibroblast cells transition into myofibroblasts that play a central role in wound contraction and collagen deposition (Apte et al. 1998; Bachem et al. 1998). In vitro CCN2 promoter activity or mRNA levels are stimulated in rat PSC in response to TGF-β1, platelet-derived growth factor, ethanol or acetaldehyde (Gao and Brigstock 2005) and, during autonomous PSC activation in culture, CCN2 is concomitantly and increasingly expressed with TGF-β1 and collagen α1(I) (Gao et al. 2004). Despite the numerous advances in this field, many aspects of CCN2 regulation and action remain poorly understood.
MicroRNAs (miRs) are a class of small noncoding RNAs. The human genome contains over 1,000 miRs (Friedman et al. 2009), each with hundreds to thousands of predicted mRNA targets. MiRs regulate expression at the translation step through Watson-Crick base pairing between the miR 5’ nucleotides 2–8 base pair “seed sequence” and the target mRNA 3’ untranslated region which results in either repression or degradation of the mRNA target (Stefani and Slack 2008). MiRs play a diverse role in many cellular processes such as proliferation (Thum et al. 2008), differentiation (Marson et al. 2008), angiogenesis (Wang et al. 2008) and morphogenesis (Harfe et al. 2005), while aberrant miR signaling occurs in numerous diseases including cancer (Shen et al. 2013), heart disease (Quiat and Olson 2013) and diabetes (Lorenzen et al. 2012) as well as fibrotic pathologies (Patel and Noureddine 2012). However, the relationship between miRs and CCN2 has not been widely studied and we therefore addressed this question in a rodent model of alcoholic CP in vivo as well as during autonomous activation of PSC during cultivation of freshly isolated PSC in vitro.
Materials and methods
Animal model
A murine model of alcoholic CP in which ethanol was chronically administered to male C57/Bl6 mice against a background of cerulein-induced acute pancreatitis was established as previously described (Charrier and Brigstock 2010). Mice were sacrificed at the fibrotic stage on Day 23 and pancreata were removed and either fixed in RNase free 4 % paraformaldehyde (pH 7.2–7.4) or processed for RNA extraction. Animal use was approved by the Institutional Animal Care and Use Committee of The Research Institute at Nationwide Children’s Hospital (Columbus, OH).
RNA isolation
Pancreata were removed and immediately immersed in RNAlater (Qiagen). RNA from homogenized pancreata tissue or lysed cultured PSC was extracted using a microRNeasy Plus Mini Kit (Qiagen) according to the manufacturer’s protocol.
MiR microarray
RNA (10 μl) from each animal (ethanol/cerulein, n = 4; water/saline, n = 4) was assessed for quality and concentration using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) and then a microarray was performed in-house (Biomedical Genomics Core, The Research Institute at Nationwide Children’s Hospital, Columbus OH) on each individual mouse RNA sample using a mouse miR Microarray release 16.0 (Agilent Technologies). Array data were combined and candidate miRs identified as being significantly up- or down-regulated were then confirmed by performing quantitative real-time PCR (qRT-PCR) on each of the above RNA samples as described below.
Reverse transcription and qRT-PCR
Aliquots of up to 1 μg of total RNA were reverse transcribed using a miScript II Reverse Transcription kit (Qiagen). Quantification of pancreatic transcripts was achieved by qRT-pCR (Eppendorf Mastercycler ep realplex2 S, Eppendorf, Hauppage, NY) using the following protocol for mRNA: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min, then 95 °C for 15 s followed by a dissociation step of 60 °C for 20 s and 95° for 15 s. mRNA PCR amplification was carried out in a 25 μl reaction containing 125 ng cDNA, 6.25 μl SYBR green PCR Master Mix (Applied Biosystems, Foster City, CA) and 0.5 μM of each primer set. The mRNA primers are shown in Table 1. Amplification conditions for miRs were: 95 °C for 15 min, followed by 40 cycles of 94 °C for 15 s and 55 °C for 30 s, then 70 °C for 30 s. miRNA PCR amplification was carried out in a 25 μl reaction containing 125 ng cDNA, 12.5 μl 2× QuantiTect Sybr Green PCR Master Mix (Qiagen), 2.5 μl 10× miScript Universal Primer (Qiagen) and 2.5 μl 10× miScript Primer Assay (Qiagen). The miRNA miScript Primer Assay primers U6, miR-21, miR-148a, miR-802, miR-199a-3p, and miR-211* were purchased from Qiagen. Each reaction was run in triplicate and all samples were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH), peptidylprolyl isomerase A (PPiA), hypoxanthine-guanine phosphoribosyl transferase 1 (HPRT1) or U6.
Table 1.
Primer sequences for qRT-PCR
| Gene | Gene accession no. | Sense (5′-3′) | Anti-sense (5′-3′) | bp |
|---|---|---|---|---|
| GAPDH (mouse) | NM_008084.2 | TGCACCACCAACTGCTTAGC | GGCATGGACTGTGGTCATGAG | 87 |
| HPRT1 (mouse) | NM_013556.2 | CCTAAGATGAGCGCAAGTTGAA | CCACAGGACTAGAACACCTGCTAA | 86 |
| PPiA (rat) (mouse) | NM_017101.1 NM_008907.1 | GCATACAGGTCCTGGCATCT | CTTCCCAAAGACCACATGCT | 121 |
| CCN2 (mouse) | NM_010217.2 | CACTCTGCCAGTGGAGTTCA | AAGATGTCATTGTCCCCAGG | 111 |
| collagen α1(I) (mouse) | NM_007742.3 | GCCCGAACCCCAAGGAAAAGAAGC | CTGGGAGGCCTCGGTGGACATTAG | 149 |
| GFP | GGACGACGGCAACTACAAGA | AAGTCGATGCCCTTCAGCTC |
In situ hybridization(ISH)
Full-length hsa-miR-21 22-mer double digoxigenin (DIG)-labeled miRCURY LNATM miRNA detection probe (TCAACATCAGTCTGATAAGCTA, RNA-Tm 83 °C) and a double DIG-labeled scramble probe (GTGTAACACGTCTATACGCCCA, RNA-TM 87 °C) as a control were purchased from Exiqon (Woburn, MA). After de-paraffinization and rehydration of mouse pancreata, sections were subjected to Proteinase-K digestion (2.5 μg/ml), (Sigma-Aldrich) for 10 min, washed in PBS and dehydrated in a series of ethanol solutions and then air-dried. Hybridization was at 55 °C for 1 h with LNA probes (50 nM) in miR ISH buffer (Exiqon). After hybridization, slides were washed in a series of SSC buffer dilutions at 55 °C, blocked in digoxygenin (DIG) blocking buffer and incubated with sheep anti-DIG-AP (1:800) (Roche) in DIG blocking buffer containing 2 % sheep serum. The slides were incubated with NBT/BCIP reaction mixture (Roche) in a humidified chamber at 37 °C for 2 h. Finally, the slides were counterstained with Nuclear Fast Red (Vector laboratories, Burlingame, CA), washed in water, ethanol, and then mounted in Eukitt mounting medium (VWR, San Dimas, CA) and examined by Axioscope 40 light microscopy (Carl Zeiss Co. Ltd, Jena, Germnay) using a 40× objective lens.
Immunohistochemistry
Fixed pancreatic tissues were embedded in paraffin, cut into 4 μm sections and stained with haemotoxylin-eosin for standard histological examination. Immunohistochemical staining for alpha-smooth muscle actin (α-SMA) was performed using monoclonal mouse-anti human α-SMA (1:1,000, Sigma-Aldrich, St. Louis, MO) followed by development with UltraTek Anti-Polyvalent, UltraTek HRP and AEC chromogenic substrate (all obtained from ScyTek Laboratories, Logan, UT). Slides were counter-stained in haemotoxylin, covered in Crystal Mount (Sigma-Aldrich) and mounted with Permaslip mounting medium (Alban Scientific, St. Louis, MO).
Cell culture and isolation
Mouse PSC were isolated by modification of previously described methods (Phillips et al. 2003). Pancreata from 10 to 15 male C57/Bl6 mice were removed, minced with scissors, and digested with 1 mg/ml pronase, 1 mg/ml DNase I and 0.5 mg/ml collagenase P (all obtained from Roche, Indianapolis, IN) in DMEM/F-12 50/50 medium. The resulting cell suspension was centrifuged in a 12 % OptiPrep (Life Technologies, Carlsbad, CA, USA) gradient at 1,400×g for 30 min. PSC separated into a hazy band just above the interface of the gradient in the aqueous buffer. This band was harvested, and the cells were washed and resuspended in DMEM/F-12 50/50 containing 20 % fetal bovine serum, 4.5 g/l l-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. Cells were plated into T-25 flasks and maintained at 37 °C in a humidified atmosphere of 5 % CO2/95 % air. Passaged or Day 8 PSC were transfected for 24–96 h in the presence of 50–200 nM syn-mmu-miR-21 mimic (Qiagen, Valencia, CA) or 50 nM CCN2 small interfering RNA (siRNA) or scramble siRNA (Ambion, Billerica, MA). Cells were then evaluated for CCN2, collagen α1(I) or miR-21 expression. Cells were not used after passage 7.
PSC transfection
All transfections were performed using Lipofectamine RNAiMax (Life Technologies) according to the manufacturer’s protocol. Briefly, PSC (at passage 3–6) were seeded 1 × 105 cells per well in a 6-well plate in DMEM/F-12 50/50 containing 10 % FBS without antibiotics 24-h prior to transfection. For transfection, siRNA, miRNA mimic, or miR-21 antagomir were diluted to the appropriate concentration in Opti-MEM. Lipofectamine RNAiMax was also pre-mixed with OPTI-MEM and then added to diluted siRNA/miRmimic and incubated for 10 min at RT. The Opti-MEM/siRNA/Lipofectamine mixtures were then added drop-wise to appropriate wells and incubated for 24–96 h.
Exosome isolation
PSC were cultured in T-175 flasks in DMEM/F-12 50/50 medium containing 10 % FBS until 90 % confluent whereupon the medium was replaced with serum free DMEM/F12-50/50 for 48 h. Medium was collected and centrifuged at 300×g for 10 min to pellet the cells. The supernatant was collected and spun at 2,000×g for 20 min and again at 10,000×g for 30 min to pellet the cell debris. The supernatant was then ultra-centrifuged at 100,000×g for 70 min from which the pellet was washed in 30 ml PBS and ultra-centrifuged again under the same conditions. In some experiments, PKH26 (Sigma-Aldrich) was added to the supernatant to fluorescently label the exosomes. All centrifugation steps were performed at 4 °C. The exosomal pellet from three T-175 flasks was resuspended in 50–100 μl PBS and examined by qRT-PCR or Western blot with anti-CD9 antibody (Lifespan Bioscience Inc., Seattle WA). Purified exosomes were analyzed for zeta potential with a ZetaPALS analyzer (Brookhaven Instruments Inc., Holtville, NY). Exosomes were allowed to settle on carbon-coated 400-mesh copper grids (Electron Microscopy Sciences, Hatfield, PA), stained with 2 % uranyl acetate, air-dried, and imaged by transmission electron microscopy (TEM) using a H-7650 microscope (Hitachi High Technologies America, Pleasanton, CA) in the Morphology Core Facility at The Research Institute at Nationwide Children’s Hospital (Columbus OH).
Isolation and functional characterization of PSC-derived exosomes
Passage 5 (P5) mouse primary PSC or the rat SAM-K PSC line (Satoh et al. 2002), were transfected by electroporation in combination with a Nucleofector Kit (Lonza, Koln, Germany) with 4 μg pCMV-CCN2-GFP (CCN2-GFP) produced as described (Hoshijima et al. 2012) or 4 μg miRNASelect pEGP-mmu-miR-21 (miR-21-GFP) expression vector containing the miR-21 stem-loop precursor cloned between BAM-H1 and NHE-1 sites flanked by its native intron sequences and under the control of the EF-1α promoter (Cell Biolabs, San Diego, CA). Cells were cultured in T-25 flasks in DMEM/F-12 50/50 medium containing 10 % FBS. After 24 h the medium was reduced to low serum (0.1 % FBS) and the cells were cultured for an additional 48 h. Medium was then concentrated using ultra-4 3 K centrifugal filter units (Ambion) at 3,000×g (25 min, 4 °C). ExoQuick solution (System Biosciences, Mountainview, CA) was added to each concentrated sample (1:1), mixed gently, and left to precipitate overnight. The precipitated exosomes were centrifuged at 3,000×g (30 min, RT) and the supernatant was removed. The pellet was centrifuged again at 3,000×g (5 min, RT), the residual supernatant was removed by aspiration, and exosomes were re-suspended in PBS and added to wells of PSC or SAM-K cells that had been previously cultured for 24 h in 12-well plates. After 24–48 h, the cells were analyzed for the presence of GFP by direct fluorescence or by qRT-PCR for GFP in RNA isolated from the cells.
Statistical analysis
All experiments were performed at least three times with triplicate measurements, with data expressed as mean ± s.e.m. RT-PCR data were analyzed by student’s t-test using Sigma plot 11.0 software (SPSS Inc., Chicago,IL) and P values < 0.05 were considered statistically significant.
Results
PSC are believed to play an essential role during fibrosis due to their ability to produce high levels of ECM proteins (e.g. collagen) in a CCN2-dependent manner following ethanol exposure. Since we previously showed that activated PSC are the main producers of collagen and CCN2 in a mouse model of CP (Charrier and Brigstock 2010), the goal of the present study was to determine the action of miRs up- or down-stream of CCN2 during PSC activation.
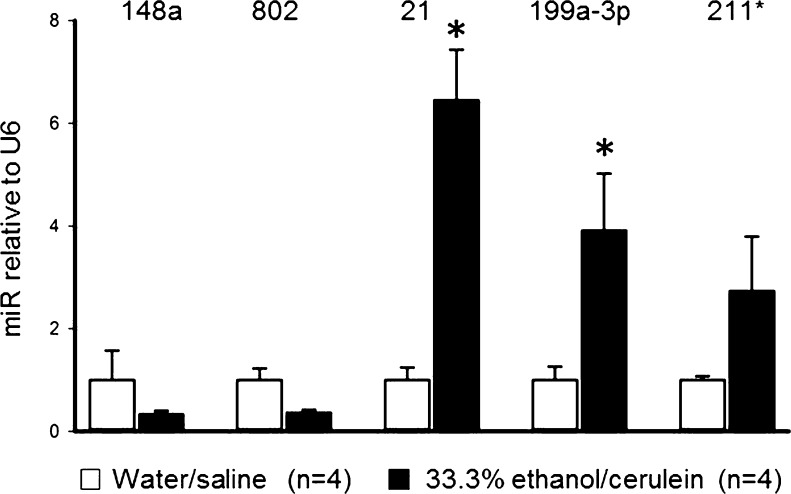
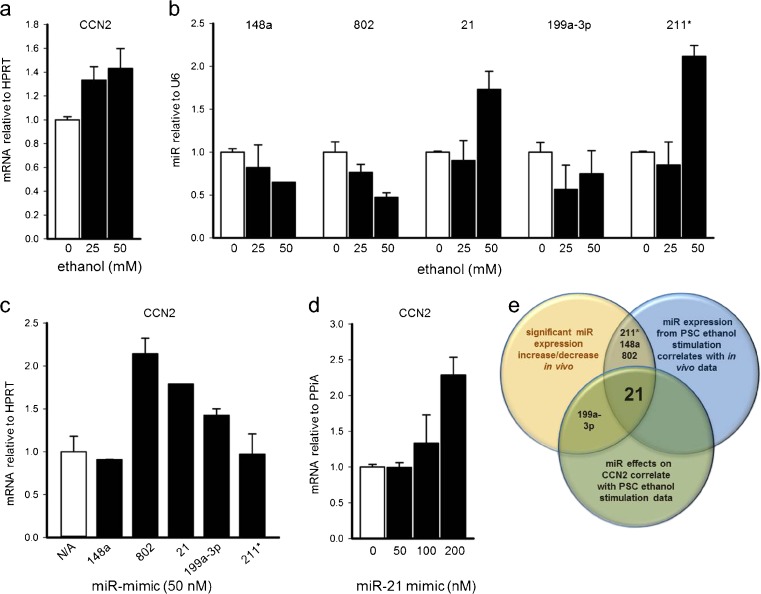
We initially performed a miR microarray on total pancreatic RNA collected from mice with ethanol-induced CP. As compared to control mice, mice with alcoholic CP exhibited significant down-regulation of miR-148 or −802 and significant up-regulation of miR-21, −199a-3p or −211* (Table 2). These findings were confirmed by qRT-PCR (Fig. 1). Since these results were from total pancreatic RNA obtained during the period of intense fibrosis and predominant PSC involvement, we next investigated if any of the same miRs were associated with ethanol-induced CCN2 production in PSC in vitro. Consistent with earlier studies (Lawrencia et al. 2009), PSC exhibited a dose-dependent stimulation of CCN2 mRNA in response to ethanol (Fig. 2a). Ethanol induction of CCN2 in PSC was accompanied by decreased expression of miR-148a, −802 or −199a-3p and increased expression of miR-211* or −21 (Fig. 2b). These patterns of expression in vitro were consistent with the in vivo model (Fig. 1) except for miR-199a-3p. Next, PSC were individually transfected with miR-mimics (pre-miRs) for miR-148a, −802, −21, −199a-3p or −211* to evaluate their respective effects on CCN2 expression. MiR-802, miR-21 or miR-199a-3p induced CCN2 levels whereas CCN2 levels were unchanged by transfection with either miR-148a or miR-211* (Fig. 2c). Also, miR-21 induced CCN2 expression in a dose-dependent manner (Fig. 2d). Taken together, of the five miRs initially detected by microarray, only miR-21 exhibited patterns of ethanol-induced expression in vivo or in vitro that were consistent with one another and with those of CCN2 as well as showing an induction of CCN2 expression that was thus consistent with the co-expression of both miR-21and CCN2. Overall, these findings suggested a causative relationship between concomitant expression of miR-21 and CCN2 in PSC downstream of ethanol exposure (Fig. 2e).
Table 2.
MiR expression changes during experimental CP assessed by miR array
| MiR | Fold change (CP v Ctrl) |
|---|---|
| mmu-miR-148a | −2.726* |
| mmu-miR-802 | −2.844* |
| mmu-miR-21 | 4.853* |
| mmu-miR-199a-3p | 2.382* |
| mmu-miR-211* | 3.25* |
Total RNA from pancreata was extracted from mice treated with ethanol/cerulein (n = 4) or water/saline (n = 4) for 23 days and subjected to miR expression array analysis. The table shows the most significant changes in miR expression for ethanol/cerulein versus water/saline treatment after the data from each microarray were combined within each treatment group. (* p < 0.05)
Fig. 1.
MiR expression changes during experimental CP assessed by qRT-PCR. QRT-PCR for miR-148a, 802, 21, 199a-3p and 211* in RNA isolated from pancreata of mice treated with ethanol/cerulein or water/saline for 23 days. (* p < 0.01)
Fig. 2.
Identification of miR-21 as being positively correlated with CCN2. Detection by qRT-PCR of a CCN2 mRNA or b miR-148a, 802, 21, 199a-3p or 211* in primary PSC treated on Day 5 of culture with 0–50 mM ethanol for 24 h. Detection by qRT-PCR of c CCN2 mRNA in PSC transfected with miR-802, 21, 199a-3p, 211* or miR-148 or d CCN2 mRNA after transfection of the cells with miR-21. e Venn diagram demonstrating the rationale for selection of miR-21 for subsequent studies
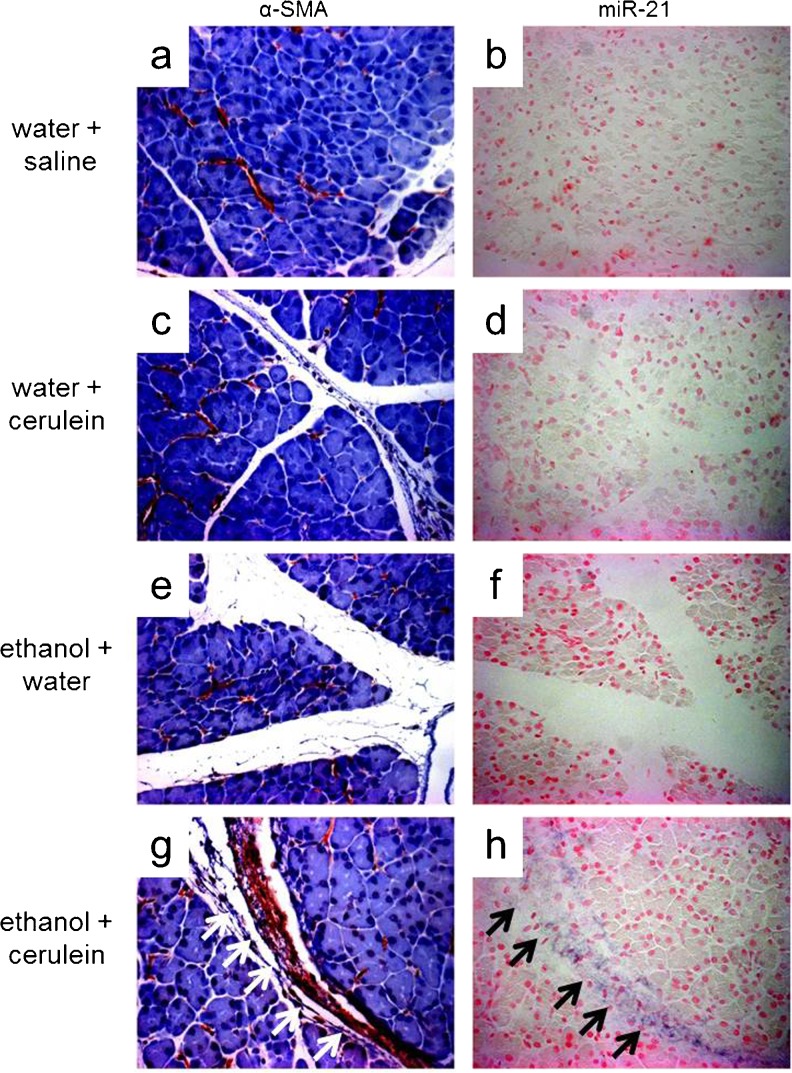
To confirm that activated PSC are the main producers of miR-21 during experimental CP, we performed in situ hybridization for miR-21 on pancreatic sections from control mice treated with cerulein alone, ethanol alone or water/saline (Fig. 3a–b, c–d, e–f), or alcoholic CP mice treated with ethanol/cerulein (Fig. 3g–h). MiR-21 was only detected in the ethanol/cerulein group (Fig. 3h) and its expression was restricted to activated PSC, which were identified by α-SMA immunoreactivity in serial sections (Fig. 3g). Thus miR-21 is up-regulated in the pancreas during experimental CP and is produced predominantly by activated PSC.
Fig. 3.
Activated PSC are the principal producer of miR-21 transcript during experimental CP. Pancreatic sections from mice treated with water/saline a–b, water alone c–d, ethanol alone e–f or ethanol/cerulein g–h were evaluated by α-SMA immunohistochemistry and miR-21 in situ hybridization
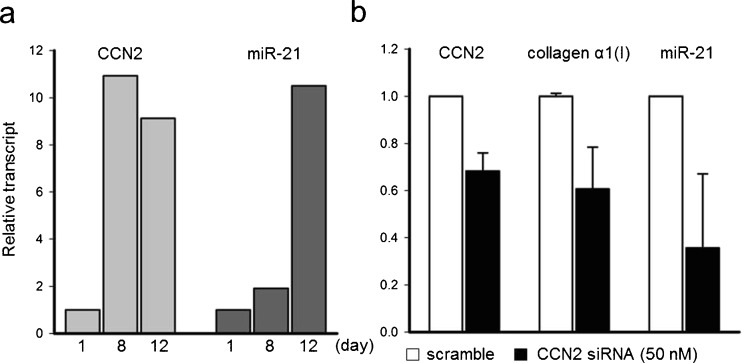
To clarify the relationship between CCN2 and miR-21 during PSC activation, primary PSC isolated from normal mouse pancreata were analyzed during their culture-induced activation in vitro. CCN2 was substantially up-regulated in PSC by Day 8 (11-fold) and remained up-regulated at Day 12 (9-fold) (Fig. 4a). MiR-21 was only slightly increased by Day 8 (2-fold) but a more robust induction was seen by Day 12 (10-fold) (Fig. 4a). To determine if the somewhat earlier increase in CCN2 expression accounted for the subsequent increase in miR-21 production, CCN2 expression was knocked down using CCN2 siRNA on Day 8 of culture. Analysis on Day 12 revealed a 35 % inhibition of CCN2 expression (Fig. 4b), a 40 % decrease in collagen α1(I) expression and a 60 % decrease in miR-21 as compared to the scramble controls (Fig. 4b). Taken together these results reveal that during PSC activation, CCN2 not only drives collagen expression as previously reported (Omary et al. 2007) but also stimulates expression of miR-21 which, in turn, can itself induce CCN2 expression (see Fig. 2c, d).
Fig. 4.
CCN2 drives miR-21 expression during autonomous PSC activation in vitro. a CCN2 and miR-21 expression during autonomous PSC activation in culture (Day 1–12) as demonstrated by qRT-PCR. b QRT-PCR for CCN2, collagen α1(I), or miR-21 in Day 8 PSC where CCN2 expression is inhibited by CCN2 siRNA
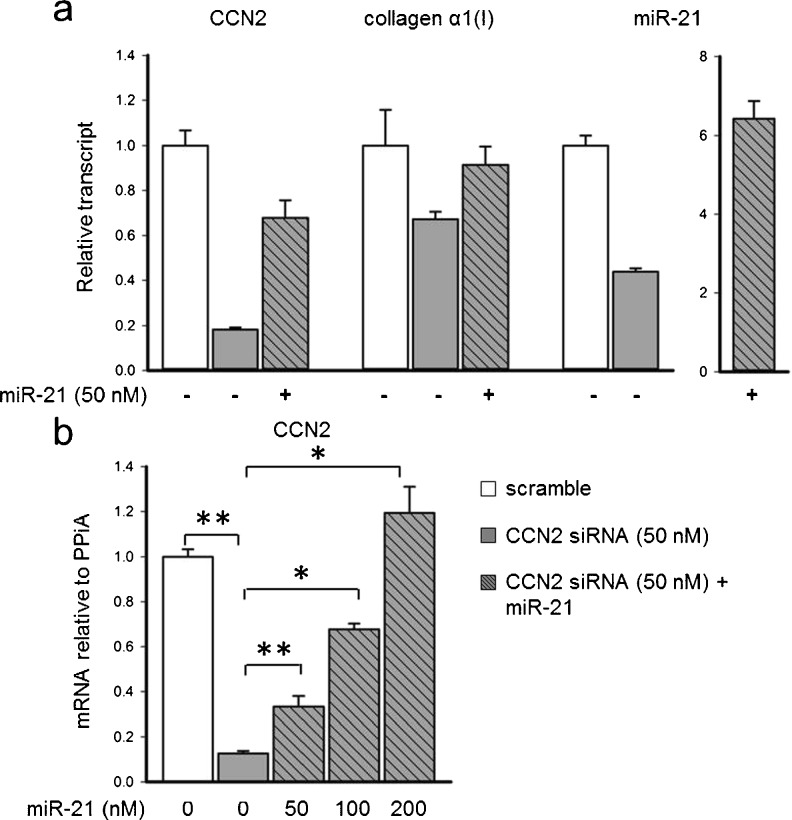
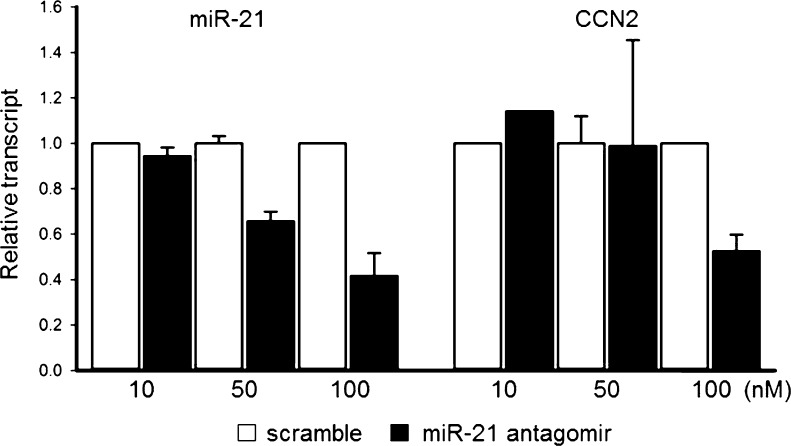
In a similar approach, the relationship between CCN2 and miR-21 was investigated in fully activated (P5) PSC. Transfection of these cells with CCN2 siRNA caused reduced expression of CCN2, collagen α1(I) or miR-21 by 80 %, 40 % or 60 %, respectively (Fig. 5a). Interestingly, the reduced levels of CCN2 or collagen α1(I) mRNA were restored towards control levels by treatment of the cells with pre-miR-21 (Fig. 5a). Strikingly, when this experiment was repeated over a 48 h period, CCN2 mRNA levels were knocked down by 90 % as compared to scramble controls, and the addition of pre-miR-21 at the time of transfection dose-dependently restored CCN2 expression to above normal levels (Fig. 5b). Collectively, these results demonstrated that not only does CCN2 drive miR-21 expression, but that miR-21 itself can stimulate CCN2 expression when the normally high levels of CCN2 activated cells are suppressed. This relationship was further demonstrated through the use of a miR-21 antagomir which inhibited expression of both miR-21 and CCN2 (Fig. 6). Thus CCN2 and miR-21 up-regulate each other and are components of a positive feedback loop that may serve as an amplification mechanism for enhanced collagen production.
Fig. 5.
CCN2 expression drives miR-21, which drives CCN2 expression in vitro. CCN2, miR-21 or collagen α1(I) expression levels as demonstrated by qRT-PCR in activated PSC (P5) transfected with CCN2 siRNA +/− pre-miR-21 for a 24 h or b 48 h. (*p < 0.05, **p < 0.001)
Fig. 6.
MiR-21 knockdown inhibits CCN2 expression in PSC in vitro. QRT-PCR for miR-21 or CCN2 mRNA in PSC transfected with 0–100 nM miR-21 antagomir
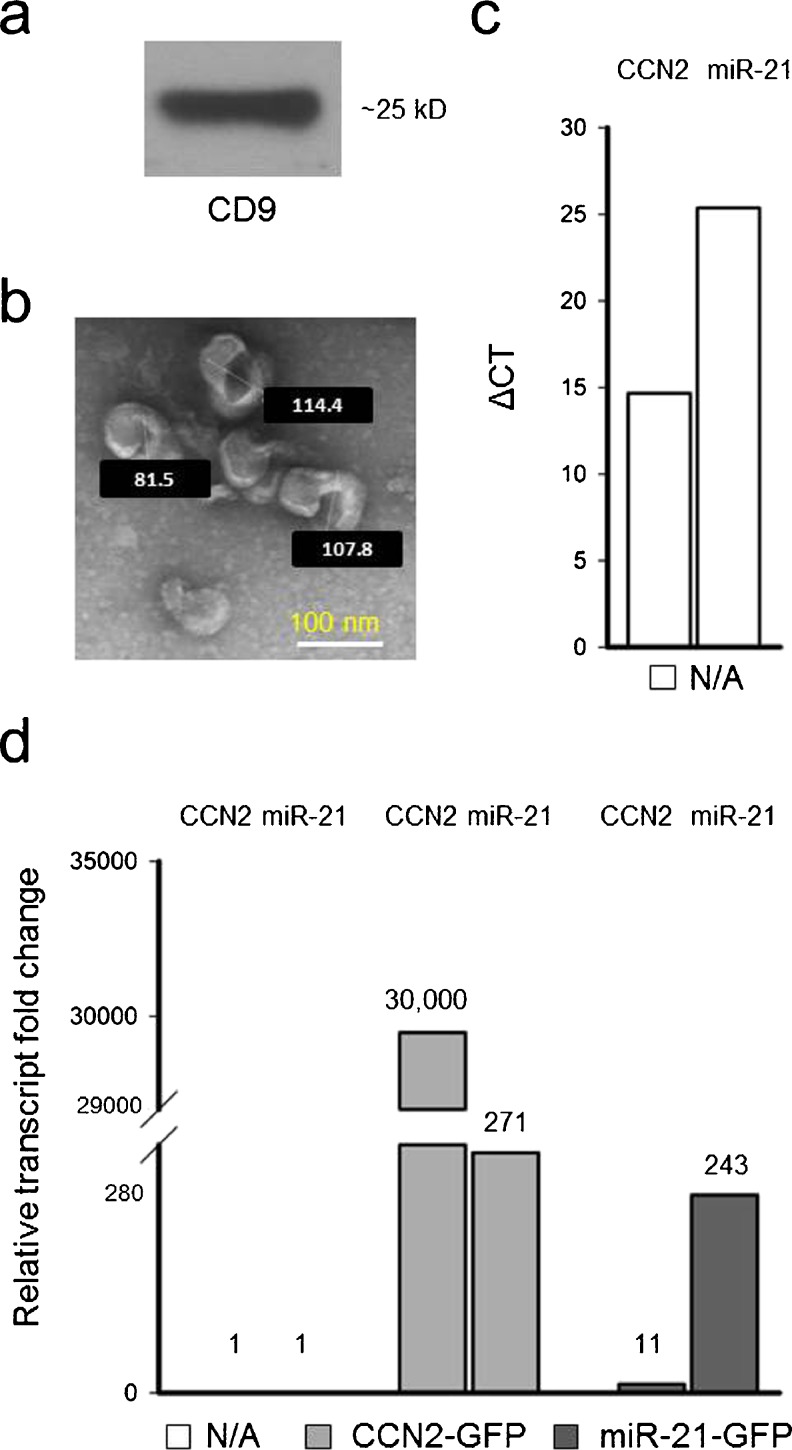
Exosomes are nano-sized vesicles that are liberated into the extracellular space and which can carry a complex molecular payload (mRNA, miRs proteins) to other cells. The ability of PSC to dynamically produce CCN2 mRNA or miR-21 prompted us to investigate the possibility that one or both of these molecules might be packaged into exosomes and delivered to other cells. Since exosome production by PSC has not previously been demonstrated, PSC conditioned medium was first analyzed to determine if exosomes were present. Indeed, PSC-derived exosomes were successfully isolated and characterized as being positive for CD9, a murine exosome marker (Fig. 7a). PSC-derived exosomes were 50–150 nm-diameter membranous vesicles as assessed by TEM (Fig. 7b) and carried a high net negative charge (−26 mV) as assessed by zeta potential (data not shown). qRT-PCR of exosomal RNA revealed the presence of CCN2 mRNA (Fig. 7c) and miR-21 (Fig. 7c). Over-expression of CCN2 in PSC led to a 30,000-fold increase of CCN2 transcript levels in isolated exosomes as well as a ~270-fold increase in miR-21 exosomal expression (Fig. 7d). Similarly, miR-21-over-expression in PSC resulted in a 243-fold increase exosomal in miR-21 levels and an 11-fold increase in exosomal CCN2 mRNA levels. These results support the hypothesis that expression of CCN2 and miR-21 in PSC is inter-dependent and further show that each transcript is incorporated into exosomes at levels reflecting their relative abundance in the producer cell.
Fig. 7.
Characterization of PSC exosomes. Exosomes isolated from the medium of cultured PSC were analyzed by a Western blot for CD9 and b TEM. qRT-PCR was performed to detect miR-21 or CCN2 mRNA in c exosomes from activated PSC or d exosomes from PSC transfected for 24 h with either CCN2-GFP or miR-21-GFP. Values above the bars in d represent the approximate fold increase in transcript level versus their respective controls which are normalized to a value of 1 and had comparable ΔCT values to those shown in c
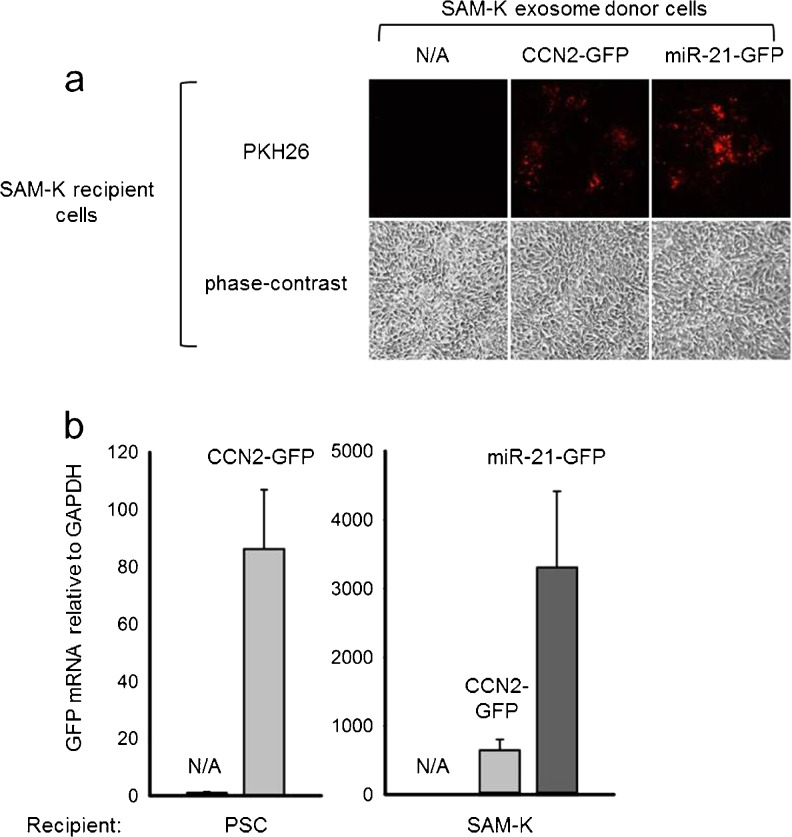
To determine if exosomal CCN2 mRNA or miR-21 could be delivered to other cells, control cultures of primary mouse PSC or rat SAM-K cells were incubated with exosomes that had been stained with PKH26 fluorescent dye after their isolation from medium of the CCN2-GFP- or miR-21-GFP-transfected cells. Within 24 h, PKH26 staining was observed in PSC receiving exosomes from either CCN2-GFP- or miR-21-GFP-transfected cells showing that exosome uptake had occurred (Fig. 8a). This was confirmed by qRT-PCR of RNA from the recipient cells which revealed a ~90- or ~800-fold increase in GFP expression in, respectively, recipient PSC or SAM-K cells receiving exosomes from CCN2-GFP-transfected donor cells or a ~3,200-fold increase in GFP expression in SAM-K cells receiving exosomes from miR-21-GFP transfected donor SAM-K cells (Fig. 8b). The overall higher levels of exosomally-transferred GFP seen in recipient SAM-K cells versus primary PSC likely reflected a higher efficiency of transfection of the GFP vectors (and subsequent incorporation into exosomes at higher levels) in the established SAM-K donor cell line as compared to the primary donor PSC (data not shown). Nonetheless, these results show that miR21 or CCN2 mRNA produced by PSC are exported from the cells in exosomes which can act as shuttles for their subsequent delivery to recipient PSC.
Fig. 8.
Exosome-mediated uptake of miR-21 or CCN2 mRNA by PSC. Primary mouse PSC or rat SAM-K cells were incubated for 24 h with PKH26-stained exosomes isolated from their CCN2-GFP- or miR-21-GFP-transfected counterparts. The figure shows a phase contrast or fluorescence microscopy of recipient SAM-K cells or b qRT-PCR for GFP in recipient PSC or SAM-K cells
Discussion
In these studies, miRs-21, 148 or −802, −199a-3p or −211* were shown to be significantly up- or down-regulated during experimental alcoholic CP through microarray analysis and independent qRT-PCR. In view of our interest in CCN2 which is produced by activated PSC and drives the fibrotic component of CP (Charrier and Brigstock 2010), our focus narrowed to miR-21 because (i) it was highly expressed in experimental CP and localized to PSC, (ii) its expression by PSC in vitro was stimulated by exposure of the cells to ethanol and (iii) its over-expression in PSC in vitro significantly induced CCN2 expression. Collectively, these finding suggested a causative relationship between expression of miR-21 and CCN2 in activated PSC. Although the relationship between miR-21 and CCN2 during CP was not anticipated, previous studies have shown that miR-21 was up-regulated in several solid tumors including those of the pancreas (Iorio et al. 2005), while fecal, serum and solid tumor levels of miR-21 were shown to be positively correlated with negative disease outcome and advanced stage (stage III and IV) in pancreatic cancer (Dillhoff et al. 2008; Link et al. 2012; Liu et al. 2012). Although our studies failed to link the significant changes in expression of miR-148 or −802, −199a-3p or −211* in alcoholic CP to that of CCN2, these miRs are nonetheless likely involved in other as yet undetermined aspects of CP.
Our data suggest that the miR-21-CCN2 axis in activated PSC cells forms a positive feedback loop that, once triggered, may exacerbate CCN2 downstream effects such as collagen production. Our findings regarding up-regulation of miR-21 in CP are consistent with several other studies of fibrosis in other organ systems. For example, miR-21 was significantly increased in fibroblasts of failing hearts in mouse or human specimens and administration of a miR-21 antagomir to mice resulted in inhibition of experimental fibrosis (Thum et al. 2008). MiR-21 was also up-regulated in the lungs of patients with idiopathic pulmonary fibrosis and in a mouse model of bleomycin-induced fibrosis in which miR-21 siRNA treatment diminished the severity of lung fibrosis (Liu et al. 2010). In both the cardiac and pulmonary models, miR-21 was up-regulated in fibroblasts and myofibroblasts (Thum et al. 2008); (Liu et al. 2010), consistent with our findings for activated PSC in CP. Finally, in a mouse model of renal fibrosis caused by unilateral ureteral obstruction, miR-21 was localized to distal tubular epithelial cells and the degree of fibrosis was reduced when miR-21 was inhibited (Zarjou et al. 2011).
In many cell types (including PSC), CCN2 acts downstream of transforming growth factor beta 1 (TGF-β1) to stimulate fibrogenic pathways. Although we have yet to study the interactions between TGF-β1 and miR-21 in PSC, lung fibroblasts have been shown to demonstrate increased miR-21 expression in response to TGF-β1 while treatment of the cells with miR-21 caused increased levels of p-Smad2 and of TGF-β1-induced fibronectin or α-SMA while inhibiting Smad7 production (Liu et al. 2010). In other studies, rat cardiac fibroblasts increasingly produced miR-21 during activation and that miR-21 directly inhibited sprouty homologue 1 (spry1) via binding to its 3’ untranslated region, causing increased extracellular signal-related kinase (ERK) mitogen activated protein (MAP) kinase signaling and increased cell proliferation (Thum et al. 2008). In a separate study, CCN2 treatment of neonatal cardiac fibroblasts resulted in inhibition of spry1 and concomitant induction of miR-21 (Adam et al. 2012); these findings are supportive of the positive association observed in the present study between CCN2 and miR-21 in PSC. Collectively, the evidence emerging from these various studies suggests that CCN2 acts as a mediator of TGF-β1-induced fibrotic pathways via miR-21 regulation. In the case of PSC, we found that CCN2 drives miR-21 expression in PSC as shown by their relative temporal expression during in vitro activation and the ability of CCN2 siRNA to block miR-21 expression. This latter effect was reversed by miR-21 overexpression, although the underlying mechanism is uncertain because new CCN2 transcripts would be expected to be inhibited by the presence of siRNA. However since, as discussed above, miR-21 promotes the expression of CCN2, TGF-β1 and stimulatory Smad2 while at the same time inhibiting expression of inhibitory Smad7, their overall combined effect in miR-21 over-expressing cells may be sufficient to overcome CCN2 siRNA-mediated knockdown, but this possibility will require further experimental investigation.
The current study also revealed that PSC produce exosomes which contain miR-21 and CCN2 transcripts which enabled their uptake by other PSC. Exosomes arise by inward budding from the limiting membranes of multivesicular bodies and are released extracellularly whereupon they traverse the intercellular space and can enter body fluids such as blood, urine, and secretions (Thery 2011). Exosomes contain a complex cargo comprising miRs, mRNA, proteins, lipids and carbohydrates and they have begun to attract attention as inter-cellular shuttles whereby some of their molecular constituents can be delivered from one cell to another. Novel modes of signaling have begun to emerge whereby exosomal surface ligands can directly stimulate target cells or in which receptors, functional proteins, infectious agents, or genetic information are transferred into target cells by endocytosis or fusion with the plasma membrane (Thery 2011). Exosomal expression of miR-21 and CCN2 were both increased by cellular over-expression of miR-21 or CCN2, thereby mirroring the changes seen in the cells themselves and reinforcing the concept of a miR-21-CCN2 positive feedback loop in the cells. Exosome-dependent delivery of CCN2 mRNA or miR-21 to PSC or SAM-K cells was shown by direct addition of purified exosomes to the cells, an approach used in other recent studies. For example addition of biliary-derived exosomes to rat cardiomyocytes resulted in increased ERK1/2 phosphorylation in cardiomyocytes and a subsequent increase in proliferation (Masyuk et al. 2010) while plasma-derived exosomes were utilized to deliver siRNA to monocytes and lymphocytes (Wahlgren et al. 2012). However, more refined techniques have been used in a few studies in which exosomal miR transfer between donor and recipient cells has been documented using co-culture systems (Rechavi et al. 2009; Pegtel et al. 2010; Yang et al. 2011; Chen et al. 2013). In the future, this type of approach may be amenable for studies of exosome trafficking between various types of pancreatic cells but this must await effective means of inhibiting exosomal production by the specific pancreatic cell types in question, especially as inhibitors used in other studies (e.g. GW4869; (Chen et al. 2013) (Gills et al. 2012)) appear to be ineffective in PSC (data not shown). While our studies suggest that PSC-derived exosomes deliver miR-21 and CCN2 to neighboring cells, we have yet to discriminate between delivery of CCN2 protein versus mRNA as GFP is detected in the recipient cells in either case. At present, attribution of altered gene expression in PSC cells to either exosomal CCN2 or miR-21 is not possible because other components of the exosomal payload likely influence expression of the same gene targets, highly specific readouts of either CCN2 or miR-21 action are lacking, and the outcome is compounded by the fact that CCN2 and miR-21 induce one another. Even so, exosomal delivery of CCN2 or miR-21 likely represents a mechanism by which cellular responses to environmental stimuli are relayed to other cells, thereby providing contextual information that allows neighboring cells to respond appropriately.
In conclusion, miR-21 expression is significantly up-regulated during experimental CP and is produced mainly by activated PSC. Up-regulation of CCN2 expression levels by activated PSC drives miR-21 induction which acts via positive feedback loop to potentiate CCN2 production, thereby amplifying collagen production in the cells. Additionally, activated PSC produce exosomes which contain CCN2 mRNA or miR-21, the levels of which are dependent on the relative expression of CCN2 and/or miR-21 by the cells. Finally, CCN2 or miR-21 in PSC-derived exosomes is taken up by other PSC, highlighting the likely existence in vivo of exosomal transport mechanisms that protects CCN2 or miR-21 from potentially hostile conditions in the extracellular environment and allows for their delivery and subsequent action in recipient cells.
Acknowledgments
Supported by NIH grant R01 AA015554 awarded to D.R.B.
Abbreviations
- α-SMA
Alpha smooth muscle actin
- CP
Chronic pancreatitis
- CCN2
Connective tissue growth factor
- ECM
Extracellular matrix
- GFP
Green fluorescent protein
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- HPRT1
Hypoxanthine-guanine phosphoribosyl transferase 1
- miR
MicroRNA
- PPiA
Peptidylprolyl isomerase A
- PSC
Pancreatic stellate cell
- qRT-PCR
Quantitative real-time PCR
- siRNA
Small interfering RNA
- TGFβ-1
Transforming growth factor beta 1
- TEM
Transmission electron microscopy
References
- Adam O, Lohfelm B, et al. Role of miR-21 in the pathogenesis of atrial fibrosis. Basic Res Cardiol. 2012;107:278. doi: 10.1007/s00395-012-0278-0. [DOI] [PubMed] [Google Scholar]
- Apte MV, Haber PS, et al. Periacinar stellate shaped cells in rat pancreas: identification, isolation, and culture. Gut. 1998;43:128–133. doi: 10.1136/gut.43.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apte MV, Haber PS, et al. Pancreatic stellate cells are activated by proinflammatory cytokines: implications for pancreatic fibrogenesis. Gut. 1999;44:534–541. doi: 10.1136/gut.44.4.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachem MG, Schneider E, et al. Identification, culture, and characterization of pancreatic stellate cells in rats and humans. Gastroenterology. 1998;115:421–432. doi: 10.1016/S0016-5085(98)70209-4. [DOI] [PubMed] [Google Scholar]
- Charrier AL, Brigstock DR. Connective tissue growth factor production by activated pancreatic stellate cells in mouse alcoholic chronic pancreatitis. Lab Investig. 2010;90:1179–1188. doi: 10.1038/labinvest.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier A, Brigstock DR. Regulation of pancreatic function by connective tissue growth factor (CTGF, CCN2) Cytokine Growth Factor Rev. 2013;24:59–68. doi: 10.1016/j.cytogfr.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Charrier A, et al. Epigenetic regulation of connective tissue growth factor by microRNA-214 delivery in exosomes from mouse or human hepatic stellate cells. Hepatology. 2013 doi: 10.1002/hep.26768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Mola FF, Friess H, et al. Connective tissue growth factor is a regulator for fibrosis in human chronic pancreatitis. Ann Surg. 1999;230:63–71. doi: 10.1097/00000658-199907000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillhoff M, Liu J, et al. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg. 2008;12:2171–2176. doi: 10.1007/s11605-008-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Brigstock DR. Connective tissue growth factor (CCN2) in rat pancreatic stellate cell function: integrin alpha5beta1 as a novel CCN2 receptor. Gastroenterology. 2005;129:1019–1030. doi: 10.1053/j.gastro.2005.06.067. [DOI] [PubMed] [Google Scholar]
- Gao R, Brigstock DR. A novel integrin alpha5beta1 binding domain in module 4 of connective tissue growth factor (CCN2/CTGF) promotes adhesion and migration of activated pancreatic stellate cells. Gut. 2006;55:856–862. doi: 10.1136/gut.2005.079178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Ball DK, et al. Connective tissue growth factor induces c-fos gene activation and cell proliferation through p44/42 MAP kinase in primary rat hepatic stellate cells. J Hepatol. 2004;40:431–438. doi: 10.1016/j.jhep.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Gills JJ, Zhang C, et al. Ceramide mediates nanovesicle shedding and cell death in response to phosphatidylinositol ether lipid analogs and perifosine. Cell Death Dis. 2012;3:e340. doi: 10.1038/cddis.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, et al. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshijima M, Hattori T, et al. Roles of heterotypic CCN2/CTGF-CCN3/NOV and homotypic CCN2-CCN2 interactions in expression of the differentiated phenotype of chondrocytes. FEBS J. 2012;279:3584–3597. doi: 10.1111/j.1742-4658.2012.08717.x. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Lawrencia C, Charrier A, et al. Ethanol-mediated expression of connective tissue growth factor (CCN2) in mouse pancreatic stellate cells. Growth Factors. 2009;27:91–99. doi: 10.1080/08977190902786319. [DOI] [PubMed] [Google Scholar]
- Link A, Becker V, et al. Feasibility of fecal microRNAs as novel biomarkers for pancreatic cancer. PLoS One. 2012;7:e42933. doi: 10.1371/journal.pone.0042933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Friggeri A, et al. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 2010;207:1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Chen X, et al. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012;58:610–618. doi: 10.1373/clinchem.2011.172767. [DOI] [PubMed] [Google Scholar]
- Lorenzen J, Kumarswamy R, et al. MicroRNAs in diabetes and diabetes-associated complications. RNA Biol. 2012;9:820–827. doi: 10.4161/rna.20162. [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masyuk AI, Huang BQ, et al. Biliary exosomes influence cholangiocyte regulatory mechanisms and proliferation through interaction with primary cilia. Am J Physiol Gastrointest Liver Physiol. 2010;299:G990–G999. doi: 10.1152/ajpgi.00093.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omary MB, Lugea A, et al. The pancreatic stellate cell: a star on the rise in pancreatic diseases. J Clin Invest. 2007;117:50–59. doi: 10.1172/JCI30082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V, Noureddine L. MicroRNAs and fibrosis. Curr Opin Nephrol Hypertens. 2012;21:410–416. doi: 10.1097/MNH.0b013e328354e559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegtel DM, Cosmopoulos K, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips PA, McCarroll JA, et al. Rat pancreatic stellate cells secrete matrix metalloproteinases: implications for extracellular matrix turnover. Gut. 2003;52:275–282. doi: 10.1136/gut.52.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiat D, Olson EN. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J Clin Invest. 2013;123:11–18. doi: 10.1172/JCI62876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi O, Erlich Y, et al. Cell contact-dependent acquisition of cellular and viral nonautonomously encoded small RNAs. Genes Dev. 2009;23:1971–1979. doi: 10.1101/gad.1789609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M, Masamune A, et al. Establishment and characterization of a simian virus 40-immortalized rat pancreatic stellate cell line. Tohoku J Exp Med. 2002;198:55–69. doi: 10.1620/tjem.198.55. [DOI] [PubMed] [Google Scholar]
- Shen J, Stass SA, et al. MicroRNAs as potential biomarkers in human solid tumors. Cancer Lett. 2013;329:125–136. doi: 10.1016/j.canlet.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Thery C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum T, Gross C, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- Wahlgren J, De LKT, et al. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012;40:e130. doi: 10.1093/nar/gks463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Aurora AB, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Chen J, et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarjou A, Yang S, et al. Identification of a microRNA signature in renal fibrosis: role of miR-21. Am J Physiol Ren Physiol. 2011;301:F793–F801. doi: 10.1152/ajprenal.00273.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]